Abstract
Objective
Biological risk factors underlying psychosis are poorly understood. Biological underpinnings of the dimension of psychosis can be derived using genetic associations with intermediate phenotypes such as sub-components of auditory event related potentials (ERPs). Various ERP sub-component abnormalities in schizophrenia (SZ) and bipolar disorder with psychosis (PBP) are heritable and expressed in unaffected relatives. Prior studies investigating genetic contributions to ERP abnormalities are limited. We used a novel parallel independent component analysis (Para-ICA) to determine which empirically-derived gene clusters are associated with data-driven ERP sub-components, assuming a complex etiology underlying psychosis.
Methods
We examined the multivariate polygenic association of ERP sub-components from 64-channel auditory oddball data in 144 SZ, 210 PBP probands and 95 healthy individuals from the multi-site BSNIP study. Data were reduced by principal component analysis to 2 target and 1 standard ERP waveforms. Multivariate association of compressed ERP waveforms with a set of 20,329 SNPs (reduced from a one million SNP array) was examined using Para-ICA. Genes associated with SNPs were further examined using pathway analysis tools.
Results
Para-ICA identified 4 ERP components that were significantly correlated with 3 genetic components. Enrichment analysis revealed complement immune response pathway and multiple processes including synaptic cell adhesion, axon guidance and neurogenesis significantly mediating ERP abnormalities in psychosis.
Conclusions
We identified three genetic components comprising multiple genes mediating ERP sub-component abnormalities in SZ and PBP. Our data suggest a possible polygenic structure comprised of genes influencing key neurodevelopmental processes, neural circuitry, brain function mediating biological pathways plausibly associated with psychoses.
Keywords: Schizophrenia, Bipolar disorder, Psychosis, Single Nucleotide Polymorphism, gene, Pathway, Event-Related Potential
Introduction
The psychosis dimension contains complex, disabling mental illnesses including schizophrenia (SZ), schizoaffective (SZA) and psychotic bipolar disorder (PBP); evidence indicates significant overlap for clinical features (1), brain function and structure (2), pharmacological treatment (3) and genetic determinants (4). Psychotic disorders demonstrate high heritability; unaffected family members are at increased risk for all diseases. Recent studies implicate shared neurobiological mechanisms of psychosis including impaired calcium channel activity (5) and synaptic function (6). Although copy number variants account for a small proportion of cases (7), under polygenic models a single gene's contribution to disease liability is small but global effects arising from combination of gene subsets are significant because together they impact specific key neural systems at multiple points (8). Endophenotypes are heritable, state-independent phenotypes intermediate between the causative genes and clinical disease (9).
The auditory oddball event related potential (ERP) is a non-invasively measured electrical brain response elicited by an external auditory stimulus. The P300 is a major ERP subcomponent visible as endogenous positive voltage deflection ∼300 ms post stimulus presentation. P300 amplitude signifies attention allocation, cognitive information processing and context updating (10) and is highly heritable (11). Additionally, auditory ERPs comprise N1, N2 and P2 sub-components generated by auditory cortices and associated with stimulus characteristics. Multiple studies report P300 amplitude abnormalities (12, 13) in both SZ and PBP, suggesting general psychosis biomarker status. Auditory P300 amplitude is a frequently-employed SZ candidate endophenotype, because it is robustly reduced in SZ and their close relatives (14-16), highly heritable, stable across the course of illness, is relatively unaffected by illness exacerbations and is elicited by a straightforward task performed easily by psychotic patients. P300 amplitude abnormalities in PBP may be proband-specific (17). Deficits in ERP subcomponents N1, P2 and N2 are found in SZ patients (18, 19) and their relatives (20). Both N1 and P2 amplitude abnormalities (13) and their endophenotypic status are less studied in PBP.
The P300 amplitude response with a parietal maximum (P3b) has been related to norepinephrine (21) and dopamine neurotransmission (22). Recent studies examining genetic associations of P300 (23, 24) adopted traditional single nucleotide polymorphism (SNP)-based univariate approaches, such as genome wide association (GWA). One polygenic association study identified several interacting SNP loci including rs1045642 in the ABCB1 gene linked to P300 ERP in healthy individuals (25), plus genes related to dopaminergic, noradrenergic and signal transduction/amplification pathways. Prior studies (26-29) documented genetic associations of P300 amplitude deficits with known SZ risk genes including DISC1, NRG1 and COMT. One study (26) identified the above-mentioned SNP (rs1045642) as a SZ risk locus.
The univariate GWA approach relates single gene variants to a unitary phenotype. While straightforward it has two main drawbacks. First, the weak individual effect of common variants prevents most from reaching genome-wide significance, due to numerous multiple-comparisons, requiring extremely large samples to gain sufficient statistical power. Second, simultaneous coupling of multiple genes is ignored. To overcome these problems, multivariate association (30) based on parallel independent component analysis (Para-ICA) has been developed to model the polygenic architecture (combinatorial gene effects) of complex psychiatric illnesses. Prior Para-ICA studies revealed known and novel gene clusters implicated in Alzheimer's disease (31) and psychoses (32) based on structural and functional imaging data respectively.
The present study sought to: 1) identify risk genes collectively associated with ERP endophenotypes in psychosis; 2) determine whether the gene clusters mediate neurophysiological abnormalities in either or both disorders. We hypothesized that Para-ICA would identify novel interacting risk gene clusters (likely including known risk genes) mediating plausible physiological pathways disturbed in psychosis.
Material and Methods
Bipolar-Schizophrenia Network on Intermediate Phenotypes (BSNIP) Study
We assessed genotype–phenotype multivariate associations by combining genetic variants and ERP from SZ, SZA, PBP probands and healthy comparison subjects (HC) from the BSNIP study (www.b-snip.org), formed to investigate intermediate phenotypes from multiple modalities and their genetic underpinnings in psychosis.
Subject Recruitment
We examined a subset of 620 probands and HC selected from the overall ∼2600-person BSNIP (33) cohort who had undergone genotyping and participated in the BSNIP auditory oddball paradigm. These comprised 144 SZ, 210 PBP and 95 HC, aged 15-65 years (see Supplementary Table ST1 for demographic and clinical information). SZA depressive and mania subtype probands were classified as SZ and PBP respectively. SZ and PBP proband groups were unmatched on age, sex or ethnicity with HC. Probands' medication is listed in supplementary Table ST2. The study was explained to all subjects and institutional review board approved written informed consent obtained at respective participating sites.
Auditory Oddball Paradigm and Electroencephalogram (EEG) Data Acquisition
EEG was collected independently at each site from identical Neuroscan equipment equipped with 64 Ag/AgCl electrodes (Quik-Cap, Compumedrics, El Paso, Texas), while subjects were performing an auditory oddball task (see supplement text). Electrode positions were defined according to the International 10-10 EEG system (see Supplementary Figure SF1).
SNP Data Collection
Blood was collected from each subject and genomic DNA extracted. Genetic variants were obtained by genotyping for ∼one million (1,140,419) target SNPs across the whole genome using the Illumina Human Omni1-Quad chip and BeadArray™ platform at Genomas Inc, Hartford Hospital.
ERP Data Processing
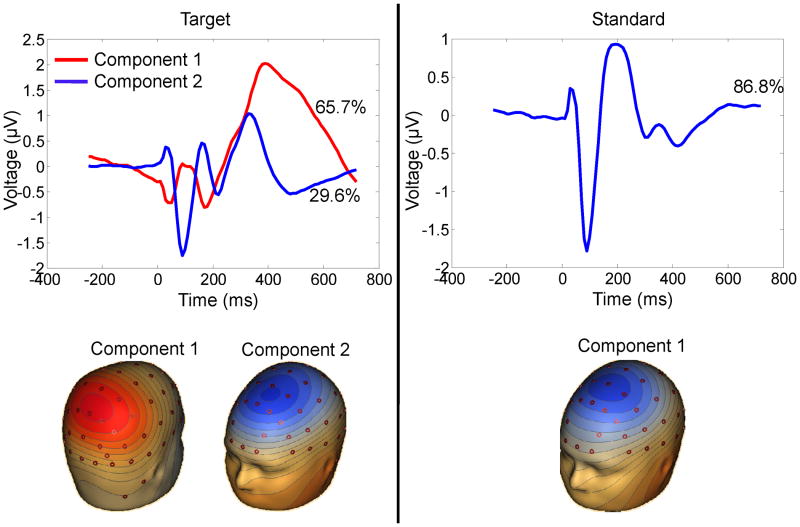
EEG data were artifact rejected and averaged ERP data were reduced to 2 targets and 1 standard component (see Figure 1) using spatial principal component analysis (PCA) (13) (see details in supplementary text).
Figure 1.
Spatial topography associated with principal components for target and standard stimulus conditions. Two targets (parietal and frontal topography) and one standard component (frontal) was derived from principal component analysis of grand average 64 channel data. The time window for each component is between 250 msec before to 720 msec after stimulus onset.
SNP Data Processing
SNP data were converted to discrete numbers and subjected to quality control process (Supplementary Figure SF2). Stratification bias was corrected using PCA (Supplementary Figure SF3). Data were reduced from million to 20,329 SNPs by selecting those significant at p<0.05 in the logistic regression (see supplementary text).
