Abstract
In the fight against heart failure, therapeutics that have the ability to increase the contractile power of the heart are urgently needed. One possible route of action to improve heart contractile power is increasing the calcium sensitivity of the thin filament. From a pharmaceutical standpoint, calcium sensitizers have the distinct advantage of not altering cardiomyocyte calcium levels and thus have lower potential for side effects. Small chemical molecules have been shown to bind to the interface between cTnC and the cTnI switch peptide and exhibit calcium sensitizing properties, possibly by stabilizing cTnC in an open conformation. Building on existing structural data of a known calcium sensitizer bound to cardiac troponin, we combined computational structure-based virtual screening drug discovery methods and solution NMR titration assays to identify a novel calcium sensitizer 4-(4-(2,5-dimethylphenyl)-1-piperazinyl)-3-pyridinamine (NSC147866) which binds to cTnC and the cTnC-cTnI147-163 complex. Its presence increases the affinity of switch peptide to cTnC by approximately a factor of two. This action is comparable to that of known levosimendan analogues.
Graphical abstract
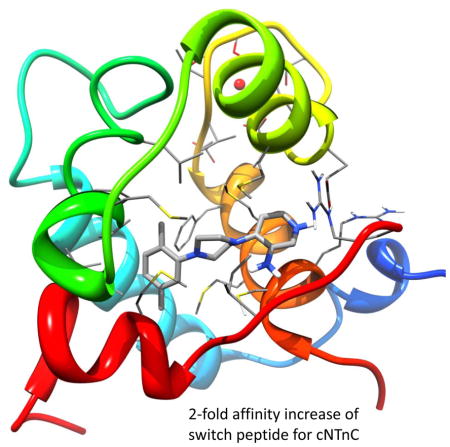
In the fight against heart failure, therapeutics that increase the calcium sensitivity of the thin filament are a promising option to improve heart contractile power. Here, we combined computational drug discovery methods and solution NMR titration assays to identify a novel calcium sensitizer which binds to cTnC and the cTnC-cTnI147-163 complex. Its presence increases the affinity of switch peptide to cTnC by approximately a factor of two, making its action comparable to that of known levosimendan analogues.
Introduction
Regular contraction of the human heart is paramount to its proper function. Human cardiomyocyte contraction is an intricate process governed by the interplay of a large number of proteins. Cross bridges between the thick (myosin) and thin (actin, tropomyosin, troponin) filament of the sarcomere produce force leading to contraction of the muscle cell. Cardiac troponin (cTn), a protein complex on the thin filament plays an important role in regulating this process. Structurally, cTn consists of three subunits: troponin C (cTnC), troponin I (cTnI) and troponin T (cTnT) which are named for their respective functions (1). It is well understood that the binding of the signaling ion, Ca2+, to the N-terminal regulatory domain of cTnC (cNTnC) results in structural and dynamic changes which initiate sarcomere contraction (2). As a consequence of calcium binding to the regulatory domain of cTnC, a hydrophobic patch on the surface of cNTnC (between helices A and B) will be exposed. The switch region of cTnI (cTnI residues 144–163, cTnI144-163) subsequently associates with this hydrophobic patch, loosening its inhibitory action on tropomyosin and actin. This process culminates in unblocking of myosin binding and contraction ensues (2, 3).
Defects in the contractile machinery can lead to heart failure. Weakened contraction of the heart will lead to diminished blood supply of the organs in the human body. Irrespective of the exact cause of the heart failure, therapeutics to increase the contractile power of the heart are urgently needed. Such drugs are generally referred to as cardiac inotropes. Various options of intervening in the contraction process exist: increasing the calcium levels in cardiomyocytes (e.g. digoxin, dobutamine, milirinone (4, 5)), interventions in cross-bridge cycling such as for example a prolonged on-time (4), and increasing the calcium sensitivity of the thin filament. Calcium sensitizers – pharmaceuticals that increase the calcium sensitivity of the thin filament – have the advantage of not altering intracellular calcium levels, an effect which can lead to arrhythmia, tachycardia and mortality. Levosimendan (Simdax) is arguably the most potent and well-known calcium sensitizing drug available so far (6). It binds to the switch peptide binding area in cNTnC and has positive inotropic function (7, 8). A few other calcium sensitizing compounds, such as pimobendan, have also entered clinical studies (9).
