Abstract
The first known outbreak of eastern equine encephalitis (EEE) in Vermont occurred on an emu farm in Rutland County in 2011. The first isolation of EEE virus (EEEV) in Vermont (VT11) was during this outbreak. Phylogenetic analysis revealed that VT11 was most closely related to FL01, a strain from Florida isolated in 2001, which is both geographically and temporally distinct from VT11. EEEV RNA was not detected in any of the 3,905 mosquito specimens tested, and the specific vectors associated with this outbreak are undetermined.
Introduction
The first isolates of Eastern equine encephalitis virus (EEEV) were obtained from the brains of encephalitic horses during an epizootic in Delaware, Maryland, Virginia and New Jersey in 1933 [1]. In 1938 EEEV was isolated from the brains of several fatal pediatric encephalitis cases and it was implicated as the causative agent [2,3]. Subsequent ecological and laboratory studies indicated that EEEV is primarily maintained in enzootic cycles in which the virus is transmitted among susceptible birds, especially passerine birds, by the ornithophilic mosquito Culiseta melanura [4–9]. Epizootic and epidemic transmission occurs only intermittently, typically during seasons of high vector population density, which are usually preceded by periods of above normal precipitation [10]. During spillover transmission, atypical hosts such as horses and humans become infected with EEEV and while these hosts are highly susceptible to EEEV, ecologically they are dead-end hosts as neither generate enough viremia to infect mosquitoes and perpetuate the transmission cycle [11]. Spillover events may include transmission by Cs. melanura, Coquillettidia perturbans, Aedes canadensis, Ae. sollicitans, Ae. taeniorhynchus and several other species [7–8,11–15]. The specific overwintering mechanism or mechanisms are currently not well understood although there is evidence of both local persistence as well as re-introduction of new strains from warmer climates [8,15].
