Abstract
Cyclospora cayetanensis is a human-specific coccidian parasite responsible for several food and water-related outbreaks around the world, including the most recent ones involving over 900 persons in 2013 and 2014 outbreaks in the USA. Multicopy organellar DNA such as mitochondrion genomes have been particularly informative for detection and genetic traceback analysis in other parasites. We sequenced the C. cayetanensis genomic DNA obtained from stool samples from patients infected with Cyclospora in Nepal using the Illumina MiSeq platform. By bioinformatically filtering out the metagenomic reads of non-coccidian origin sequences and concentrating the reads by targeted alignment, we were able to obtain contigs containing Eimeria-like mitochondrial, apicoplastic and some chromosomal genomic fragments. A mitochondrial genomic sequence was assembled and confirmed by cloning and sequencing targeted PCR products amplified from Cyclospora DNA using primers based on our draft assembly sequence. The results show that the C. cayetanensis mitochondrion genome is 6274 bp in length, with 33% GC content, and likely exists in concatemeric arrays as in Eimeria mitochondrial genomes. Phylogenetic analysis of the C. cayetanensis mitochondrial genome places this organism in a tight cluster with Eimeria species. The mitochondrial genome of C. cayetanensis contains three protein coding genes, cytochrome (cytb), cytochrome C oxidase subunit 1 (cox1), and cytochrome C oxidase subunit 3 (cox3), in addition to 14 large subunit (LSU) and nine small subunit (SSU) fragmented rRNA genes.
Introduction
C. cayetanensis is an emerging human pathogen causing gastrointestinal disease in humans around the world and is acquired through food and waterborne transmission, often by consumption of contaminated fresh produce [1–4]. In the United States, Cyclospora was the etiological agent of recent food-borne outbreaks affecting more than nine hundred people in 2013 and 2014 (http://www.cdc.gov/parasites/cyclosporiasis/outbreaks/index.html). Cyclospora belongs to the phylum Apicomplexa, which is a large group of protists, related to dinoflagellates and ciliates [5]. Other members of Apicomplexa include Plasmodium, Babesia, Theileria, Toxoplasma, Eimeria, and Cryptosporidium.
Mitochondria are organelles essential for cellular processes such as energy metabolism, signaling, cellular differentiation and cell death [6]. Mitochondria contain their own genomes referred to as mitochondrial (mt) genomes [7,8]. Most apicomplexan species possess mt genomes of ~6kb in size with considerable variability in structure and organization [9]. The genomes of apicomplexan mitochondria encode three mt protein coding genes (cytochrome c oxidase subunits, cox1 and cox3, and cytochrome b, cob), in addition to highly fragmented rRNA genes. These mt genomes exist as either monomeric or concatemeric linear forms [10]. Among the apicomplexan organisms, Eimeria is the phylogenetically closest genus to Cyclospora based on 18S gene sequences [11] [12]. The complete mitochondrial genome sequences for 13 Eimeria species have been reported [13–15], all of which range from 6.1 to 6.4 kb in size with highly conserved content and structure [13].
Despite the significant clinical and public health importance of C. cayetanensis, very little sequence information is available for this organism. Lack of a laboratory culture method for Cyclospora [16] has hampered the ability to obtain large numbers of highly purified oocysts and sufficient Cyclospora DNA for genomics studies. Nevertheless, because of recent advances in next generation sequencing technologies, we performed whole genome sequencing (WGS) of C. cayetanenesis using genomic DNA samples extracted from oocysts purified from human fecal samples. The sequence output was metagenomic in nature, but we were able to obtain contigs containing Eimeria-like mitochondrial, apicoplastic and some chromosomal genomic fragments by bioinformatically filtering out the reads of non-coccidian origin sequences.
Here we report the sequence, gene organization, and structure of the C. cayetanensis mt genome from Cyclospora originating in Nepal. Our data show that the C. cayetanensis mitochondrial genome is organized as a concatemeric linear 6.3 kb molecule which is closely related to the mt genomes of Eimeria species.
