SUMMARY
Progenitor differentiation requires remodeling of genomic expression; however, in many tissues, such as epidermis, the spectrum of remodeled genes and the transcription factors (TFs) that control them are not fully defined. We performed kinetic transcriptome analysis during regeneration of differentiated epidermis and identified gene sets enriched in progenitors (594 genes), in early (159 genes), and in late differentiation (387 genes). Module mapping of 1,046 TFs identified MAF and MAFB as necessary and sufficient for progenitor differentiation. MAF:MAFB regulated 393 genes altered in this setting. Integrative analysis identified ANCR and TINCR lncRNAs as essential upstream MAF:MAFB regulators. ChIP-seq analysis demonstrated MAF:MAFB binding to known epidermal differentiation TF genes whose expression they controlled, including GRHL3, ZNF750, KLF4, and PRDM1. Each of these TFs rescued expression of specific MAF:MAFB target gene subsets in the setting of MAF:MAFB loss, indicating they act downstream of MAF:MAFB. A lncRNA-TF network is thus essential for epidermal differentiation.
Graphical abstract
INTRODUCTION
In self-renewing tissues, progenitor differentiation commonly involves induction of a well-orchestrated genetic program that engages cell-cycle exit followed by cellular conversion to perform tissue-specialized functions. Progenitor differentiation has been studied in multiple tissues, however, the genetic interactions that occur in this intricate process are not fully understood in any setting. Among somatic tissues, the self-renewing interfollicular epidermis of skin consists of a basal layer of proliferating progenitor cells that migrate outward as they undergo cell-cycle arrest and enter the terminal differentiation pathway. Recent work identified epigenetic regulators such as DNMT1, the BAF complex, and histone modifiers such as EZH2 (Sen et al., 2010; Bao et al., 2013; Mulder et al., 2012) that repress differentiation and maintain the epidermal progenitor state. Pro-differentiation factors in epidermis also have been identified. For example, NOTCH signaling mediates well-known pro-differentiation effects (Okuyama et al., 2004; Blanpain et al., 2006). Additionally, cooperation between transcription factors (TFs) and other factors have been shown to promote differentiation, as in the case of GRHL3 and trithorax proteins (Yu et al., 2006; Hopkin et al., 2012) as well as ZNF750 and KLF4, RCOR1, and KDM1A (Boxer et al., 2014; Sen et al., 2012). It is likely that additional important regulators of epidermal differentiation remain to be identified.
Members of the AP-1 family of TFs play an important role in multiple processes, including proliferation, differentiation, and death. The MAF subfamily of TFs consists of both large and small MAFs, all of which share a C-terminal bZIP domain and leucine-rich protein-binding domain, with the large MAFs containing an N-terminal transactivation domain absent in the small MAFs (Blank and Andrews, 1997; Eychène et al., 2008). The large MAFs have been implicated in controlling cell states in various cells types. For example, MAF has been shown to promote differentiation of immune cells, osteoclasts, and most recently development of mechanosensory neurons (Hedge et al., 1998; Kroenke et al., 2012; Matsuoka et al., 2003; Nishikawa et al., 2010a Rutz et al., 2011). Similarly, MAFB has been shown to promote differentiation of macrophages, β-islet cells, osteoclasts, and glomerular epithelial cells (Kelly et al., 2000; Nishikawa et al., 2010b; Artner et al., 2007; Sadl et al., 2002; Sarrazin et al., 2009; Sieweke et al., 1996). Interestingly, double knockout of MAF and MAFB in macrophages allowed for self-renewal of differentiated monocytes, demonstrating they promote differentiation by linking cell-cycle inhibition to terminal differentiation (Aziz et al., 2009). While small MAFs have been implicated in differentiation of forestomach (Motohashi et al., 2004), a role for MAF proteins in epidermal stratified epithelia has not been defined. Thus, MAFs appear to be important regulators of cell fate in various cell types, yet the full scope of their action is not yet characterized.