Since 2005, there has been increased detection of EEEV activity in northeastern US [16–18] with activity reported in New Hampshire [19], Massachusetts [18], Maine [20], New York [16], Connecticut [19] and recently in Vermont [21,22]. The EEEV activity in Vermont is particularly intriguing because despite well documented EEEV activity in all the surrounding states and territories [8,15–19,23], there was no documented detection of EEEV activity in Vermont until the fall of 2010 [21,22]. In this manuscript we describe the first outbreak of EEE in Vermont and the phylogenetic relationships of the first EEEV isolate from this state.
Materials and Methods
Sample collection and Virus isolation
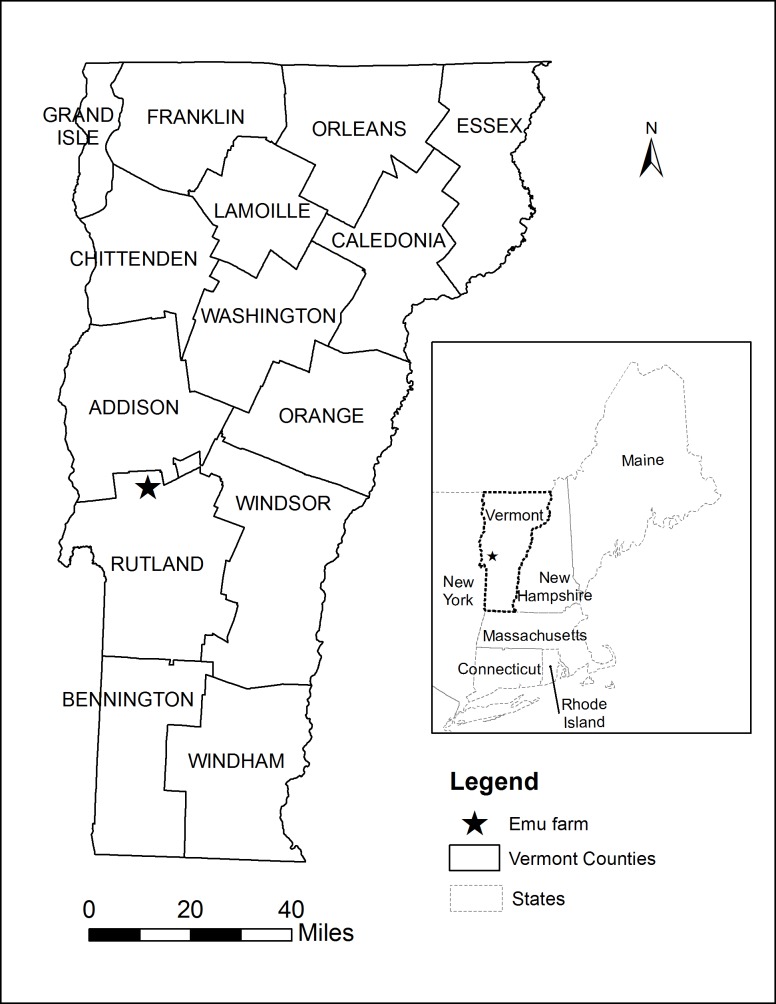
On September 21st, 2011, a local veterinarian performed a necropsy on one of the dead emus from a farm in Rutland County (Fig 1). Brain tissue was collected and sent to the New Hampshire Department of Health’s Public Health Laboratory for PCR testing for the presence of EEEV RNA. On September 22nd, 2011, the lab confirmed that the PCR test was positive for EEEV. The sample was then sent to the Centers for Disease Control in Fort Collins, CO for additional characterization.
Fig 1. A map of Vermont showing the location of the emu farm in Rutland County where the EEE outbreak occurred in 2011.
Upon arrival at the CDC laboratory in Fort Collins, CO, the virus, Rutland_Co_VT2011_Emu (VT11, Genbank Accession number: JX2623861), was isolated from the brain homogenate through a single passage in Vero cells. Viral RNA was extracted using the QIAamp viral RNA protocol (Qiagen, Valencia, CA). The OneStep RT-PCR protocol (Qiagen, Valencia, CA) was utilized to generate PCR amplicons for classical sequencing using virus-specific primers (Table 1) designed within the Primer Select program (DNASTAR, Madison, WI). Appropriate amplicons were cleaned using the MinElute Gel Extraction Kit (Qiagen, Valencia, CA). The DNA was subsequently sequenced using virus-specific primers and the Big Dye v3.1 kit on an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA). The VT11 sequence was assembled and analyzed for sequence quality and genome coverage using Lasergene suite software (DNASTAR, Madison, WI).
Table 1. The oligonucleotides used for EEEV full genome sequencing.
| Name | Use | Sequence | Tm (C) |
|---|---|---|---|
| EEE 40 FWD | RT-PCR and sequencing | GGCAACCACCCTATTTCCACCTA | 70 |
| EEE 626 FWD | Sequencing | GGCGCCTACCCTACATACA | 60 |
| EEE 1087 FWD | Sequencing | ATGACCGGGATACTGGCGACTGAC | 76 |
| EEE 1349 REV | Sequencing | GGCGGGCACCTTCTTAATAGTTTG | 72 |