Materials and Methods
Cyclospora
Human stool samples containing C. cayetanensis oocysts were obtained from the Microbiology and Public Health Research Laboratory at Tribhuvan University Teaching Hospital in Kathmandu, Nepal and the University of Georgia in Athens, Georgia, USA (This study was reviewed and approved by Institutional review Board of FDA, RIHSC- ID#10-095F). Cyclospora oocysts were purified by a method similar to the one used for Cryptosporidium [17](Arrowood and Donaldson 1996). Briefly, Cyclospora oocysts were recovered from sieved fecal samples by differential sucrose and cesium chloride gradient centrifugations. Cyclospora oocysts were counted using a haemocytometer and a Zeiss Axio Imager D1 microscope with an HBO mercury short arc lamp and a UV filter (350 nm excitation and 450 nm emission).
Sequencing
Genomic DNA was isolated from purified C. cayetanensis oocyst preparations using the ZR Fecal DNA MiniPrep kit (Zymo Research, Irvine, CA) following the manufacturer’s instructions. DNA concentration was measured with a Qubit 1.0 Fluorimeter using the Qubit dsDNA HS Assay Kit (Life Technologies, Grand Island, NY). Shotgun sequencing of genomic DNA isolated from three oocyst preparations purified from two different fecal samples was performed on the Illumina MiSeq platform (Illumina, San Diego, CA) using the Nextera XT library preparation kit (Illumina, San Diego, CA). Approximately 12 pmol of each library was paired-end sequenced (2X 250 cycles) on the MiSeq.
Bioinformatic analysis
Metagenomic profiling of the shotgun datasets was carried out by MetaPhlAn (PMID: 2688413) using default parameters [18]. A local database of apicomplexan and dinoflagellates genomes from Cryptosporidium, Perkinsus and Eimeria was created using sequences downloaded from NCBI (PMID: 25398906) and used for alignment with Bowtie2 [19]. This strategy filtered sequence reads specific to Apicomplexa from the metagenomic runs by collecting those sequences that mapped to the target apicomplexan genome database. CLCworkbench 6 (http://www.clcbio.com/products/clc-genomics-workbench/) was used to generate de novo assembly of sequences to obtain a final set of contigs of various lengths, and to map the filtered reads back to the apicomplexan genomic regions.
Confirmation of the mitochondrial genome structure via PCR amplification and re-sequencing
PCR reactions were performed using the Platinum PCR SuperMix High Fidelity kit (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instructions. Genomic DNA extracted from C. cayetanensis oocysts was used (100 pg) as the template. The PCR primers (Table 1) were designed to cover the entire sequence of the longest mitochondrial contig (6.3 kb) generated. The PCR products were gel purified using the QIAGEN Gel Extraction kit, and sequenced either directly or after cloning using the TOPO TA cloning kit with One Shot TOP10 Chemically Competent E.coli (Invitrogen, USA). DNA sequences were trimmed and assembled using the CodonCode Aligner version 5.0 (CodonCode Corporation, Centerville, MA). WebACT tool [20], a web version of Artemis genome comparison tool [21] was used to visualize the mappings of PCR products to the genome assemblies generated by NCBI BLAST.
Table 1. Primer sequences used for PCR.
| Primers | Sequence (5’-3’) |
|---|---|
| 1F | GGTACTACATCAGCTTCTATGG |
| 1R | GCGTGGAAAGGTTCCCGTGAG |
| 3Fb | GTTGGAGCTCAATTACCTCAAGAAG |
| 3Rb | TTTGTATGGATTTCACGGTCAACTC |
| 4Fb | CCATTTCGTACTATCTCTTGGTGCG |
| 4Rb | CTAATACAGTGAGCAAGAATGGTGA |
| 4B-F | GCTGGAAGACGGAATCGTTAC |
| 4B-R | CAAATAGTAGTAACTCAGTAC |
| 2F-begin | GACAAAGTCCCAGCAGTAGC |
| 2R | CTTTATGGTTTGCAGCAGTTAG |
| 5F | CAGGAGCTCTGATCCTTGAATTCTAT |
| 5R | GCACAAGCACTCAGCAAGTTAAGAGAATG |
| End6R | CACATGATGCTCCAGTAGCATGTAGG |
| 2Fb | CATTCATGCAGGACGGAATTTACC |
| End5R | GCAATCGGATCGTGTTGGCTAGGTGTAC |
| End1F | GGCGTGTAGAGCATCTTTATCTAGGTAG |
| End2F | CTGTCCGAAACAATAACCTTAGGC |
| End3F | CTACCAAAGCATCCATCTACAGCTG |
| End4F | GCATCCATCTACAGCTGCGGAAACTG |
Mitochondrial sequence analysis
The mt genome sequences for thirteen Eimeria species and P. falciparum as an outlier were downloaded from NCBI and used for comparative analysis. The accession numbers of the sequences used are as follows: E. acervulina-HQ702479; E. meleagridis-KJ608418; E. meleagrimitis-KJ608414; E. dispersa-KJ608416; E. adenoeides-KJ608415; E. gallopavonis-KJ608413; E. magna-KF419217; E. mitis-JN864949; E. tenella-HQ702484; E. praecox-HQ702483; E. necatrix-HQ702482; E. maxima-HQ702481; E. brunetti- HQ702480; and P. falciparaum NF54-AJ27684.