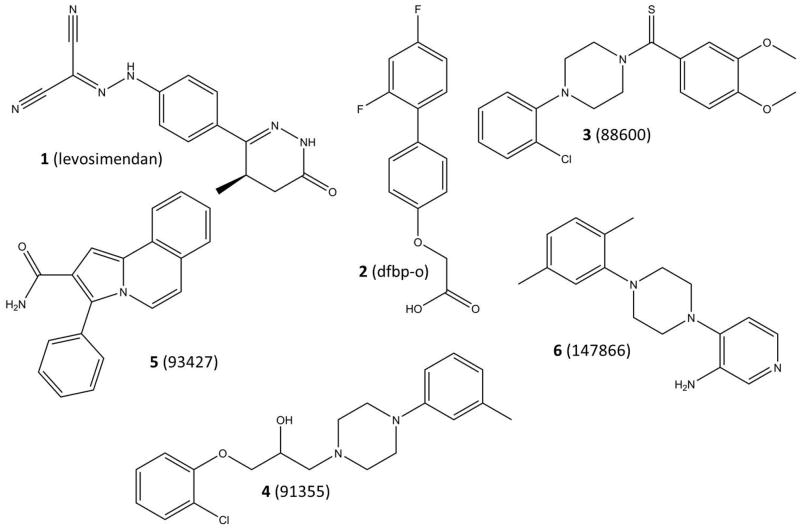
Currently, no atomic structure of levosimendan (1, Figure 1) bound to cNTnC exists. Modeling of the interaction suggested that levosimendan-binding stabilizes the hydrophobic patch of cTnC in a semi-open conformation and thus increases the binding affinity of the cTnI switch peptide to this area of cTnC (10, 11). In a previous study we have determined the structures of two more stable levosimendan analogues - 2′,4′-difluoro(1,1′-biphenyl)-4-yl acetic acid (dfbp) and 2′,4′-difluorobiphenyl-4-yloxy acetic acid (dfbp-o; 2, Figure 1) (12). It was shown that the fluoride containing analogue dfbp-o bound to cTnC both in the presence and absence of the cTnI switch peptide and increased the switch peptide affinity towards cTnC. Here we used the structure of dfbp-o bound to the cNTnC•Ca2+-cTnI144-163 complex as a starting point for computer-aided explorations of novel calcium sensitizers targeting the cardiac troponin complex. Using the relaxed complex scheme – a combination of molecular dynamics (MD) simulations and virtual screening – and NMR titration assays we identified a novel calcium sensitizer 4-(4-(2,5-dimethylphenyl)-1-piperazinyl)-3-pyridinamine (NSC147866), whose action is comparable to that of the levosimendan analogues.
Figure 1.
Structures of the known calcium sensitizers: levosimendan (1) and dfbp-o (2), as well as structure of experimentally validated cTnC binders: NSC88600 (3), NSC91355 (4), NSC93427 (5), and NSC147866 (6).
Methods and Materials
Molecular Dynamics simulations of dfbp-o bound cNTnC•Ca2+-cTnI144-163 complex and cluster analysis
The system prepared for simulations was based on the recent NMR structure of calcium sensitizer 2′,4′-difluorobiphenyl-4-yloxy acetic acid (dfbp-o) bound to the complex of cNTnC and cTnI (cTnI144-163) (PDB-ID 2L1R, (12)). Model 1 was chosen for the simulations since it was the best representative conformer. The system was in the sensitizer-bound, site II calcium-bound state. Tleap (13) neutralized the system by adding Na+ counter ions and solvated it using a TIP3P water box. The fully solvated sensitizer-bound system contained 26214 atoms. The entire dfbp-o ligand was geometry optimized using the B3LYP/6-31G(d) basis set in Gaussian 03 and then the minimized conformation was parameterized using Antechamber and RESP in Amber Tools 11 with the General AMBER force field (GAFF) (14, 15). After building up the system, minimization using SANDER (13) was carried out in two stages: 1000 steps of minimization of solvent and ions with the protein and sensitizer restrained using a force constant of 500 kcal/mol/Å2, followed by a 2500 step minimization of the entire system. A short initial 20 ps MD simulation with weak restraints (10 kcal/mol/Å2) on the protein and sensitizer atoms was used to heat the system to a temperature of 300K. Subsequently, 100 ns of MD simulations were performed. The MD simulations were performed under the NPT ensemble at 300 K using AMBER (13) and the ff99SBildn force field (16, 17). Periodic boundary conditions were used, along with a non-bonded interaction cutoff of 10 Å for Particle Mesh Ewald (PME) long-range electrostatic interaction calculations. Bonds involving hydrogen atoms were constrained using the SHAKE algorithm (18), allowing for a time step of 2 fs. Structures representing the conformational variability of the dfbp-o binding site during the simulation were extracted using clustering. For clustering, frames every 8 ps were extracted from the MD trajectory. Alignment was based on all Cα atoms within 10 Å of the sensitizer in the sensitizer-bound starting structure. Subsequent clustering was performed by RMSD using GROMOS++ conformational clustering (19). A RMSD cutoff of 1.5 Å was chosen, resulting in 7 clusters that represented at least 90% of the trajectory. The central members of each of these clusters were chosen to represent the protein conformations within the cluster and thereby the conformations sampled by the trajectory.
Pocket-volume calculations
To quantify the variability of the sensitizer binding pocket within the chosen clusters, the volume of the dfbp-o binding pocket was calculated for 2L1R model 1 (representative model) and all seven cluster centers. POVME (20) was used for pocket-volume calculations. The coordinates of the following atoms were used as centers for POVME inclusion spheres: CAO, CAS, CAR, CAP, CAL, and CAN. Coordinates of these atoms were extracted from each pdb individually. Points were generated in POVME with a grid spacing of 1 Å using inclusion spheres of 5 Å radius around the atom positions. The volume was calculated using the contiguous option (with contiguous seed spheres of radius 4 Å centered at the same coordinates as the inclusion spheres).