Long non-coding RNAs (lncRNAs) influence many biological processes. LncRNAs promote lineage commitment in neurons and erythroid and cardiovascular cells, and influence myogenesis and adipogenesis (Hu et al., 2011; Ramos et al., 2013; Ng et al., 2012; Klattenhoff et al., 2013; Grote et al., 2013; Wang et al., 2013; Sun et al., 2013). In epidermis, two lncRNAs, ANCR and TINCR, are essential for homeostasis. The anti-differentiation lncRNA, ANCR, promotes progenitor maintenance and recently has been shown to prevent differentiation in osteoblasts by interacting with EZH2 to suppress gene activation (Kretz et al., 2012; Zhu and Xu, 2013). In contrast, the terminal differentiation-induced lncRNA, TINCR, promotes terminal differentiation through a mechanism involving direct RNA:RNA interactions and recruitment of STAU1 protein to stabilize differentiation-specific mRNAs (Kretz et al., 2013). The epidermal target genes of these lncRNAs are being identified and integration of these datasets with those of other epidermal regulators may permit identification of the genetic networks that control epidermal progenitor differentiation.
Here we have employed a kinetic approach to define the transcriptional landscape during regeneration of differentiated epidermal tissue from epidermal progenitors. We have identified three distinct gene sets whose enhanced expression distinguishes between epidermal progenitors and their progeny undergoing early and late differentiation. Expression module mapping of the kinetic epidermal transcriptome with 1,046 expressed TFs predicted MAF and MAFB as key regulators of differentiation. Altering the function of MAF and MAFB in organotypic epidermal tissue and in vivo knockout tissue demonstrated an essential requirement for MAF:MAFB in epidermal differentiation, where MAF:MAFB induces cell-cycle arrest and terminal differentiation genes. Integrative analysis of epidermal regulator target gene sets suggested upstream control of MAF:MAFB by two specific lncRNAs. The ANCR lncRNA targeted the EZH2 Polycomb protein to both MAF and MAFB genes and repressed their expression in progenitors. The TINCR lncRNA was required for normal mRNA stability of MAF and MAFB, and was essential for increased MAF:MAFB expression during differentiation. Characterization of MAF:MAFB genomic binding by ChIP-seq linked MAF:MAFB to four TFs essential for epidermal differentiation, including GRHL3, ZNF750, PRDM1, and KLF4. These data characterize MAF and MAFB as essential mediators of epidermal progenitor differentiation, and demonstrate that they reside within a network between the ANCR and TINCR lncRNAs and a set of canonical pro-differentiation TFs.
RESULTS
Kinetic Transcriptome Analysis during Regeneration of Differentiated Epidermal Tissue
To characterize genomic expression during epidermal differentiation, we profiled gene expression during regeneration of differentiated organotypic epidermal tissue from undifferentiated, progenitor-containing keratinocyte populations. Progenitor keratinocytes were seeded on native human dermal mesenchymal tissue and followed daily over a 7-day time course in a process that culminated in the production of a fully stratified epithelium, expressing both early and late differentiation markers (Figure S1A). This defined setting captured dynamic changes in the process of regenerating a differentiated tissue. For example, representative differentiation markers for the spinous layer (keratin 1) first appeared on day 3, whereas markers for the outer granular layer of the skin, such as filaggrin and loricrin, were detected later in the time course at days 4 and 5, respectively (Figure S1A). This regeneration time course therefore recapitulated the earlier and later induction of differentiation markers characteristic of epidermis.
We next cataloged the dynamic changes occurring during differentiation at the transcriptome level. Using microarray analysis, we defined three distinct gene sets whose expression was enriched in either the progenitor state (day 0, 594 genes), early differentiation (days 1–4, 159 genes), or late differentiation (days 5–7, 387 genes) (Figure S1B; Table S1). The progenitor gene signature was characterized by genes that decreased over the time course and were enriched for GO terms related to cell-cycle and cell division (Figure S1C). Genes that rapidly increased from the progenitor state (day 0) through day 4 and gradually decreased over the remainder of the time course represented the early differentiation signature and were enriched for GO terms involved in cell migration and motility (Figure S1D), consistent with the development of tissue morphology evident at those time points. Finally, genes that gradually increased over the time course and peaked at days 5–7 characterized the late differentiation signature, which was enriched for epidermis development and keratinocyte differentiation GO terms (Figure S1E), consistent with the production of the terminally differentiated outer epidermis in that time frame. Global GO term changes spanning this kinetic process were consistent with progenitor exit from cellular replication into the epidermal differentiation pathway (Figure S1F).