The 5’ and 3’ ends of the C. cayetanensis mitochondrial sequences were manually curated by comparing the initial assembly with complete mitochondrial genomes from Eimeria as reference to obtain the 6,274 bp long complete sequence. The Eimeria and Cyclospora mt sequences were aligned with MEGA6 suite [22] using ClustalW [23]. A phylogenetic tree was built using using default parameters in Maximum Likelihood algorithm options for the 14 genomes. Mitochondrial genome from P.falciparum was initially used as an outgroup. Test of phylogeny was conducted on Mega 6 using 500 replicates for bootstrap analysis. The 6274 bp long mitochondrial genome from C. cayatenensis was annotated using Genbank record KJ608417 submitted to NCBI and assigned accession number KP231180.
Results and Discussion
Assembly of C. cayetanensis mitochondrial genome sequence
Metaphlan analysis of the quality-filtered data revealed that more than 98% of our reads mapped to fecal bacterial sequences. To identify the apicomplexan sequences, we first designed a custom database with genome sequences from Eimeria, Perkinsus and other related apicomplexans, and mapped our sequence reads against this database identifying 47,000 apicomplexan-specific reads that assemble into 482 contigs. From these, we identified a 6.3 kb long sequence which is highly similar to those mt sequences from Eimeria (average BLASTn score 91–93%).
The complete sequence of C. cayatenensis mitochondrial genome
By aligning the initial Cyclospora 6.3 kb contig assembly with the E. tenella mt genome, we were able to delineate the terminal bases of the molecule in silico. Next, we sequenced the PCR products amplified from Cyclospora oocysts using seven primer sets covering the whole 6.3 kb-long assembled sequence (Fig 1 and Table 2), confirming and closing the complete C. cayatenensis mt genome (Acc: KP231180). When compared with Eimeria mt sequences using ClustalW, the Cyclospora mt genome aligned well with 13 available Eimerid mitochondrial genomes in sequence and organization. In particular, a front block of about 200 bases and a large stretch of sequences towards the end are highly conserved in Eimeria and Cyclospora genomes, suggesting a common ancestral mt genome (S1 File).