Redocking of dfbp-o
Before performing a virtual screen using structures derived from a molecular dynamics simulation of a dfbp-o-bound cNTnC•Ca2+-cTnI144-163 complex (PDB-ID 2L1R), we evaluated the ability of the Glide SP and XP docking functions to dock the known troponin sensitizer dfbp-o into the representative NMR structure. The sensitizer sdf file was downloaded from the protein data bank and ligand model 5 was chosen as starting model, deliberately different from the protein model (model 1) chosen for the docking analysis. The sensitizer input file was further prepared using LigPrep, which added missing hydrogen atoms, generated all possible ionization states, as well as tautomers. The representative sensitizer-bound NMR structure was prepared with the Receptor Grid Generation tool. Docking was performed with the Glide SP and XP scoring functions.
Virtual screen of NCI diversity set II
The virtual screen was performed using the National Cancer Institute (NCI) diversity set II, a subset of the full NCI compound database. Ligands were prepared using LigPrep, adding missing hydrogen atoms, generating all possible ionization states, as well as tautomers. The final set used for virtual screening contained 1541 compounds. Docking simulations were performed with Glide (21–23), using the SP scoring function. All seven cluster centers from the molecular dynamics trajectory were screened. For each ligand, the best scoring of the seven poses was added to a consensus list over all seven receptors and the top scoring 21 compounds were chosen for experimental verification.
Experimental inhibition assays
Sample preparation
Recombinant human cardiac [15N]-cNTnC (cTnC residues 1–89) with the mutations C35S and C84S was used in this study. The expression and purification of [15N]-cNTnC in E. coli were as described previously (24). 21 portions (0.5–2 mgs) of solid [15N]-cNTnC were dissolved separately into 500μL NMR buffer containing 100 mM KCl, 10 mM imidazole in 90% H2O/10% D2O. The sample concentrations range from 50 to 300 μM. Protein concentration was determined by integrating 1D 1H and 2D (1H,15N)-HSQC NMR spectroscopy. To each sample, 5 μL of 1M CaCl2 was added to ensure that the protein was Ca2+-saturated and the pH was adjusted by 1M NaOH and 1M HCl to 6.7. The synthetic peptide, cTnI147-163, acetyl-RISADAMMQALLGARAK-amide, was purchased from GL Biochem Ltd. (Shanghai, China). Peptide quality was verified by HPLC and ESI Mass Spectrometry. Solid peptide is only marginally soluble in aqueous solutions, thus was dissolved in d6-DMSO to make a stock solution of ~5 mM, as determined by integrating 1D 1H NMR spectrum using DSS as an internal standard. 21 NCI compounds were kindly provided by National Cancer Institute in NIH. The purity and structure of the drug were verified by 1D 1H NMR spectroscopy. Stock solutions of the compounds, in d6-DMSO, were prepared and the vials containing the solutions were wrapped in aluminum foil to protect the molecules from light catalyzed degradation. Gilson Pipetman P (model P2 and P10) was used to deliver the drug or peptide solutions for all titrations.
Titrations
Each compound was titrated to a NMR sample containing [15N]-cNTnC•Ca2+, 4 (NSC88600, NSC93427, NSC147866, and NSC91355) out of 21 were found to induce backbone chemical shift changes on cNTnC. We then titrated the 4 shortlisted compounds to a cNTnC-cTnI chimera construct (cNTnC-C35S-cTnI144-173•Ca2+), only NSC147866 was found to induce chemical shift changes. Thus, we focused on NSC147866.
A. Titration of [15N]-cNTnC•Ca2+ with cTnI147-163
This titration has been done many times previously in our laboratory and the results have been reproducible (25). The results were used here for the purpose of comparison.
B. Titration of [15N]-cTnC•Ca2+•NSC147866 with cTnI147-163
To a 500 μL NMR sample containing a 0.16 mM [15N]-cTnC•Ca2+•NSC147866 complex, aliquots of 1, 2, 5, 5, 5, 10, 7 μL of 5 mM cTnI147-163 in d6-DMSO were added consecutively. The sample was mixed thoroughly with each addition. The total volume increase was 35 μL and the change in protein concentration due to dilution was taken into account for data analyses. The pH decrease from cTnI147-163 addition was compensated by 1M NaOH. Both 1D 1H and 2D (1H, 15N)-HSQC spectra were acquired at every titration point.
C. Titration of [15N]-cNTnC•Ca2+ with NSC147866
To a 500 μL NMR sample containing a 0.17 mM [15N]-cTnC•Ca2+, aliquots of 0.5, 5, 5, 5 μL of 59 mM NSC147866 in d6-DMSO were added consecutively. The sample was mixed thoroughly with each addition. The pH increase from NSC147866 was compensated by 1M HCl. Another 5 μL addition resulted in a small amount of brown precipitate. This precipitate is likely unbound NSC147866, which is insoluble in aqueous solution. This titration point was not used in data analysis. The total volume increase was 15.5 μL and the change in protein concentration due to dilution was taken into account for data analyses. Both 1D 1H and 2D (1H, 15N)-HSQC NMR spectra were acquired at every titration point.
