Abstract
Luminal A and B breast cancers are the most prevalent forms of breast cancer diagnosed in women. Compared to luminal A breast cancer patients, patients with luminal B breast cancers experience increased disease recurrence and lower overall survival. The mechanisms that regulate the luminal B subtype remain poorly understood. The chemokine CCL2 is overexpressed in breast cancer, correlating with poor patient prognosis. The purpose of this study was to determine the role of CCL2 expression in luminal B breast cancer cells. Breast tissues, MMTV-PyVmT and MMTV-Neu transgenic mammary tumors forming luminal B-like lesions, were immunostained for CCL2 expression. To determine the role of CCL2 in breast cancer cells, CCL2 gene expression was silenced in mammary tumor tissues and cells using TAT cell-penetrating peptides non-covalently cross linked to siRNAs (Ca-TAT/siRNA). CCL2 expression was examined by ELISA and flow cytometry. Cell growth and survival were analyzed by flow cytometry, immunocytochemistry, and fluorescence microscopy. CCL2 expression was significantly increased in luminal B breast tumors, MMTV-PyVmT and MMTV-Neu mammary tumors, compared or normal breast tissue or luminal A breast tumors. Ca-TAT delivery of CCL2 siRNAs significantly reduced CCL2 expression in PyVmT mammary tumors, and decreased cell proliferation and survival. CCL2 gene silencing in PyVmT carcinoma cells or BT474 luminal B breast cancer cells decreased cell growth and viability associated with increased necrosis and autophagy. CCL2 expression is overexpressed in luminal B breast cancer cells and is important for regulating cell growth and survival by inhibiting necrosis and autophagy.
Keywords: CCL2, siRNA, TAT cell-penetrating peptide, Luminal breast cancer, Necrosis, Autophagy
Introduction
With approximately 174,000 cases per year, luminal breast cancers are the most common forms of breast cancer diagnosed in the US and Canada [1, 2]. In contrast to Her2-overexpressing breast cancers, or triple negative breast cancers, which lack expression of estrogen receptor (ER), Progesterone receptor, and Her2, luminal breast cancers frequently express hormone and Her2 receptors, rendering these luminal breast cancers responsive to anti-hormonal and Her2-targeted therapies [2–4]. Gene profiling studies have revealed that a subset of luminal breast cancers show high expression of cell proliferation genes. This poorly understood subset, classified as luminal B, is significantly associated with lower patient survival, and is less responsive to anti-hormonal therapy compared to luminal A breast cancers [1, 3, 5]. Understanding the mechanisms that drive luminal B breast cancers would contribute to improved treatments tailored specifically to this disease.
CCL2 (MCP-1) is a chemotactic cytokine (8 kDa), which binds to CCR2 G-protein-coupled receptors to regulate macrophage recruitment during normal processes such as wound healing [6–13], and during inflammatory diseases such as rheumatoid arthritis [14, 15]. CCL2 is overexpressed in breast cancer tumors [16–18]. CCL2 expression in the stroma correlates with macrophage recruitment and poor patient prognosis [18, 19]. Antibody neutralization of CCL2 in tumor-bearing mice inhibits breast tumor growth and metastasis, and reduces the number of M2 polarized macrophages in primary tumors [19, 20]. Macrophage depletion through clodronate treatment or knockout of the CSF-1 Receptor in tumor-bearing mice inhibits tumor growth and metastasis, associated with a reduction in angiogenesis, cell proliferation, and survival [19, 21]. These studies demonstrate an important role for CCL2-mediated macrophage recruitment in breast cancer progression.
Recent studies showed that CCL2 was highly expressed in tumor epithelium, and signaled directly to breast cancer cells, to regulate cell survival, migration, and invasion through CCR2-dependent mechanisms [22, 23]. With few analytical tools, the role of epithelial CCL2 expression has not been well studied. Neutralizing antibodies block CCL2 activity non-specifically in tissues [19, 20, 24]. Small interfering RNAs (siRNAs) could selectively inactivate critical oncogenes if coupled to a carrier that targeted carcinoma cells [25, 26]. The HIV-1 derived trans-activating transcriptor peptide (TAT49–57: RKKRRQRRR) exhibits unusual properties by efficiently penetrating cell membranes, independent of temperature or cell surface receptor expression [27]. In recent studies, TAT peptides calcium cross linked to siRNAs or plasmids (Ca-TAT/siRNA or Ca-TAT/plasmid) formed stable nanoparticles that transfected basal and lung carcinoma cells, with lower toxicity and higher efficiency than conventional polymeric carriers or TAT peptides alone [28, 29]. Thus, Ca-TAT peptides may be used to block CCL2 expression and activity in breast carcinoma cells.
The relevance of CCL2 expression in specific subtypes has remained unclear. We show for the first time that CCL2 expression is overexpressed in luminal B breast cancer. Using Ca-TAT/siRNA complexes to knockdown CCL2 expression in ex vivo tissue models and in breast cancer cell lines, we demonstrate that epithelial-specific CCL2 positively regulates carcinoma cell growth, and negatively regulates necrosis and autophagy. These studies reveal novel mechanisms for the regulation of luminal B breast cancers, and identify a potentially important therapeutic target for this breast cancer subtype.
Methods
Tissue microarrays
Paraffin-embedded breast cancer tissue microarrays were obtained from the National Cancer Institute Diagnostics Program. The arrays were composed of 24 cores of normal breast tissue 1 mm in diameter (6 cores/slide), and 400 cores of invasive breast ductal carcinoma 0.6 mm in diameter (100 cores/slide). Using clinical criteria previously validated [2, 30, 31], luminal A breast cancers were identified as ER+ and/or PR+, Her2−, with a mitotic score of 1. Luminal B breast cancers were identified as ER and/or PR+, Her2+ or Her2−, with a mitotic score of 2+.
Tissue harvesting
MMTV-PyVmT positive animals were generated as described [32], and were maintained in accordance with AAALAC and University of Kansas Medical Center guidelines. Late-stage carcinomas were harvested from PyVmT transgenic females 14–16 weeks of age for ex vivo cultures, or were fixed in 10 % neutral formalin buffer (10 % NBF) for immunohistochemistry, as described previously [33]. MMTV-Neu tumors derived from FVB mice 8–10 weeks of age were kindly provided by Harold L. Moses, M.D (Vanderbilt University Medical Center). Normal mammary glands were harvested from age-matched wild type FVB females.
Immunohistochemistry
Five micron sections were stained for CCL2 expression as described [22]. Briefly, sections were heated in 1 M urea, blocked in PBS containing 5 % rabbit serum, and incubated with antibodies (1:100) to murine or human CCL2 (Santa Cruz Biotechnology), overnight at 4 °C. CCL2 expression was detected using secondary goat biotinylated antibodies (Vector Laboratories, 1:1000 dilution), conjugated to streptavidin peroxidase (Vector Laboratories). Reactions were catalyzed with 3,3′-Diaminobenzidine (DAB) substrate (Dako).
