Abstract
Symmetry is an important feature of protein tertiary and quaternary structure that has been associated with protein folding, function, evolution and stability. Its emergence and ensuing prevalence has been attributed to gene duplications, fusion events, and subsequent evolutionary drift in sequence. This process maintains structural similarity and is further supported by this study. To further investigate the question of how internal symmetry evolved, how symmetry and function are related, and the overall frequency of internal symmetry, we developed an algorithm, CE-Symm, to detect pseudosymmetry within the tertiary structure of protein chains. Using a large manually curated benchmark of 1007 protein domains, we show that CE-Symm performs significantly better than previous approaches. We use CE-Symm to build a census of symmetry among domain superfamilies in SCOP and note that 18% of all superfamilies are pseudo-symmetric. Our results indicate that more domains are pseudo-symmetric than previously estimated. We establish a number of recurring types of symmetry–function relationships and describe several characteristic cases in detail. Using the Enzyme Commission classification, symmetry was found to be enriched in some enzyme classes but depleted in others. CE-Symm thus provides a methodology for a more complete and detailed study of the role of symmetry in tertiary protein structure.
Availability
CE-Symm can be run from the web at http://source.rcsb.org/jfatcatserver/symmetry.jsp. Source code and software binaries are also available under the GNU Lesser General Public License (v. 2.1) at https://github.com/rcsb/symmetry. An interactive census of domains identified as symmetric by CE-Symm is available from: http://source.rcsb.org/jfatcatserver/scopResults.jsp.
Keywords: structural biology, protein evolution, pseudo-symmetry, tertiary structure, algorithm, structural alignment, symmetry detection, evolution, protein function
Introduction
Many proteins have a high degree of symmetry in both their tertiary and quaternary structures. This observation dates back to the determination of the quaternary structure of hemoglobin in 1960 [1], which was discovered to contain symmetric pairs of subunits. Subsequently, symmetry has been found to be important for understanding protein evolution [2], DNA binding [3, 4], allosteric regulation [5, 6], cooperative enzyme effects [7], and folding [8]. The relationships between protein symmetry, evolution, and function are reviewed in [9, 10, 11, 7, 12].
Symmetry is characterized by an alignment between equivalent substructures. In the case of quaternary symmetry, these substructures are defined by the inherent equivalence of interactions between identical chains, and often can be determined from the space group of the crystal for X-ray structures. However, this equivalence can be relaxed to allow for evolutionary divergence, revealing pseudo-symmetric arrangements within individual polypeptide chains (internal symmetry) or that span two or more non-identical chains. Figure 1 contains examples of proteins with such symmetry within a single chain. This study will focus on internal pseudo-symmetry.
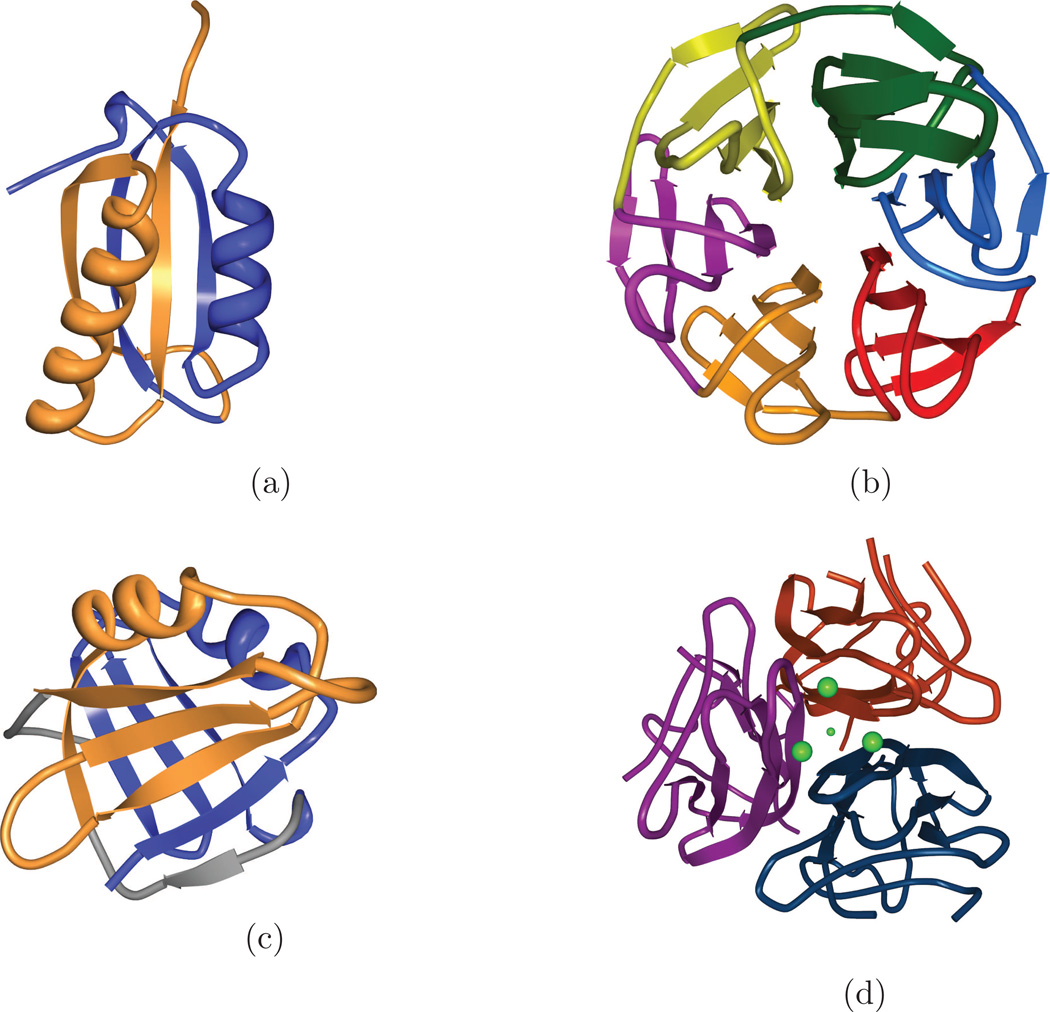
Figure 1.
Several protein domains with internal symmetry that CE-Symm detects. Coloring is by symmetry unit. (a) A ferredoxin-like fold with two-fold symmetry. (SCOP ID: d2j5aa1) (b) A 6-bladed β propeller. Each blade contains a Kelch sequence motif [69], which is also found in some 7-bladed β-propellers (SCOP ID: d1u6dx_) (c) A single DNA clamp domain of a human proliferating cell nuclear antigen (PCNA). The full biological assembly contains 6 of these domains arranged with six-fold symmetry as a trimer of PCNA chains (SCOP ID: d1vyma1) (d) Adiponectins normally assemble into homotrimers of 3 single-domain chains. Shown here (PDB ID: 4DOU) is a designed single-chain three-fold symmetric repeat of an adiponectin globular domain that folds much like an adiponectin trimer [28]. The construct was found to increase insulin sensitivity in mice [29].
Symmetry and protein evolution
Considering all proteins in the Protein Data Bank (PDB) [13, 14] that contain at least two chains in the annotated biological assembly, we find that approximately 80% of all protein complexes contain quaternary structural symmetry (unpublished, see http://www.rcsb.org). Large symmetric oligomers are thought to have been present in primordial life [7, 15], and symmetry continues to be an important feature of proteins.
One model explaining the evolution of internal symmetry has been described by Andrade et al. [16] and Abraham et al. [17]. They proposed gene duplication and fusion as a model for the emergence of symmetric protein chains from complexes with quaternary symmetry. These architectures are then subject to evolutionary drift, but their overall symmetric architectures are preserved. An alternative hypothesis, the emergent architecture model, posits that symmetric architectures arise primarily via convergent evolution [18]. Most likely both mechanisms are correct for different protein families. Another possible driving force for the evolution of symmetry could be random chance, driven by negative selection against destabilizing mutations [19].
Well-known cases of symmetry include TIM barrels, β-trefoils, β-propellers, ferredoxin-like proteins, pentein propellers, and immunoglobulin proteins.
TIM barrels consist of eight pairs of alternating α-helices and β-sheets that interact in parallel to form a cylinder. The TIM barrel fold is extremely versatile and supports a wide diversity of enzymatic reactions [20]. Canonical TIM barrels have eight-fold symmetry around the central channel. However, the overall structure is robust to changes in the (βα)8 sequence: functional TIM barrels are known with single anti-parallel sheets, with deleted (βα) subunits [21, 22], and even as a dimer of (βα)4 chains [23].