D. Titration of [15N]-cNTnC•Ca2+•cTnI147-163 with NSC147866
A NMR sample of [15N]-cNTnC•Ca2+•cTnI147-163 was made by dissolving solid [15N]-cNTnC and cTnI147-163 to NMR buffer. To a 500 μL NMR sample contains ~50 μM [15N]-cNTnC and ~180 μM cTnI147-163, aliquots of 0.5, 1, 1.5, 2, 3 μL of 5.22 mM NSC147866 in d6-DMSO were added for the first 5 titration points, and aliquots of 0.5, 1.5, 3, 5, 5 μL of 52.2 mM NSC147866 in d6-DMSO were added for the next 5 titration points. The sample was mixed thoroughly with each addition. The pH increase from NSC147866 was compensated by 1M HCl. The total volume increase was 23 μL and the change in protein concentration due to dilution was taken into account for data analyses. Both 1D 1H and 2D (1H, 15N)-HSQC NMR spectra were acquired at every titration point.
NMR Spectroscopy
All NMR experiments were run on either a Varian Inova 500 MHz spectrometer or a Unity 600 MHz spectrometer. All data were collected at 30°C. Both spectrometers are equipped with a triple resonance 1H13C15N probe and z-pulsed field gradients. The NMR chemical shift changes in each titration were used to calculate the dissociation constant (KD). The binding of NSC147866 and cTnI147–163 to the target molecules or complexes were fit with a 1:1 stoichiometry. The dissociation constants were calculated by averaging the normalized individual chemical shifts as a function of the ligand to protein ratios and fitting was done using xcrvfit (www.bionmr.ualberta.ca/bds/software/xcrvfit). The amide resonances that were perturbed larger than the mean plus one standard deviation were chosen for the KD calculation. The KD was determined by fitting the data to the equation
Results and Discussion
Molecular Dynamics simulation generates a wide range of binding pocket conformations
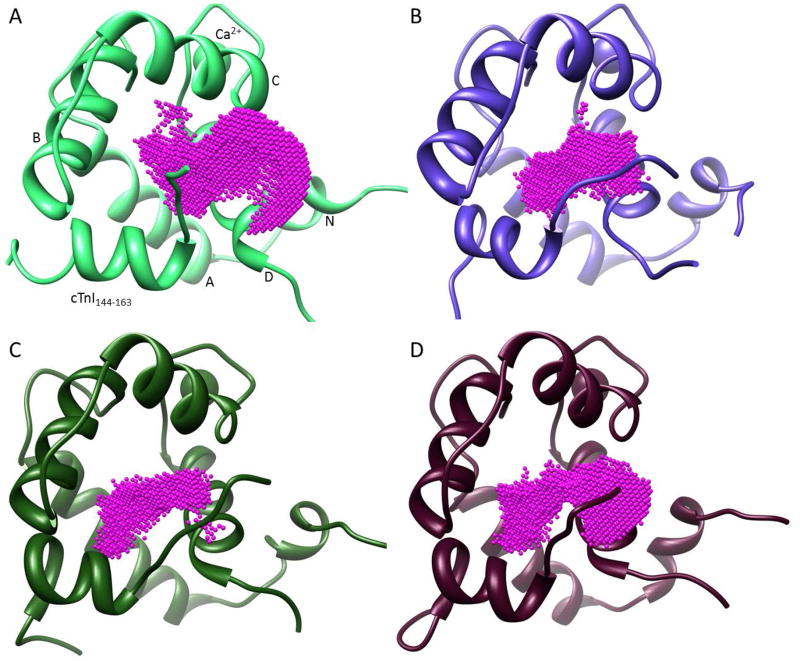
To account for troponin flexibility in the sensitizer binding region, a 100 ns molecular dynamics simulation of the cNTnC•Ca2+-cTnI144-163 complex was evaluated. It is widely accepted that receptor flexibility plays a crucial role in docking of small molecules to proteins (26). The relaxed complex scheme is a computational approach that utilizes molecular dynamics to generate conformational ensembles to serve as multiple receptors in virtual screening studies – thereby accounting for receptor flexibility (27, 28). The sensitizer-bound simulation was clustered with respect to variation in residues surrounding the sensitizer binding site. Seven representative structures characterizing the conformational flexibility of this region were extracted from the simulation and used as receptor structures for virtual screening. For all seven structures and the representative structure from the 2L1R NMR model, the volume of the binding site was calculated. Interestingly the experimental NMR model exhibits the largest volume, 308 Å3. All representative structures from the MD simulation have smaller binding site volumes. The observed volumes of the seven cluster centers range from 108 Å3 to 241 Å3. Visual inspection of the trajectory showed the sensitizer moved deeper into the pocket (towards the helix A-B interface), which is accompanied by a closing of the solvent accessible end of the pocket (towards the C-terminal part of helix D). This explains the smaller pocket volumes compared to the experimental structure. Figure 2 shows the pockets for the 2L1R NMR structure and three of the representative cluster centers extracted from the trajectory. The range in observed pocket volumes and shapes also illustrates nicely how different snapshots from the MD trajectory can prove valuable in virtual screening.