| EEE 2059 REV | RT-PCR and sequencing | GCATCCCCTTTCTTCACGCACTTC | 74 |
| EEE 1824 FWD | RT-PCR and sequencing | CGCGCAGGACGATACAA | 54 |
| EEE 2586 FWD | Sequencing | CTCGGCGATGCACTAAGAC | 60 |
| EEE 3030 FWD | Sequencing | CATGGCGAAAATACTTGAGAC | 60 |
| EEE 3438 FWD | Sequencing | ATAACCCGCTAATAAATGT | 50 |
| EEE 4035 REV | RT-PCR and sequencing | TTATGTCGCCGCGCACCACTCTAT | 74 |
| EEE 3836 FWD | RT-PCR and sequencing | GCAGTCGCCCGCTCATTCA | 62 |
| EEE 4619 FWD | Sequencing | CCGAGGGCAAGGTGTATT | 56 |
| EEE 4956 REV | Sequencing | GCCGGGGGTACAGTGCCAGAGA | 74 |
| EEE 5102 FWD | Sequencing | TGCAAGAATCCCCAGCCCTCCAT | 72 |
| EEE 5742 REV | RT-PCR and sequencing | GTTTGCATTGCCGCGTAGATTTTT | 68 |
| EEE 5568 FWD | RT-PCR and sequencing | GAGCCGCAGCGCAGACACGAT | 70 |
| EEE 6252 FWD | Sequencing | ATTTGCGGCCTGAGATACG | 58 |
| EEE 6562 REV | Sequencing | ACATCCCGTTTAAGGTCCATC | 62 |
| EEE 7081 FWD | Sequencing | CGCAGCATTTATCGGCGACGACAA | 74 |
| EEE 7776 REV | RT-PCR and sequencing | CGTTTGGCGGGCGGTCCTG | 66 |
| EEE 7509 FWD | RT-PCR and sequencing | CCATAACCCTCTACGGCTGACCTAAAT | 65 |
| EEE 8352 FWD | Sequencing | AAAGGGGGTTACAGTCAAAGATAC | 68 |
| EEE 8666 REV | Sequencing | GCATGCGCATCCCCTCTGACTTC | 74 |
| EEE 9403 REV | RT-PCR and sequencing | TGGTCCGGGTGCAGGTGTAAAATC | 74 |
| EEE 9153 FWD | RT-PCR and sequencing | CGAGCGGCGCCCAAGTGAAATA | 70 |
| EEE 9536 FWD | Sequencing | CCGGAGAAGGGTTGGAGT | 58 |
| EEE 9877 REV | Sequencing | GGATAAGCGTCTGCATCCAG | 64 |
| EEE 10466 FWD | Sequencing | ATGGTGAAACTCCCGCGAAAATAG | 70 |
| EEE 11183 REV | Sequencing | TCGCCGACGTAAAGGATTC | 58 |
| T25 REV | RT-PCR and sequencing | T(25)V | 78 |
Phylogenetic analysis
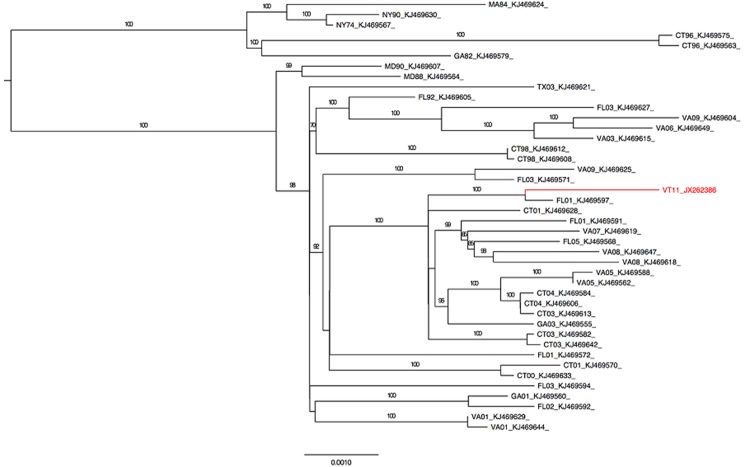
Since there were no other sequences of EEEV from Vermont, studies were undertaken to determine genetic relationships with other strains from the US. Sequences were selected from GenBank (Fig 2) to represent isolates from diverse counties in multiple US states. Sequences for the E2 glycoprotein gene and the full coding sequences were aligned using MUSCLE on the Cipres Science Gateway [24,25]. Maximum likelihood inference was performed using RAxML 7.06 on the Cipres Science Gateway [26]. 1000 replicates of bootstrapping resampling were utilized to assess the accuracy of tree topologies. Output trees were generated for publication using FigTree v1.4.