Para-ICA based SNP-ERP Association
Para-ICA is a multivariate association analysis that links linearly associated functional ERP activity with synergistic gene clusters by relating the hidden structures from both ERP and SNP data. Data were organized as a matrix of (i) subjects by SNPs (449 × 20329) and (ii) subjects by ERP component waveforms (449 × 294 (3×98 time points)) and were reduced to 11 and 8 components or factors respectively. ERP and SNP data were jointly processed by Para-ICA (25, 30) (see Supplementary Figure SF4 and supplementary text).
Correlation and Statistical Analysis
Bi-modal feature association using Para-ICA was assessed by correlations between SNP and ERP loading coefficients (LC). Significance levels were adjusted through partial-correlation by regressing effects of age, sex and site on LCs. Correlations for all component pair combinations (8×11=88) were evaluated and significance levels adjusted using Bonferroni multiple comparison correction number of combinations (p<0.05/88). ERP and SNP LCs of significant component pairs were examined for group differences using t-tests. Dominant SNPs and ERP features in the components contributing to the association were identified with a threshold of |Z| = 2.5 for both modalities. Supplementary analyses included correlation of ERP LCs with intelligence quotient (IQ) scores and chlorpromazine (CPZ) equivalents.
ERP-Structural Magnetic Resonance Imaging (sMRI) Association Analysis
Brain structures (see Supplementary methods for sMRI data collection) associated with the ERP components were identified by partial correlation (accounting for age, sex and site) of regional volumetric measures with ERP loading coefficients. P-values were corrected for multiple comparisons using false discovery rate.
Pathway Analysis
Genes corresponding to SNPs selected from the genetic component were entered into MetaCore annotation software GeneGo (Thompson Reuters, New York, NY) to determine the biological pathways associated with ERP abnormalities (see supplementary text).
Results
Gene-ERP Multivariate Linkage
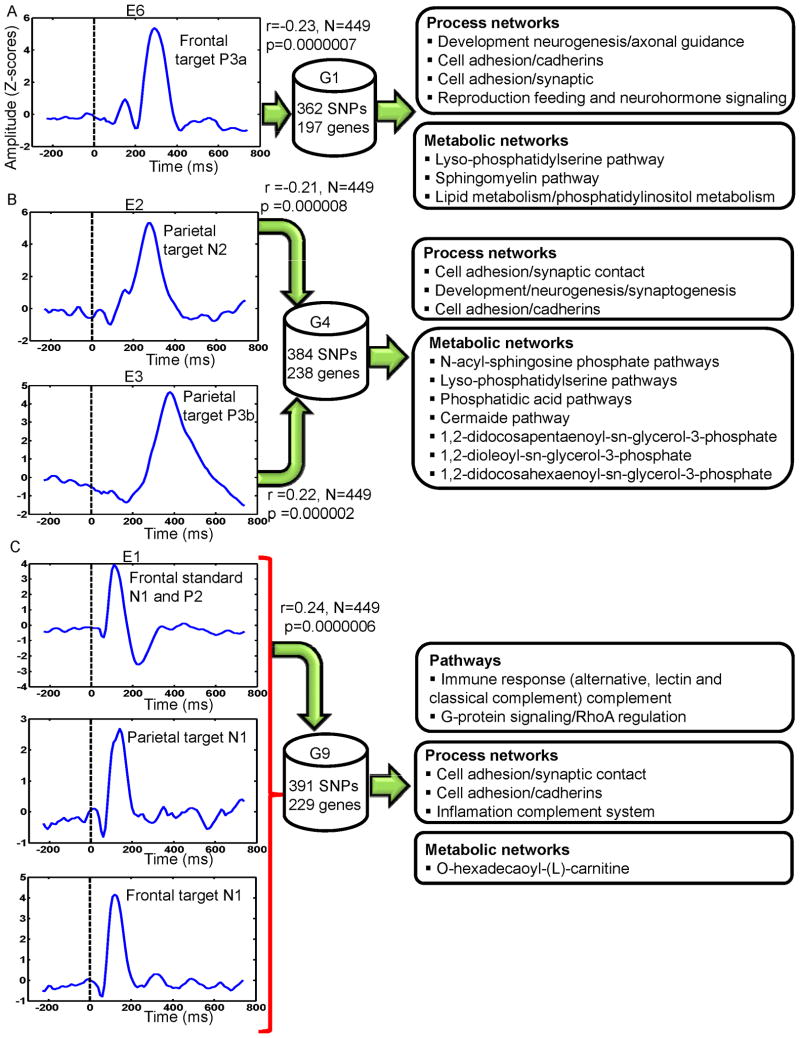
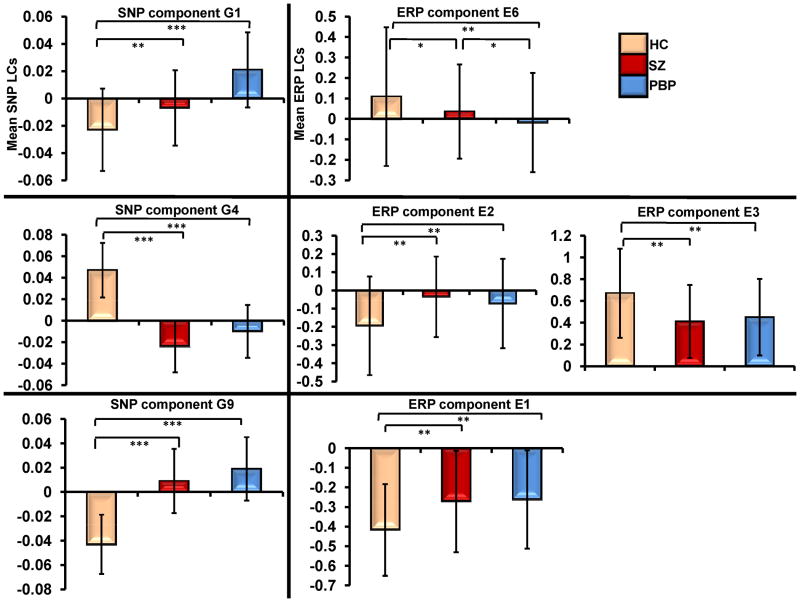
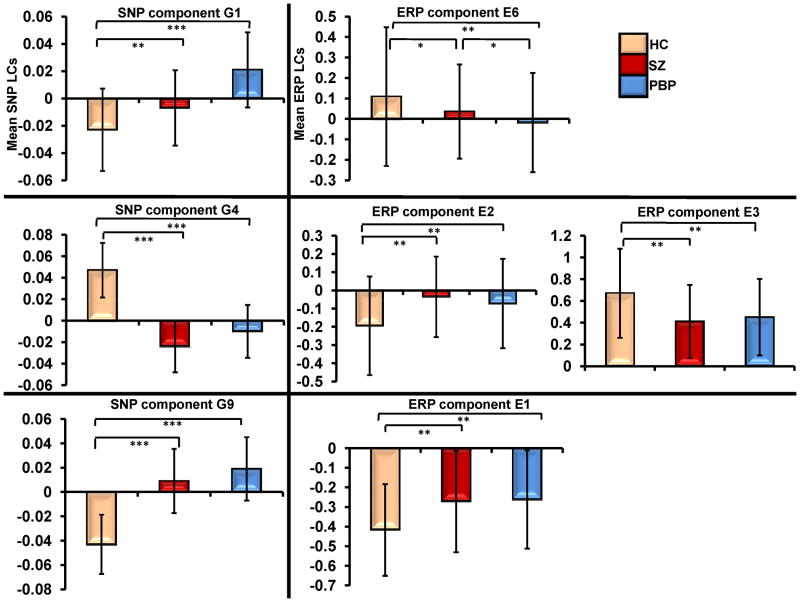
Four (E1, E2, E3 and E6) of the 8 independent ERP components were significantly connected to 3 (G1, G4 and G9) of the 11 genetic/SNP components. ERP component E6 (frontal target P3a) was correlated negatively with G1 (r=-0.23, N=449, p<0.0000007) (Figure 2A); reflecting increased loading on G1 was related to diminished frontal target P3a. E1 was positively associated with G9 (r=0.24, N=449, p<0.0000006) (Figure 2C); indicating increased loading on G9 related to increased ERP features in E1 (frontal standard N1, P2, frontal and parietal target N1). Both E2 (parietal target N2) and E3 (parietal target P3b) components were negatively (r=-0.21, N=449, p<0.000008) and positively (r=0.22, N=449, p<0.000002) correlated (see Figure 2B) with the same gene component G4 respectively (see scatter plot in Figure 3). Although IQ differed between groups and was negatively correlated (r= -0.166, N=446, p<0.0004) with component E3, controlling its effects did not alter the ERP-SNP correlation. ERP component E2 positively correlated (r= 0.179, N=215, p= 0.009) with CPZ equivalents, reflecting higher CPZ dosage associated with increased abnormality. Post-hoc comparisons revealed significant differences in ERP and SNP LCs between HC and both SZ and PBP for all significantly correlated components (Figure 4). ERP component E6 (frontal target P3a) differed between SZ and PBP, while other ERP components failed to differentiate two proband groups. Reliability test by leave-one-out cross-validation revealed an average within modality correlation > 0.85, indicating stable ERP and SNP components.