The β-trefoil fold has three-fold symmetry and similarly spans a wide range of functions. Several studies have investigated the role of symmetry in β-trefoils by creating β-trefoils with perfect three-fold symmetry [10, 2, 18]. Both studies found that perfect trimeric β-trefoils are highly stable. One of these constructs—a synthetic glycosidase carbohydrate binding domain—not only retained its function, but was found to have increased binding activity. However, a similar construct of an FGF-1 protein showed none of its normal binding activity. This suggests that exact symmetry improves the function of some proteins, while the normal function of other proteins requires imperfect symmetry.
Adiponectin is a hormone involved in metabolic regulation [24] whose normal functioning has been associated with increased insulin sensitivity [25, 26, 27, 28]. The protein normally assembles as a homotrimer with threefold crystallographic symmetry (PDB ID: 1C3H). Ge et al. [29] constructed a single-chain repeat of an Adiponectin globular domain (Figure 1d), which folded into a perfectly three-fold symmetric monomer with a structure similar to that of its multimeric counterpart. Expression of the protein construct increased insulin sensitivity in mice and is hoped to be useful in the treatment of diabetes. Given the contribution of symmetry to protein stability, symmetry may become important in protein design, similar to the increased importance of circular permutations [30].
Algorithms that detect symmetry
The examples described in the previous section provide a compelling reason to accurately establish and classify symmetry in protein tertiary structure. Many symmetry-detection algorithms have been developed, including COSEC2 [31, 12], DAVROS [32], OPAAS [33, 34], Swelfe [35], RQA [36], GANGSTA+ [37], and SymD [38].
Some of the early methods are based on the alignment of secondary structure elements. These are sensitive to secondary structure assignment, which limits their power to detect some cases of pseudo-symmetry. Moreover, several of these approaches are no longer available. One algorithm, SymD, is still being actively developed. It aligns proteins at the residue level, detecting symmetry by systematically performing a structural alignment for all possible circular permutations of a protein. This results in the determination of protein symmetry, including the detection of multiple axes of symmetry for some cases. Using SymD, Kim et al. [38] estimated that 10–15% of known protein domains are symmetric.
Symmetry detection using structural alignment
We have previously developed the Combinatorial Extension (CE) algorithm for global three-dimensional protein structure alignment [39, 40] and integrated it into the RCSB PDB as part of the Protein Comparison Tool [41]. CE is a well-established protein structure comparison algorithm that has been used in a number of benchmarks as one of the reference methods in terms of alignment accuracy [42, 43, 44]. Here, the intention is to use our experience in performing protein structure alignments using CE and employ it to detect symmetry in protein tertiary structure using a new variation of CE, called CE-Symm.
With several algorithms for the detection of symmetry available, it is surprising that no reference benchmark to evaluate and compare the quality of these algorithms has been introduced previously. Here we present a manually curated benchmark containing 1007 protein domains.
In the following sections we describe CE-Symm and the benchmark, and we use both to demonstrate that CE-Symm is currently the leading method for the detection of symmetry. Finally, we systematically apply CE-Symm to establish a census of symmetry found in superfamilies as defined by SCOPe 2.03 (formerly SCOP 1.75C) [45, 46, 47].
Results
To evaluate the accuracy of CE-Symm and competing methods overall, a total of 1100 SCOP superfamilies from SCOPe 2.01 (SCOP 1.75A)1 were initially sampled at random, with one domain arbitrarily selected as the representative structure. Sampling superfamilies rather than domains was intended to reduce the effect of bias in the PDB towards easily crystallized or heavily studied proteins. Repeated motifs were classified as cyclic symmetry, dihedral symmetry, linear repeats, helical symmetry, or superhelical. For explanations of these types of symmetry, see Detailed evaluation.
The presence and type of symmetry for each of these domains was determined manually, resulting in a table of SCOP IDs with their corresponding space groups presented in Supplemental Table S1. When testing algorithms against the benchmark, we considered only cyclic and dihedral symmetry to be cases of symmetry.
Evaluating CE-Symm
CE-Symm performed well on the benchmark set, and fared particularly well at higher thresholds for specificity (fewer false positives). While maintaining a false-positive rate (FPR) of just 3.3%, it correctly identified 86% of the symmetric domains in the benchmark set. Among true-positive results, CE-Symm determined the correct order of symmetry 83% of the time. In 96% of cases, it reported either the correct order or an integral multiple or divisor of it.
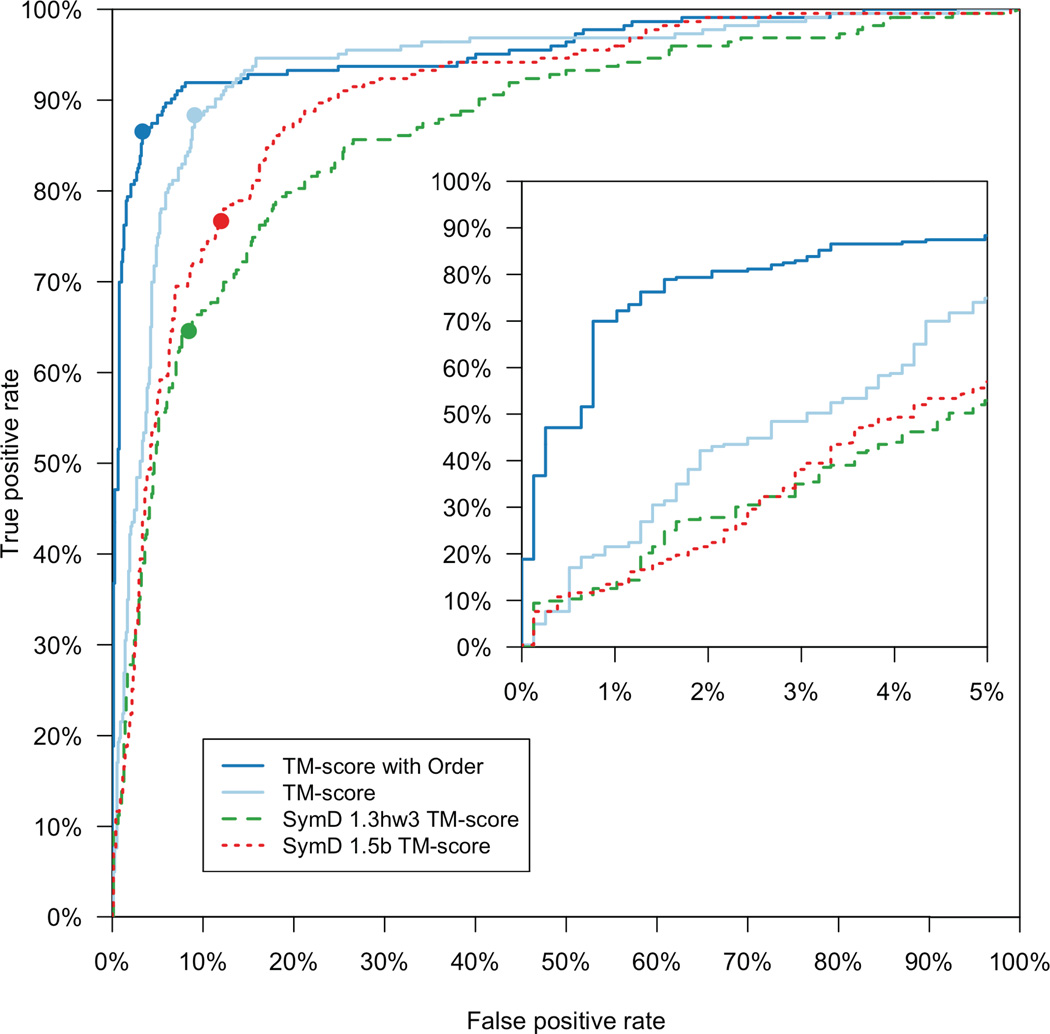
To compare CE-Symm against what we considered the best previously available method, we also ran results from SymD (version 1.3hw3) against our benchmark set. Kim et al. [38] provided us with a copy of an unpublished update to SymD (version 1.5b), which we also benchmarked. For comparison, SymD 1.3hw3 found only 39% of symmetric domains while maintaining the same FPR of 3.3%. The two algorithms are compared in the receiver operating characteristic (ROC) curves shown in Figure 2.
Figure 2.
Receiver Operating Characteristic curves for CE-Symm and SymD on a benchmark set of 1007 SCOP domains. Two curves for CE-Symm are shown: using only TM-score for scoring (light blue), and using TM-score and the order-method described in Methods (dark blue, solid). Two curves for SymD are shown, one for SymD 1.3hw3 (green), and one for the unpublished version 1.5b (red). The thresholds used for determining symmetry (refer to the footnotes in Table 2) are indicated with circles.
The ROC curve for CE-Symm (dark blue) results had an area under curve (AUC) of 0.95, and this value was 0.87 for SymD (version 1.3hw3; orange). The difference between these values was determined to be highly statistically significant (P-value = 2.2 × 10−5) using StAR [48]. Therefore, overall CE-Symm performs much better than SymD. We also benchmarked an alternate scoring system for CE-Symm (light blue).