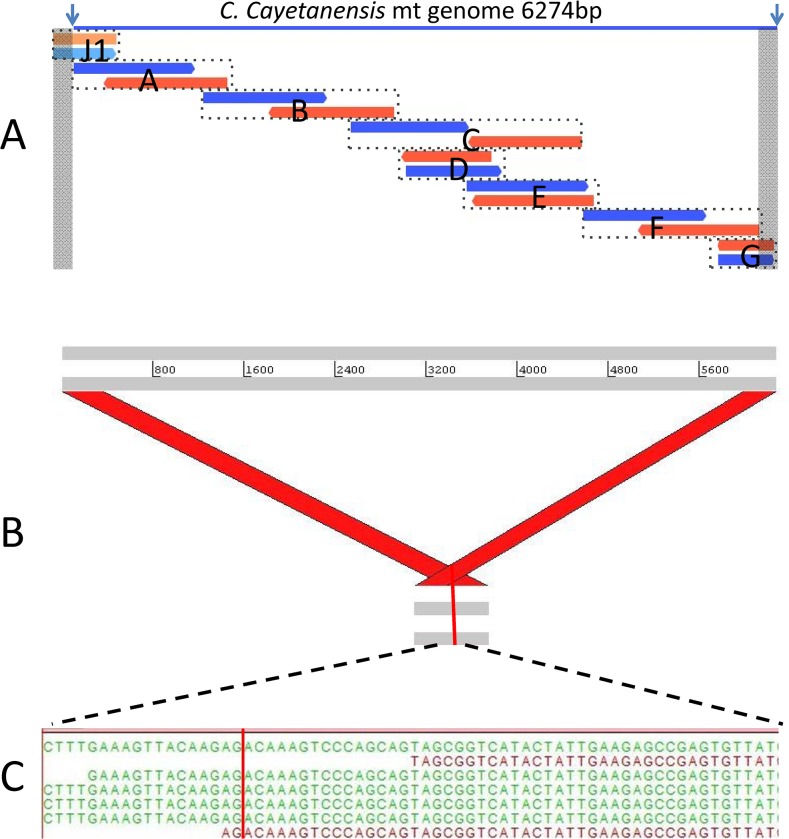
Fig 1. Confirmation of the C. Cayetanensis mt genome sequence.
A) Overlapping PCR fragments used to confirm the C. cayetanenesis mt genome sequence and concatemeric structure. PCR fragments are shown with dotted lines. Forward sequence reads are shown in blue or orange and reverse reads are shown in red or light blue. Terminal grey bands indicate identical sequence regions. B) A 659 bp contig constructed using overlapping junction PCR fragment sequences (see Table 2) is represented in gray. Diagonal red bars represent portions of the junction PCR product mapping to the ends of the complete C. cayetanensis mt genome. C) WGS reads spanning the junction region in the initial mt assembly. The vertical red lines mark the tail:head junction.
Table 2. PCR primer sets and amplicons.
| Full length mt genome PCR | |||
| Amplicons | Forward Primer | Reverse Primer | Amplicon size (bp) |
| A | 2F-Begin | 2R | 1393 |
| B | 3Fb | 3Rb | 1609 |
| C | 4Fb | 4Rb | 1984 |
| D | 5F | 5R | 901 |
| E | 4B-F | 4B-R | 1022 |
| F | 1F | 1R | 1507 |
| G | 2Fb | End5-R | 458 |
| Junction PCR | |||
| J1 | End1-F | End6-R | 634 |
| J2 | 2Fb | End5-R | 458 |
| J3 | End2-F | End6-R | 603 |
| J4 | End3-F | End6-R | 541 |
| J5 | End4-F | End6-R | 535 |
In the phylum Apicomplexa, mitochondrial genomes have either concatemeric or monomeric linear structures as represented by Eimeria, and Piroplasms, respectively [10]. To determine the C. cayetanensis mt genome structure, we amplified, sequenced and assembled the PCR products designed to span the tail to head junction (Table 2, Fig 1A). The resulting 659 bp sequence mapped to the terminal ends of the C. cayatenensis mt genome as expected for a structure with tail to head junction (Fig 1B). We were able to identify a few apicomplexan-specific metagenomic reads that spanned across this junction region as shown in the Fig 1C. Our results are consistent with a concatemeric structure that is the hallmark of the closely related Eimeria mt genomes; however, we cannot rule out a circular mt genome structure.
The complete C. cayetanensis mt genome is 6,274 bp, with a base composition of A (30%), T (36%), C (16%), and G (17%). Based on the available annotations in the NCBI for Eimeria gallopavonis, Cyclospora mitochondrian genome was annotated after aligning the two genomes. The C. cayetanensis mt genome encodes three protein-coding genes; cytb, 1080 bp [128 bp-1207 bp], ATG start codon; cox1, 1443 bp [1248 bp- 2690 bp], GTT start codon; and cox3, 780 bp [4226 bp- 5005 bp], ATT start codon. In addition to these three protein-coding genes, twelve large-subunit (LSU), and seven small-subunit (SSU) fragmented rRNA genes are present in the mt genome (Fig 2). Pairwise amino acid sequence alignments between the individual protein coding genes of C. cayetanensis mt genome, and the corresponding coding genes of seventeen other published Eimeria mt genomes revealed sequence identities that ranged from 90–97% for cytbB, 93–97% for cox1, and 83–93% for cox3.