Figure 2.
Event-related potential (ERP) sub-components and associated gene components derived from the multivariate parallel independent component analysis. A E6-G1 component pair, B E2-G4 and E3-G4 component pairs, C E1-G9 component pair E6 comprised frontal target P3a sub-component. E2 and E3 comprised parietal target N2 and P3b subcomponents respectively. E1 comprised standard N1 and P2, parietal target N1 and frontal target N1 subcomponents. Peak amplitude within predefined time periods post-stimulus was used to identify the ERP component. The polarity of the ERP waveforms for N1, N2 and P2 subcomponents is reversed by independent component analysis. Biological pathways and process networks associated with each gene component from enrichment analysis are also displayed.
Figure 3.
Scatter plots of loading coefficients (LC) for the three groups including schizophrenia (SZ), psychotic bipolar disorder (PBP) probands and healthy comparison subjects (HC). The overall association between the LCs across three groups is displayed as the linear fit. ERP=event-related potential; SNP=single nucleotide polymorphism
Figure 4.
Mean loading coefficients (LC) for significantly associated event-related potential (ERP) and gene components for schizophrenia (SZ) (N=144), psychotic bipolar probands (PBP) (N=210) and healthy comparison subjects (HC) (N=95). Error bars represent standard deviation. Post-hoc comparisons included pairwise t-tests between HCs and SZ and PBP probands. *** P≪0.000000001 ** p<0.0001 * p<0.05 SNP=single nucleotide polymorphism
ERP-Structural Volume Associations
ERP component E1 (frontal standard N1, P2, frontal and parietal target N1) was associated with volume reduction in gray matter regions including pars opercularis, posterior cingulate, insula, caudal middle frontal, middle temporal and supramarginal gyrus (see Table 1). E3 (P3b) was associated with volume reduction in left entorhinal and hippocampal regions.
Table 1.
Association of event-related potential (ERP) with cortical and subcortical structural volumes. 82 brain volumes were extracted for N=369 subjects out of the 449 who had ERP data using Freesurfer. The association was evaluated using partial correlation adjusting for age, sex and data collection site. ERP component E1 was correlated with six regional volumes (after false discovery rate correction (FDR)), while E3 was related to 2 regional volumes. ERP components E2 and E6 did not yield significant correlations after FDR correction.
| Brain Regions (volume) | E1 | |
|---|---|---|
| R | P | |
| L. caudal middle frontal | -0.158 | 0.00238 |
| L. pars opercularis | -0.173 | 0.00086 |
| L. posterior cingulate | -0.153 | 0.00324 |
| L. insula | -0.158 | 0.00236 |
| R. middle temporal | -0.152 | 0.00351 |
| R. supramarginal | -0.148 | 0.00433 |
| E3 | ||
| L. entorhinal | 0.171 | 0.00098 |
| L. Hippocampus | 0.167 | 0.00125 |
L, Left; R, Right
Pathway Analysis
Prominent process networks associated (see Supplementary Table ST3) with G1 (related to ERP component E6) included (but not limited to) neurogenesis-axon guidance and cell adhesion (cadherins, synaptic contact). Major metabolic components included Lyso-phosphatidylserine and sphingomyelin pathways. Functional properties for the top 20 genes from all the three genetic components are summarized in Table 2.
Table 2.
Functional attributes and gene-specific information for top 20 unique genes in genetic components G1, G4 and G9 from multivariate association analysis. Only the single nucleotide polymorphism (SNP) with maximal loading within each gene is listed in the table, (most genes were represented by several SNPs). Relative weights indicate normalized Z-scores (ZS) (absolute Z-score divided by the maximum absolute SNP weight). SNP locations are based on GRCh37/hg19 assembly.
| Gene networks | ||||||
|---|---|---|---|---|---|---|
| Gene (G1) | SNP | Chromosome | Position | Z-score | Relative weights | Functional Attribute |
| DCC* | rs16956411I | 18q21.2 | 50775428 | -9.93 | 1 | Axon guidance, neuronal migration |
| BOC* | rs775228I | 3q13.2 | 112997554 | -4.84 | 0.487 | CA and synaptic development |
| SEC14L2* | rs4820845I | 22q12.2 | 30800338 | -4.5 | 0.461 | Lipid binding |
| PDLIM5* | rs13121500I | 4q22.3 | 95577290 | 4.41 | 0.444 | Regulates protein kinase C activity; synapse & dendritic spine morphogenesis |
| HDAC9* | rs12699994I | 7p21.1 | 18895297 | -4.31 | 0.434 | Neocortical neuron development by transcriptional regulation |
| MAML3* | rs7678266I | 4q31.1 | 140668374 | -4.23 | 0.426 | Unknown |
| B3GNTL1* | rs1001865I | 17q25.3 | 80914988 | 4.1 | 0.417 | Unknown |
| TBCD* | rs3785520I | 17q25.3 | 80895745 | -4.00 | 0.402 | Captures & stabilizes beta-tubulin |
| SNAP91* | rs1546977I | 6q14.2 | 84314423 | -3.96 | 0.399 | Formation of clathrin coated vesicles at presynaptic membranes |
| LY9 | rs574610Utr-3 | 1q23.3 | 160797684 | -3.91 | 0.384 | Unknown |
| CD244 | rs485618Utr-3 | 1q23.3 | 160800480 | -3.86 | 0.389 | Mediates non-major HC restricted killing and modulates natural killer-cell cytolysis |
| CDKAL1* | rs7758129I | 6p22.3 | 20609241 | 3.85 | 0.388 | Unknown |
| ESRRG* | rs3929399I | 1q41 | 216718378 | 3.84 | 0.387 | CP |
| MCTP2* | rs1655455I | 15q26.2 | 94858686 | -3.83 | 0.386 | Ca2+ ion binding; intracellular signal transduction |
| PIP4K2A | rs7071450I | 10p12.2 | 22867451 | 3.79 | 0.381 | CPD |
| TMEFF2* | rs10185068I | 2q32.3 | 192924987 | -3.78 | 0.381 | Unknown |
| ZC3H18* | rs12445653I | 16q24.2 | 88677230 | -3.75 | 0.378 | Unknown |
| VWA3B* | rs10211067Utr-5 | 2q11.2 | 98703659 | -3.73 | 0.376 | Unknown |
| RASGRP3 | rs11687777I | 2p22.3 | 33764709 | -3.66 | 0.368 | Signal transduction |
| ANKIB1 | rs721015I | 7q21.2 | 91925256 | -3.59 | 0.361 | Unknown |
| (G4) | ||||||
| MSRA* | rs7459532I | 8p23.1 | 10261068 | -7.96 | 1 | Enzymatic reduction of methionine sulfoxide & repairs oxidative damage |
| XKR6* | rs2409691I | 8p23.1 | 10943276 | -7.3 | 0.917 | Unknown |
| RP1L1 | rs7386213I | 8p23.1 | 10503525 | -6.47 | 0.813 | Binds to microtubules; regulates microtubule polymerization |
| BLK | rs2618451I | 8p23.1 | 11376266 | -6.32 | 0.794 | CPD |
| TNKS* | rs7840706I | 8p23.1 | 9535056 | -5.39 | 0.677 | Unknown |
| TPO* | rs2276702I | 2p25.3 | 1426621 | -5.06 | 0.635 | Iodination of tyrosine residues in thyroglobulin |
| IL1F10* | rs6761276M | 2q13 | 113832312 | -5.04 | 0.632 | Regulates innate immune responses |
| MFHAS1* | rs4841044I | 8p23.1 | 8664940 | 5.00 | 0.628 | Unknown |
| ADAMTS16* | rs270178I | 5p15.32 | 5165415 | 4.65 | 0.584 | Unknown |
| DOCK8* | rs10967788I | 9p24.3 | 282180 | - 4.62 | 0.58 | Interacts with Rho GTPases, intracellular signaling networks |
| TAF8 | rs6917299I | 6p21.1 | 42025058 | -4.04 | 0.508 | Involved in transcription factors |
| GSG1L* | rs1645362I | 16p12.1 | 27891473 | -4.04 | 0.508 | Unknown |
| ODZ3* | rs957053I | 4q35.1 | 183248389 | -4.04 | 0.508 | Unknown |
| VAT1L* | rs9933953I | 16q23.1 | 77909692 | 4.02 | 0.507 | Unknown |
| USH2A* | rs17025267I | 1q41 | 215903907 | 3.97 | 0.495 | Development & homeostasis of inner ear |
| COL2A1* | rs1793923I | 12q13.11 | 48384122 | 3.95 | 0.498 | Unknown |
| INPP5K* | rs1109303I | 17p13.3 | 1403477 | 3.92 | 0.496 | Regulates actin cytoskeleton |
| NRXN3* | rs10782463I | 14q24.3 | 78978482 | - 3.92 | 0.492 | Receptors and synaptic CAM in CNS |
| PNPLA1* | rs12197079M | 6p21.31 | 36274153 | 3.85 | 0.483 | Lipid metabolism, (lipolytic & acyltransferase) |
| PKP3 | rs7105848I | 11p15.5 | 396546 | 3.83 | 0.481 | Links cadherins to intermediate cytoskeleton filaments |
| (G9) | ||||||
| ME1* | rs1170348I | 6q14.2 | 84020324 | -6.90 | 1 | Fatty acid biosynthesis and brain Co2 fixation |
| PEMT* | rs11078389I | 17p11.2 | 17478352 | 5.60 | 0.812 | Synthesizes membrane phospholipids:affects choline levels |
| GPC6* | rs4369513I | 13q31.3 | 94958719 | 5.42 | 0.786 | Cell growth, cell division & CNS synapse formation |
| SNAP91* | rs217291I | 6q14.2 | 84393942 | 5.38 | 0.78 | Refer to table G1 |
| DLGAP1* | rs1465947I | 18p11.31 | 3973509 | 4.76 | 0.69 | Molecular organization of synapses & neuronal cell signaling |
| PRSS35 | rs592911I | 6q14.2 | 84223563 | -4.47 | 0.648 | Unknown |
| PCSK5* | rs2842467I | 9q21.13 | 78938628 | -4.32 | 0.626 | Mediates posttranslational endoproteolytic processing for several integrin alpha subunits |