Based on these results, suggested thresholds for the binary decision of symmetric/asymmetric using CE-Symm were established (Table 2). Thresholds for SymD are included for reference.
Table 2.
Folds with known symmetry
| ID | Fold | No.1 | CE-Symm (%) | SymD (%) | GANG (%) | |||
|---|---|---|---|---|---|---|---|---|
| Ord2 | TM3 | Z84 | Z105 | 1.5b6 | FSAR7 | |||
| d.58 | Ferredoxin-like | 59 | 73 | 73 | 19 | 5.0 | 43 | 23 |
| b.l | Immunoglobulin-like | 28 | 61 | 61 | 8.9 | 0.54 | 26 | 8.4 |
| b.42 | β-trefoil | 8 | 98 | 98 | 100 | 95 | 98 | 56 |
| a.24 | Four-helical bundle | 24 | 60 | 71 | 51 | 25 | 56 | 25 |
| d.131 | DNA clamp | 1 | 100 | 100 | 91 | 73 | 96 | 64 |
| b.69 | 7-bladed β-propeller | 14 | 94 | 100 | 100 | 100 | 100 | 37 |
| c.1 | TIM barrel | 33 | 70 | 88 | 83 | 69 | 70 | 3.7 |
| b.11 | γ-Crystallin-like | 1 | 92 | 92 | 75 | 58 | 92 | 83 |
Percentage of domains determined to be symmetric according to different decision methods. Data for SymD 1.3hw3 and GANGSTA+, in addition to the list of SCOP domains, is taken from supplemental 3 of Kim et al. [38]. The best-performing methods for each fold are in bold.
The number of superfamilies in the fold
CE-Symm using TM-score ≥ 0.4 and requiring order ≥ 2
CE-Symm using TM-score ≥ 0.4
SymD using Z-score ≥ 8 (recommended by authors)
SymD using Z-score ≥ 10 (recommended by authors)
The unpublished SymD version 1.5b using TM-score ≥ 0.4
GANGSTA+ using FSAR(fraction of sequentially aligned residues) ≥ 0.8, which the authors recommend [37]
Folds with well-known symmetry
In the interest of continuing the benchmark by Kim et al. [38], which compared SymD against the secondary structure-based symmetry detection algorithm GANGSTA+, we ran CE-Symm on a set of 8 SCOP folds that are known to be symmetric (Table 2). This evaluation is useful to compare CE-Symm with GANGSTA+, and CE-Symm with SymD for selected cases; however, we emphasize that this table contains only a limited and arbitrary choice of folds compared to the more comprehensive benchmark described above. CE-Symm was at least as likely to classify a domain as symmetric than either SymD and GANGSTA+ in 7 of 8 cases. It was 6 times as likely to find symmetry among immunoglobulin-like β-sandwiches than SymD, and 23 times as likely as GANGSTA+ to find symmetry among TIM barrels.
Detailed evaluation
We analyzed a number of cases where CE-Symm determined symmetry correctly but SymD did not, and vice versa. Generally, we found that CE-Symm was more robust to insertions and small structural differences than SymD. For example, CE-Symm correctly identified C2 symmetry in the Ferredoxin-like domain d1r0bl1 and C8 symmetry in the β/α barrel domain d2i5ia1.
One strength of SymD is its superior order-detection capabilities, due to its systematic consideration of all circular permutation points. The order-detection methods used by CE-Symm are useful for eliminating many asymmetric cases and for estimating the order of symmetry. However, the methods are heuristics and sometimes incorrectly report the order, particularly among structures with order greater than 8 or those whose order has no small factors (Supplemental Table S6). The order-detection heuristic can also fail for proteins with variable-length subunits, such as some β-barrels. For example, CE-Symm’s order-detection incorrectly reports C1 for the autotransporter domain (SCOP ID: d1uyox_), but CE-Symm is able to correctly classify it as symmetric based on TM-Score alone. A complete listing of predictions on the benchmark set by CE-Symm and SymD is available in S4.
CE-Symm and SymD were found to have comparable computation times. Both SymD 1.3hw3 and CE-Symm with order-detection completed in about 2 seconds per domain when run on the benchmark set in a single-threaded environment on a 64-bit Mac OS system with a 2.8Ghz Intel Core i7 processor and 16GB RAM. On the same system, SymD 1.5b required about 4 seconds per domain; however, we note that this version has not been released publicly.
Symmetry order
The types of symmetry identified in the benchmark set are given in Table 4). We found that 23.9% of the superfamilies sampled contained some form of structural repeat. Of these, cyclic symmetry was by far the most common (91.3%). Two-fold symmetry was the most common type of cyclic symmetry (75.5%), followed by eight-fold cyclic symmetry.
Table 4.
Benchmark symmetry by order
| Order | Superfamilies | Example Folds | |
|---|---|---|---|
| Asymmetric | |||
| 766 | 76.1% | ||
| Rotational | |||
| 2 | 166 | 16.5% | Immunoglobulin-like, Ferredoxin-like, Rossmann, γ-crystallin-like, DNA clamp, up-down 4-helical bundle |
| 3 | 10 | 1.0% | β-Trefoil, β-Prism, Flavodoxin-like |
| 4 | 2 | 0.2% | 4-bladed β-propeller, Streptavidin-like, Prealbumin-like, OMPA-like |
| 5 | 3 | 0.3% | 5-bladed β-propeller, Pentein β/α-propeller, PT-barrel |
| 6 | 9 | 0.9% | 6-hairpin glycosidases, 6-bladed β-propeller, autotransporter |
| 7 | 9 | 0.9% | 7-bladed β-propeller, 7-bladed α/α-toroid, 7-hairpin glycosidase |
| 8 | 21 | 2.1% | TIM barrel, 8-bladed β-propeller |
| Dihedral | |||
| 2 | 2 | 0.2% | Transmembrane β-barrels, Streptavidin-like |
| 4 | 1 | 0.1% | Streptavidin-like |
| Helical | |||
| 2 | 9 | 0.9% | Leucine-rich repeat, β-helix, α-α superhelix |
| 3 | 2 | 0.2% | α-α superhelix, β-helix |
| Non-integral | 2 | 0.2% | α-α superhelix, Triple-stranded β-helix |
| Superhelical | 2 | 0.2% | α-α superhelix |
| Translational | |||
| 3 | 0.3% | Ankyrin repeat, β-helix, Bacteriochlorophyll A protein | |
Types of symmetry found in the benchmark.
Dihedral symmetry, helical symmetry, and translational repeats accounted for the remainder, about 2.1%. Linear repeats have translational symmetry, which is given by the repeated application of a translation but no rotation. In most helically symmetric structures, rotating by 360°/k for some integer k is equivalent to no rotation; such a structure is said to have helical symmetry of order k. For some structures, no such integer exists; we labeled this type of symmetry “non-integral helical”. Superhelical symmetry is the unusual symmetry seen in domains such as in Leucine-rich repeats.
A census of symmetry in SCOP
A census of symmetry in the tertiary structure of domains was created by running CE-Symm on every domain in each superfamily in SCOPe 2.03 [45, 46]. This version of SCOP is an update by John-Marc Chandonia, Naomi K. Fox, and Steven E. Brenner; it is available at http://scop.berkeley.edu.
SCOPe 2.03 contains 1766 superfamilies over 5 main classes: all-α, all- β, α/β, α + β, and trans-membrane. We constructed a census of symmetry over these superfamilies by running CE-Symm (with order detection enabled) on every domain in each superfamily and normalizing by the number of domains per superfamily. We found that 18.0% of these superfamilies are symmetric. This percentage of symmetric superfamilies is slightly higher than the percentage of symmetric domains in SCOP among ASTRAL 40 representatives [49] found by SymD, which was 10–15% [38]. Figure 1 shows some examples of symmetric proteins identified by CE-Symm.
Interestingly, symmetric α + β superfamilies are disproportionately rare (Table 3). α + β folds consist of α and β regions that are physically separated in sequence; we hypothesize that this separation limits the number of viable symmetric architectures. In contrast, all-β proteins are enriched for symmetry. This class contains a number of common symmetric folds, such as β-barrels and β-propellers. The extended hydrogen-bonding networks in β-sheets may also contribute to this enrichment, as planar structures are inherently more likely to be symmetric due to their reduced dimensionality.
Table 3.
Symmetry by SCOP class
| Class | Total Number | % Symmetric |
|---|---|---|
| α | 507 | 18.5% |
| β | 354 | 24.6% |
| α/β | 244 | 16.8% |
| α+β | 551 | 14.3% |
| Multi-domain1 | 66 | 4.5% |
| Membrane | 109 | 23.8% |
| Overall | 1831 | 18.0% |
Percentage of superfamilies identified as symmetric by CE-Symm. Note that, to maintain a low false-discovery rate, CE-Symm underestimates the number of symmetric superfamilies in SCOP by about 27% (see Figure 2).