Module Mapping Identifies MAF and MAFB in Epidermal Differentiation
Given these dynamic transcriptional changes, we used expression module mapping to identify transcriptional regulators of epidermal differentiation. Chosen for their possession of the GO term “transcription factor” and positive expression in skin, we used Genomica to query the expression patterns of 1,046 potential TFs for correlation with expression modules generated from our time-course data. We performed 100 permutations of the analysis and generated 100 predicted regulators whose mRNA expression correlates with differential gene expression patterns during differentiation. Of these, 36 are previously known regulators of epidermal homeostasis (p value = 5.6 × 10−33), including ID3, CEBPA, HOPX, UHRF1, ETS1, and KLF4 (Figure 1A; Table S2). Unexpectedly, given that a role for MAF proteins in epidermal differentiation has not been described, MAF was the TF most frequently correlated with epidermal differentiation expression modules, as was its close family member, MAFB, a predicted regulator observed in about half of all tested module maps (Figure 1A). The expression modules predicted to be regulated by MAF and MAFB (MAF:MAFB) represent both up- and downregulated genes, suggesting activating and repressing roles for MAF:MAFB in differentiation (Figures 1B, S2A, and S2B).
Figure 1. MAF and MAFB Are Induced in Epidermal Differentiation.
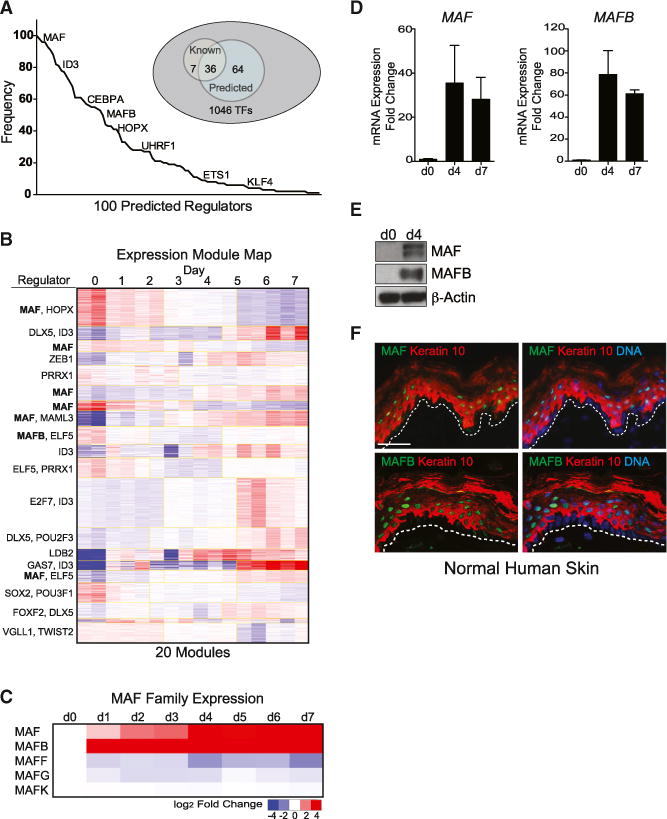
(A) Expression module mapping of time-course microarrays performed in Genomica against a set of 1,046 potential regulators expressed in skin with GO term “transcription factor.” Graph represents frequency of predicted regulators in 100 permutations of module mapping with select TFs displayed.
(B) Representative output of expression module map showing high frequency of MAF in predicted module regulation. Yellow boxes demarcate modules.
(C–E) (C) Gene expression changes in MAF TF family members during regeneration of differentiated epidermis. Induction of MAF and MAFB (D) mRNA and (E) protein expression during calcium-induced keratinocyte differentiation in vitro. Mean ± SEM; n = 2 biological replicates.