| FRK* | rs12662901I | 6q22.1 | 116305637 | 4.24 | 0.615 | Tyrosine kinase role in cell cycle & growth suppression |
| KCTD8* | rs2020159I | 4p13 | 44330067 | 4.23 | 0.613 | Unknown |
| PDLIM1* | rs11593722I | 10q24.1 | 97006105 | -4.13 | 0.599 | Regulates actin cytoskeleton dynamics & neutrite growth |
| CYP2C19* | rs10786172I | 10q23.33 | 96581094 | 4.08 | 0.591 | |
| ANO2* | rs1035066I | 12p13.31 | 5756592 | -4.08 | 0.591 | Unknown |
| SRRM4 | rs1405050I | 12q24.23 | 119443713 | -4.05 | 0.586 | Unknown |
| YSK4* | rs4953941I | 2q21.3 | 135727531 | -4.03 | 0.584 | Unknown |
| CNTNAP2* | rs700281I | 7q35 | 146192877 | 4.03 | 0.584 | CNS Cell to cell interaction |
| PRDM16* | rs1798246I | 16q24.2 | 3080855 | -4.02 | 0.583 | Unknown |
| TNNI1 | rs3767548I | 1p36.32 | 201388148 | -3.98 | 0.577 | Regulate Ca2+ sensitivity of muscle myofibril contractile apparatus |
| GLT1D1* | rs516034I | 12q24.33 | 129459959 | 3.98 | 0.576 | Unknown |
| ESRRG* | rs1833036I | 1q41 | 216707012 | -3.86 | 0.359 | Refer to table for G1 |
| ZNF385B* | rs10432487I | 2q31.2 | 180512804 | 3.85 | 0.558 | Unknown |
indicates multiple SNP occurrences (>2) of the gene within the genetic network.
CA, cell adhesion; CA2+, Calcium ion, CAM, cell adhesion molecule, CNS; Central nervous system; CO2, carbon dioxide; CP, cell proliferation; CPD, cell proliferation and differentiation; HC, histocompatibility complex; I, Intronic; M, missense; Utr-3, three prime untranslated region; Utr-5, five prime untranslated region
Process networks (collection of coordinated genes controlling a biological processes) associated with G4 were cell adhesion (synaptic contact, cadherins and amyloid proteins) and development neurogenesis (synaptogenesis). Several metabolic networks enriched for genes in G4 included (but were not limited to) N-acyl-sphingosine phosphate, Lyso-phosphatidylserine, phosphatidic acid and ceramide pathways. Gene component G9 (related to ERP component E1) was significantly (after false discovery rate correction) enriched with immune response genes (classical, alternative and lectin-induced complements) and G-protein signaling (RhoA regulation). Primary process networks related to G9 were cell adhesion (synaptic contact, cadherins) and the inflammation-complement system. Pathways, known diseases and major human brain regions from Allen Brain Atlas (www.brain-map.org) (assessed based on expression Z-scores) associated with the top 20 genes are listed in supplementary Table ST4.
Discussion
Etiological mechanisms for SZ and PBP are complex, but one approach to gain insight is to examine biological measures such as ERP that are putatively close to the genetic variation. Currently, the neural substrates or genetic basis for eliciting ERP components are unknown. Various ERP sub-components demonstrate significant heritability, suggesting strong genetic influences. As a preliminary step, we investigated polygenic sources underlying abnormal ERP sub-components in the psychosis dimension from a large multi-site dataset including all probands, based on multivariate association. Additionally we compared SZ and PBP probands against HC based on DSM criteria.
ERP Components
ERP component E1 (a linear combination of standard N1, P2, frontal and parietal target N1) showed overall increased loadings (less change from baseline) in SZ and PBP compared to controls reflecting deficits in early sensory processing, consistent with prior SZ (18, 20, 34) and PBP (13) studies. The primary source of N1 and P2 is the auditory cortex (35, 36) (Heschl's gyrus); we identified reduction in gray matter regions surrounding this structure. Parietal target N2 in E2 was significantly increased (reduced change from baseline) in probands indexing abnormal stimulus classification (37). Psychosis probands exhibited decreased frontal target P3a in E6 and parietal target P3b in E3, consistent with prior psychosis literature (12). The role of hippocampus in P3b generation is debated (38); however, our finding of P3b amplitude reduction associated with lesser hippocampus volume is consistent with prior evidence (39).
Gene Components
Para-ICA identified gene groups comprising interacting genes with a weight associated to each linkage-contributing gene. We discuss here the genetic underpinnings of ERP abnormalities based on the functionality of top ranking and frequently appearing genes in each component and biological properties associated with the gene clusters. Of all unique genes pooled across the 3 genetic components found in this study (N=376), 76 have previously been identified as risk genes for SZ, PBP or both, while we report 300 new genes associated with ERP alterations in psychosis
G1
Top Genes and Those Identified Repeatedly
The highest-ranking gene in G1 was DCC, encoding a netrin-1 (40) receptor and a novel SZ candidate risk gene (41). Multiple (27 occurrences in the top 50) intronic SNPs within DCC were associated with frontal target P3a in E6. The dominant SNP loading was negative with a negative correlation between G1 and E6, indicating decreased minor allele frequency associated with increased frontal target P3a. DCC plays a critical role in neuronal circuitry modulating synaptic connectivity by mediating brain development via axon guidance and dendrite growth (42) with roles in functional reorganization of mesocortical dopamine circuitry (43). DCC is also involved in development of human brain lateralization (44), abnormal in SZ (45).
Several intronic SNPs within the BOC gene were found in G1. BOC is the receptor for the molecule Sonic Hedgehog, whose critical role in spatial specificity of synapse formation (46) guide the formation of cortical microcircuits (47) and dorsoventral axon patterning (48). Multiple SNPs within PDLIM5 encoding the LIM domain protein were detected in G1. PDLIM5 is a candidate risk gene for SZ, PBP and major depression and interacts with brain neuronal N-type calcium channel and protein kinase-C (49). Postsynaptic density proteins containing PDZ domain, interact with glutamate receptors to regulate synaptic plasticity (50, 51). The HDAC9 gene encoding the histone deacetylase 9 enzyme involved in transcriptional regulation and cell cycle progression is a known SZ risk gene (52), pivotal in neocortical neuron development through transcription regulation via histone modification and regulates dendritic growth in developing cortical neurons (53). HDAC inhibitors modulate cell lineage differentiation during brain development (52) and are direct targets of valproic acid, a mood stabilizer used to treat PBP (54); and may impact antipsychotic response (55). Several SNPs within DISC1, a known SZ risk gene were found in G1 but not ranked in the top 20.
Pathways, Process and Metabolic Networks
Neurogenesis (in particular axon guidance) and cell adhesion (cadherins and synaptic contact) were the enriched functional processes associated with gene clusters in G1 engaged in modulating frontal target P3a abnormality common to psychosis, indicating that these processes are general psychosis risk mediators. Adult neurogenesis likely plays essential roles in both brain function (56) and pathophysiology of psychiatric illnesses (57, 58). The primary neurogenesis-axon guidance associated genes are PCSK5, CACNA1C, DCC, SLIT3, UNC5C, SLIT1, DISC1, NCAM1, NTRK3, ROBO2, GDA and PLCB1: of these several are implicated in psychosis risk.
Another key process identified was cell-adhesion mediated by cadherins and synaptic contact. Cadherins depend on calcium ion function to facilitate cell adhesion involved in intracellular signaling, memory-formation, neuronal migration, synapse-formation and maturation (59), associated with neuropsychiatric disorders. Genes associated with cadherin-based cell adhesion were CTNND2, PTPRJ, PCDH15, PDZK3, CTNNA2, CDH12, VAV2, CDH13 and WNT5B. Several of these are implicated in multiple psychiatric disorders; in particular CDH12 is associated psychosis. Synaptic cell adhesion molecules mediate neural connections and synapse development. Such genes were SYT9, NRXN3, CTNND2, GRIN2B, CTNNA2, NCAM1 and OBCAM, of which NRXN3 is implicated in SZ, NCAM1 and GRIN2B in both SZ and PBP. Sphingomyelin is a signal transduction system, impacting neural tissue and neuronal membrane integrity, suggesting that sphingolipid metabolism may play a key role in pathogenesis of neuropsychiatric disorders (60).