These are large protein chains that have only been observed in their entirety.
Symmetry is also disproportionately frequent among membrane superfamilies, in agreement with previous observations [50, 51]. Membrane proteins often contain additional quaternary symmetry in addition to the internal symmetry within individual domains. The axis of symmetry is typically perpendicular to the membrane plane, although some cases are known with the axis of symmetry parallel to the plane [7]. The symmetric arrangement of subunits in membrane proteins minimizes the lipid interface for each subunit, and the gap formed at the axis of symmetry often forms the channel for membrane transporters.
Sequence conservation
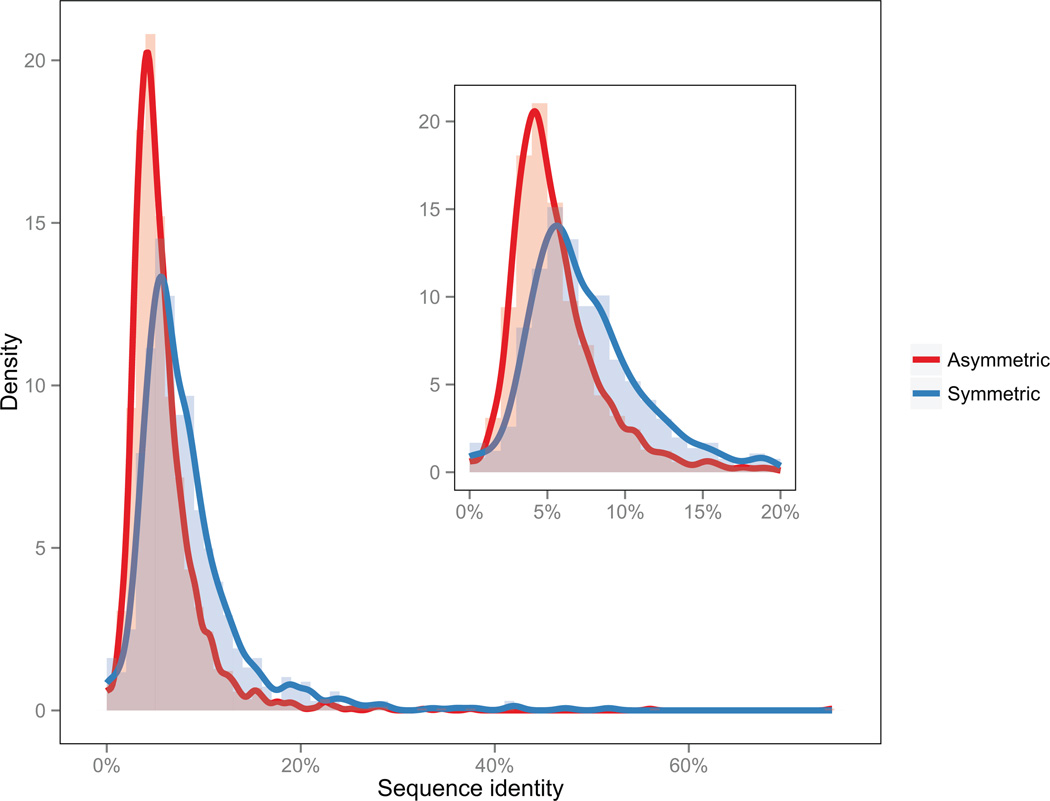
Using all superfamilies in the census, we calculated the percentage identity of the alignment given by CE-Symm. In the case of self-alignments given by CE-Symm, the percentage identity is defined as the percentage of amino acids that are conserved when the domain is superimposed on itself following a rotation about the axis of symmetry. Percent identity was graphed separately for symmetric and asymmetric superfamilies (Figure 3).
Figure 3.
Sequence identity between symmetry units. Distribution of sequence identity between aligned subunits for symmetric superfamilies (blue). For comparison, the distribution of percentage identity among asymmetric superfamilies (red). Most CE-Symm alignments of asymmetric proteins represent random alignments, although a few examples contain translational repeats or helical symmetry.
Surprisingly, the distributions in Figure 3 are very similar. Indeed, the mean %id among symmetric results is 8.2%, not substantially higher than the mean %id among asymmetric results, 5.8%. Moreover, there are few symmetric domains with greater than 16 %id. Considering amino acid similarity rather than identity produces similar results (see Supplemental Figure S1). This lack of sequence conservation between the structural units that give rise to the symmetry (symmetry units) could indicate (a) that the majority of internally symmetric superfamilies arose following ancient duplication events, (b) that convergent evolution between subunits is a more significant contributor to internally symmetric proteins than previously thought, or (c) that the relationship between sequence and structural motifs is relatively flexible, making it difficult to detect sequence similarities based on structure-based methods such as CE-Symm. A similar observation has also been made by Wright et al. [52], where a low sequence identity between proteins might be associated with the inhibition of misfolding and aggregation of proteins in the crowded environment of a living cell.
Enzyme function
To investigate the relationship between symmetry and protein function, we grouped symmetric superfamilies by their Enzyme Commission (EC) numbers [53]). Consistent with our methodology for the census, we normalized by the number of domains per superfamily to mitigate bias in the PDB. A superfamily was assigned an EC number if it contained a domain having that EC number, meaning that multiple enzyme classes can be assigned to a single superfamily.
Analysis of the top-level EC classes proved difficult due to the breadth of structures which provide scaffolds for each type of reaction. Isomerases were enriched for internal symmetry (24% symmetric), while oxidoreductases and ligases contained fewer symmetric domains than average (each 15%; see Supplemental Figure S2). Oxidoreductases span a broad range of evolutionarily and structurally disparate folds (148 in the analysis), and the distribution of folds and the distribution of superfamilies over these folds are both diffuse. Therefore, the low level of symmetry cannot be ascribed to the class having a constrained set of viable folds.
Considering second-level EC subclasses allows the relationship between symmetry and function to be more clearly established. The number of symmetric superfamilies for selected EC subclasses is given in Table 1 and is fully detailed in Supplemental Table S2. Although the number of superfamilies annotated with each subclass is fairly small, enrichment for symmetry also could not be explained by a lack of structural diversity in enzymes with each function.
Table 1.
Percentage of superfamilies found to be symmetric for selected second-level Enzyme Commission numbers, restricted to the most and least symmetric 5 EC subclasses containing at least 20 superfamilies. See Table S2 for the complete list.
| EC | Description | %S1 | NSf2 |
|---|---|---|---|
| 5.1 | Isomerases: racemases and epimerases | 38 | 21 |
| 5.3 | Isomerases: intramolecular oxidoreductases | 26 | 34 |
| 4.1 | Lyases: carbon-carbon lyases | 26 | 57 |
| 2.5 | Transferases: transferring alkyl or aryl groups, other than methyl groups | 23 | 31 |
| 3.4 | Hydrolases: acting on peptide bonds (peptide hydrolases) | 21 | 95 |
| 6.3 | Ligases: forming carbon-nitrogen bonds | 11 | 74 |
| 1.8 | Oxidoreductases: acting on a sulfur group of donors | 10 | 29 |
| 4.2 | Lyases: carbon-oxygen lyases | 10 | 79 |
| 1.10 | Oxidoreductases: acting on diphenols and related substances as donors | 10 | 20 |
| 1.4 | Oxidoreductases: acting on the CH-NH(2) group of donors | 8.3 | 24 |
Percentage of superfamilies that are symmetric
The number of superfamilies
One of the most enriched subclasses for symmetry is that of the racemases and epimerases (EC 5.1). While perfect symmetry would be unexpected in racemase active sites based on their need to bind multiple sterioisomers equally well, pseudosymmetric scaffolds may be amenable to these types of function [54]. Many racemases exhibit quaternary symmetry, in addition to the internal symmetry considered for the census. Several oxidoreductase subclasses are significantly below average for symmetry. Oxidoreductases often contain multiple cofactors for electron transport, which may be less easily supported by symmetric protein scaffolds. Thus, certain enzymatic reactions may support or preclude symmetry.
Discussion
To further investigate potential relationships between symmetry and protein function, we analyzed a large number of proteins to ascertain their symmetry–function relationships. Based on this analysis, we identified recurring types of symmetry–function relationships.
Symmetry around ligand-binding sites
Symmetry around ligand-binding sites is the most basic symmetry–function relationship. For example, glyoxalase I (Figure 4a) is a two-fold symmetric protein with a metal-binding site at its center. [55]. Searching systematically in our census and counting only one domain per superfamily, we found that 22% of symmetric, ligand-containing domains contained a ligand within 5Å from the centroid of the domain. Unaligned residues, such as insertions, were excluded from the calculation of the centroid.