(F) MAF and MAFB protein expression (green) in differentiated layers, represented by keratin 10 staining (orange), of normal adult skin tissue (dotted white line demarcates the epidermal basement membrane). Scale bar, 50 mm (see also Figures S1 and S2).
The MAF TF family is composed of large and small MAFs, both of which are expressed in epidermis based on our microarray data. However, only the large MAFs, MAF:MAFB, are upregulated during differentiation, whereas the small MAFs are largely downregulated (Figures 1C and S2C). MAFA and NRL were not expressed. MAF:MAFB mRNA and protein were both dramatically upregulated during differentiation (Figures 1D and 1E). Furthermore, immunostaining of MAF:MAFB in adult human skin revealed MAF and MAFB in the differentiated suprabasal layers of the epidermis (Figures 1F, S2D, and S2E), supporting a possible role for these TFs in epidermal differentiation.
MAF:MAFB Is Necessary and Sufficient to Drive Epidermal Progenitor Differentiation
To examine MAF and MAFB function in differentiation, we first depleted MAF:MAFB mRNA expression in organotypic epidermal tissue using small interfering RNAs (siRNAs) (Figures S3A–S3C). Knockdown of either MAF or MAFB alone decreased expression of differentiation marker genes KRT1, FLG, and LOR, with combined knockdown of both MAF:MAFB demonstrating even stronger effects (Figures S3A and S3D), indicating that MAF:MAFB is essential for differentiation gene induction, but not stratification. Such stratification with impaired differentiation marker expression has been observed previously, for example, in ZNF750-deficient epidermal tissue (Sen et al., 2012). In addition to impaired differentiation gene induction, we observed increased expression of basal markers, such as CCNB1, CDK1, and DNMT1, with decreased expression of cell-cycle inhibitor CDKN1A (Figure S3E).
To rule out simple delay in differentiation due to the limited siRNA-mediated depletion time frame, we employed CRISPR/Cas9-mediated gene ablation of MAF and MAFB to assess differentiation dynamics over a longer time course. Recently, a similar approach using CRISPR/Cas9 was implemented in primary human hematopoietic stem cells (Mandal et al., 2014); however, this is the first example of such gene-targeting in human primary epithelial cells to generate three-dimensional human knockout tissue. We used two independent single guide RNAs (sgRNAs) to target MAF or MAFB in primary keratinocytes and generated pools of keratinocytes with double MAF:MAFB gene ablation (crMAF:crMAFB) (Figures 2A–2C). These gene-targeted primary keratinocytes were then placed in organotypic culture for 4 or 7 days (Figure 2A) or grafted onto mice to generate in vivo human knockout tissue (Figures 2B and S3F). Strikingly, expression of differentiation markers was severely impaired in crMAF:crMAFB tissue (Figures 2A and 2B). Consistent with results observed with siRNA-mediated depletion, crMAF:crMAFB tissue displayed impaired induction of multiple differentiation genes (Figure 2D). Furthermore, the mitotic index of crMAF:crMAFB tissue, as measured by Ki67 staining, was increased compared to control (Figure S3G). Moreover, TUNEL staining showed decreased apoptotic index in crMAF:crMAFB tissue (Figure S3H). These results, along with the prolonged differentiation gene expression defects, suggest an important role for MAF:MAFB in the maintenance of epidermal homeostasis.
Figure 2. MAF:MAFB Is Necessary and Sufficient for Differentiation Gene Induction and Progenitor Compartment Exit.