G4
Top Genes and Those Identified Repeatedly
The most significant G4 candidate gene (associated with parietal target N2 and P3b) was MSRA, a gene that shields against oxidative stress and is associated with SZ susceptibility (61-63). Oxidative stress is important in SZ pathophysiology (64), associated with hypoactive N-methyl-D-aspartate glutamate receptors (65). The chromosome region 8p23.1, in particular SNP D8S542 within MSRA is strongly associated with both SZ and PBP (66, 67).
Pathways, Process and Metabolic Networks
Developmental neurogenesis, synaptogenesis and cell adhesion was the significant process network mediating psychosis risk through parietal P3b and N2 abnormalities. Metabolic processes including Lyso-phosphatidylserine, phosphatidic acid and ceramide networks regulated P3b and N2 abnormalities in SZ and PBP. Phospholipids constitute cell membranes regulating signal transduction and acting as key secondary messengers. Abnormal phospholipid distribution occurs in SZ prefrontal cortex (68). Both calcium independent and dependent Phospholipase A2 enzymes are abnormal in SZ (69, 70). Ceramides are lipid signaling molecules guiding cellular differentiation, proliferation and apoptosis altered in both first-episode schizophrenia (71) and depression.
G9
Top Genes and Those Identified Repeatedly
Multiple SNPs were identified in the ME1 gene encoding the cytosolic, NADP-dependent malic enzyme involved in metabolic pathways. No direct evidence from prior studies suggests ME1's role in SZ and PBP, except for increased postmortem expression in unmedicated bipolar brain (72) and, linkage signals in SZ in the vicinity of ME1 (73). The PEMT gene encodes for an enzyme synthesizing membrane phospholipids associated with SZ (74). This gene helps mediate Arachidonic acid signaling, neuronal differentiation and neurite growth (75). The GPC6 gene belongs to the glypicans that promote glutamate receptor clustering, receptivity and induce postsynaptic synapse formation (76). SNAP91 encodes for clathrin coat assembly protein 180, enriched in presynaptic neuronal terminals (77). SNAP91 is involved in intracellular signaling (78) associated with the WNT pathway (79) and is implicated in PBP (80).
Pathways, Process and Metabolic Networks
Multiple immune response pathways were associated with psychosis risk mediated by frontal, parietal target N1 and standard N1 and P2 ERP abnormalities in probands, suggesting immune system's role in psychosis pathogenesis (81), consistent with prior SZ studies (82, 83). The classical complement pathway acts on synapse remodeling, pruning, neuronal plasticity and neurodevelopmental processes via immune system cells and molecules (C3) (84) and implicated in the pathophysiology of neurodegenerative disorders (85, 86). This study implicated immune system CR2 and C3 genes; the latter is a SZ risk gene (87). C3 is involved in synapse development and regulates neuronal connectivity (88). Gene component G9 was associated with cell adhesion regulated by cadherins and synaptic contact, relevant to SZ and PBP risk.
Replication analyses were not conducted to validate the current findings; however several candidate genes and processes found in this study are previously implicated in SZ and PBP, supporting the neurodevelopmental hypothesis of SZ (89) and affective disorders (90). Findings from the current study point to converging evidence from pathway-based studies that identified similar biological mechanisms including axon guidance (91-93), cell adhesion (94, 95), inflammation and immune system (96) associated with SZ risk. A recent BSNIP resting fMRI-genetic study (32) using Para-ICA identified similar processes including developmental neurogenesis, axon guidance, synaptic and cadherin cell adhesion, immune response, nervous system development, ceramide and sphingosine pathways mediating aberrant default mode activity in psychosis. These data help confirm the current findings and validate the approach undertaken to merge functional and genetic data to dissect the complex mechanisms mediating biological phenotypes in these disorders. A complete overlap in pathways and processes across diverse phenotypes is unlikely as they probably probe different domains of neurophysiology in these disorders.
Strengths and Limitations
Advantages of the present study include our use of high density spatial ERP data in a large multi-site psychosis sample. Limitations included unmatched age, sex and sample sizes between groups, adjusted for by controlling their effects through partial correlation between the SNP and ERP LCs: multi-site effects were accounted for by regressing data collection site. Examined ERP phenotypes may be medication-influenced; it was not straightforward to control for medication effects because probands were on numerous medications, combined with varying chronicity, severity and illness duration. The current findings could not be validated due to lack of replication sample. Para-ICA was optimized by selecting nominally disease-associated SNPs from univariate analysis. Thus, other genetic netrowks that may be weakly associated with the ERP sub-components were overlooked.
Conclusions
As hypothesized, we derived both novel interacting candidate genes and known SZ and PBP risk genes mediating ERP abnormalities in psychosis. In general, our data suggests a strong multifactorial genetic component comprised of brain-relevant genes playing a key role in neural functions, specifically those controlling neuronal circuits and neurodevelopmental processes. We identified one genetic cluster enriched with genes for neurogenesis related axon guidance mediating psychosis risk via frontal target P3a abnormalities in probands. The second genetic component was enriched with genes involved in developmental neurogenesis-synaptogenesis, ceramide and phospholipid networks, influencing P3b abnormalities in psychosis. The third genetic component comprised genes coding for the complement immune response pathway associated with SZ and PBP risk through standard N1, P2, frontal and parietal target N1 abnormalities. All three genetic components mediated ERP abnormalities across the psychosis dimension. Cell adhesion mediated by synaptic contact and cadherins was the prominent network process driving all the ERP abnormalities in psychoses.
Supplementary Material
Supplementary Figure SF1 Schematic of 64-channel EEG montage used for data collection.
Supplementary Figure SF2 Schematic depiction of the processing pipeline for quality control (QC) of single nucleotide polymorphism (SNP) data.
The QC process involved two stages: individual based followed by SNP-based QC.
Poorly genotyped individuals and bad SNPs were removed from the final regression analysis.
SNP data were corrected for stratification bias by correcting for top 3 Eigen factors associated with self-reported ethnicity.
LD= linkage disequilibrium
Supplementary Figure SF3 Q-Q plot of theoretical and empirical p-values from logistic regression for (A) schizophrenia (SZ) vs healthy comparison subjects (HC) and (B) psychotic bipolar disorder (PBP) vs HC.
Logistic regression was applied to individual markers in case-control fashion for both SZ and PBP groups.
Supplementary Figure SF4 Schematic illustration of the parallel independent component analysis (Para-ICA) for genetic association of event-related potential (ERP) data.
Data were constructed as a matrix of subjects by SNP (K=449 × M=20,329) and subjects by reduced ERP waveforms (K=449 × P=294 (3×98 time points)). The number of components extracted from Para-ICA for the ERP and SNP data were N1 = 8 and N2 = 11 respectively.
LC=loading coefficient; SNP= single nucleotide polymorphism
Supplementary Table ST1: Demographic information for schizophrenia, psychotic bipolar probands and comparison subjects.
Supplementary Table ST2: Medication information for probands
Supplementary Table ST3: Results of enrichment analysis including GeneGo pathway maps, process networks, metabolic networks and gene ontology processes associated with contributing genes from components G1, G4 and G9. Bold P-values indicate significant (p<0.05) after false discovery rate (FDR) correction.
Supplementary Table ST4: Pathways, disease risk and brain regions associated with top 20 genes from gene components G1, G4 and G9. Expression Z-scores from Allen brain atlas database was used to identify brain regions. Other mental disorders associated with these genes from prior studies are also provided (references in online supplementary text). Gene information was obtained from Dbsnp and Genecard.
Acknowledgments
We thank the study participants for their contributions. This study was supported by funding from National Institute of Mental Health through linked R01 MH077851 to Drs. Godfrey Pearlson, MH078113, Carol Tamminga, MH077945 Matcheri Keshavan, MH077862, John Sweeney and MH077852, Dr. Gunavant Thaker. Additional support includes 2R44 MH075481 to Dr. Gualberto Ruaño and P20GM1034672 and R01EB006841 to Dr. Vince Calhoun.
Funding: This study was supported by funding from National Institute of Mental Health through linked R01 MH077851 to Drs. Godfrey Pearlson, MH078113 to Carol Tamminga, MH077945 to Matcheri Keshavan, MH077862 to John Sweeney and MH077852 to Dr. Gunavant Thaker. Additional support includes 2R44 MH075481 to Dr. Gualberto Ruaño and P20GM1034672 and R01EB006841 to Dr. Vince Calhoun.
Dr. John Sweeney has received support from Takeda, BMS, Lilly, Roche and Janssen. Dr. Matcheri Keshavan has received support from Sunovion. Dr. Carol Tamminga has received funding from Astellas, Eli Lilly, Intracellular Therapies, Lundback and Pure Tech Ventures.
Footnotes
Conflict of Interest: Other authors declare no financial interest in relation to the work described in this manuscript other than the grant funding.