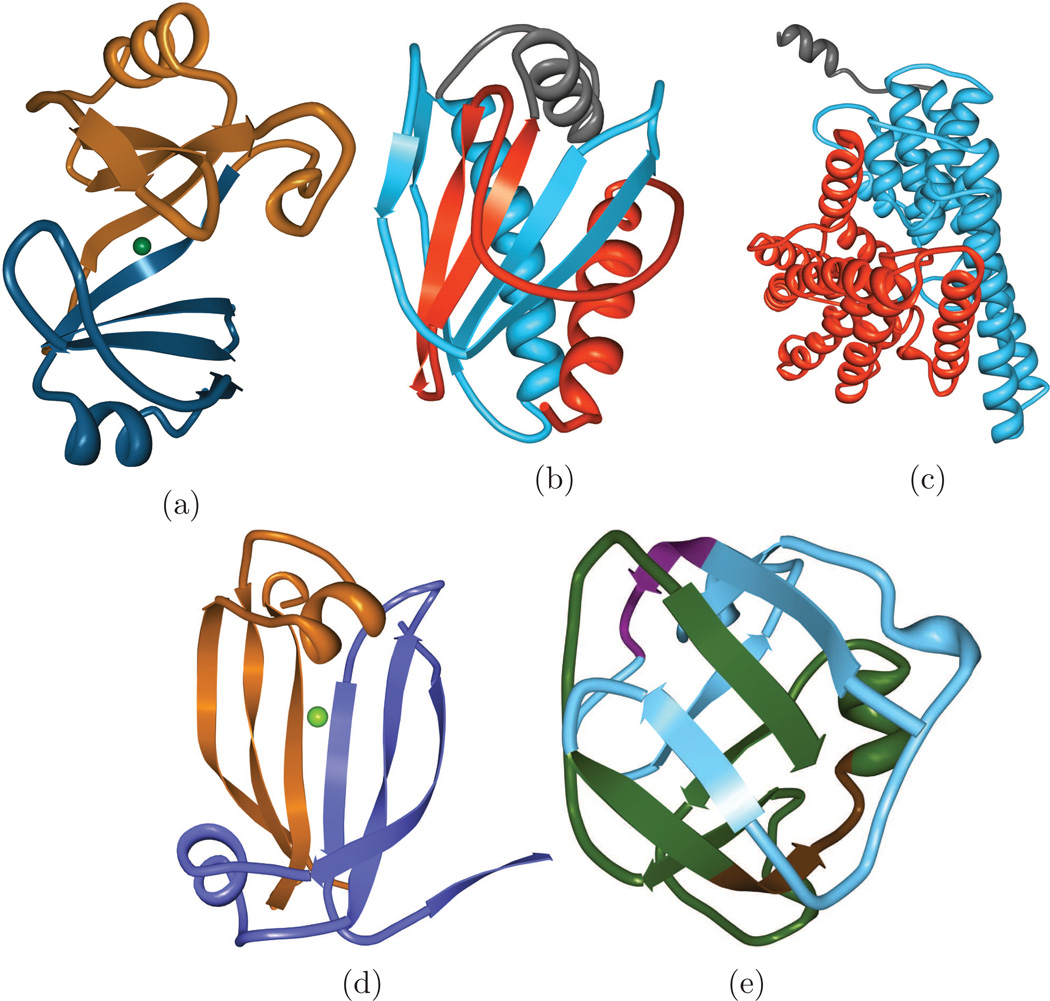
Figure 4.
Examples of proteins with symmetry and function relationships. (a) Glyoxalase I contains a duplication around the nickel-binding active site (PDB ID: 3HDP). (b) CheX protein contains two identical active sites (PDB ID: 1SQU). (c) CLC-ec1 chloride carrier, where ions are thought to flow along its symmetric interface (PDB ID: 2FEE). (d) A chorismate lyase-like protein with a two-fold symmetry that is not clearly related to its little-understood function (PDB ID: 3DDV). (e) PTSIIA/GutA-like domain (PDB ID: 2F9H). Both subdomains of the symmetry contain the same 8-amino-acid sequence (residues 9–16, shown in purple and 67–74, shown in brown).
Function along symmetric interfaces
Many symmetric proteins have function at the interface between symmetry units, the repeated structural units that describe the symmetry. This differs from symmetry around a ligand–binding site, described above, in that the functional site can occur anywhere along the axis of symmetry. An example of this relationship is the chloride channel, in which the symmetric interface between the two symmetry units forms a gate at the core of the channel [56]. Interestingly, the chloride channel is thought to be moderately rigid compared to other channels, such as potassium ion channels or bacterial leucine transporters, both of which are activated by the rotation of subunits relative to each other [56, 57]. Currently, it seems that only the movement of one side chain at the core of the gate is responsible for letting Cl− ions pass. Using the same systematic, preliminary analysis we applied to find ligands near the centroids of domains, we found that 63% of symmetric, ligand-containing domains contained a ligand within 5ÅA from the axis of symmetry. This number was 37% within a mere 1ÅA of the axis.
Duplication of ligand-binding sites
Duplication of ligand-binding sites is another common feature of symmetric proteins. For example, it occurs in the chemotaxis protein CheC (Figure 4b), which is a globular α/β protein that functions in bacterial chemotaxis and is involved in flagella movement. The protein is two-fold symmetric. Each of the two units of symmetry contains a dephosphorylation center comprising asparagine and glutamate residues. Gene duplication followed by domain swapping has been proposed as an evolutionary process for the emergence of CheC [58].
Unknown functions
Besides examples such as those listed above, there are many symmetric domains with no obvious relationship between their symmetry and their function. The chorismate lyase-like protein (Figure 4d) consists of a two-fold internally symmetric domain. Its biologically active form is a dimer such as PhnF from E. coli (PDB ID: 2FA1) or YurK from B. subtilis (PDB ID: 2IKK).
Conserved sequence motifs
In some cases we can identify conserved sequence motifs shared between symmetry units. The PTSIIA/GutA-like domain is an antiparallel β-barrel fold with highly conserved two-fold symmetry. The overall sequence identity of this symmetry is 16%. Little is known about this protein structure since it is a novel fold and does not have an associated publication. Similarly, not much is known about its sequence, with Uniprot only listing a manuscript that describes the larger genomic region covering the gene encoding this structure. However, by investigating the symmetric alignment, we can identify a motif that corresponds to equivalent residues in the structure and that is observable in the Pfam domain (PF03829) [59], which contains a conserved [IV]XX[IV]GXX[VA] motif at the corresponding positions (Figure 4e). Sequence homology between the subunits can be established using the protein sequence alone. However, the analysis of symmetry reveals structural homology and shows that the two types of homology correspond. Based on this correspondence, we postulate that these residues are important functionally, and that they can serve as a guide for further experimental analysis.
Relationship between tertiary and quaternary symmetry
We also suggest that there is a relationship between symmetry of proteins and their biological assemblies. It has been speculated that this can be related to mono- and oligomerization events during evolution that keep the biologically active assembly essentially unmodified [17]. We can confirm this finding and identify several domains with complex relationships between symmetry in the biological assembly and internal symmetry in tertiary structure. An example of this is the DNA clamp. In eukaryotes (PDB ID: 1VYM), it exists as a three-chain symmetric biological assembly. Each chain consists of two protein domains, which in turn have two-fold symmetry (Figure 1c). Thus, the overall assembly has six-fold pseudo-symmetry. The overall symmetry is highly conserved in the bacterial DNA clamp, which has only two chains in the biological assembly, but with each chain consisting of three internally symmetric domains (PDB ID: 1MMI; Kelman and O’Donnell [60]).
Another example with an interesting relationship between the biological assembly and internal pseudo-symmetry is the vitamin B12 transporter BtuCD-F (PDB ID: 4FI3; Korkhov et al. [61]). It consists of three components: BtuC, BtuD, and BtuF. BtuC and BtuD are present as a dimer and bound to BtuF, which is a monomer in the biological assembly. However, BtuF has has internal pseudo-symmetry, giving the whole complex pseudo-twofold symmetry. For a classification of symmetry in structural complexes of proteins see [62].
Types of symmetry CE-Symm identifies
The modifications described in the Methods section enable CE-Symm to detect rotational pseudo-symmetry within protein backbones. It can also detect non-rotational repeats, such as linear repeats, helical proteins, and β- helices. Rotational symmetry can be easily distinguished from other repeats using geometric criteria (see Materials and Methods).
Because CE-Symm uses dynamic programming, it is limited to finding alignments that contain at most a single circular permutation. Types of symmetry that contain more than one axis of symmetry (dihedral, tetrahedral, octahedral, or icosahedral) require multiple changes in sequence topology to align. In such cases, CE-Symm typically will identify one axis of rotation, though additional axes may be found by rerunning CE-Symm on just one of the symmetric domains identified by the first run.