Cell culture
Sum225 cells originated from Asterand. Raw 264.7, BT474, MCF10A, and MCF-7 cell lines were obtained from the American tissue culture collection. PyVmT carcinoma cells were isolated from transgenic mice (Jackson Laboratories), as described [23, 34]. All cell lines were cultured as commercially specified. Normal fibroblasts were isolated from mouse mammary tissue, validated, and cultured as described [33].
siRNA reagents
Sense and anti-sense oligonucleotides were synthesized and annealed by Dharmacon Fisher. The following siRNA targeting sequences were designed: enhanced green fluorescent protein (eGFP) [35] as a negative control: 5′-GCUGACCCUGAAGUUCAUC-3′, mCCL2si: 5′-AAACCUGGAUCGGAACCAA-3′, huCCL2si1: 5′-ACCUGCUGUUAUAACUUCA-3′, huCCL2si2: 5′-CAGCAAGUGUCCCAAAGAA-3′.
Preparation of Ca-TAT complexes
The following formula was used to determine the amount of TAT peptide needed for a specific N/P ratio per μg of DNA or siRNA: μg of TAT = 0.446 × (N/P ratio) + 0.116. TAT peptides were mixed with siRNA or plasmid DNA in 45 μl sterile deionized water containing: 37.5, 75, or 112.5 mM CaCl2. The solution was pipetted 20 times and incubated on ice for 20 min. For in vitro studies, these complexes were added directly to cells. For ex vivo cultures, 25 μl of 10 % glucose was added to Ca-TAT complexes. Ca-TAT complexes were diluted in PBS to a total volume of 100 μl, before injection.
Luciferase assay
40,000 cells/well were plated in duplicate onto 24-well plates, and transfected with Ca-TAT peptides complexed to 2 μg MSCV-luciferase plasmid(Addgene) for 48 h. Cells were analyzed for luciferase activity using the Promega Luciferase Assay System, and a Veritas Microplate Luminometer.
ELISA
40,000 cells/well were seeded in duplicate onto 24-well plates, and incubated in 400 μl/well serum-free DMEM for 24 h. Conditioned media were analyzed by murine or human CCL2 ELISA kit (Peprotech). Reactions were catalyzed using tetramethylbenzidine substrate (RnD systems). Absorbance was read at OD 450 nM using a BioTek Microplate Reader.
Ex vivo culture
Using a scalpel, tumors were partitioned into 0.125 cm3 sizes, placed in mesh filters (70 micron pores) fitted to 6-well plates, and cultured in DMEM/10 % FBS with antibiotics for 24 h. Ca-TAT peptides were complexed to 10 μg siRNA, and injected into four different areas of the tumor at 25 μl increments using a 27-gage needle. Tissues were incubated for 48 h, before analysis.
Flow cytometry
PyVmT mammary tumors were digested into single-cell suspensions, as described [36], and then fixed in 10 % NBF overnight at 4 °C. Cells were permeabilized with 0.1 % Triton-X-100 at 37 °C for 15 min, washed in PBS, incubated with the following antibodies overnight at a 1:50 dilution in PBS/2 % BSA: murine CCL2 (Santa Cruz Biotechnology), Ki67 (Santa Cruz Biotechnology), HMGB1 (Cell Signaling Technology) or LC3B (Invitrogen), CD24 (Santa Cruz Biotechnology), Fsp1 (Abcam) or Cd11b-FITC (BD Biosciences). CCL2 or CD24 proteins were detected by anti-goat-Alexa-488 (Life Technologies, 1: 500 dilution), on ice for 1 h. Ki67, HMGB1, LC3B, or Fsp1 proteins were detected by anti-rabbit-Alexa-647 (Life Technologies, 1: 500 dilution). Cells were washed with PBS three times, before analysis on an LSRII Flow cytometer.
Propidium iodide assay
500,000 cells/well were seeded in 6-cm dishes for 24 h. Cells were transfected with Ca-TAT peptides complexed to 6 μg siRNA for 48 h. Cells were detached by 5 mM EDTA treatment and stained with 1 μg/ml of PI for 15 min at room temperature. Cells were assayed using an LSRII Flow cytometer.
Immunocytochemistry
10,000 cells/well were seeded in triplicate onto 96-well plates for 24 h. Cells were transfected with Ca-TAT peptides complexed to 500 ng siRNA for 48 h. Cells were fixed in 10 %NBF, methanol permeabilized, blocked with PBS/3 % FBS, and immunostained overnight with antibodies to Ki67 (Santa Cruz Technology, 1:50 dilution), HMGB1 (Cell Signaling Technology, 1:100 dilution), LC3B expression (Cell Signaling Technology, 1:100 dilution), and conjugated to rabbit-biotinylated antibodies (1:1000 dilution). Expression was detected by incubation with streptavidin-peroxidases and DAB substrate. Three fields per sample were captured using a Motic AE31 inverted microscope. Protein expression was quantified by Image J [23], and normalized to hematoxylin counterstain.
Viability/cell count growth assay
8000 cells/well were seeded in triplicate onto 96-well plates for 24 h. Cells were transfected with Ca-TAT complexed to 500 ng siRNA for up to 48 h, washed with PBS, detached by trypsin and stained with 0.1 % trypan blue. The number of trypan blue-positive cells was counted by hemocytometer and normalized to the total number of cells per field. Brightfield images were captured at ×10 magnification using a Motic microscope. Growth was measured by the total number of viable cells at 24 and 48 h.
Autophagy assay
100,000 cells were seeded onto coverslips coated with 0.1 % gelatin. Cells were transfected with Ca-TAT complexed to 5 μg siRNA for 48 h. Cells were incubated with Cyto-ID® green dye uptake as described (Enzo Life Sciences). Images were captured at ×20 magnification using a Motic microscope. Uptake was quantified by Image J [23], and normalized to DAPI counterstain.
Statistical analysis
Experiments were performed in a minimum of triplicate. Data are expressed as mean ± SE of the mean (SEM). Statistical analysis was performed using Mann–Whitney two-sample test or ANOVA with Bonferroni post-hoc comparisons using Graphpad Software. Statistical significance was determined by p < 0.05. *p < 0.001, **p < 0.01, ***p < 0.05, ****p > 0.05.