Fig 2. A maximum likelihood tree for 42 full genome sequences of EEEV isolates isolated from eastern US.
VT11 is highlighted in red print. Numbers at nodes are bootstrap values based on 1000 replicates.
Mosquito collection and processing
The Vermont Department of Health (VDH) and the Vermont Agency of Agriculture, Food and Markets (VAAFM) routinely conduct statewide mosquito based surveillance at 77 trap sites. They monitor CDC gravid traps, resting traps and CDC CO2-baited light traps at all the sites. Mosquitoes were collected once a week, beginning in mid-June and ending in mid-October, and identified to species by using the keys of Darsie and Ward [27]. Mosquitoes of the same species and sex were pooled into groups of 50 or less and screened for EEEV antigen using the VecTest (Medical Analysis Systems, Inc., Camarillo, CA). During the outbreak, additional traps were placed at several locations close to the emu farm to capture mosquitoes at the outbreak site with the aim of incriminating the vector(s) associated with the outbreak.
Results
Outbreak description
On September 21, 2011, the VDH was notified by the Assistant State Veterinarian at the VAAFM about an emu flock in Rutland County (Fig 1) that had suffered multiple fatalities from illness starting on September 15th. By September 21st, 14 emus had died with symptoms of hemorrhagic gastroenteritis. Additional emus showed symptoms of illness, including bloody discharges, weakness and inability to stand. In total, 19 emus died over a 10 day period. All ages were affected: 3 of 44 emus less than 6 months old, 9 of 27 emus 16–18 months old, and 7 of 22 breeder stock birds which were over 4 years old. Two additional birds became ill but survived resulting in an overall attack rate of 22.6%.
Phylogenetic analysis
Despite the limited availability of full-length coding sequences from recent isolates, phylogenetic analyses using sequences from the range of North American strains revealed that VT11 has the highest identity with an isolate from Florida in 2001 (FL01 KJ469597) with a pairwise identity of 99.8% across the coding regions (Fig 2). In all analyses performed (using the range of sequences available), FL01 was the only strain that shared a common ancestor with VT11. However, VT11 also showed high similarity (6 or fewer nucleotide differences) to other viruses within the observable polytomy including strains from Connecticut, New Hampshire, Virginia, Florida, and Georgia [CT01 (KJ468628), NH05 (KJ469631 and KJ469556), VA08 (KJ469647 and KJ469618), FL05 (KJ469568), CT03 (KJ469642 and KJ469613), VA05 (KJ469588), FL01 (KJ469591), VA07 (KJ469619), and GA03 (KJ469555)]. Given the low degree of sequence variation, all analyses had low bootstrap values and poor basal resolution, as expected.
Mosquito surveillance
In 2011, 42,129 mosquitoes belonging to 8 genera and 30 species were captured between June and October. Of these, 3,905 (9.27%) (Table 2) were screened for EEEV antigen. No positive results were obtained, which was possibly due to the small test sample size or the lack of sensitivity of the assay. However, the mosquito trapping did indicate a strong presence of species previously associated with EEEV transmission in North America. For example, there were 1,070 Cs. melanura [8], 7,758 Cq. perturbans [8], 429 Ae. canadensis [8], 17,291 Aedes vexans [8], and 279 Culiseta morsitans) [8] which comprised 2.54%, 18.42%, 1.02%, 41.08% and 0.66% of the total collections, respectively. Although EEEV antigen was not detected in any of the 2011 specimens tested, these species are capable of both epizootic and epidemic transmission of EEEV [8,11–15]. In subsequent years in Vermont, a total of 40 EEEV positive mosquito pools were detected from Cs. melanura, Cq. perturbans, Cs. morsitans and Ae. canadensis (Table 1) which were abundant species during the 2011 outbreak and were the most likely vectors for that outbreak.