CE-Symm is also limited to returning the single highest-scoring alignment. This may not correspond to the smallest rotational symmetry present in the protein. For instance, in proteins with four-fold pseudo-symmetry, the alignment corresponding to the 180° rotation may score higher than the 90° or 270° alignments. This sometimes leads to the protein being identified as containing two-fold pseudo-symmetry, which incompletely describes the relationships within the protein. More broadly, accurate detection of order of symmetry is a current limitation in CE-Symm which we expect to rectify in a future version.
Conclusions
In this study we introduced a new method for determining pseudo-symmetry in protein structure and used it to build a census of symmetry over domains in SCOP. We also established a reliable benchmark set containing SCOP domains for which both presence and type of symmetry was determined manually. We used this benchmark set to compare our algorithm and previously published symmetry-detection algorithms and demonstrated that our algorithm is more suitable than other methods for detecting symmetry at high specificity. The benchmark set can be used to verify the accuracy of results from other methods for symmetry detection or classification.
By systematically applying CE-Symm on many protein domains we found that more proteins contain internal symmetry than previously estimated. The symmetry of most domains lacks any sequence signal that CE-Symm readily detects. However, clear sequence signals were found for certain folds, such as β-propellers [63].
We also found symmetry to be more associated with some types of enzymatic activity than with others and suggest that certain enzymatic functions preclude or hinder symmetry. We note that in several cases there is a clear relationship between protein symmetry and function, which may explain why certain domains are symmetric.
The analysis of symmetry and pseudo-symmetry in protein structures leads to a deeper understanding of protein function and evolution. Besides detecting pseudo-symmetry in protein structures, CE-Symm allows also the detection of conserved sequence motifs in symmetry units. This can provide insight useful for further analysis of a protein. This is particularly important if the function or active sites of the protein are unknown.
Materials and Methods
CE-Symm Algorithm
The Combinatorial Extension (CE) algorithm operates by using a geometric distance score to evaluate the local structural similarity between two proteins around each residue [39]. Dynamic programming is used to identify high-scoring paths in the dynamic programming matrix, corresponding to regions of local structural similarity. An iterative algorithm then heuristically combines local fragments to identify a high-scoring global superposition of the two proteins.
Building on the CE concept, and to identify self-similar regions within a protein, CE-Symm compares a protein structure to itself. It runs CE to compare two copies of the input protein, with the following modifications:
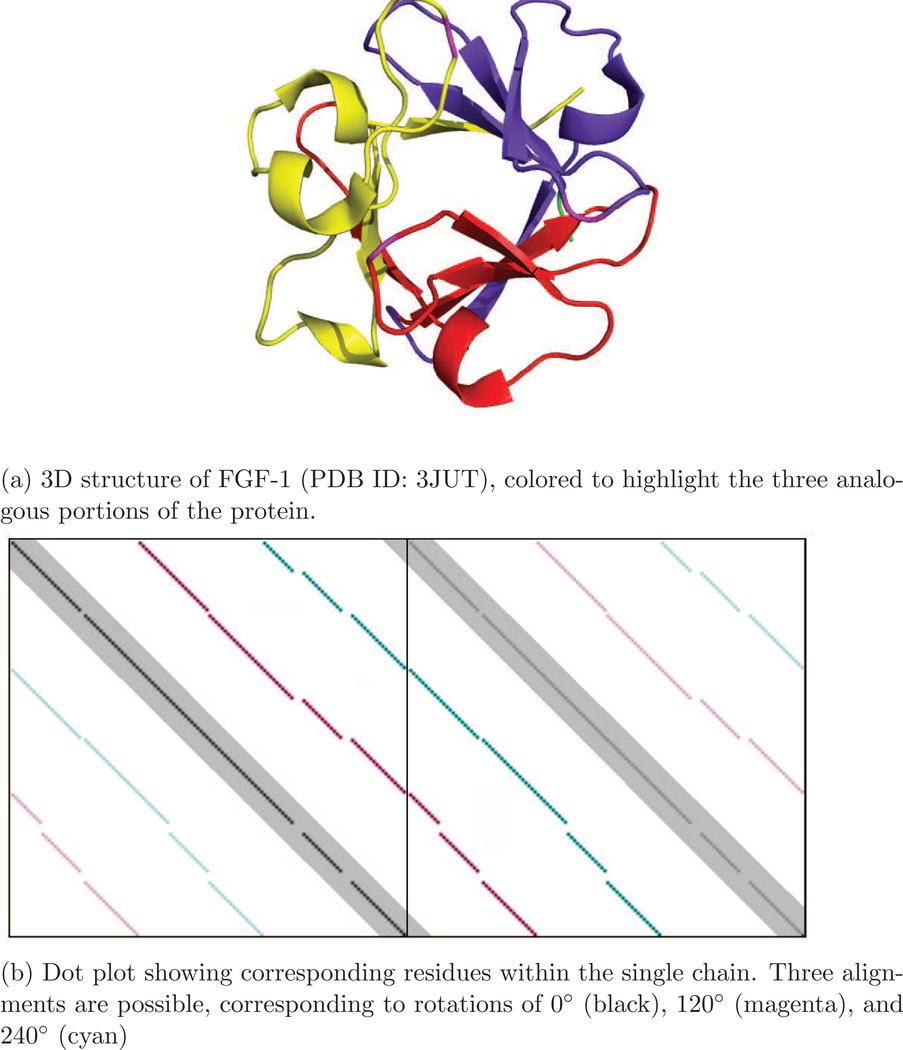
Prohibit alignments near the diagonal. To prevent the algorithm from finding trivial identity similarity, the distance score between residues less than δ residues apart was defined as infinity, preventing the optimal path from traversing the region near the diagonal in the dynamic programming matrix (black line in Figure 5). δ = 8 performed well in practice.
Figure 5.
Self-similarity in FGF-1, a three-fold symmetric protein.
Allow circular permutations. When comparing a protein to a rotated copy of itself, the aligned sequence of the rotated copy will appear to be circularly permuted relative to the original protein. This can be seen in Figure 5b as discontinuities in the magenta and cyan alignments. To detect circular permutations we apply an approach similar to Uliel et al. [64]. The dynamic programming matrix is duplicated in one direction (see Figure 5) and CE is run normally. This allows the full length of a symmetric protein to be aligned. The results are then post-processed to map the alignment back onto the single protein. While it is possible with this technique that single residues may be aligned twice, this is rare in practice. In cases where it does occur, alignment length is used as a heuristic to choose which residues to include in the final alignment.
Identifying symmetry order
CE-Symm identifies self-similar structures within a protein. Rotational symmetry is the most abundant form of structural repeat, but linear repeats with high self-similarity can also be found (concentric turns of β-helices, for example). To filter out such cases, we developed an algorithm to estimate the symmetry order of a self-alignment. Proteins with order 1 (no rotational symmetry) were removed from the results.
The algorithm considers a self-alignment to be a function from the set of residues in a protein to itself. We say that f(x) = y if CE-Symm aligned residues x and y. If CE-Symm identified rotational symmetry within the protein, then the repeated function composition fk(x) corresponds to repeated rotations. When the function is applied a number of times equal to the order of the underlying CE-Symm alignment, k*, then fk* (x) ≈ x, corresponding to a rotation by 360°. To identify the order of a self-alignment, successively larger values of k are tried and the root mean squared deviation (RMSD) found for each according to the formula:
The correct order is determined by identifying large decreases in RMSD. In practice, a threshold of 40% decreases was found to correctly identify the order in most cases. If no such drops are identified for k of 8 or less, an order of 1 (no rotational symmetry) is assumed.
We also employed a secondary method to determine order based on the angle between aligned subunits. The rotation axis and angle of rotation is first calculated based on the procedure in Kim et al. [38]. We then compare the angle of rotation, θ, to the ideal angles for proteins with low orders of rotational symmetry.
If this angle is below a threshold, τ, we label the protein as symmetric with order k. For this study we used a stringent threshold of τ = 1°.
Initial tests found two methods to be complementary. Method 1 is more robust to geometrical distortions, while method 2 is more robust to inaccuracies in the alignment. Thus, proteins were classified as symmetric if either method determined the symmetry order to be greater than one.
Scoring schemes
Several alternate scoring schemes were considered, both for optimizing the alignment and for detecting the presence of symmetry. By default, the CE scoring scheme is used to judge the quality of alignments [39]. This is a purely structural scoring which attempts to maximize the alignment length while maintaining a low RMSD. We also implemented an alternate score that incorporates sequence similarity in addition to the structural alignment. Sequence similarity is quantified using the structure-derived substitution matrix [65], which is optimized for the alignment of distantly related proteins. The relative weight of structure and sequence scores can be adjusted with a configuration parameter.