Results
Expression patterns of CCL2 in luminal breast cancer
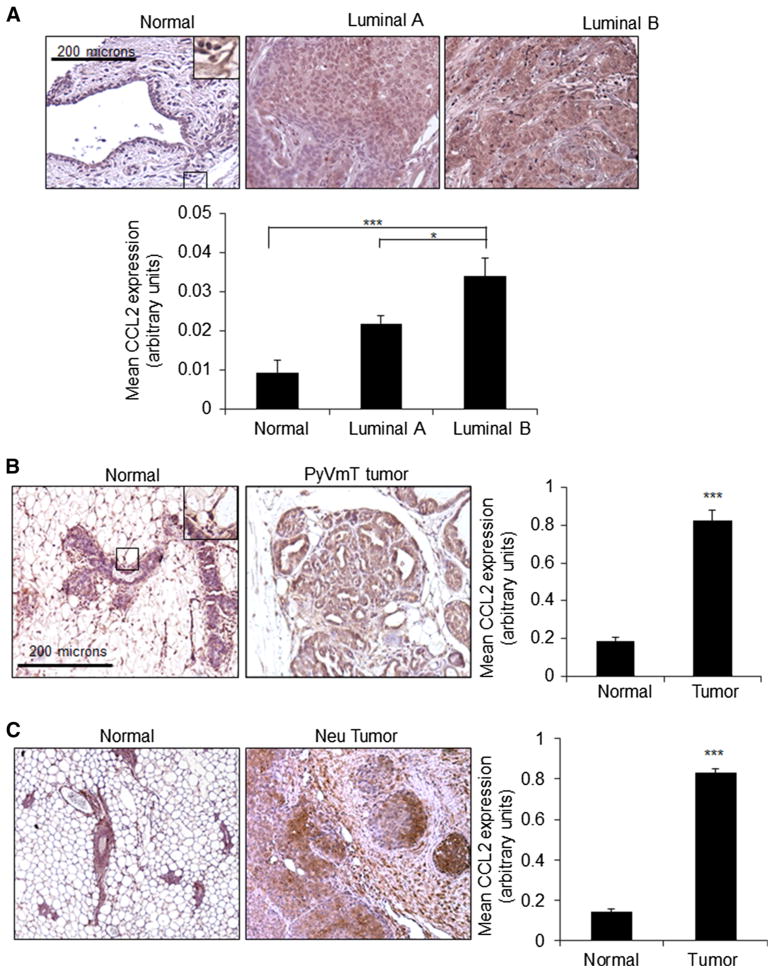
To determine the relevance of CCL2 expression in luminal breast cancer, we analyzed patient samples for CCL2 expression by immunohistochemistry. CCL2 expression was lowly expressed in normal breast epithelium, and in some stromal cells including fibroblasts and macrophages. Luminal B breast tumors showed the highest levels of CCL2 expression, compared to luminal A breast tumors and normal breast tissues (Fig. 1a). Animal models are vital for the testing of new molecular therapies [37, 38]. Therefore, we analyzed CCL2 expression in mouse mammary tumors induced by polyoma virus middle T antigen (PyVmT) or Neu overexpression, under the control of the mouse mammary tumor virus promoter (MMTV). PyVmT and Neu transgenic mice form invasive mammary tumors histologically similar to breast ductal carcinomas [34, 35], and are of luminal B classification [39, 40]. Similar to patient samples, CCL2 was more highly expressed in MMTV-PyVmT and MMTV-Neu mammary tumors compared to normal tissues (Fig. 1b, c). These studies demonstrate increased CCL2 expression in luminal B breast cancer.
Fig. 1.
CCL2 expression is increased in luminal B breast cancer.
a Immunohistochemistry staining was performed for CCL2 expression in patient samples of normal breast tissue (n = 24), luminal A (n = 173) or luminal B (n = 81) breast tumor tissues.
b Immunohistochemistry staining was performed for CCL2 expression in normal mouse mammary gland (n = 11) or PyVmT mammary tumor tissues (n = 17).
c Immunohistochemistry staining was performed for CCL2 expression in normal mammary gland (n = 7) or Neu-overexpressing mammary tumor tissues (n = 7). Magnified inset shows low-level expression in normal epithelium and stromal cells. Expression was quantified by Image J. Statistical analysis was performed by One Way ANOVA with Bonferroni post hoc comparison (A) or Mann–Whitney two-sample test (B). Statistical significance was determined by p value < 0.05. *p < 0.05, ***p < 0.001. Values are shown as Mean ± SEM
Calcium chloride and charge ratios modulate transfection efficiency to mammary carcinoma cells
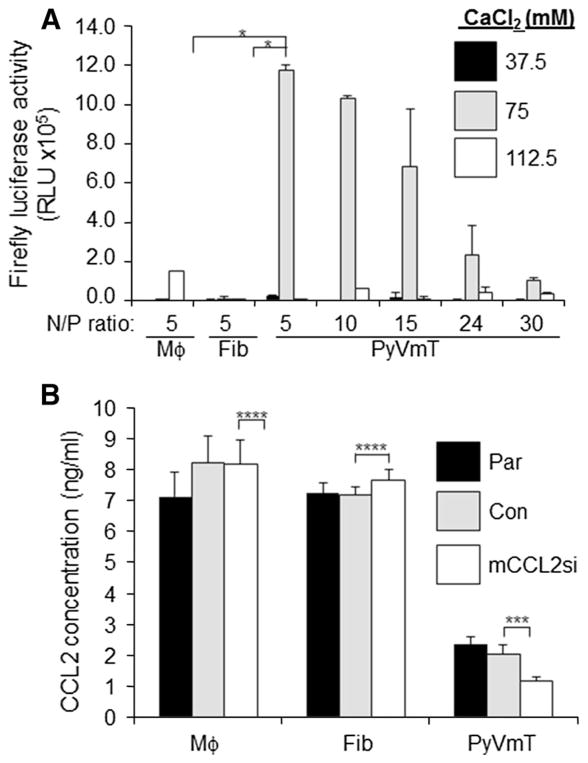
To study the functional role of CCL2 in luminal B breast cancer, we designed a strategy to silence CCL2 expression in mammary carcinoma cells using Ca-TAT peptides. Studies have indicated that transfection efficiency of Ca-TAT/siRNA or Ca-TAT/plasmid complexes depends on cell type, CaCl2 concentration, and charge ratio (N/P) ratio of amino groups to phosphate groups [28, 29]. To optimize transfection conditions for mammary carcinoma cells, PyVmT cells were transfected with Ca-TAT/luciferase plasmid complexes, formulated at varying CaCl2 concentrations and N/P ratios. Ca-TAT/plasmid complexes formulated at N/P = 5 in 75 mM CaCl2 induced the highest level of luciferase activity in PyVmT mammary carcinoma cells (Fig. 2a). Mesenchymal cells such as mammary fibroblasts and macrophages, showed minimal luciferase activity at this formulation. These data indicate that Ca-TAT/plasmid complexes preferentially transfected mammary carcinoma cells.
Fig. 2.
Calcium concentrations and N/P ratios modulate transfection efficiency of Ca-TAT complexes to PyVmT mammary carcinoma cells. a Mammary fibroblasts (Fib), Raw 264.7 macrophages (Mϕ) or PyVmT mammary carcinoma cells, were transfected with Ca-TAT peptides complexed to MSCV-luciferase plasmids at the indicated CaCl2 concentrations and N/P ratios. Luciferase activity was measured 48 h post-transfection. RLU relative light units. b Ca-TAT peptides were complexed to control siRNA (Con) or CCL2 siRNAs (mCCL2si), and transfected into the indicated cell lines. CCL2 expression was measured in parental (Par) or transfected cells by ELISA 48 h later. Statistical analysis was performed by One-Way ANOVA, followed by Bonferroni post-hoc comparisons. Statistical significance was determined by p value < 0.05. *p < 0.001,***p < 0.05, ****p > 0.05. Values are shown as Mean ± SEM
Using this formulation, PyVmT cells were transfected with Ca-TAT peptides complexed to control or CCL2 siRNAs (mCCL2si) complexes. Ca-TAT/mCCL2si complexes significantly knocked down CCL2 expression in PyVmT cells, but not in cultured macrophages or fibroblasts (Fig. 2b). While cultured PyVmT cells showed lower levels of CCL2 compared to stromal cells, flow cytometry analysis revealed that carcinoma cells comprised the bulk (75%) of PyVmT tumor tissues (Supplemental Fig. 1). These studies indicate that Ca-TAT/plasmid or Ca-TAT/siRNA complexes formulated at N/P = 5 and 75 mM CaCl2 optimally transfect mammary carcinoma cells, a major source for CCL2 expression in tumor tissues.