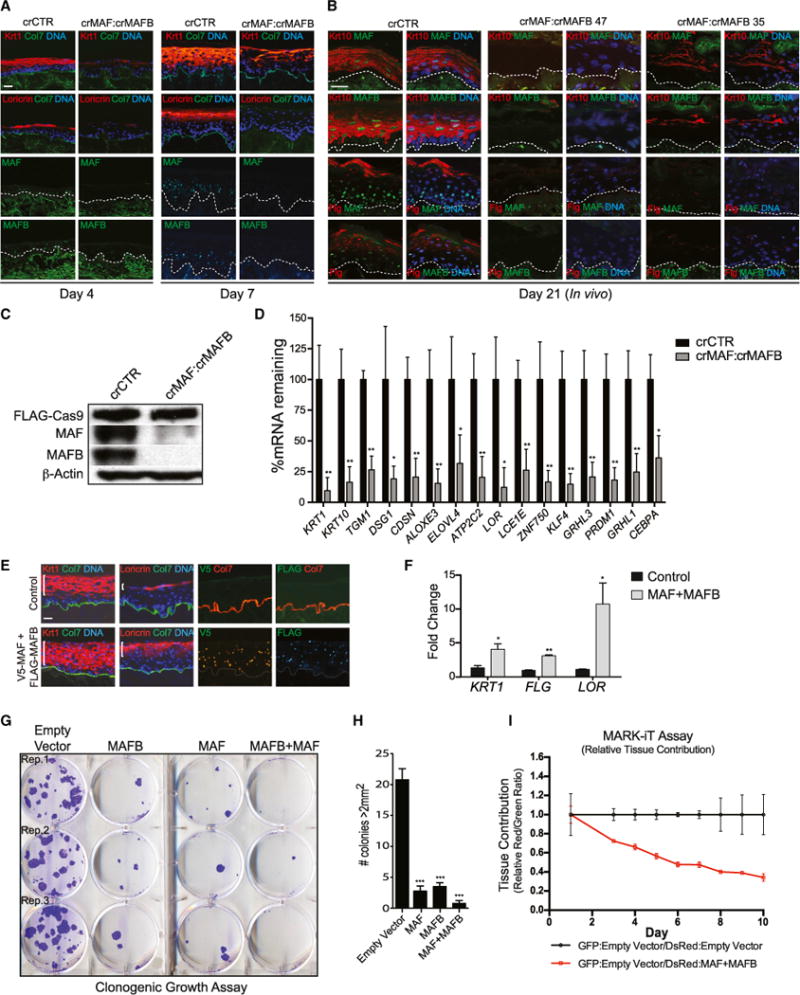
(A) Organotypic culture of epidermis generated by CRISPR/Cas9-mediated genetic ablation of MAF and MAFB (crMAF:crMAFB) across day 4 and day 7 of differentiation, with staining for differentiation markers (top two rows) keratin 1 and loricrin (orange), collagen VII (green), and nuclei (blue) or staining for (bottom two rows) MAF or MAFB (green). Note loss of differentiation proteins upon MAF:MAFB loss. Scale bar, 50 μm.
(B) In vivo skin xenografts generated from CRISPR/Cas9-targeted primary keratinocytes with a control sgRNA or two different combinations of sgRNAs targeting MAF and MAFB (crMAF:crMAFB 47 or crMAF:MAFB 35), harvested 21 days post-seeding, and immunostained for differentiation markers keratin 10 and filaggrin (red) and MAF or MAFB (green). Scale bar, 50 μm.
(C) Western blots for Cas9, MAF, and MAFB in CRISPR/Cas9-targeted primary keratinocytes.
(D) Quantification of differentiation gene expression levels in crMAF:crMAFB day 7 organotypic tissue. Mean ± SEM; n = 3 biological replicates; *p < 0.05, **p < 0.01.
(E) Immunostaining of overexpressed epitope-tagged V5-MAF and FLAG-MAFB in organotypic epidermis and differentiation markers as in (A, top); white brackets indicate increased distribution of differentiation protein expression in MAF:MAFB compared to control (left); epitope tag immunostaining shows MAF and MAFB overexpression throughout the tissue (right). Scale bar, 50 μm.
(F) Quantification of mRNA levels of differentiation markers in tissue overexpressing MAF:MAFB. Mean ± SEM; n = 2 biological replicates; *p < 0.05, **p < 0.01.
(G) Clonogenic assay of progenitor keratinocytes overexpressing MAF and/or MAFB over a 2-week time span.
(H) Quantification of colonies > 2 mm2. Mean ± SEM; n = 4; ***p < 0.001.