Fig 2. Gene arrangement of the C. cayetanensis mt genome.
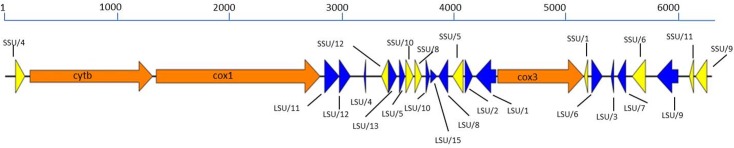
Predicted genes on the Cyclospora mitochondrial genomes were derived based on ClustalW alignment with the Eimeria gallopavonis (KJ608417). Orange indicates coding genes, blue indicates fragments of LSU rRNA, and yellow indicates fragments of SSU rRNA. Transcriptional direction is indicated by arrowed end.
Phylogeny
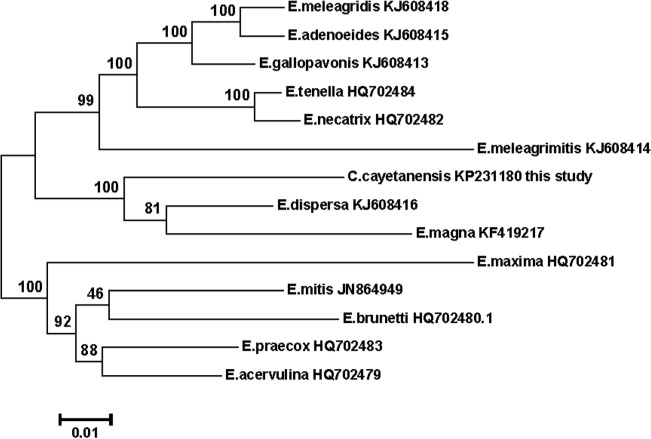
Mt genomes from thirteen species of Eimeria and from a strain NF54 of P. falciparum were aligned with the C. cayetenensis mt genome for Maximum Likelihood (ML) phylogenetic reconstruction analysis (Fig 3). The mt genome nucleotide phylogeny of C. cayetenensis supports a closer relationship to Eimeria species E. magna and E. dispersa that infect rabbits and turkeys, respectively. C. cayetenensis appears to infect only humans [16] and, notably, we found that the C. cayetenensis mt sequences fall into a clade that contains the rabbit infecting E. magna (Fig 3). Avian infecting Eimeria species in our comparison clustered into two other distinct clades (Fig 3). The cladistic distribution of the two groups of Eimeria spp. followed mostly the pattern described in Barta 2014 [15]. The ‘turkey Eimerids’ that infect the caeca of turkeys (E. meleagridis, E. meleagrimitis, E. adenoeides, and E. gallopavonis) formed a clade that included E. tenella and E. necatrix (chicken Eimerids that infect the caeca) as sister groups. The other ‘chicken Eimerids’ (E. mitis, E. praecox, E.acervulina, E.maxima and E. brunette) grouped together. C. cayatenensis clustered together with rabbit Eimerid, E.magna, and a turkey infecting strain, E. dispersa. Previous studies using 18S rRNA [24–26]and ITS-1 sequences [27]place C. cayetanensis into a clade with chicken eimerids. We constructed a phylogenetic tree using currently available Eimeria spp.18S rRNA gene sequences, and C. cayetanensis 18S rRNA gene (S1 Fig). In our 18S rRNA bootstrap analysis, C. cayetanensis appears to cluster with E. meleagrimitis. Mammalian coccidians E. magna and E. bovis are found to be closely related to each other, but located in a clade different from C. cayetanensis tree location. The monophyletic grouping of mammalian Coccidians (Cyclospora and E. magna) in mitochondrial phylogenetic tree warrants further analysis, though beyond the scope of this work, preferably using apicomplexan apicoplastic and chromosomal genome sequences.
Fig 3. Phylogenetic relationships among Eimeria species and C. cayetanensis based on mitochondrial genome sequences.