Table 2. The number and percentage of mosquitoes collected and screened for EEEV in Vermont from 2011 to 2014.
| Year | Number of Mosquitoes Collected | Number of Mosquitoes Tested | EEEV Positive Pools | EEEV Positive Species** |
|---|---|---|---|---|
| 2011* | 42,129 | 3,905 (9.27%) | 0 | |
| 2012 | 10,498 | 4,676 (44.27%) | 10 | Cs. melanura (10) |
| 2013 | 32,727 | 16,729 (51.12%) | 22 | Cs. melanura (20) |
| Cq. perturbans (1) | ||||
| Cx. pipiens/restuans (1) | ||||
| 2014 | 67,335 | 41,700 (61.93%) | 8 | Cs. melanura (5) |
| Cs. morsitans (2) | ||||
| Ae. canadensis (1) |
* Tested using the VecTest. 2012–2014 samples were tested using RT-PCR (27)
** Number in parenthesis indicates number of positive pools.
Discussion
The first known outbreak of EEE in Vermont occurred on an emu farm in Rutland County in September 2011. The outbreak was restricted to emus on only one farm and no other farm animals, domestic animals, or humans were infected. The outbreak was intriguing because there were no detections of EEEV activity in the state before 2010 [21,22] yet EEEV activity had been previously detected in all surrounding states and territories including Quebec, Canada, directly north of Vermont [8,15–21,28]. The detection of EEEV antibodies in free-ranging deer and moose in Vermont in 2010 [21,22] in concert with the detection of human infections in 2012 and detection of EEEV RNA in field-collected mosquitoes in 2012, 2013 and 2014 (Table 1) leads to at least two possibilities for EEEV presence in the state. Either EEEV was only recently introduced into Vermont and the period from 2010 to 2014 represents the first major emergence events, or EEEV had been circulating enzootically in Vermont but was simply undetected until 2010.
Based on phylogenetic analysis (Fig 2) and pairwise sequence differences, VT11 is most closely related to a strain from Florida in 2001 (KJ469597), which is both geographically and temporally distinct from VT11. The isolate was also found to be highly similar to other virus strains from northeastern US including those from Connecticut (2001–2004) and Virginia (2005–2008) (Fig 2). It is difficult to draw conclusions regarding the importance of geographically similar viruses due to the general lack of temporal overlap; however the observed clustering is consistent with what is known about North American EEEV ecology. Maintenance by avian hosts that allows for wide geographic distribution and the introduction of genotypes from remote geographical locations, particularly along seasonal flyway patterns, contributes to limited genetic diversity. This is a likely explanation as to why the VT11 sequence has such a high degree of homology with a geographically disparate isolate such as FL01. However, examples of regionally confined evolution of EEEV with occasional introductions from geographically distant locations has been observed multiple times [19,23,29–31], and there is much evidence to support the presence of and continued spread of enzootic EEEV in the North eastern states [19–23]. So while it is probable that this particular epizootic was engendered from a novel introduction from Florida, the potential for future epizootic spillover from the recently identified enzootic foci in Vermont deer and moose is also a concern [21,22].
Studies of in vivo infection of emus with EEEV, have shown that they develop a high-titered viremia [greater than 9.0 log10 (PFU/ml)], and shed virus in secretions and excretions increasing the risk of transmission to adjacent animals and human caretakers [32]. Such a high-titered viremia could have increased the likelihood of virus acquisition and transmission by resident vector populations, especially less competent mosquitoes or atypical vectors, which could have potentially acted as bridge vectors.
These finding highlight the potential for endemic EEEV to emerge in Vermont and cause disease in the human population, especially given the established presence of EEEV in local wildlife [21,22]. Future studies examining the seroprevalence of EEEV in humans living in endemic areas will further elucidate the risk to human populations and aid development of preventative measures and targeted surveillance. Similarly, in depth vector prevalence and blood-meal identification studies will enhance our knowledge of likely transmission dynamics.