Figure 2.
Ligand binding pockets as calculated by POVME. Pockets for the 2L1R NMR structure (A) and three of the representative MD cluster centers (B–D) are shown. Ligands were removed before the actual pocket calculation. Calculated pocket volumes are 309 Å3 (A), 145 Å3 (B), 108 Å3 (C), and 241 Å3 (D). The structural components of cTnC and cTnI are labeled in panel A.
Experimental troponin sensitizer pose is recovered by Glide SP docking
Before running virtual screens on the cardiac troponin complex to find novel calcium sensitizers, it is important to assess the ability of the docking algorithm and scoring function to correctly dock known calcium sensitizers. For this work we picked Schroedinger’s Glide as the docking program of choice. We assessed its ability of correctly finding the docked pose of a known calcium sensitizer by docking dfbp-o into the representative model (model 1) of the 2L1R NMR structure. The standard precision (SP, (21)) and extra precision (XP, (22)) scoring functions were used and their results compared. Figure 3 shows the experimentally determined conformation of the dfbp-o ligand in the cNTnC•Ca2+-cTnI144-163 complex, as well as the docked poses obtained with Glide SP and XP. Glide SP determined a docking score of −6.83 kcal/mol and docked the ligand with an RMSD of 1.80 Å with respect to the NMR structure. The RMSD came almost entirely from a translation of the docked compound with respect to the experimental pose. The actual conformational RMSD was only 0.5 Å (as determined by RMSD calculation allowing for rigid body movements). Surprisingly, Glide XP did not perform as well. The RMSD of the ligand compared to the experimental pose was 2.53 Å, whereas the docking score was −6.24 kcal/mol. An RMSD calculation allowing for rigid body movements yielded 0.98 Å. The higher RMSD is entirely driven by the incorrect position of the terminal carboxyl group. Based on these results, we decided to use Glide SP as docking function for the virtual screen.
Figure 3.
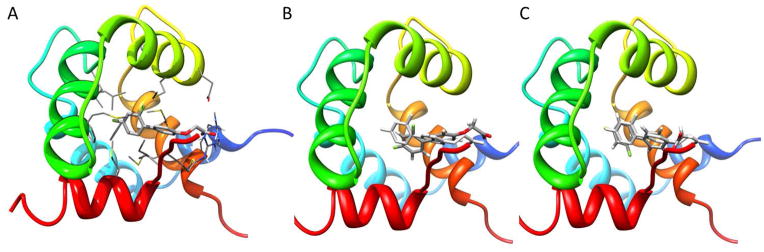
Experimental and docked poses of troponin calcium sensitizer dfbp-o. A) NMR conformation of dfbp-o bound to cTnC-cTnI144-163 interface. The backbone of cTnC and cTnI is represented as ribbons and colored in rainbow. Side chains interacting with the ligand, as well as the ligand itself, are colored by element type. Glide SP (B) and Glide XP (C) docked poses of dfbp-o. The protein backbone is shown in rainbow. The experimental binding pose is colored in light grey, while the docked pose is colored by element type.
Virtual screen of NCI diversity set II
Seven representative structures from a MD simulation of a calcium sensitizer-bound cNTnC•Ca2+-cTnI144-163 complex were used as receptors for the relaxed complex scheme docking protocol using the Glide SP scoring function. The NCI diversity set II was used as screening library. The Glide SP docking results were ranked according to the predicted docking score and subsequently the best scoring of the seven poses for each ligand was added to a consensus list over all seven receptors. The top 21 compounds from this list had docking scores ranging from −10.1 kcal/mol to −9.17 kcal/mol (corresponding to the respective top scoring receptor conformation) and were selected for experimental investigation.
Experimental results
We performed solution NMR titration assays which monitor chemical shift changes indicative of compound binding and protein-ligand interactions. We utilized 2D (1H, 15N) HSQC NMR spectra to follow perturbations in the chemical environment at each 15N-labeled amide nucleus. When a ligand binds to a protein, the amide resonances of residues in direct contact with the bound ligand will experience a change in both 1H and 15N chemical shift. These changes may also occur by ligand induced-conformational changes in the protein. In either scenario, the chemical shift change reflects the ligand effect and can be used to determine the stoichiometry and affinity for protein-ligand interactions.