References
- 1.Hill SK, Reilly JL, Harris MS, Rosen C, Marvin RW, Deleon O, Sweeney JA. A comparison of neuropsychological dysfunction in first-episode psychosis patients with unipolar depression, bipolar disorder, and schizophrenia. Schizophr Res. 2009;113:167–175. doi: 10.1016/j.schres.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chai XJ, Whitfield-Gabrieli S, Shinn AK, Gabrieli JD, Nieto Castanon A, McCarthy JM, Cohen BM, Ongur D. Abnormal medial prefrontal cortex resting-state connectivity in bipolar disorder and schizophrenia. Neuropsychopharmacology. 2011;36:2009–2017. doi: 10.1038/npp.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter WT, Bustillo JR, Thaker GK, van Os J, Krueger RF, Green MJ. The psychoses: cluster 3 of the proposed meta-structure for DSM-V and ICD-11. Psychol Med. 2009;39:2025–2042. doi: 10.1017/S0033291709990286. [DOI] [PubMed] [Google Scholar]
- 4.Bramon E, Sham PC. The common genetic liability between schizophrenia and bipolar disorder: a review. Curr Psychiatry Rep. 2001;3:332–337. doi: 10.1007/s11920-001-0030-1. [DOI] [PubMed] [Google Scholar]
- 5.Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, Nurnberger JI, Ripke S, Santangelo S, Sullivan PF. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eastwood SL, Harrison PJ. Hippocampal synaptic pathology in schizophrenia, bipolar disorder and major depression: a study of complexin mRNAs. Mol Psychiatry. 2000;5:425–432. doi: 10.1038/sj.mp.4000741. [DOI] [PubMed] [Google Scholar]
- 7.Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK, St Clair DM, Young AH, Ferrier N, Farmer AE, McGuffin P, Holmans PA, Owen MJ, O'Donovan MC, Craddock N. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2010;67:318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 10.Polich J. Updating P300: an integrative theory of P3a and P3b. Clin Neurophysiol. 2007;118:2128–2148. doi: 10.1016/j.clinph.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Beijsterveldt CE, van Baal GC, Molenaar PC, Boomsma DI, de Geus EJ. Stability of genetic and environmental influences on P300 amplitude: a longitudinal study in adolescent twins. Behav Genet. 2001;31:533–543. doi: 10.1023/a:1013389226795. [DOI] [PubMed] [Google Scholar]
- 12.O'Donnell BF, Vohs JL, Hetrick WP, Carroll CA, Shekhar A. Auditory event-related potential abnormalities in bipolar disorder and schizophrenia. Int J Psychophysiol. 2004;53:45–55. doi: 10.1016/j.ijpsycho.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Ethridge LE, Hamm JP, Shapiro JR, Summerfelt AT, Keedy SK, Stevens MC, Pearlson G, Tamminga CA, Boutros NN, Sweeney JA, Keshavan MS, Thaker G, Clementz BA. Neural activations during auditory oddball processing discriminating schizophrenia and psychotic bipolar disorder. Biol Psychiatry. 2012;72:766–774. doi: 10.1016/j.biopsych.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winterer G, Egan MF, Raedler T, Sanchez C, Jones DW, Coppola R, Weinberger DR. P300 and genetic risk for schizophrenia. Arch Gen Psychiatry. 2003;60:1158–1167. doi: 10.1001/archpsyc.60.11.1158. [DOI] [PubMed] [Google Scholar]
- 15.Turetsky BI, Cannon TD, Gur RE. P300 subcomponent abnormalities in schizophrenia: III. Deficits In unaffected siblings of schizophrenic probands. Biol Psychiatry. 2000;47:380–390. doi: 10.1016/s0006-3223(99)00290-5. [DOI] [PubMed] [Google Scholar]
- 16.Bestelmeyer PE, Phillips LH, Crombie C, Benson P, St Clair D. The P300 as a possible endophenotype for schizophrenia and bipolar disorder: Evidence from twin and patient studies. Psychiatry Res. 2009;169:212–219. doi: 10.1016/j.psychres.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 17.Schulze KK, Hall MH, McDonald C, Marshall N, Walshe M, Murray RM, Bramon E. Auditory P300 in patients with bipolar disorder and their unaffected relatives. Bipolar Disord. 2008;10:377–386. doi: 10.1111/j.1399-5618.2007.00527.x. [DOI] [PubMed] [Google Scholar]
- 18.Salisbury DF, Collins KC, McCarley RW. Reductions in the N1 and P2 auditory event-related potentials in first-hospitalized and chronic schizophrenia. Schizophr Bull. 2010;36:991–1000. doi: 10.1093/schbul/sbp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Donnell BF, Shenton ME, McCarley RW, Faux SF, Smith RS, Salisbury DF, Nestor PG, Pollak SD, Kikinis R, Jolesz FA. The auditory N2 component in schizophrenia: relationship to MRI temporal lobe gray matter and to other ERP abnormalities. Biol Psychiatry. 1993;34:26–40. doi: 10.1016/0006-3223(93)90253-a. [DOI] [PubMed] [Google Scholar]
- 20.Foxe JJ, Yeap S, Snyder AC, Kelly SP, Thakore JH, Molholm S. The N1 auditory evoked potential component as an endophenotype for schizophrenia: high-density electrical mapping in clinically unaffected first-degree relatives, first-episode, and chronic schizophrenia patients. Eur Arch Psychiatry Clin Neurosci. 2011;261:331–339. doi: 10.1007/s00406-010-0176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieuwenhuis S, Aston-Jones G, Cohen JD. Decision making, the P3, and the locus coeruleus-norepinephrine system. Psychol Bull. 2005;131:510–532. doi: 10.1037/0033-2909.131.4.510. [DOI] [PubMed] [Google Scholar]
- 22.Pogarell O, Padberg F, Karch S, Segmiller F, Juckel G, Mulert C, Hegerl U, Tatsch K, Koch W. Dopaminergic mechanisms of target detection - P300 event related potential and striatal dopamine. Psychiatry Res. 2011;194:212–218. doi: 10.1016/j.pscychresns.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Mulert C, Juckel G, Giegling I, Pogarell O, Leicht G, Karch S, Mavrogiorgou P, Moller HJ, Hegerl U, Rujescu D. A Ser9Gly polymorphism in the dopamine D3 receptor gene (DRD3) and event-related P300 potentials. Neuropsychopharmacology. 2006;31:1335–1344. doi: 10.1038/sj.npp.1300984. [DOI] [PubMed] [Google Scholar]
- 24.Tsai SJ, Yu YW, Chen TJ, Chen MC, Hong CJ. Association analysis for dopamine D3 receptor, dopamine D4 receptor and dopamine transporter genetic polymorphisms and P300 event-related potentials for normal young females. Psychiatr Genet. 2003;13:51–53. doi: 10.1097/00041444-200303000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Kiehl KA, Pearlson G, Perrone-Bizzozero NI, Eichele T, Calhoun VD. Genetic determinants of target and novelty-related event-related potentials in the auditory oddball response. Neuroimage. 2009;46:809–816. doi: 10.1016/j.neuroimage.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Decoster J, De Hert M, Viechtbauer W, Nagels G, Myin-Germeys I, Peuskens J, van Os J, van Winkel R. Genetic association study of the P300 endophenotype in schizophrenia. Schizophr Res. 2012;141:54–59. doi: 10.1016/j.schres.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Bramon E, Dempster E, Frangou S, Shaikh M, Walshe M, Filbey FM, McDonald C, Sham P, Collier DA, Murray R. Neuregulin-1 and the P300 waveform--a preliminary association study using a psychosis endophenotype. Schizophr Res. 2008;103:178–185. doi: 10.1016/j.schres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 28.Gallinat J, Bajbouj M, Sander T, Schlattmann P, Xu K, Ferro EF, Goldman D, Winterer G. Association of the G1947A COMT (Val(108/158)Met) gene polymorphism with prefrontal P300 during information processing. Biol Psychiatry. 2003;54:40–48. doi: 10.1016/s0006-3223(02)01973-x. [DOI] [PubMed] [Google Scholar]
- 29.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Demirci O, Calhoun VD. A parallel independent component analysis approach to Investigate genomic influence on brain function. IEEE Signal Process Lett. 2008;15:413–416. doi: 10.1109/LSP.2008.922513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meda SA, Narayanan B, Liu J, Perrone-Bizzozero NI, Stevens MC, Calhoun VD, Glahn DC, Shen L, Risacher SL, Saykin AJ, Pearlson GD. A large scale multivariate parallel ICA method reveals novel imaging-genetic relationships for Alzheimer's disease in the ADNI cohort. Neuroimage. 2012;60:1608–1621. doi: 10.1016/j.neuroimage.2011.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meda SA, Ruaño G, Windemuth A, O'Neil K, Berwise C, Dunn S, Boccaccio L, Narayanan B, Kocherla M, Sprooten E, Keshavan M, Tamminga CA, Sweeney JA, Clementz BA, Calhoun VD, Pearlson GD. Multivariate analysis reveals genetic associations of the resting state network in psychotic bipolar disorder and schizophrenia. Proc Natl Acad Sci U S A in Press. 2014 doi: 10.1073/pnas.1313093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamminga CA, Ivleva EI, Keshavan MS, Pearlson GD, Clementz BA, Witte B, Morris DW, Eliot A, Thaker GK, Sweeney JA. Clinical Phenotypes of Psychosis in the Bipolar and Schizophrenia Network on Intermediate phenotypes (B-SNIP) American Journal of psychiatry. 2012 doi: 10.1176/appi.ajp.2013.12101339. [DOI] [PubMed] [Google Scholar]