Acknowledgments
We thank the New Hampshire Department of Health’s Public Health Laboratory for PCR confirmatory testing of the emu brain and for forwarding the brain sample to CDC. We also thank Dr. Keely Henderson, who reported the outbreak to the Vermont State Veterinarian.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was conducted doing the normal duties at the CDC (United States Centers for Disease Control and Prevention) and the state of Vermont. The authors received no specific funds for this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Giltner LT, Shahan MS (1933) The immunological relationship of eastern and western strains of equine encephalomyelitis virus. Science 78: 587–588. [DOI] [PubMed] [Google Scholar]
- 2. Forthergill LD, Dingle JH, Farber S, Connerley ML (1938) Human encephalitis caused by the virus of the eastern variety of equine encephalomyelitis. N Engl J Med 219: 411. [Google Scholar]
- 3. Webster LT, Wright FH (1938) Recovery of equine encephalitis virus from brain tissue of human cases of fatal encephalitis in Massachusetts. Science 257: 305–306. [DOI] [PubMed] [Google Scholar]
- 4. Sudia WD, Chamberlain RW, Coleman PH (1968) Arbovirus isolations from mosquitoes collected in South Alabama, 1959–1963, and serologic evidence of human infection. Am J Epidemiol 87: 112–116. [DOI] [PubMed] [Google Scholar]
- 5. Chamberlain RW, Sudia WD, Coleman PH, Johnston JG Jr., Work TH (1969) Arbovirus isolations from mosquitoes collected in Waycross, Georgia, 1963, during an outbreak of equine encephalitis. Am J Epidemiol 89: 82–88. [DOI] [PubMed] [Google Scholar]
- 6. Bigler WJ, Lassing EB, Buff EE, Prather EC, Beck EC,Hoff GL (1976) Endemic eastern equine encephalomyelitis in Florida: a twenty-year analysis, 1955–1974. Am J Trop Med Hyg 25: 884–890. [DOI] [PubMed] [Google Scholar]
- 7. Crans WJ, McNelly J, Schulze TL, Main A (1986) Isolation of eastern equine encephalitis virus from Aedes sollicitans during an epizootic in southern New Jersey. J Am Mosq Control Assoc 2: 68–72. [PubMed] [Google Scholar]
- 8. Morris CD (1988) Eastern equine encephalomyelitis In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. Boca Raton, FL: CRC Press; pp. 1–20. [Google Scholar]
- 9. Komar N, Dohm DJ, Turell MJ, Spielman A (1999) Eastern equine encephalitis virus in birds: relative competence of European starlings (Sturnus vulgaris). Am J Trop Med Hyg 60: 387–391. [DOI] [PubMed] [Google Scholar]
- 10. Hayes RO, Hess AD (1964) Climatilogical conditions associated with outbreaks of eastern encephalitis. Am J Trop Med Hyg 60: 851–858. [DOI] [PubMed] [Google Scholar]
- 11.Weaver SC (2005) Host range, amplification and arboviral disease emergence. Arch Virol Suppl: 33–44. [DOI] [PubMed]
- 12. Andreadis TG, Anderson JF, Tirrell-Peck SJ (1998) Multiple isolations of eastern equine encephalitis and highlands J viruses from mosquitoes (Diptera: Culicidae) during a 1996 epizootic in southeastern Connecticut. J Med Entomol 35: 296–302. [DOI] [PubMed] [Google Scholar]
- 13. Turell MJ (1998) Effect of salt concentration in larval rearing water on susceptibility of Aedes Mosquitoes (Diptera: Culicidae) to eastern equine and Venezuelan equine encephalitis viruses. J Med Entomol 35: 670–673. [DOI] [PubMed] [Google Scholar]
- 14. Turell MJ, Beaman JR, Neely GW (1994) Experimental transmission of eastern equine encephalitis virus by strains of Aedes albopictus and A. taeniorhynchus (Diptera: Culicidae). J Med Entomol 31: 287–290. [DOI] [PubMed] [Google Scholar]
- 15. Scott TW, Weaver SC (1989) Eastern equine encephalomyelitis virus: epidemiology and evolution of mosquito transmission. Adv Virus Res 37: 277–328. [DOI] [PubMed] [Google Scholar]
- 16.USDA (2009) Summary of Eastern Equine Encephalitis Cases in the United States. Available at: http://wwwaphisusdagov/vs/nahss/equine/ee/.