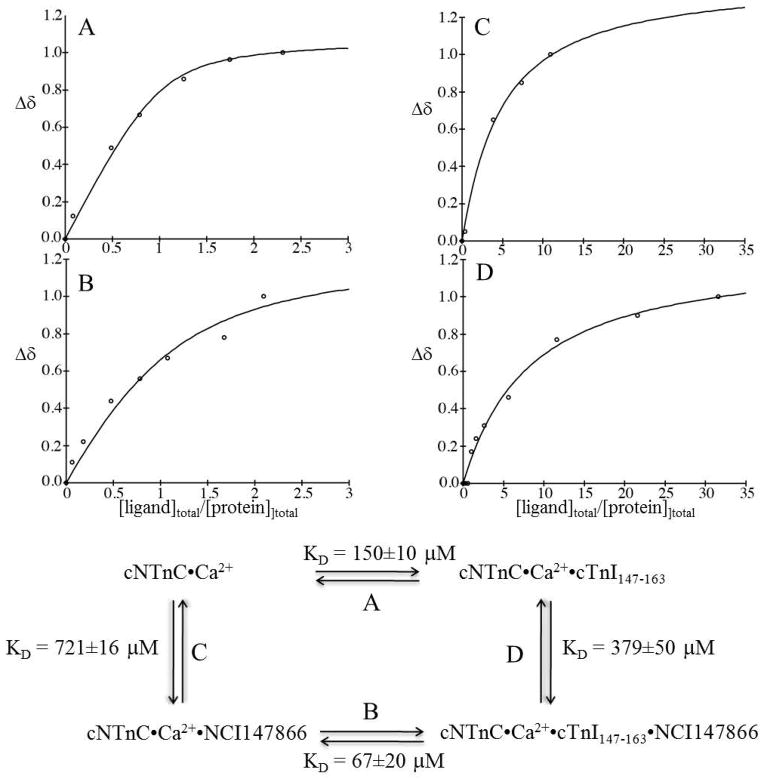
We titrated the initial 21 NCI compounds to 21 cNTnC•Ca2+ NMR samples. For each titration we first dissolved the compounds in DMSO to generate concentrated stock solutions. The compounds are all soluble in DMSO. When we titrated the drug stock to an aqueous NMR sample, the drugs that did not interact with the protein precipitated right away (the drugs are not soluble in aqueous solution). If the drug binds to the protein no precipitation is observed since the complex is soluble. Some of the compounds precipitated after first additions. Others did not precipitate but induced no spectral changes on cNTnC•Ca2+. Four out of 21 compounds (NSC88600 (3, Figure 1), NSC93427 (5, Figure 1), NSC147866 (6, Figure 1), and NSC91355 (4, Figure 1)) induced chemical shift changes in the 2D (1H, 15N) HSQC NMR spectra of cNTnC•Ca2+. In all 4 cases, the chemical shift changes fell into the fast exchange limit on the NMR time scale. The linear movement of the cross-peaks indicated that only two species existed in the interaction between the drug and the protein. The position of each cross-peak corresponded to the weighted average of the bound and free chemical shifts of cNTnC. This phenomenon has been observed many times in our earlier studies, e.g. dfbp-o binding to cNTnC•Ca2+•cTnI147-163 (12), and is indicative of 1:1 stoichiometry. We then titrated the 4 compounds to a construct of cNTnC linked to the switch region of cTnI (cNTnC-C35S•Ca2+-cTnI144-163 chimera). Only NSC147866 was found to cause chemical shift perturbations. We noticed that among the 4 compounds (NSC88600, NSC93427, NSC147866, and NSC91355) that bind to cNTnC•Ca2+, NSC88600, NSC147866, and NSC91355 each contains a piperazine group. However, both NSC88600 and NSC91355 consist of other bulkier groups as compared with NSC147866. This may be why only NSC147866 bound to cNTnC•Ca2+ in the presence of cTnI. We subsequently focused on NSC147866 and probed NSC147866’s ability to alter affinity of cNTnC for cTnI147-163; specifically NSC147866 was titrated into cNTnC•Ca2+ and cNTnC•Ca2+•cTnI147-163 complex, respectively. We found that NSC147866 bound to cNTnC•Ca2+ with a dissociation constant of 721±16 μM; albeit weakly, this affinity was enhanced ~2-fold in the presence of cTnI147-163 (379±50 μM). We then titrated cTnI147-163 to the cNTnC•Ca2+•NSC147866 complex and found that NSC147866 also enhanced the affinity of cTnI147-163 for cNTnC•Ca2+ by ~2-fold: 150±10 μM to 67±20 μM. This enhancement is comparable to the calcium sensitization of dfbp-o (12). The experimentally determined dissociation constants are summarized in Figure 4. Encouragingly, NSC147866 has very drug-like properties (MW = 282 Da, logP = 2.8, 4 hydrogen bond acceptors, 2 hydrogen bond donors) and does not violate a single of Lipinski’s rules of five (29). Due to its small size it is suitable to lead improvement.
Figure 4.
(A) Titration of cTnI147–163 into cNTnC•Ca2+. (B) Titration of cTnI147–163 into cNTnC•Ca2+•NSC147866 (~20 fold excess of NSC147866). (C) Titration of NSC147866 into cNTnC•Ca2+. (D) Titration of NSC147866 into cNTnC•Ca2+•cTnI147–163 (~3 fold excess of cTnI147–163). A summary of the experimentally determined dissociation constants is shown at the bottom.