NCBI accession numbers are included. The evolutionary relationship between the Eimeria (13), Cyclospora(1) and P.falciparum (1) mitochondrial genomes was inferred using the Maximum Likelihood method with 500 replications for bootstrap analysis. The confidence value for clustering is given as percentage above the branches. The scale bar points to the number of substitutions per site. After analysis, the outgroup branch was removed for clarity. Default parameters in MEGA6 were retained for phylogenetic analysis and tree building.
Conclusions
Our data suggest that the C. cayetanensis mt genome is a 6.3 kb linear molecule with a concatemeric structure. The content and order of protein-coding, and rRNA genes are highly conserved with those found in Eimera spp. The Cyclospora mt genome shows a close phylogenetic relationship with that of mammalian-infecting E. magna. Our study potentially opens the way for studies aimed toward development of organellar genome single nucleotide polymorphism (SNP) based trace-back assays for investigation of Cyclospora outbreaks, an approach successfully employed for the epidemiology of P. falciparum [28–30].
Supporting Information
18S rRNA sequence files from NCBI were used to infer evolutionary relationship between 22 Eimeria spp. and C. cayatenensis strain. The bootstrap analysis was carried out using the Maximum Likelihood method with 500 replications. The confidence value for the resultant clusters is given as percentage above the branches. The scale bar points to the number of substitutions per site. The analysis and the tree building were carried out using default parameters in MEGA 6 software.
(TIF)
87 bp in some of the Eimeria sequences were cut and appended to the 3’ end of the respective sequence files. ClustalW identified blocks of highly conserved 5’ and 3’ ends of Eimeria and Cyclospora mitochondrial sequences.
(TXT)
Acknowledgments
Authors thank Ben Tall and Hulusi Cinar for their comments and valuable suggestions on the manuscript. We also thank Ynes Ortega for helpful discussions regarding oocyst purification methods.
Data Availability
All relevant data are within the paper and its Supporting Information files. Cyclospora cayetanensis complete mitochondrial genome data are deposited at NCBI database (accession number KP231180).
Funding Statement
The authors received no specific funding for this research.
References
- 1. Ortega YR, Sterling CR, Gilman RH, Cama VA, Diaz F (1993) Cyclospora species—a new protozoan pathogen of humans. N Engl J Med 328: 1308–1312. 10.1056/NEJM199305063281804 [DOI] [PubMed] [Google Scholar]
- 2. Ortega YR, Gilman RH, Sterling CR (1994) A new coccidian parasite (Apicomplexa: Eimeriidae) from humans. J Parasitol 80: 625–629. [PubMed] [Google Scholar]
- 3. Sterling CR, Ortega YR (1999) Cyclospora: an enigma worth unraveling. Emerg Infect Dis 5: 48–53. 10.3201/eid0501.990106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ortega YR, Sanchez R (2010) Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin Microbiol Rev 23: 218–234. 23/1/218; 10.1128/CMR.00026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fast NM, Xue L, Bingham S, Keeling PJ (2002) Re-examining alveolate evolution using multiple protein molecular phylogenies. J Eukaryot Microbiol 49: 30–37. [DOI] [PubMed] [Google Scholar]
- 6. McBride HM, Neuspiel M, Wasiak S (2006) Mitochondria: more than just a powerhouse. Curr Biol 16: R551–R560. S0960-9822(06)01781-7; 10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- 7. Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. [DOI] [PubMed] [Google Scholar]
- 8. Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767–1780. gkc319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gray MW, Lang BF, Burger G (2004) Mitochondria of protists. Annu Rev Genet 38: 477–524. 10.1146/annurev.genet.37.110801.142526 [DOI] [PubMed] [Google Scholar]
- 10. Hikosaka K, Kita K, Tanabe K (2013) Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol 188: 26–33. S0166-6851(13)00031-5; 10.1016/j.molbiopara.2013.02.006 [DOI] [PubMed] [Google Scholar]
- 11. Relman DA, Schmidt TM, Gajadhar A, Sogin M, Cross J, Yoder K et al. (1996) Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J Infect Dis 173: 440–445. [DOI] [PubMed] [Google Scholar]