- 34.Leavitt VM, Molholm S, Ritter W, Shpaner M, Foxe JJ. Auditory processing in schizophrenia during the middle latency period (10-50 ms): high-density electrical mapping and source analysis reveal subcortical antecedents to early cortical deficits. J Psychiatry Neurosci. 2007;32:339–353. [PMC free article] [PubMed] [Google Scholar]
- 35.Liem F, Zaehle T, Burkhard A, Jancke L, Meyer M. Cortical thickness of supratemporal plane predicts auditory N1 amplitude. Neuroreport. 2012;23:1026–1030. doi: 10.1097/WNR.0b013e32835abc5c. [DOI] [PubMed] [Google Scholar]
- 36.Naatanen R, Picton T. The N1 wave of the human electric and magnetic response to sound: a review and an analysis of the component structure. Psychophysiology. 1987;24:375–425. doi: 10.1111/j.1469-8986.1987.tb00311.x. [DOI] [PubMed] [Google Scholar]
- 37.Naatanen R, Picton TW. N2 and automatic versus controlled processes. Electroencephalogr Clin Neurophysiol Suppl. 1986;38:169–186. [PubMed] [Google Scholar]
- 38.Polich J, Criado JR. Neuropsychology and neuropharmacology of P3a and P3b. Int J Psychophysiol. 2006;60:172–185. doi: 10.1016/j.ijpsycho.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 39.Dutt A, Ganguly T, Shaikh M, Walshe M, Schulze K, Marshall N, Constante M, McDonald C, Murray RM, Allin MP, Bramon E. Association between hippocampal volume and P300 event related potential in psychosis: support for the Kraepelinian divide. Neuroimage. 2012;59:997–1003. doi: 10.1016/j.neuroimage.2011.08.067. [DOI] [PubMed] [Google Scholar]
- 40.Mehlen P, Furne C. Netrin-1: when a neuronal guidance cue turns out to be a regulator of tumorigenesis. Cell Mol Life Sci. 2005;62:2599–2616. doi: 10.1007/s00018-005-5191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant A, Fathalli F, Rouleau G, Joober R, Flores C. Association between schizophrenia and genetic variation in DCC: a case-control study. Schizophr Res. 2012;137:26–31. doi: 10.1016/j.schres.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 42.Zhang C, Gao J, Zhang H, Sun L, Peng G. Robo2--slit and Dcc--netrin1 coordinate neuron axonal pathfinding within the embryonic axon tracts. J Neurosci. 2012;32:12589–12602. doi: 10.1523/JNEUROSCI.6518-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grant A, Hoops D, Labelle-Dumais C, Prevost M, Rajabi H, Kolb B, Stewart J, Arvanitogiannis A, Flores C. Netrin-1 receptor-deficient mice show enhanced mesocortical dopamine transmission and blunted behavioural responses to amphetamine. Eur J Neurosci. 2007;26:3215–3228. doi: 10.1111/j.1460-9568.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- 44.Srour M, Riviere JB, Pham JM, Dube MP, Girard S, Morin S, Dion PA, Asselin G, Rochefort D, Hince P, Diab S, Sharafaddinzadeh N, Chouinard S, Theoret H, Charron F, Rouleau GA. Mutations in DCC cause congenital mirror movements. Science. 2010;328:592. doi: 10.1126/science.1186463. [DOI] [PubMed] [Google Scholar]
- 45.Shirakawa O, Kitamura N, Lin XH, Hashimoto T, Maeda K. Abnormal neurochemical asymmetry in the temporal lobe of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2001;25:867–877. doi: 10.1016/s0278-5846(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 46.Courchet J, Polleux F. Sonic hedgehog, BOC, and synaptic development: new players for an old game. Neuron. 2012;73:1055–1058. doi: 10.1016/j.neuron.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Harwell CC, Parker PR, Gee SM, Okada A, McConnell SK, Kreitzer AC, Kriegstein AR. Sonic hedgehog expression in corticofugal projection neurons directs cortical microcircuit formation. Neuron. 2012;73:1116–1126. doi: 10.1016/j.neuron.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connor RM, Allen CL, Devine CA, Claxton C, Key B. BOC, brother of CDO, is a dorsoventral axon-guidance molecule in the embryonic vertebrate brain. J Comp Neurol. 2005;485:32–42. doi: 10.1002/cne.20503. [DOI] [PubMed] [Google Scholar]
- 49.Maeno-Hikichi Y, Chang S, Matsumura K, Lai M, Lin H, Nakagawa N, Kuroda S, Zhang JF. A PKC epsilon-ENH-channel complex specifically modulates N-type Ca2+ channels. Nat Neurosci. 2003;6:468–475. doi: 10.1038/nn1041. [DOI] [PubMed] [Google Scholar]
- 50.Remedios R, Subramanian L, Tole S. LIM genes parcellate the embryonic amygdala and regulate its development. J Neurosci. 2004;24:6986–6990. doi: 10.1523/JNEUROSCI.0001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jelen F, Oleksy A, Smietana K, Otlewski J. PDZ domains - common players in the cell signaling. Acta Biochim Pol. 2003;50:985–1017. [PubMed] [Google Scholar]
- 52.Lang B, Alrahbeni TM, Clair DS, Blackwood DH, McCaig CD, Shen S. HDAC9 is implicated in schizophrenia and expressed specifically in post-mitotic neurons but not in adult neural stem cells. Am J Stem Cells. 2011;1:31–41. [PMC free article] [PubMed] [Google Scholar]
- 53.Sugo N, Oshiro H, Takemura M, Kobayashi T, Kohno Y, Uesaka N, Song WJ, Yamamoto N. Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur J Neurosci. 2010;31:1521–1532. doi: 10.1111/j.1460-9568.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- 54.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 55.Kurita M, Holloway T, Garcia-Bea A, Kozlenkov A, Friedman AK, Moreno JL, Heshmati M, Golden SA, Kennedy PJ, Takahashi N, Dietz DM, Mocci G, Gabilondo AM, Hanks J, Umali A, Callado LF, Gallitano AL, Neve RL, Shen L, Buxbaum JD, Han MH, Nestler EJ, Meana JJ, Russo SJ, Gonzalez-Maeso J. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neurosci. 2012;15:1245–1254. doi: 10.1038/nn.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Bostrom E, Westerlund I, Vial C, Buchholz BA, Possnert G, Mash DC, Druid H, Frisen J. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisch AJ, Cameron HA, Encinas JM, Meltzer LA, Ming GL, Overstreet-Wadiche LS. Adult neurogenesis, mental health, and mental illness: hope or hype? J Neurosci. 2008;28:11785–11791. doi: 10.1523/JNEUROSCI.3798-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toro CT, Deakin JF. Adult neurogenesis and schizophrenia: a window on abnormal early brain development? Schizophr Res. 2007;90:1–14. doi: 10.1016/j.schres.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 59.Stanescu L, Georgescu G, Zaharescu J, Giosanu M, Fica V. Dynamic aspects of blood gastrin after antral stimulation. Rev Med Interna Neurol Psihiatr Neurochir Dermatovenerol Med Interna. 1978;30:311–318. [PubMed] [Google Scholar]
- 60.Keshavan MS, Mallinger AG, Pettegrew JW, Dippold C. Erythrocyte membrane phospholipids in psychotic patients. Psychiatry Res. 1993;49:89–95. doi: 10.1016/0165-1781(93)90032-c. [DOI] [PubMed] [Google Scholar]
- 61.Walss-Bass C, Soto-Bernardini MC, Johnson-Pais T, Leach RJ, Ontiveros A, Nicolini H, Mendoza R, Jerez A, Dassori A, Chavarria-Siles I, Escamilla MA, Raventos H. Methionine sulfoxide reductase: a novel schizophrenia candidate gene. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:219–225. doi: 10.1002/ajmg.b.30791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X, Deng W, Liu X, Li M, Chen Z, He Z, Wang Y, Wang Q, Hu X, Collier DA, Li T. A genome-wide association study for quantitative traits in schizophrenia in China. Genes Brain Behav. 2011;10:734–739. doi: 10.1111/j.1601-183X.2011.00712.x. [DOI] [PubMed] [Google Scholar]
- 63.Yao JK, Keshavan MS. Antioxidants, redox signaling, and pathophysiology in schizophrenia: an integrative view. Antioxid Redox Signal. 2011;15:2011–2035. doi: 10.1089/ars.2010.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flatow J, Buckley P, Miller BJ. Meta-Analysis of Oxidative Stress in Schizophrenia. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stahl SM. Beyond the dopamine hypothesis to the NMDA glutamate receptor hypofunction hypothesis of schizophrenia. CNS Spectr. 2007;12:265–268. doi: 10.1017/s1092852900021015. [DOI] [PubMed] [Google Scholar]
- 66.Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, Zambuto CT, Schmitt K, Matise TC, Harkavy Friedman JM, Hampe C, Lee H, Shore D, Wynne D, Faraone SV, Tsuang MT, Cloninger CR. NIMH Genetics Initiative Millenium Schizophrenia Consortium: linkage analysis of African-American pedigrees. Am J Med Genet. 1998;81:282–289. [PubMed] [Google Scholar]