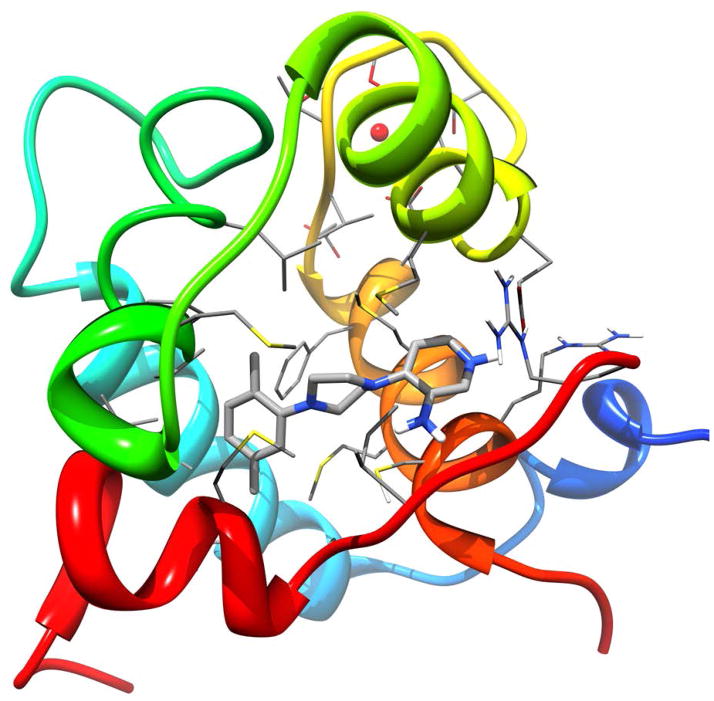
Interestingly, NSC147866 has been identified by docking into the conformation representing the cluster with the smallest binding pocket (4th most populated of the seven cluster centers) with a volume of 108 Å3 as seen in panel C of Figure 2. This pocket is the most different from the experimental structure, underlining the power of the relaxed complex scheme. Figure 5 shows the docked pose of NSC147866. It is also interesting to note that Omecamtiv mecarbil (a cardiac myosin activator) (30), trifluoperazine (calcium sensitizer in muscle contraction) (31, 32), and ranolazine (calcium sensitivity modulator in diastolic cardiac dysfunction) (33) all contain a piperazine group. This supports the notion that a piperazine group might be the key pharmacophore in the sensitization of cardiac muscle contraction.
Figure 5.
Docked pose of identified calcium sensitizer NSC147866 in molecular dynamics cluster 4. The backbone of cTnC and cTnI is represented as ribbons and colored in rainbow. Side chains interacting with the ligand, as well as the ligand itself, are colored by element type.
Conclusions
Calcium sensitizers are compounds that increase the calcium sensitivity of the thin filament. That is in the presence of the compound a stronger contractile force is observed at a set calcium concentration. One possible target for calcium sensitization drugs is the interface between cTnC and the cTnI switch region. In this study we combined molecular dynamics, structure-based drug discovery methods and sensitive solution NMR titration assays to identify a novel calcium sensitizer 4-(4-(2,5-dimethylphenyl)-1-piperazinyl)-3-pyridinamine (NSC147866) which binds to cNTnC and the cNTnC-cTnI147-163 complex. Its presence increases the affinity of switch peptide to cNTnC by approximately a factor of two. This action is comparable to that of known levosimendan analogues and a great starting point for future follow-up work to improve the binding affinity of the compound, which needs to be higher for any pharmaceutical applications. We identified a piperazine group as a possible key pharmacophore in the sensitization of cardiac muscle contraction. Building on this finding is of interest to researchers working on development of drugs for calcium sensitization.
Acknowledgments
We thank Peter Kekenes-Huskey for interesting discussions concerning the cardiac troponin complex, as well as other members of the McCammon group, for useful discussions. This work was supported by the National Institutes of Health, the National Science Foundation, the Howard Hughes Medical Institute, the National Biomedical Computation Resource, the NSF Supercomputer Centers, and the Canadian Institutes of Health Research (grant 37769 to BDS). Computational resources were supported, in part, by the National Science Foundation grant PHY-0822283 and the Center for Theoretical Biological Physics. S. L. was supported by the American Heart Association (12POST11570005) and the Center for Theoretical Biological Physics.