CCL2 gene silencing inhibits cell growth and survival in PyVmT mammary tumor tissues
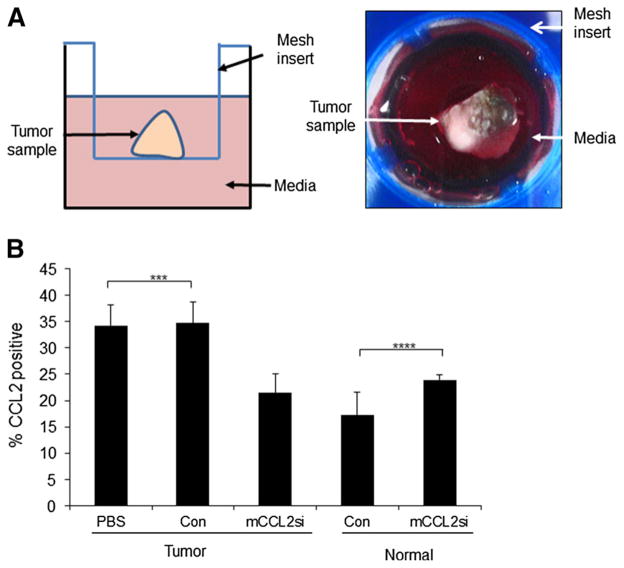
To determine the functional relevance of CCL2 expression in the context of tumor tissues, we used a modified ex vivo tissue culture system [41]. This efficient and cost-effective system allowed us to control for the size of tissue samples and dosage of Ca-TAT/siRNA delivery, minimizing potential variations in treatment. PyVmT mammary tumors were cultured in suspension (Fig. 3a), injected with Ca-TAT/siRNA complexes, and analyzed by flow cytometry. Injecting tumor samples with Ca-TAT/mCCL2si complexes significantly reduced CCL2 protein expression by 40 % (Fig. 3b). CCL2 expression was not affected in normal mammary tissue (Fig. 3b), indicating a preference for Ca-TAT/siRNA complexes in targeting mammary tumor tissues.
Fig. 3.
Ca-TAT/siRNA complexes selectively target CCL2 gene expression in mammary tumor tissues cultured ex vivo. a Left panel Diagram of PyVmT mammary tumors cultured in suspension ex vivo. Ca-TAT complexes are injected directly into the tumor tissue. Right panel representative image of a tumor sample shown in an overhead view. b Flow cytometry analysis was performed to measure overall CCL2 expression in normal mammary tissues or PyVmT tumors injected with phosphate buffered saline (PBS vehicle control), or Ca-TAT peptides complexed to control siRNA (Con) or CCL2 siRNA (mCCL2si). CCL2 expression levels were normalized to secondary antibody only control, and are relative to whole cell population in either the tumor or normal group. N = 6 per group. Statistical analysis was performed using One-Way ANOVA with Bonferroni post-hoc comparison. Statistical significance was determined by p value < 0.05. ***p < 0.05, ****p > 0.05. Values are shown as Mean ± SEM
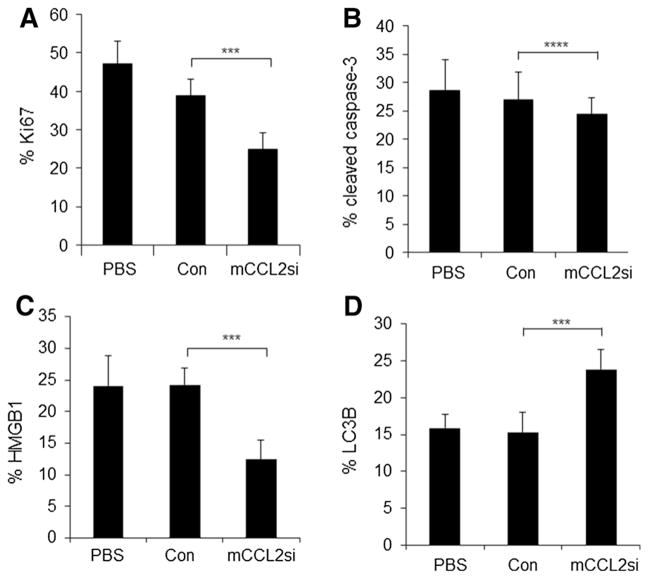
We analyzed the effects of CCL2 knockdown on expression of markers associated with cell proliferation or cell death. Compared to control siRNA-treated tumors, tumors injected with Ca-TAT/mCCL2si complexes showed a 30 % decrease in Ki67 expression (Fig. 4a), indicating decreased cell proliferation [2]. CCL2 knockdown did not significantly affect expression of cleaved caspase-3 (Fig. 4b), a marker for apoptosis [41]. CCL2 knockdown significantly decreased intracellular HMGB1 expression by 33 % (Fig. 4c), indicating increased necrosis [42, 43]. CCL2 knockdown increased expression of LC3B by 30 % (Fig. 4d), demonstrating increased autophagy [44]. These studies indicate that CCL2 knockdown inhibited PyVmT tumor cell proliferation and survival in ex vivo cultures.
Fig. 4.
Ca-TAT-mediated delivery of CCL2 siRNAs to PyVmT ex vivo cultures lead to decreased cell proliferation and increased necrosis and autophagy. PyVmT tumor tissues were cultured for 24 h before injection with: PBS vehicle control, or Ca-TAT complexed to control siRNA (Con) or CCL2 siRNA (mCCL2si). Samples were analyzed by flow cytometry 48 h post-injection, for expression of the following: a Ki67, b cleaved caspase-3, c HMGB1, or d LC3B. Expression levels were normalized to samples stained with secondary antibody only. Statistical analysis was performed using One-Way ANOVA followed by Bonferroni post-hoc comparisons. Statistical significance was determined by p value < 0.05, ***p < 0.05, ****p > 0.05. Values are expressed as Mean ± SEM. N = 6 per group
CCL2 gene silencing enhances necrosis and autophagy in cultured PyVmT mammary carcinoma cells
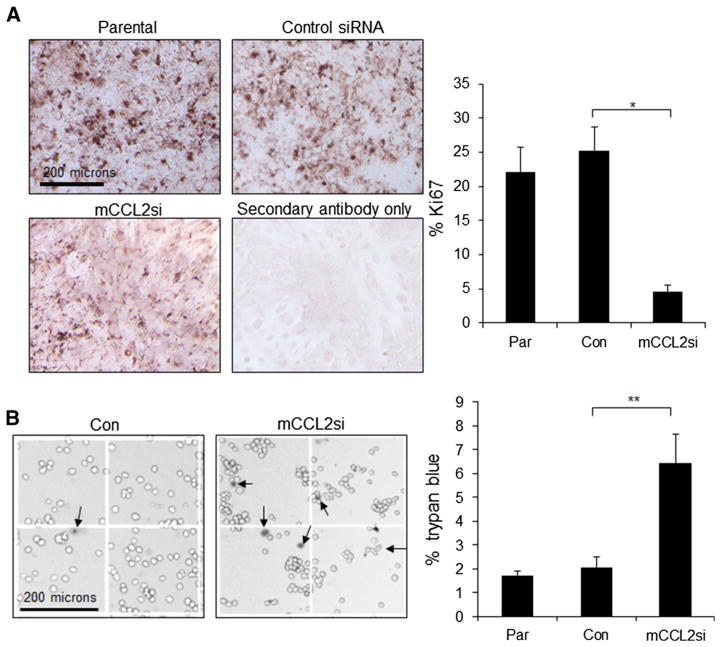
We next determined which cell types were most affected by CCL2 knockdown, using cultured cells. Ca-TAT/mCCL2si complexes significantly decreased cell proliferation in PyVmT cells, as determined by Ki67 expression (Fig. 5a), and cell count assay (Supplemental Fig. 2). Moreover, CCL2 knockdown significantly increased trypan blue staining in PyVmT cells (Fig. 5b). Ca-TAT/siRNA complexes did not affect cell proliferation or viability of macrophages or fibroblasts (Supplemental Fig. 3). These studies indicate that Ca-TAT/mCCL2si complexes selectively inhibited PyVmT carcinoma cell proliferation and survival.