- 67.Ophoff RA, Escamilla MA, Service SK, Spesny M, Meshi DB, Poon W, Molina J, Fournier E, Gallegos A, Mathews C, Neylan T, Batki SL, Roche E, Ramirez M, Silva S, De Mille MC, Dong P, Leon PE, Reus VI, Sandkuijl LA, Freimer NB. Genomewide linkage disequilibrium mapping of severe bipolar disorder in a population isolate. Am J Hum Genet. 2002;71:565–574. doi: 10.1086/342291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsumoto J, Sugiura Y, Yuki D, Hayasaka T, Goto-Inoue N, Zaima N, Kunii Y, Wada A, Yang Q, Nishiura K, Akatsu H, Hori A, Hashizume Y, Yamamoto T, Ikemoto K, Setou M, Niwa S. Abnormal phospholipids distribution in the prefrontal cortex from a patient with schizophrenia revealed by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal Bioanal Chem. 2011;400:1933–1943. doi: 10.1007/s00216-011-4909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Law MH, Cotton RG, Berger GE. The role of phospholipases A2 in schizophrenia. Mol Psychiatry. 2006;11:547–556. doi: 10.1038/sj.mp.4001819. [DOI] [PubMed] [Google Scholar]
- 70.Ross BM, Hughes B, Kish SJ, Warsh JJ. Serum calcium-independent phospholipase A2 activity in bipolar affective disorder. Bipolar Disord. 2006;8:265–270. doi: 10.1111/j.1399-5618.2006.00299.x. [DOI] [PubMed] [Google Scholar]
- 71.Smesny S, Schmelzer CE, Hinder A, Kohler A, Schneider C, Rudzok M, Schmidt U, Milleit B, Milleit C, Nenadic I, Sauer H, Neubert RH, Fluhr JW. Skin Ceramide Alterations In First-Episode Schizophrenia Indicate Abnormal Sphingolipid Metabolism. Schizophr Bull. 2012 doi: 10.1093/schbul/sbs058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 73.Lee BD, Walss-Bass C, Thompson PM, Dassori A, Montero PA, Medina R, Contreras S, Armas R, Ramirez M, Pereira M, Salazar R, Leach RJ, Quezada P, Raventos H, Escamilla MA. Malic enzyme 2 and susceptibility to psychosis and mania. Psychiatry Res. 2007;150:1–11. doi: 10.1016/j.psychres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, Zhang H, Ju G, Zhang X, Xu Q, Liu S, Yu Y, Shi J, Boyle S, Wang Z, Shen Y, Wei J. A study of the PEMT gene in schizophrenia. Neurosci Lett. 2007;424:203–206. doi: 10.1016/j.neulet.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 75.Paoletti L, Elena C, Domizi P, Banchio C. Role of phosphatidylcholine during neuronal differentiation. IUBMB Life. 2011;63:714–720. doi: 10.1002/iub.521. [DOI] [PubMed] [Google Scholar]
- 76.Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yao PJ, Coleman PD, Calkins DJ. High-resolution localization of clathrin assembly protein AP180 in the presynaptic terminals of mammalian neurons. J Comp Neurol. 2002;447:152–162. doi: 10.1002/cne.10217. [DOI] [PubMed] [Google Scholar]
- 78.Cho JH, Oh DY, Kim HJ, Park SY, Choi HJ, Kwon SJ, Lee KS, Han JS. The TSP motif in AP180 inhibits phospholipase D1 activity resulting in increased efficacy of anticancer drug via its direct binding to carboxyl terminal of phospholipase D1. Cancer Lett. 2011;302:144–154. doi: 10.1016/j.canlet.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 79.Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 80.Goes FS, Hamshere ML, Seifuddin F, Pirooznia M, Belmonte-Mahon P, Breuer R, Schulze T, Nothen M, Cichon S, Rietschel M, Holmans P, Zandi PP, Craddock N, Potash JB. Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl Psychiatry. 2012;2:e180. doi: 10.1038/tp.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mayilyan KR, Weinberger DR, Sim RB. The complement system in schizophrenia. Drug News Perspect. 2008;21:200–210. doi: 10.1358/dnp.2008.21.4.1213349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Havik B, Le Hellard S, Rietschel M, Lybaek H, Djurovic S, Mattheisen M, Muhleisen TW, Degenhardt F, Priebe L, Maier W, Breuer R, Schulze TG, Agartz I, Melle I, Hansen T, Bramham CR, Nothen MM, Stevens B, Werge T, Andreassen OA, Cichon S, Steen VM. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol Psychiatry. 2011;70:35–42. doi: 10.1016/j.biopsych.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 83.Consortium SWGotPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stephan AH, Barres BA, Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- 85.Maier M, Peng Y, Jiang L, Seabrook TJ, Carroll MC, Lemere CA. Complement C3 deficiency leads to accelerated amyloid beta plaque deposition and neurodegeneration and modulation of the microglia/macrophage phenotype in amyloid precursor protein transgenic mice. J Neurosci. 2008;28:6333–6341. doi: 10.1523/JNEUROSCI.0829-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weksler ME, Gouras G, Relkin NR, Szabo P. The immune system, amyloid-beta peptide, and Alzheimer's disease. Immunol Rev. 2005;205:244–256. doi: 10.1111/j.0105-2896.2005.00264.x. [DOI] [PubMed] [Google Scholar]
- 87.Rudduck C, Beckman L, Franzen G, Lindstrom L. C3 and C6 complement types in schizophrenia. Hum Hered. 1985;35:255–258. doi: 10.1159/000153555. [DOI] [PubMed] [Google Scholar]
- 88.Boulanger LM. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64:93–109. doi: 10.1016/j.neuron.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Owen MJ, O'Donovan MC, Thapar A, Craddock N. Neurodevelopmental hypothesis of schizophrenia. Br J Psychiatry. 2011;198:173–175. doi: 10.1192/bjp.bp.110.084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Green MF, Gur RE, Gur RC, Hardiman G, Kelsoe JR, Leonard S, Light GA, Nuechterlein KH, Olincy A, Radant AD, Schork NJ, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Freedman R, Braff DL. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2011;168:930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenwood TA, Light GA, Swerdlow NR, Radant AD, Braff DL. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PLoS One. 2013;7:e29630. doi: 10.1371/journal.pone.0029630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O'Dushlaine C, Kenny E, Heron E, Donohoe G, Gill M, Morris D, Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 95.Juraeva D, Haenisch B, Zapatka M, Frank J, Witt SH, Muhleisen TW, Treutlein J, Strohmaier J, Meier S, Degenhardt F, Giegling I, Ripke S, Leber M, Lange C, Schulze TG, Mossner R, Nenadic I, Sauer H, Rujescu D, Maier W, Borglum A, Ophoff R, Cichon S, Nothen MM, Rietschel M, Mattheisen M, Brors B. Integrated pathway-based approach identifies association between genomic regions at CTCF and CACNB2 and schizophrenia. PLoS Genet. 2014;10:e1004345. doi: 10.1371/journal.pgen.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jia P, Wang L, Meltzer HY, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure SF1 Schematic of 64-channel EEG montage used for data collection.
Supplementary Figure SF2 Schematic depiction of the processing pipeline for quality control (QC) of single nucleotide polymorphism (SNP) data.
The QC process involved two stages: individual based followed by SNP-based QC.
Poorly genotyped individuals and bad SNPs were removed from the final regression analysis.
SNP data were corrected for stratification bias by correcting for top 3 Eigen factors associated with self-reported ethnicity.
LD= linkage disequilibrium
Supplementary Figure SF3 Q-Q plot of theoretical and empirical p-values from logistic regression for (A) schizophrenia (SZ) vs healthy comparison subjects (HC) and (B) psychotic bipolar disorder (PBP) vs HC.
Logistic regression was applied to individual markers in case-control fashion for both SZ and PBP groups.
Supplementary Figure SF4 Schematic illustration of the parallel independent component analysis (Para-ICA) for genetic association of event-related potential (ERP) data.
Data were constructed as a matrix of subjects by SNP (K=449 × M=20,329) and subjects by reduced ERP waveforms (K=449 × P=294 (3×98 time points)). The number of components extracted from Para-ICA for the ERP and SNP data were N1 = 8 and N2 = 11 respectively.
LC=loading coefficient; SNP= single nucleotide polymorphism
Supplementary Table ST1: Demographic information for schizophrenia, psychotic bipolar probands and comparison subjects.
Supplementary Table ST2: Medication information for probands
Supplementary Table ST3: Results of enrichment analysis including GeneGo pathway maps, process networks, metabolic networks and gene ontology processes associated with contributing genes from components G1, G4 and G9. Bold P-values indicate significant (p<0.05) after false discovery rate (FDR) correction.
Supplementary Table ST4: Pathways, disease risk and brain regions associated with top 20 genes from gene components G1, G4 and G9. Expression Z-scores from Allen brain atlas database was used to identify brain regions. Other mental disorders associated with these genes from prior studies are also provided (references in online supplementary text). Gene information was obtained from Dbsnp and Genecard.