- 12. Pieniazek NJ, Herwaldt BL (1997) Reevaluating the molecular taxonomy: is human-associated Cyclospora a mammalian Eimeria species? Emerg Infect Dis 3: 381–383. 10.3201/eid0303.970319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hikosaka K, Nakai Y, Watanabe Y, Tachibana S, Arisue N, Palacpac NM et al. (2011) Concatenated mitochondrial DNA of the coccidian parasite Eimeria tenella. Mitochondrion 11: 273–278. S1567-7249(10)00187-X; 10.1016/j.mito.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 14. Lin RQ, Qiu LL, Liu GH, Wu XY, Weng YB, Xie WQ et al. (2011) Characterization of the complete mitochondrial genomes of five Eimeria species from domestic chickens. Gene 480: 28–33. S0378-1119(11)00094-1; 10.1016/j.gene.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 15. Ogedengbe ME, El-Sherry S, Whale J, Barta JR (2014) Complete mitochondrial genome sequences from five Eimeria species (Apicomplexa; Coccidia; Eimeriidae) infecting domestic turkeys. Parasit Vectors 7: 335 1756-3305-7-335; 10.1186/1756-3305-7-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eberhard ML, Ortega YR, Hanes DE, Nace EK, Do RQ, Robl MG et al. (2000) Attempts to establish experimental Cyclospora cayetanensis infection in laboratory animals. J Parasitol 86: 577–582. 10.1645/0022-3395(2000)0862.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 17. Arrowood MJ, Donaldson K (1996) Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol 43: 89S [DOI] [PubMed] [Google Scholar]
- 18. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9: 811–814. nmeth.2066; 10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. nmeth.1923; 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abbott JC, Aanensen DM, Bentley SD (2007) WebACT: an online genome comparison suite. Methods Mol Biol 395: 57–74. 1-59745-514-8:57. [PubMed] [Google Scholar]
- 21. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J (2005) ACT: the Artemis Comparison Tool. Bioinformatics 21: 3422–3423. bti553; 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. mst197; 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgins DG, Sharp PM (1988) CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244. 0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 24. Relman DA, Schmidt TM, Gajadhar A, Sogin M, Cross J, Yoder K et al. (1996) Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J Infect Dis 173: 440–445. [DOI] [PubMed] [Google Scholar]
- 25. Lopez FA, Manglicmot J, Schmidt TM, Yeh C, Smith HV, Relman DA (1999) Molecular characterization of Cyclospora-like organisms from baboons. J Infect Dis 179: 670–676. JID980940; 10.1086/314645 [DOI] [PubMed] [Google Scholar]
- 26. Eberhard ML, da Silva AJ, Lilley BG, Pieniazek NJ (1999) Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg Infect Dis 5: 651–658. 10.3201/eid0505.990506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olivier C, van de Pas S, Lepp PW, Yoder K, Relman DA (2001) Sequence variability in the first internal transcribed spacer region within and among Cyclospora species is consistent with polyparasitism. Int J Parasitol 31: 1475–1487. S0020-7519(01)00283-1. [DOI] [PubMed] [Google Scholar]
- 28. Conway DJ (2003) Tracing the dawn of Plasmodium falciparum with mitochondrial genome sequences. Trends Genet 19: 671–674. S0168-9525(03)00299-3; 10.1016/j.tig.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 29. Conway DJ (2007) Molecular epidemiology of malaria. Clin Microbiol Rev 20: 188–204. 20/1/188; 10.1128/CMR.00021-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Preston MD, Campino S, Assefa SA, Echeverry DF, Ocholla H, Amambua-Ngwa A et al. (2014) A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat Commun 5: 4052 ncomms5052; 10.1038/ncomms5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
18S rRNA sequence files from NCBI were used to infer evolutionary relationship between 22 Eimeria spp. and C. cayatenensis strain. The bootstrap analysis was carried out using the Maximum Likelihood method with 500 replications. The confidence value for the resultant clusters is given as percentage above the branches. The scale bar points to the number of substitutions per site. The analysis and the tree building were carried out using default parameters in MEGA 6 software.
(TIF)
87 bp in some of the Eimeria sequences were cut and appended to the 3’ end of the respective sequence files. ClustalW identified blocks of highly conserved 5’ and 3’ ends of Eimeria and Cyclospora mitochondrial sequences.
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Cyclospora cayetanensis complete mitochondrial genome data are deposited at NCBI database (accession number KP231180).