Fig. 5.
CCL2 gene silencing in PyVmT mammary carcinoma cells leads to decreased cell proliferation and viability. PyVmT mammary carcinoma cells were transfected with Ca-TAT peptides complexed to control (Con) or CCL2 siRNAs (mCCL2si). Parental cells (Par) or transfected cells were analyzed for changes in the following: a Ki67 expression by immunocytochemistry staining, followed by hematoxylin counterstain. Expression was quantified by Image J, and normalized to hematoxylin staining. b Cell viability by trypan blue staining. Arrows in magnified sample images point to positive staining. Statistical analysis was performed using One-Way ANOVA with Bonferroni post-hoc comparisons. Statistical significance was determined by p value < 0.05.
*p < 0.001,**p < 0.01. Values are shown as Mean ± SEM
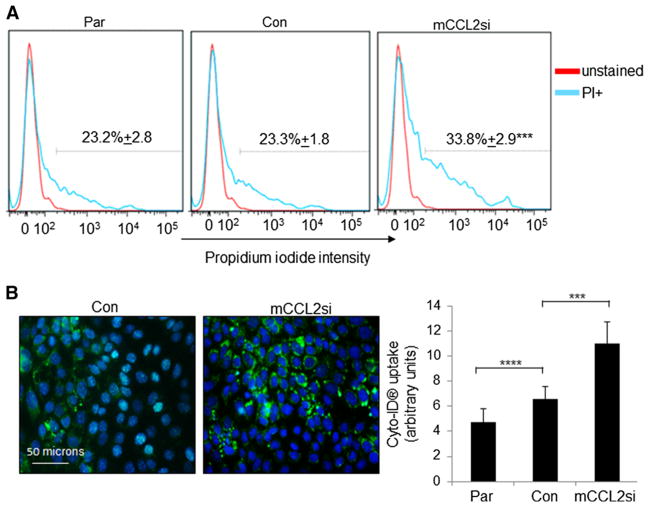
We further examined the effects of CCL2 silencing on necrosis and autophagy in cultured cells. PyVmT mammary carcinoma cells were examined for changes in propidium iodide (PI) staining, which labels cells undergoing cellular necrosis [45]. Consistent with decreased HMGB1 expression in ex vivo cultures, CCL2 knockdown significantly increased the number of PI+ cells, indicating increased cellular necrosis (Fig. 6a). To examine the role of CCL2 expression on cellular autophagy, PyVmT cells were transfected with Ca-TAT/siRNA complexes, and examined for live cell uptake of Cyto-ID® substrate, which labels lysosomal/autophagic vacuoles. CCL2 knockdown significantly increased the uptake of Cyto-ID® (Fig. 6b), which corresponded to increased trypan blue staining (Fig. 5b). In summary, CCL2 knockdown in PyVmT mammary carcinoma cells enhances necrosis and cellular autophagy, ultimately decreasing cell viability.
Fig. 6.
CCL2 gene silencing in PyVmT mammary carcinoma cells leads to increased necrosis and autophagy. PyVmT cells were transfected with Ca-TAT peptides complexed to control (Con) or CCL2 siRNAs (mCCL2si). 48 h later, transfected cells or parental cells (Par) were analyzed for the following: a Propidium iodide (PI) staining by flow cytometry. b Autophagosome formation by uptake of Cyto-ID® green dye. LC3B expression was quantified by Image J. Levels were normalized to DAPI stain. Statistical analysis was performed using One-Way ANOVA with Bonferroni post-hoc comparisons. Significance was determined by p < 0.05. ***p < 0.05, ****p > 0.05. For (A), ***p < 0.05 versus Control siRNA group. Values are shown as Mean ± SEM
CCL2 gene silencing enhances necrosis and autophagy of luminal B breast cancer cells
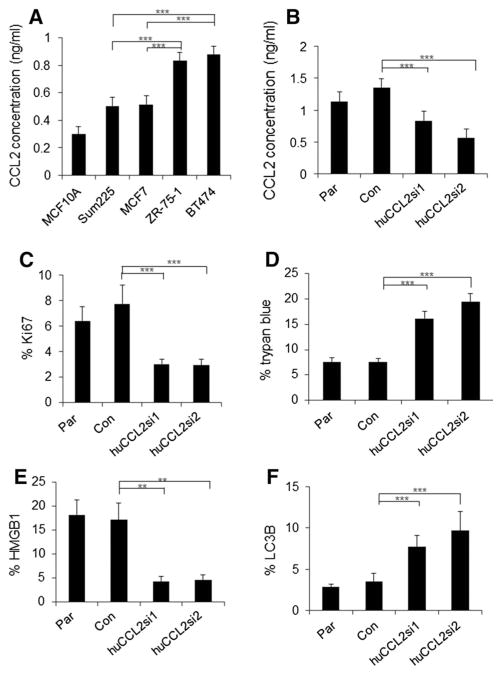
To determine the relevance of CCL2 expression in human breast cancer cells, we examined: BT474 and Zr-75-1, luminal B breast cancer cell lines, MCF-7 and Sum225, luminal A cell lines and MCF10A, a benign breast epithelial cell line [46]. The molecular subtypes of the breast cancer cell lines were characterized in studies on gene expression profiling, and ER, PR, and Her2 expression patterns of breast cancer cell lines [30, 47–49]. By ELISA, BT474 and Zr-75-1 conditioned medium showed the highest levels of CCL2 expression (Fig. 7a). Next, we examined the effects of CCL2 knockdown on BT474 cell growth and survival using human-specific siRNA sequences (huCCL2si1 and huCCL2si2), which targeted two non-overlapping regions of CCL2 (Supplemental Fig. 4). Ca-TAT delivery of huCCL2si1 or huCCL2si2 to BT474 cells significantly reduced CCL2 expression, compared to control siRNA delivery (Fig. 7b). CCL2 knockdown using either siRNA decreased cell proliferation as indicated by decreased Ki67 expression (Fig. 7c), and cell count assay (Supplemental Fig. 5). Moreover, CCL2 knockdown increased expression of HMGB1 and LC3B proteins, corresponding to increased trypan blue staining (Fig. 7d–f). These studies indicate that CCL2 positively regulates cell growth and negatively regulates necrosis and autophagy in BT474 luminal B breast cancer cells.
Fig. 7.
Ca-TAT delivery of CCL2 siRNAs inhibits CCL2 expression, cell growth, and survival in BT474 breast cancer cells. a The indicated breast epithelial cell lines were analyzed for CCL2 expression by ELISA. b–f BT474 breast cancer cells were transfected with Ca-TAT peptides complexed to control (Con) or CCL2 siRNAs (huCCL2si1, huCCL2si2). Parental (Par) or transfected cells were analyzed for CCL2 expression by ELISA (b), Ki67 expression (c), trypan blue staining (d), HMGB1 expression (e), and LC3B expression (f). Statistical analysis performed by One-Way ANOVA followed by Bonferroni post hoc comparisons. Statistical significance determined by p value < 0.05. **p < 0.01, ***p < 0.05. Values are shown as Mean ± SEM
Discussion
The role of CCL2 in regulating macrophage recruitment during breast cancer progression is well recognized [14, 15, 19, 20]. Here, we demonstrate that CCL2 is important for promoting luminal B breast cancer cell proliferation and negatively regulating necrosis and autophagy independent of macrophage recruitment. We previously demonstrated an important role for paracrine CCL2 signaling in regulating migration and apoptosis, but not autophagy, necrosis, or proliferation of breast cancer cells [22]. These results suggest a unique role for autocrine CCL2 signaling in breast cancer cells in modulating breast cancer cell growth and survival.