References
- 1.Farah CS, Reinach FC. The troponin complex and regulation of muscle contraction. Faseb J; 1995;9:755–67. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- 2.Li MX, Wang X, Sykes BD. Structural based insights into the role of troponin in cardiac muscle pathophysiology. Journal of Muscle Research and Cell Motility; 2004;25:559–79. doi: 10.1007/s10974-004-5879-2. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu Rev Physiol; 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 4.Hasenfuss G, Teerlink JR. Cardiac inotropes: current agents and future directions. Eur Heart J; 2011 doi: 10.1093/eurheartj/ehr026. [DOI] [PubMed] [Google Scholar]
- 5.Teerlink JR, Metra M, Zaca V, Sabbah HN, Cotter G, Gheorghiade M, Cas LD. Agents with inotropic properties for the management of acute heart failure syndromes. Traditional agents and beyond. Heart failure reviews; 2009;14:243–53. doi: 10.1007/s10741-009-9153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasenfuss G, Pieske B, Castell M, Kretschmann B, Maier LS, Just H. Influence of the novel inotropic agent levosimendan on isometric tension and calcium cycling in failing human myocardium. Circulation; 1998;98:2141–7. doi: 10.1161/01.cir.98.20.2141. [DOI] [PubMed] [Google Scholar]
- 7.Haikala H, Kaivola J, Nissinen E, Wall P, Levijoki J, Linden IB. Cardiac troponin C as a target protein for a novel calcium sensitizing drug, levosimendan. J Mol Cell Cardiol; 1995;27:1859–66. doi: 10.1016/0022-2828(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 8.Haikala H, Nissinen E, Etemadzadeh E, Levijoki J, Linden IB. Troponin C-mediated calcium sensitization induced by levosimendan does not impair relaxation. J Cardiovasc Pharmacol; 1995;25:794–801. doi: 10.1097/00005344-199505000-00016. [DOI] [PubMed] [Google Scholar]
- 9.Perrone SV, Kaplinsky EJ. Calcium sensitizer agents: a new class of inotropic agents in the treatment of decompensated heart failure. Int J Cardiol; 2005;103:248–55. doi: 10.1016/j.ijcard.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Sorsa T, Pollesello P, Solaro RJ. The contractile apparatus as a target for drugs against heart failure: interaction of levosimendan, a calcium sensitiser, with cardiac troponin c. Mol Cell Biochem; 2004;266:87–107. doi: 10.1023/b:mcbi.0000049141.37823.19. [DOI] [PubMed] [Google Scholar]
- 11.Pollesello P, Ovaska M, Kaivola J, Tilgmann C, Lundstrom K, Kalkkinen N, Ulmanen I, Nissinen E, Taskinen J. Binding of a new Ca2+ sensitizer, levosimendan, to recombinant human cardiac troponin C. A molecular modelling, fluorescence probe, and proton nuclear magnetic resonance study. J Biol Chem; 1994;269:28584–90. [PubMed] [Google Scholar]
- 12.Robertson IM, Sun YB, Li MX, Sykes BD. A structural and functional perspective into the mechanism of Ca2+-sensitizers that target the cardiac troponin complex. J Mol Cell Cardiol; 2010;49:1031–41. doi: 10.1016/j.yjmcc.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of computational chemistry; 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model; 2006;25:247–60. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem; 2004;25:1157–74. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 16.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins; 2006;65:712–25. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins; 2010;78:1950–8. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics; 1977;23:327–41. [Google Scholar]
- 19.Christen M, Hunenberger PH, Bakowies D, Baron R, Burgi R, Geerke DP, Heinz TN, Kastenholz MA, Krautler V, Oostenbrink C, Peter C, Trzesniak D, van Gunsteren WF. The GROMOS software for biomolecular simulation: GROMOS05. Journal of computational chemistry; 2005;26:1719–51. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- 20.Durrant JD, de Oliveira CA, McCammon JA. POVME: an algorithm for measuring binding-pocket volumes. J Mol Graph Model; 2011;29:773–6. doi: 10.1016/j.jmgm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem; 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 22.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem; 2006;49:6177–96. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 23.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem; 2004;47:1750–9. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 24.Li MX, Saude EJ, Wang X, Pearlstone JR, Smillie LB, Sykes BD. Kinetic studies of calcium and cardiac troponin I peptide binding to human cardiac troponin C using NMR spectroscopy. European biophysics journal : EBJ. 2002;31:245–56. doi: 10.1007/s00249-002-0227-1. [DOI] [PubMed] [Google Scholar]
- 25.Li MX, Spyracopoulos L, Sykes BD. Binding of cardiac troponin-I147-163 induces a structural opening in human cardiac troponin-C. Biochemistry; 1999;38:8289–98. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 26.Sinko W, Lindert S, McCammon JA. Accounting for Receptor Flexibility and Enhanced Sampling Methods in Computer-Aided Drug Design. Chem Biol Drug Des; 2013;81:41–9. doi: 10.1111/cbdd.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaro R, Baron R, McCammon J. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. Journal of Computer-Aided Molecular Design; 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J Am Chem Soc; 2002;124:5632–3. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 29.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev; 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 30.Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, Brejc K, et al. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science; 2011;331:1439–43. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurebayashi N, Ogawa Y. Increase by trifluoperazine in calcium sensitivity of myofibrils in a skinned fibre from frog skeletal muscle. J Physiol; 1988;403:407–24. doi: 10.1113/jphysiol.1988.sp017256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silver PJ, Pinto PB, Dachiw J. Modulation of vascular and cardiac contractile protein regulatory mechanisms by calmodulin inhibitors and related compounds. Biochem Pharmacol; 1986;35:2545–51. doi: 10.1016/0006-2952(86)90052-3. [DOI] [PubMed] [Google Scholar]
- 33.Lovelock JD, Monasky MM, Jeong EM, Lardin HA, Liu H, Patel BG, Taglieri DM, Gu L, Kumar P, Pokhrel N, Zeng D, Belardinelli L, Sorescu D, Solaro RJ, Dudley SC., Jr Ranolazine improves cardiac diastolic dysfunction through modulation of myofilament calcium sensitivity. Circ Res; 2012;110:841–50. doi: 10.1161/CIRCRESAHA.111.258251. [DOI] [PMC free article] [PubMed] [Google Scholar]