(I) MARK-iT cell competition assay over 10 days showing relative tissue contribution detected by red:green fluorescence ratios of GFP:empty vector/DsRed:empty vector mixed samples compared to GFP:empty vector/DsRed:MAF+MAFB mixed samples; note progressive decrease in the proportion of MAF:MAFB-expressing cells over time. Mean ± SEM; n = 3 biological replicates (see also Figure S3).
We next asked if enforced expression of MAF:MAFB in progenitor populations, where they are not normally expressed, can drive epidermal differentiation. Progenitor keratinocytes were transduced with V5-MAF and FLAG-HA-MAFB via lentivirus and seeded in organotypic culture (Figure 2E). Compared to control, MAF:MAFB increased mRNA and protein expression of differentiation genes in tissue (Figures 2F and S3I). Remarkably, we observed expression of keratin 1 into the basal layer and aberrant expression of loricrin in the spinous layer of MAF:MAFB-overexpressing tissue (Figure 2E, white bracket), supporting the sufficiency of MAF and MAFB to drive ectopic differentiation in less differentiated epidermal cells. In contrast, overexpression of DNA-binding-deficient MAF:MAFB mutants did not result in premature differentiation (Figures S3I and S3J). MAF:MAFB-driven ectopic differentiation was partially recovered by day 7 of culture, suggesting that cells overexpressing MAF:MAFB are lost over time (Figures S3I and S3J). Consistent with these data, a decreased mitotic with concomitant increased apoptotic index were observed in tissue expressing wild-type, but not mutant, MAF:MAFB (Figures S3I, S3K, and S3L).
MAF:MAFB Promotes Epidermal Progenitor Cell-Cycle Exit
We next investigated if MAF:MAFB suppresses epidermal progenitor self-renewal. Expression of MAF:MAFB in epidermal progenitors at levels found in differentiating cells impaired clonogenic growth, a measure of progenitor proliferative capacity in vitro over multiple cell divisions (Figures 2G and 2H). Consistent with this, enforced MAF:MAFB expression also impaired proliferation in epidermal progenitor-containing populations (Figures S3M and S3N). Furthermore, a decrease in basal gene expression and increase in the cell-cycle inhibitor p21 also were observed in MAF:MAFB-overexpressing progenitor cells (Figure S3O). In contrast, overexpression of DNA-binding-deficient mutants of MAF:MAFB did not alter cell-cycle profiles (Figure S3M); however, a slight increase in basal gene expression was observed, suggesting a potential dominant-negative effect (Figure S3O).
To determine if MAF:MAFB also impairs epidermal progenitor self-renewal in tissue, we developed the MARK-iT mosaic progenitor cell competition assay. In this assay, two populations of cells labeled with differing marker genes (GFP or DsRed) are mixed together at a 1:1 ratio, then used to regenerate organotypic epidermal tissue, and followed over time to determine if the altered progenitor population contributes to tissue formation more or less effectively than control, as measured by fluorescence emission from intact tissue in real time. Comparable to the clonogenic assay, the MARK-iT assay assesses long-term progenitor maintenance over multiple cell divisions in tissue. We performed this assay by measuring DsRed:GFP fluorescence intensity ratios of GFP-expressing empty vector cells mixed with DsRed-expressing empty vector cells for control tissue, and compared to GFP-expressing empty vector cells mixed with DsRed-expressing MAF:MAFB cells over 10 days (Figure 2I). MAF:MAFB-expressing progenitor populations failed to compete to sustain tissue in a comparable fashion to marked control cells. A similar failure of MAF:MAFB-expressing progenitor population was observed when the marker genes were reversed, indicating this is not due to differential marker gene toxicity (Figure S3P). These impacts on tissue regeneration and maintenance were not observed with expression of either MAF or MAFB alone, suggesting that combined MAF:MAFB action is necessary to impair progenitor function. Thus, in addition to being essential for induction of differentiation, MAF and MAFB oppose proliferative self-renewal, consistent with their role in epidermal differentiation.