Previous studies demonstrated that paracrine CCL2 signaling positively regulating cell migration and negatively regulated apoptosis through Smad3- and MAPK-dependent mechanisms. However, it is not likely that these same pathways are involved in autocrine CCL2-mediated proliferation, autophagy and necrosis, breast cancer cells alone expressed low to undetectable levels of these phosphorylated proteins [22]. In prostate cancer cells, recombinant CCL2 treatment inhibited autophagic cell death and enhanced cell proliferation and migration through PI3-K-, AKT-, and AMPK-dependent mechanisms [50–52]. It is possible that autocrine CCL2 signaling modulates breast cancer cell proliferation and autophagy through similar mechanisms. In addition, necrosis is modulated by a number of pathways, including RIPK1/3 and poly(ADP-ribose) polymerase to inhibit glycolysis and deplete ATP levels [53, 54]. Studies are currently being conducted to understand how autocrine CCL2 signaling integrates cell proliferation with survival in breast cancer cells.
We show that CCL2 knockdown in luminal B breast cancer cells enhanced necrosis. Breast tumors frequently exhibit central necrosis, a factor associated with poor prognosis [55, 56]. Functional studies indicate that aggressive tumor growth contributes to necrosis, as proliferative cells consume the existing pool of nutrients and promote a hypoxic environment [57–59]. Hypoxia induces angiogenesis and macrophage recruitment through HIF-1-dependent mechanisms [60, 61]. Necrosis itself benefits tumor progression by releasing HMGB1 from cells into the extracellular space. HMGB1 acts as a pro-inflammatory cytokine to enhance the recruitment of M2 polarized macrophages [43]. Anti-cancer drug treatment also enhances necrosis in solid tumors. However, these drugs also inhibit cell proliferation, suppressing tumor growth [54]. These studies indicate that cellular necrosis may be tumor suppressive when accompanied by decreased tumor cell proliferation. As CCL2 knockdown enhances necrosis and decreases breast cancer cell proliferation, CCL2 functions as a tumor promoter in luminal B breast cancer.
We showed that CCL2 knockdown enhanced autophagy of breast cancer cells. While autophagy enhances cancer cell survival to conserve energy during metabolic stress, cytotoxic drugs such as the mTOR inhibitor RAD001 and sorafenib induce and sustain autophagy, leading to cell death in cancers including: glioma, hepatocellular carcinoma, and ovarian cancer [62]. In prostate cancer, CCL2 protects against autophagic cell death induced by serum deprivation and rapamycin [52]. Here, CCL2 silencing in breast cancer cells enhanced LC3B expression correlating with decreased survival. These results indicate that CCL2 expression in breast cancer cells may protect against autophagic cell death. Alternatively, CCL2 may enhance autophagy as an initial protective response to necrosis signals. To distinguish these possibilities, it would be necessary to conduct further biochemical studies on CCL2-mediated autophagy and necrosis in breast cancer cells.
Tissue specificity and penetration are major challenges in nanomedicine [63, 64]. Here, we demonstrated that Ca-TAT/mCCL2si complexes penetrated PyVmT mammary tumors preferentially over normal tissues to significantly reduce CCL2 expression and inhibit mammary tumor cell proliferation and survival. Net ionic charge and nanoparticle size are two factors modulating specificity of Ca-TAT. Complexing TAT peptides with siRNAs with CaCl2 results in a net positive charge of Ca-TAT complexes [28]. CaCl2 also condenses TAT/siRNA complexes to nanoparticle sizes. These positively charged nanocomplexes may interact with negatively charged phospholipids to form micelle structures, which facilitate entry into the cell [28, 65, 66]. Furthermore, differences in cell membrane organization could render tumor cells more easily penetrable to Ca-TAT complexes, compared to normal cells [67].
Future studies using in vivo models will further determine the contribution of CCL2 signaling in epithelial cells versus macrophages in breast cancer progression. Here, we show that CCL2 expression is associated with luminal B breast cancers, and plays an important role in regulating growth and survival of this breast cancer subtype. Inducing necrosis and autophagic cell death provides a potential therapeutic advantage where tumors acquire resistance to drug-induced apoptosis [68]. Moreover, Ca-TAT/siRNA complexes may be used as a therapeutic targeting agent to control expression and activity of molecules in selected cell types [25]. By understanding the role of CCL2 signaling in luminal B breast cancer progression, we could design alternative therapies to more effectively treat this disease.
Supplementary Material
Acknowledgments
This work was supported by grants from: the KUCC Pilot Grants Program, Kansas Bioscience Authority, NIH/NCI (CA127357) and American Cancer Society (RSG-13-182-01-CSM).
Abbreviations
- Ca-TAT
Calcium crosslinked TAT
- CCL2
Chemokine (C–C motif) ligand 2
- siRNA
Small interfering RNA
- MMTV
Mouse mammary tumor virus
- PyVmT
Polyoma virus middle T
- HMGB1
High mobility group box 1
- LC3B
Light chain 3B
- TNF-a
Tumor necrosis alpha
- TRAIL
TNF-related apoptosis inducing ligand
- RIPK
Receptor interacting protein kinase
- FBS
Fetal bovine serum
- BSA
Bovine serum albumin
- NBF
Neutral formalin buffer
- SEM
Standard error of the mean
- ANOVA
Analysis of variance
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-015-3324-4) contains supplementary material, which is available to authorized users.
Ethical standards The authors declare that the experiments comply with the current laws of the country in which they were performed.
Conflict of interest C. Berkland has a patent (US 20110287547 A1) on Ca-TAT peptides. These studies were not solicited or funded by any pharmaceutical company.
Contributor Information
Wei Bin Fang, Department of Pathology and Laboratory, University of Kansas Medical Center, 3901 Rainbow Boulevard, Wahl Hall East 1020, Kansas, KS 66160, USA.
Min Yao, Department of Pathology and Laboratory, University of Kansas Medical Center, 3901 Rainbow Boulevard, Wahl Hall East 1020, Kansas, KS 66160, USA.
Iman Jokar, Department of Pathology and Laboratory, University of Kansas Medical Center, 3901 Rainbow Boulevard, Wahl Hall East 1020, Kansas, KS 66160, USA.
Nabil Alhakamy, Department of Pharmaceutical Chemistry, University of Kansas, Lawrence, KS 66045, USA.
Cory Berkland, Department of Pharmaceutical Chemistry, University of Kansas, Lawrence, KS 66045, USA.
Jin Chen, Department of Cancer Biology, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Dana Brantley-Sieders, Department of Cancer Biology, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Nikki Cheng, Email: ncheng@kumc.edu, Department of Pathology and Laboratory, University of Kansas Medical Center, 3901 Rainbow Boulevard, Wahl Hall East 1020, Kansas, KS 66160, USA.