- 17. Armstrong PM, Andreadis TG (2013) Eastern equine encephalitis virus—old enemy, new threat. N Eng J Med 368: 1670–1673. 10.1056/NEJMp1213696 [DOI] [PubMed] [Google Scholar]
- 18. Silverman MA, Misasi J, Smole S, Feldman HA, Cohen AB, Santagata S, et al. (2013) Eastern equine encephalitis in children, Massachusetts and New Hampshire,USA, 1970–2010. Emerg Infect Dis 19: 194–201; quiz 352. 10.3201/eid1902.120039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Armstrong PM, Andreadis TG, Anderson JF, Stull JW, Mores CN (2008) Tracking eastern equine encephalitis virus perpetuation in the northeastern United States by phylogenetic analysis. Am J Trop Med Hyg 79: 291–296. [PubMed] [Google Scholar]
- 20. Gibney KB, Robinson S, Mutebi JP, Hoenig DE, Bernier BJ, Webber L, et al. (2011) Eastern equine encephalitis: an emerging arboviral disease threat, Maine, 2009. Vector Borne Zoonotic Dis 11: 637–639. 10.1089/vbz.2010.0189 [DOI] [PubMed] [Google Scholar]
- 21. Berl E, Eisen RJ, MacMillan K, Swope BN, Saxton-Shaw KD, Graham AC, et al. (2013) Serological evidence for eastern equine encephalitis virus activity in white-tailed deer, Odocoileus virginianus, in Vermont, 2010. Am J Trop Med Hyg 88: 103–107. 10.4269/ajtmh.2012.12-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mutebi JP, Swope BN, Saxton-Shaw KD, Graham AC, Turmel JP, Berl E, et al. (2012) Eastern equine encephalitis in moose (Alces americanus) in northeastern Vermont. J Wildl Dis 48: 1109–1112. 10.7589/2012-03-076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Young DS, Kramer LD, Maffei JG, Dusek RJ, Backenson PB, Mores CN, et al. (2008) Molecular epidemiology of eastern equine encephalitis virus, New York. Emerg Infect Dis 14: 454–460. 10.3201/eid1403.070816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; 2010; New Orleans, LA.
- 26. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 27. Darsie RF Jr., Ward RA (2005) Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico Gainesville: University Press of Florida. [Google Scholar]
- 28. Chenier S, Cote G, Vanderstock J, Macieira S, Laperle A, Helie P. 2010. An eastern equine encephalomyelitis (EEE) outbreak in Quebec in the fall of 2008. Can Vet J 51: 1011–1015. [PMC free article] [PubMed] [Google Scholar]
- 29. Arrigo NC, Adams AP, Weaver SC (2010) Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J Virol 84: 1014–1025. 10.1128/JVI.01586-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brault AC, Powers AM, Chavez CL, Lopez RN, Cachon MF, Gutierrez LF, et al. (1999) Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. Am J Trop Med Hyg 61: 579–586. [DOI] [PubMed] [Google Scholar]
- 31. Weaver SC, Hagenbaugh A, Bellew LA, Gousset L, Mallampalli V, Holland JJ, et al. (1994) Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol 68: 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tengelsen LA, Bowen RA, Royals MA, Campbell GL, Komar N, Craven RB, (2001) Response to and efficacy of vaccination against eastern equine encephalomyelitis virus in emus. J Am Vet Med Assoc 218: 1469–1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.