A number of features were considered for classifying proteins as either rotationally symmetric or asymmetric, including RMSD, TM-score, Z-score (as reported by CE), alignment length, and sequence identity. Of these, the TM-score gave the best performance on the ROC curves. A variant of TM-score that incorporates order information was also evaluated, in which 1.0 was added to the TM-score if either method for determining symmetry order determined an order of symmetry greater than 1. This ensures that rotationally symmetric structures always have scores strictly greater than asymmetric ones, reducing false positives especially from helical symmetry and translational repeats. To classify the structure as symmetry or asymmetric, a threshold of ≥ 1.4 is applied to the sum. This last method was yielded the best performance and is recommended by the authors.
Supplementary Material
Acknowledgments
We would like to thank Jean-Pierre Changeux for inspiring discussions about protein symmetry and Chin-Hsien (Emily) Tai for providing access to SymD.
Funding: The RCSB PDB is managed by two members of the RCSB: Rutgers and UCSD, and it is funded by National Science Foundation (NSF), National Institute of General Medical Sciences, Department of Energy (DOE), National Library of Medicine, National Cancer Institute, National Institute of Neurological Disorders and Stroke and National Institute of Diabetes and Digestive and Kidney Diseases. The RCSB PDB is a member of the ww-PDB. This work was supported by the RCSB PDB grant NSF DBI 0829586. SB was also supported by a grant from the National Institutes of Health, USA (NIH grants T32GM8806). Computation provided in part by the Open Science Grid [66, 67]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The census (described in the preceding paragraph) originally used SCOPe 2.01 but was updated to SCOPe 2.03 when that version was released. The benchmark was fixed at SCOPe 2.01. No differences at the level of superfamily or higher exist between the two versions.
References
- 1.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North AC. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960 Feb;185(4711):416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee Jihun, Blaber Michael. Experimental support for the evolution of symmetric protein architecture from a simple peptide motif. PNAS. 2011 Jan;108(1):126–130. doi: 10.1073/pnas.1015032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, Dickerson RE. How proteins recognize the TATA box. J Mol Biol. 1996 Aug;261(2):239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 4.Waldrop Grover L. The role of symmetry in the regulation of bacterial carboxyltransferase. BioMolecular Concepts. 2011;2(1–2) doi: 10.1515/bmc.2011.009. [DOI] [PubMed] [Google Scholar]
- 5.Monod Jacque, Wyman Jeffries, Changeux Jean-Pierre. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 6.Changeux Jean-Pierre, Edelstein Stuart J. Allosteric mechanisms of signal transduction. Science. 2005 Jun;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 7.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 8.Wolynes PG. Symmetry and the energy landscapes of biomolecules. PNAS. 1996 Dec;93(25):14249–14255. doi: 10.1073/pnas.93.25.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraldo Jesus, Ciruela Francisco., editors. Progress in Molecular Biology and Translational Science. Academic Press; 2013. May, Oligomerization in Health and Disease. [Google Scholar]
- 10.Broom Aron, Doxey Andrew C, Lobsanov Yuri D, Berthin Lisa G, Rose David R, Howell P Lynne, McConkey Brendan J, Meiering Elizabeth M. Modular evolution and the origins of symmetry: reconstruction of a three-fold symmetric globular protein. Structure. 2012 Jan;20(1):161–171. doi: 10.1016/j.str.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Matthews Jacqueline M., editor. Protein Dimerization and Oligomerization in Biology - Google Books. Landes Bioscience and Springer Science+ Business Media, LLC; 2012. May, [Google Scholar]
- 12.Kinoshita K, Kidera A, Go N. Diversity of functions of proteins with internal symmetry in spatial arrangement of secondary structural elements. Protein Sci. 1999 Jun;8(6):1210–1217. doi: 10.1110/ps.8.6.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne Philip E. The Protein Data Bank. Nucleic Acids Res. 2000 Jan;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose PW, Bi C, Bluhm Wolfgang F, Christie CH, Dimitropoulos D, Dutta S, Green RK, Goodsell DS, Prlic A, Quesada M, Quinn GB, Ramos AG, DWestbrook J, Young J, Zardecki C, Berman HM, Bourne Philip E. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 2012 Dec;41(D1):D475–D482. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshland DE. The evolution of function in enzymes. Fed. Proc. 1976 Aug;35(10):2104–2111. [PubMed] [Google Scholar]
- 16.Andrade Miguel A, Perez-Iratxeta Carolina, Ponting Chris P. Protein Repeats: Structures, Functions, and Evolution. J Struct Biol. 2001 May;134(2-3):117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 17.Abraham Anne-Laure, Pothier Joël, Rocha Eduardo PC. Alternative to homo-oligomerisation: the creation of local symmetry in proteins by internal amplification. J Mol Biol. 2009 Dec;394(3):522–534. doi: 10.1016/j.jmb.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 18.Blaber Michael, Lee Jihun, Longo Liam. Emergence of symmetric protein architecture from a simple peptide motif: evolutionary models. Cell. Mol. Life Sci. 2012 Jul;69:3999–4006. doi: 10.1007/s00018-012-1077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bershtein Shimon, Mu Wanmeng, Wu Wanmeng, Shakhnovich Eugene I. Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. PNAS. 2012 Mar;109(13):4857–4862. doi: 10.1073/pnas.1118157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagano Nozomi, Orengo Christine A, Thornton Janet M. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002 Aug;321(5):741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 21.Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001 Apr;134(2-3):167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 22.Sadreyev Ruslan I, Kim Bong-Hyun, Grishin Nick V. Discretecontinuous duality of protein structure space. Curr Opin Struct Biol. 2009 Jun;19(3):321–328. doi: 10.1016/j.sbi.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fortenberry Carie, Bowman Elizabeth Anne, Proffitt Will, Dorr Brent, Combs Steven, Harp Joel, Mizoue Laura, Meiler Jens. Exploring symmetry as an avenue to the computational design of large protein domains. J Am Chem Soc. 2011 Nov;133(45):18026–18029. doi: 10.1021/ja2051217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hug Christopher, Lodish Harvey F. The role of the adipocyte hormone adiponectin in cardiovascular disease. Curr Opin Pharmacol. 2005 Apr;5(2):129–134. doi: 10.1016/j.coph.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Maeda Norikazu, Shimomura Iichiro, Kishida Ken, Nishizawa Hitoshi, Matsuda Morihiro, Nagaretani Hiroyuki, Furuyama Naoki, Kondo Hidehiko, Takahashi Masahiko, Arita Yukio, Komuro Ryutaro, Ouchi Noriyuki, Kihara Shinji, Tochino Yoshihiro, Okutomi Keiichi, Horie Masato, Takeda Satoshi, Aoyama Toshifumi, Funahashi Tohru, Matsuzawa Yuji. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat. Med. 2002 Jul;8(7):731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 26.Shklyaev Stanislav, Aslanidi George, Tennant Michael, Prima Victor, Kohlbrenner Eric, Kroutov Vadim, Campbell-Thompson Martha, Crawford James, Shek Eugene W, Scarpace Philip J, Zolotukhin Sergei. Sustained peripheral expression of transgene adiponectin offsets the development of diet-induced obesity in rats. PNAS. 2003 Nov;100(24):14217–14222. doi: 10.1073/pnas.2333912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Combs Terry P, Pajvani Utpal B, Berg Anders H, Lin Ying, Jelicks Linda A, Laplante Mathieu, Nawrocki Andrea R, Rajala Michael W, Parlow Albert F, Cheeseboro Laurelle, Ding Yang-Yang, Russell Robert G, Lindemann Dirk, Hartley Adam, Baker Glynn RC, Obici Silvana, Deshaies Yves, Ludgate Marian, Rossetti Luciano, Scherer Philipp E. A transgenic mouse with a deletion in the collagenous domain of adiponectin displays elevated circulating adiponectin and improved insulin sensitivity. Endocrinology. 2004 Jan;145(1):367–383. doi: 10.1210/en.2003-1068. [DOI] [PubMed] [Google Scholar]
- 28.Min Xiaoshan, Lemon Bryan, Tang Jie, Liu Qiang, Zhang Richard, Walker Nigel, Li Yang, Wang Zhulun. Crystal structure of a single-chain trimer of human adiponectin globular domain. FEBS Lett. 2012 Mar;586(6):912–917. doi: 10.1016/j.febslet.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 29.Ge Hongfei, Xiong Yumei, Lemon Bryan, Lee Ki Jeong, Tang Jay, Wang Ping, Weiszmann Jennifer, Hawkins Nessa, Laudemann John, Min Xiaoshan, Penny David, Wolfe Tom, Liu Qiang, Zhang Richard, Yeh Weh-Chen, Shen Wenyan, Lindberg Richard, Wang Zhulun, Sheng Jackie, Li Yang. Generation of novel long-acting globular adiponectin molecules. J Mol Biol. 2010 May;399(1):113–119. doi: 10.1016/j.jmb.2010.03.062. [DOI] [PubMed] [Google Scholar]