References
- 1.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zong Y, Zhu L, Wu J, Chen X, Huang O, Fei X, He J, Chen W, Li Y, Shen K. Progesterone receptor status and Ki-67 index may predict early relapse in luminal B/HER2 negative breast cancer patients: a retrospective study. PLoS One. 2014;9(8):e95629. doi: 10.1371/journal.pone.0095629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanagawa M, Ikemot K, Kawauchi S, Furuya T, Yamamoto S, Oka M, Oga A, Nagashima Y, Sasaki K. Luminal A and luminal B (HER2 negative) subtypes of breast cancer consist of a mixture of tumors with different genotype. BMC Res Notes. 2012;5:376. doi: 10.1186/1756-0500-5-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran B, Bedard PL. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011;13(6):221. doi: 10.1186/bcr2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, Ransohoff RM. The roles of chemokine CXCL12 in embryonic and brain tumor angiogenesis. Semin Cancer Biol. 2009;19(2):111–115. doi: 10.1016/j.semcancer.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Bruserud O, Kittang AO. The chemokine system in experimental and clinical hematology. Curr Top Microbiol Immunol. 2009;341:3–12. doi: 10.1007/82_2010_18. [DOI] [PubMed] [Google Scholar]
- 8.Hemmerich S, Paavola C, Bloom A, Bhakta S, Freedman R, Grunberger D, Krstenansky J, Lee S, McCarley D, Mulkins M, Wong B, Pease J, Mizoue L, Mirzadegan T, Polsky I, Thompson K, Handel TM, Jarnagin K. Identification of residues in the monocyte chemotactic protein-1 that contact the MCP-1 receptor, CCR2. Biochemistry. 1999;38(40):13013–13025. doi: 10.1021/bi991029m. [DOI] [PubMed] [Google Scholar]
- 9.Han KH, Green SR, Tangirala RK, Tanaka S, Quehenberger O. Role of the first extracellular loop in the functional activation of CCR2. The first extracellular loop contains distinct domains necessary for both agonist binding and transmembrane signaling. J Biol Chem. 1999;274(45):32055–32062. doi: 10.1074/jbc.274.45.32055. [DOI] [PubMed] [Google Scholar]
- 10.Monteclaro FS, Charo IF. The amino-terminal extracellular domain of the MCP-1 receptor, but not the RANTES/MIP-1alpha receptor, confers chemokine selectivity. Evidence for a two-step mechanism for MCP-1 receptor activation. J Biol Chem. 1996;271(32):19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 11.Zabel BA, Zuniga L, Ohyama T, Allen SJ, Cichy J, Handel TM, Butcher EC. Chemoattractants, extracellular proteases, and the integrated host defense response. Exp Hematol. 2006;34(8):1021–1032. doi: 10.1016/j.exphem.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Zimmermann HW, Tacke F. Modification of chemokine pathways and immune cell infiltration as a novel therapeutic approach in liver inflammation and fibrosis. Inflamm Allergy Drug Targets. 2011;10(6):509–536. doi: 10.2174/187152811798104890. [DOI] [PubMed] [Google Scholar]
- 13.Raman D, Sobolik-Delmaire T, Richmond A. Chemokines in health and disease. Exp Cell Res. 2011;317(5):575–589. doi: 10.1016/j.yexcr.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Paepe B, Creus KK, De Bleecker JL. Chemokines in idiopathic inflammatory myopathies. Front Biosci. 2008;13:2548–2577. doi: 10.2741/2866. [DOI] [PubMed] [Google Scholar]
- 15.Koelink PJ, Overbeek SA, Braber S, de Kruijf P, Folkerts G, Smit MJ, Kraneveld AD. Targeting chemokine receptors in chronic inflammatory diseases: an extensive review. Pharmacol Ther. 2009 doi: 10.1016/j.pharmthera.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Chavey C, Bibeau F, Gourgou-Bourgade S, Burlinchon S, Boissiere F, Laune D, Roques S, Lazennec G. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007;9(1):R15. doi: 10.1186/bcr1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, Koike M, Inadera H, Matsushima K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6(8):3282–3289. [PubMed] [Google Scholar]
- 18.Saji H, Koike M, Yamori T, Saji S, Seiki M, Matsushima K, Toi M. Significant correlation of monocyte chemoattractant protein-1 expression with neovascularization and progression of breast carcinoma. Cancer. 2001;92(5):1085–1091. doi: 10.1002/1097-0142(20010901)92:5<1085::aid-cncr1424>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto H, Sangai T, Ishii G, Ikehara A, Nagashima T, Miyazaki M, Ochiai A. Stromal MCP-1 in mammary tumors induces tumor-associated macrophage infiltration and contributes to tumor progression. Int J Cancer. 2009;125(6):1276–1284. doi: 10.1002/ijc.24378. [DOI] [PubMed] [Google Scholar]
- 20.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193(6):727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P, Cheng N. CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J Biol Chem. 2012;287(43):36593–36608. doi: 10.1074/jbc.M112.365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang WB, Jokar I, Chytil A, Moses HL, Abel T, Cheng N. Loss of one Tgfbr2 allele in fibroblasts promotes metastasis in MMTV: polyoma middle T transgenic and transplant mouse models of mammary tumor progression. Clin Exp Metastasis. 2011;28(4):351–366. doi: 10.1007/s10585-011-9373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haringman JJ, Gerlag DM, Smeets TJ, Baeten D, van den Bosch F, Bresnihan B, Breedveld FC, Dinant HJ, Legay F, Gram H, Loetscher P, Schmouder R, Woodworth T, Tak PP. A randomized controlled trial with an anti-CCL2 (anti-monocyte chemotactic protein 1) monoclonal antibody in patients with rheumatoid arthritis. Arthritis Rheum. 2006;54(8):2387–2392. doi: 10.1002/art.21975. [DOI] [PubMed] [Google Scholar]
- 25.Makley LN, Gestwicki JE. Expanding the number of ‘druggable’ targets: non-enzymes and protein–protein interactions. Chem Biol Drug Des. 2013;81(1):22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T, Altman RB. Identifying druggable targets by protein microenvironments matching: application to transcription factors. CPT. 2014;3:e93. doi: 10.1038/psp.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272(25):16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 28.Baoum A, Xie SX, Fakhari A, Berkland C. “Soft” calcium crosslinks enable highly efficient gene transfection using TAT peptide. Pharm Res. 2009;26(12):2619–2629. doi: 10.1007/s11095-009-9976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickel L, Matsuzuka T, Doi C, Ayuzawa R, Maurya DK, Xie SX, Berkland C, Tamura M. Overexpression of angiotensin II type 2 receptor gene induces cell death in lung adenocarcinoma cells. Cancer Biol Ther. 2010;9(4):277. doi: 10.4161/cbt.9.4.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO, Perou CM. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal a breast cancer. J Clin Oncol. 2013;31(2):203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aleskandarany MA, Green AR, Benhasouna AA, Barros FF, Neal K, Reis-Filho JS, Ellis IO, Rakha EA. Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res. 2012;14(1):R3. doi: 10.1186/bcr3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bugge TH, Lund LR, Kombrinck KK, Nielsen BS, Holmback K, Drew AF, Flick MJ, Witte DP, Dano K, Degen JL. Reduced metastasis of Polyoma virus middle T antigen-induced mammary cancer in plasminogen-deficient mice. Oncogene. 1998;16(24):3097–3104. doi: 10.1038/sj.onc.1201869. [DOI] [PubMed] [Google Scholar]