- 30.Bliven Spencer E, Prlić Andreas. Circular permutation in proteins. PLoS Comput Biol. 2012 Mar;8(3):e1002445. doi: 10.1371/journal.pcbi.1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuguchi K, Go N. Comparison of spatial arrangements of secondary structural elements in proteins. Protein Eng. 1995 Apr;8(4):353–362. doi: 10.1093/protein/8.4.353. [DOI] [PubMed] [Google Scholar]
- 32.Murray Kevin B, Taylor William R, Thornton Janet M. Toward the detection and validation of repeats in protein structure. Proteins: Structure, Function, and Bioinformatics. 2004 Nov;57(2):365–380. doi: 10.1002/prot.20202. [DOI] [PubMed] [Google Scholar]
- 33.Shih Edward SC, Hwang Ming-Jing. Alternative alignments from comparison of protein structures. Proteins: Structure, Function, and Bioinformatics. 2004 Aug;56(3):519–527. doi: 10.1002/prot.20124. [DOI] [PubMed] [Google Scholar]
- 34.Shih Edward SC, Gan Ruei-chi R, Hwang Ming-Jing. OPAAS: a web server for optimal, permuted, and other alternative alignments of protein structures. Nucleic Acids Res. 2006 Jul;34(Web Server issue):W95–W98. doi: 10.1093/nar/gkl264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham Anne-Laure, Rocha Eduardo PC, Pothier Joël. Swelfe: a detector of internal repeats in sequences and structures. Bioinformatics. 2008 Jul;24(13):1536–1537. doi: 10.1093/bioinformatics/btn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Hanlin, Huang Yanzhao, Xiao Yi. A simple method of identifying symmetric substructures of proteins. Comput Biol Chem. 2009 Feb;33(1):100–107. doi: 10.1016/j.compbiolchem.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 37.Guerler Aysam, Wang Connie, Knapp Ernst Walter. Symmetric structures in the universe of protein folds. J Chem Inf Model. 2009 Sep;49(9):2147–2151. doi: 10.1021/ci900185z. [DOI] [PubMed] [Google Scholar]
- 38.Kim Changhoon, Basner Jodi, Lee Byungkook. Detecting internally symmetric protein structures. BMC Bioinformatics. 2010;11:303. doi: 10.1186/1471-2105-11-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shindyalov IN, Bourne Philip E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998 Aug;11(9):739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 40.Jia Yuting, Dewey T Gregory, Shindyalov Ilya N, Bourne Philip E. A new scoring function and associated statistical significance for structure alignment by CE. J Comput Biol. 2004;11(5):787–799. doi: 10.1089/cmb.2004.11.787. [DOI] [PubMed] [Google Scholar]
- 41.Prlić Andreas, Bliven Spencer E, Rose Peter W, Bluhm Wolfgang F, Bizon Chris, Godzik Adam, Bourne Philip E. Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics. 2010 Dec;26(23):2983–2985. doi: 10.1093/bioinformatics/btq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayr Gabriele, Domingues Francisco S, Lackner Peter. Comparative analysis of protein structure alignments. BMC Struct Biol. 2007;7:50. doi: 10.1186/1472-6807-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Yang, Skolnick Jeffrey. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye Yuzhen, Godzik Adam. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003 Oct;19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 45.Fox Naomi K, Brenner Steven E, Chandonia John-Marc. SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014 Jan;42(1):D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murzin Alexey G, Brenner Steven E, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995 Apr;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 47.Andreeva Antonina, Howorth Dave, Chandonia John-Marc, Brenner Steven E, Hubbard Tim JP, Chothia Cyrus, Murzin Alexey G. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008 Jan;36(Database issue):D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vergara Ismael A, Norambuena Tomás, Ferrada Evandro, Slater Alex W, Melo Francisco. StAR: a simple tool for the statistical comparison of ROC curves. BMC Bioinformatics. 2008;9:265. doi: 10.1186/1471-2105-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandonia John-Marc, Hon Gary, Walker Nigel S, Lo Conte Loredana, Koehl Patrice, Levitt Michael, Brenner Steven E. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32(Database issue):D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klingenberg M. Membrane protein oligomeric structure and transport function. Nature. 1981 Apr;290(5806):449–454. doi: 10.1038/290449a0. [DOI] [PubMed] [Google Scholar]
- 51.Choi Sungwon, Jeon Jouhyun, Yang Jae-Seong, Kim Sanguk. Common occurrence of internal repeat symmetry in membrane proteins. Proteins: Structure, Function, and Bioinformatics. 2008 Apr;71(1):68–80. doi: 10.1002/prot.21656. [DOI] [PubMed] [Google Scholar]
- 52.Wright Caroline F, Teichmann Sarah A, Clarke Jane, Dobson Christopher M. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005 Dec;438(7069):878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 53.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000 Jan;28(1):304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitman CP, Hegeman GD, Cleland WW, Kenyon GL. Symmetry and asymmetry in mandelate racemase catalysis. Biochemistry. 1985 Jul;24(15):3936–3942. doi: 10.1021/bi00336a020. [DOI] [PubMed] [Google Scholar]
- 55.Bergdoll M, Eltis LD, Cameron AD, Dumas P, Bolin JT. All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci. 1998 Aug;7(8):1661–1670. doi: 10.1002/pro.5560070801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dutzler Raimund, Campbell Ernest B, MacKinnon Roderick. Gating the selectivity filter in ClC chloride channels. Science. 2003 Apr;300(5616):108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 57.Forrest Lucy R, Rudnick Gary. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009 Dec;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park Sang-Youn, Chao Xingjuan, Gonzalez-Bonet Gabriela, Beel Bryan D, Bilwes Alexandrine M, Crane Brian R. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol Cell. 2004 Nov;16(4):563–574. doi: 10.1016/j.molcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 59.Punta Marco, Coggill Penny C, Eberhardt Ruth Y, Mistry Jaina, Tate John, Boursnell Chris, Pang Ningze, Forslund Kristoffer, Ceric Goran, Clements Jody, Heger Andreas, Holm Liisa, Sonnhammer Erik LL, Eddy Sean R, Bateman Alex, Finn Robert D. The Pfam protein families database. Nucleic Acids Res. 2012 Jan;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelman Z, O’Donnell M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995 Sep;23(18):3613–3620. doi: 10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korkhov Vladimir M, Mireku Samantha A, Locher Kaspar P. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature. 2012 Oct;490(7420):367–372. doi: 10.1038/nature11442. [DOI] [PubMed] [Google Scholar]
- 62.Levy Emmanuel D, Pereira-Leal Jose B, Chothia Cyrus, Teichmann Sarah A. 3D complex: a structural classification of protein complexes. PLoS Comput Biol. 2006 Nov;2(11):e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaudhuri Indronil, Söding Johannes, Lupas Andrei N. Evolution of the β-propeller fold. Proteins. 2008;71(2):795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 64.Uliel S, Fliess A, Amir A, Unger R. A simple algorithm for detecting circular permutations in proteins. Bioinformatics. 1999 Nov;15(11):930–936. doi: 10.1093/bioinformatics/15.11.930. [DOI] [PubMed] [Google Scholar]
- 65.Prlic A, Domingues FS, Sippl MJ. Structure-derived substitution matrices for alignment of distantly related sequences. Protein Eng Des Sel. 2000 Aug;13(8):545–550. doi: 10.1093/protein/13.8.545. [DOI] [PubMed] [Google Scholar]
- 66.Sfiligoi I, Bradley DC, Holzman B, Mhashilkar P, Padhi S, Wurthwein F. The Pilot Way to Grid Resources Using glideinWMS. Computer Science and Information Engineering, 2009 WRI World Congress on. 2009;2:428–432. [Google Scholar]
- 67.Pordes Ruth, Petravick Don, Kramer Bill, Olson Doug, Livny Miron, Roy Alain, Avery Paul, Blackburn Kent, Wenaus Torre, Würthwein Frank, Foster Ian, Gardner Rob, Wilde Mike, Blatecky Alan, McGee John, Quick Rob. The open science grid. J. Phys.: Conf. Ser. 2007 Aug;78(1):012057. [Google Scholar]
- 68.Zhang Yang, Skolnick Jeffrey. Scoring function for automated assessment of protein structure template quality. Proteins: Structure, Function, and Bioinformatics. 2004 Dec;57(4):702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 69.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000 Jan;10(1):17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 70.Webb EC, IUBMB . Enzyme nomenclature 1992. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. Academic Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.