- 33.Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R, Arteaga CL, Neilson EG, Hayward SW, Moses HL. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene. 2005;24(32):5053–5068. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maglione JE, Moghanaki D, Young LJ, Manner CK, Ellies LG, Joseph SO, Nicholson B, Cardiff RD, MacLeod CL. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61(22):8298–8305. [PubMed] [Google Scholar]
- 35.Guy C, Cardiff R, Muller W. Induction of mammary tumors by expression a polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hembruff SL, Jokar I, Yang L, Cheng N. Loss of transforming growth factor-beta signaling in mammary fibroblasts enhances CCL2 secretion to promote mammary tumor progression through macrophage-dependent and -independent mechanisms. Neoplasia. 2010;12(5):425–433. doi: 10.1593/neo.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graeber TG, Sawyers CL. Cross-species comparisons of cancer signaling. Nat Genet. 2005;37(1):7–8. doi: 10.1038/ng0105-7. [DOI] [PubMed] [Google Scholar]
- 38.Budhu S, Wolchok J, Merghoub T. The importance of animal models in tumor immunity and immunotherapy. Curr Opin Genet Dev. 2014;24:46–51. doi: 10.1016/j.gde.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD, Cushing RC, Seagroves TN. Hypoxia-inducible factor 1 alpha promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 2012;14(1):R6. doi: 10.1186/bcr3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Usary J, Zhao W, Darr D, Roberts PJ, Liu M, Balletta L, Karginova O, Jordan J, Combest A, Bridges A, Prat A, Cheang MC, Herschkowitz JI, Rosen JM, Zamboni W, Sharpless NE, Perou CM. Predicting drug responsiveness in human cancers using genetically engineered mice. Clin Cancer Res. 2013;19(17):4889–4899. doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirnia F, Frese S, Gloor B, Hotz MA, Luethi A, Gugger M, Betticher DC, Borner MM. Ex vivo assessment of chemotherapy-induced apoptosis and associated molecular changes in patient tumor samples. Anticancer Res. 2006;26(3A):1765–1772. [PubMed] [Google Scholar]
- 42.Guerriero JL, Ditsworth D, Fan Y, Zhao F, Crawford HC, Zong WX. Chemotherapy induces tumor clearance independent of apoptosis. Cancer Res. 2008;68(23):9595–9600. doi: 10.1158/0008-5472.CAN-08-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418(6894):191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 44.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36(12):2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabakov AE, Kudryavtsev VA, Gabai VL. Determination of cell survival or death. Methods Mol Biol. 2011;787:231–244. doi: 10.1007/978-1-61779-295-3_17. [DOI] [PubMed] [Google Scholar]
- 46.Miller FR. Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia. 2000;5(4):379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 47.Riaz M, Elstrodt F, Hollestelle A, Dehghan A, Klijn JG, Schutte M. Low-risk susceptibility alleles in 40 human breast cancer cell lines. BMC Cancer. 2009;9:236. doi: 10.1186/1471-2407-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jonsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong P, Oredsson S, Ringner M, Hoglund M, Borg A. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosom Cancer. 2007;46(6):543–558. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 50.Loberg RD, Day LL, Harwood J, Ying C, John LNS, Giles R, Neeley CK, Pienta KJ. CCL2 is a potent regulator of prostate cancer cell migration and proliferation. Neoplasia. 2006;8(7):578–586. doi: 10.1593/neo.06280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roca H, Varsos ZS, Pienta KJ. CCL2 is a negative regulator of AMP-activated protein kinase to sustain mTOR complex-1 activation, survivin expression, and cell survival in human prostate cancer PC3 cells. Neoplasia. 2009;11(12):1309–1317. doi: 10.1593/neo.09936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roca H, Varsos Z, Pienta KJ. CCL2 protects prostate cancer PC3 cells from autophagic death via phosphatidylinositol 3-kinase/AKT-dependent survivin up-regulation. J Biol Chem. 2008;283(36):25057–25073. doi: 10.1074/jbc.M801073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15(2):135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 54.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13(24):7271–7279. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 55.Carlomagno C, Perrone F, Lauria R, de Laurentiis M, Gallo C, Morabito A, Pettinato G, Panico L, Bellelli T, Apicella A, et al. Prognostic significance of necrosis, elastosis, fibrosis and inflammatory cell reaction in operable breast cancer. Oncology. 1995;52(4):272–277. doi: 10.1159/000227472. [DOI] [PubMed] [Google Scholar]
- 56.Yu L, Yang W, Cai X, Shi D, Fan Y, Lu H. Centrally necrotizing carcinoma of the breast: clinicopathological analysis of 33 cases indicating its basal-like phenotype and poor prognosis. Histopathology. 2010;57(2):193–201. doi: 10.1111/j.1365-2559.2010.03601.x. [DOI] [PubMed] [Google Scholar]
- 57.Tomes L, Emberley E, Niu Y, Troup S, Pastorek J, Strange K, Harris A, Watson PH. Necrosis and hypoxia in invasive breast carcinoma. Breast Cancer Res Treat. 2003;81(1):61–69. doi: 10.1023/A:1025476722493. [DOI] [PubMed] [Google Scholar]
- 58.Rundqvist H, Johnson RS. Tumour oxygenation: implications for breast cancer prognosis. J Intern Med. 2013;274(2):105–112. doi: 10.1111/joim.12091. [DOI] [PubMed] [Google Scholar]
- 59.Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol. 2012;4(9):a008763. doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murdoch C, Giannoudis A, Lewis CE. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood. 2004;104(8):2224–2234. doi: 10.1182/blood-2004-03-1109. [DOI] [PubMed] [Google Scholar]
- 61.Ahluwalia A, Tarnawski AS. Critical role of hypoxia sensor–HIF-1alpha in VEGF gene activation. Implications for angiogenesis and tissue injury healing. Curr Med Chem. 2012;19(1):90–97. doi: 10.2174/092986712803413944. [DOI] [PubMed] [Google Scholar]
- 62.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, Pan H. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khurana B, Goyal AK, Budhiraja A, Arora D, Vyas SP. siRNA delivery using nanocarriers - an efficient tool for gene silencing. Curr Gene Ther. 2010;10(2):139–155. doi: 10.2174/156652310791111010. [DOI] [PubMed] [Google Scholar]
- 64.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269(14):10444–10450. [PubMed] [Google Scholar]
- 66.Thoren PE, Persson D, Isakson P, Goksor M, Onfelt A, Norden B. Uptake of analogs of penetratin, Tat(48-60) and oligoarginine in live cells. Biochem Biophys Res Commun. 2003;307(1):100–107. doi: 10.1016/s0006-291x(03)01135-5. [DOI] [PubMed] [Google Scholar]
- 67.Prinetti A, Prioni S, Loberto N, Aureli M, Nocco V, Illuzzi G, Mauri L, Valsecchi M, Chigorno V, Sonnino S. Aberrant glycosphingolipid expression and membrane organization in tumor cells: consequences on tumor-host interactions. Adv Exp Med Biol. 2011;705:643–667. doi: 10.1007/978-1-4419-7877-6_34. [DOI] [PubMed] [Google Scholar]
- 68.Fulda S. Tumor resistance to apoptosis. Int J Cancer. 2009;124(3):511–515. doi: 10.1002/ijc.24064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.