Transcriptome Analysis of MAF:MAFB-Regulated Genes
To identify the specific MAF:MAFB target genes among the global gene sets identified in epidermal regeneration, we profiled MAF:MAFB-depleted organotypic epidermal tissue versus control. Biologic tissue replicate microarray analysis identified 393 highly concordant genes significantly changed by MAF:MAFB loss, with 315 genes downregulated and 78 genes upregulated (Figure 3A; Table S3). We did not observe such widespread impacts with depletion of either MAF or MAFB alone, implying that they functionally compensate for each other. MAFi+ MAFBi-downregulated genes, representing positively regulated MAF:MAFB targets, were enriched for GO terms associated with epidermal and ectodermal development (Figure S4A). In contrast, GO term analysis of the MAFi+MAFBi-upregulated genes, representing MAF:MAFB-repressed targets, were enriched for cell-cycle regulation (Figure S4B). MAF:MAFB-regulated genes were enriched in phenotypic terms related to human genetic skin diseases characterized by incomplete epidermal differentiation (Figure 3B). Gene set enrichment analysis (GSEA) comparing the entire MAF:MAFB gene set to a published calcium-differentiated keratinocyte gene set (Sen et al., 2010) identified an enrichment and correlation between MAF:MAFB-downregulated genes and those induced in calcium-differentiated keratinocytes (Figure 3C). Genes downregulated with MAF:MAFB loss encompassed 20% of the late differentiation gene signature (p value = 1.3 × 10−58) (Figure 3D), indicating that MAF:MAFB is required for induction of a substantial portion of the differentiation gene set. The identity of MAF:MAFB-regulated genes is consistent with an essential role for MAF:MAFB in promoting epidermal differentiation.
Figure 3. MAF:MAFB Regulates Epidermal Differentiation Genes.
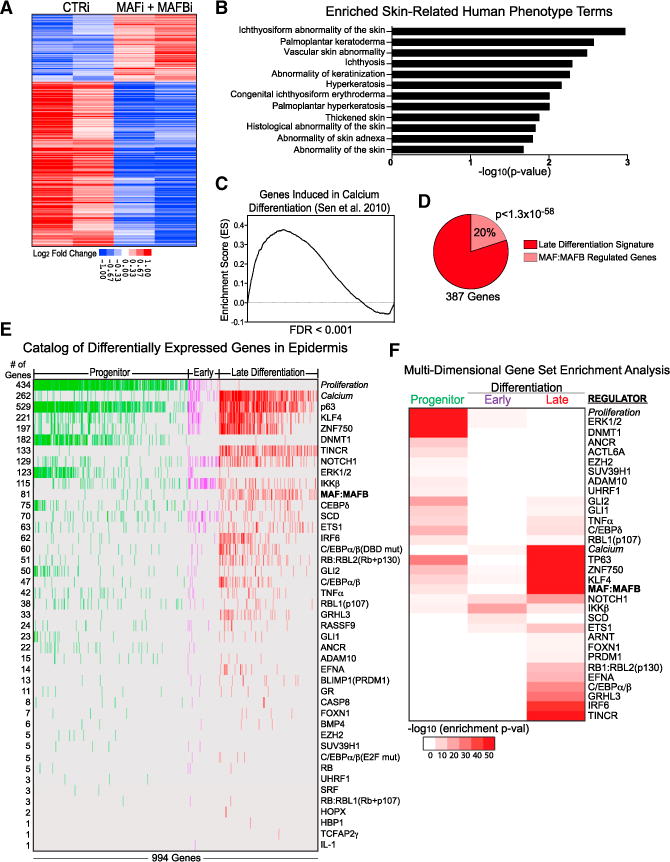
(A) Epidermal differentiation genes regulated by MAF:MAFB. Heatmap of duplicate, mean-centered microarray analysis of MAF:MAFB depleted tissue showing genes significantly changed compared to control siRNA.
(B) Skin-related human disease phenotype terms of MAF:MAFB target genes.
(C) GSEA of MAF:MAFB depleted tissue against a calcium-differentiated keratinocyte gene set (Sen et al., 2010) ranked by MAF:MAFB genes downregulated to upregulated (left to right).
(D) Quantification of the overlap of MAF:MAFB downregulated genes with the late differentiation signature.
(E) Catalog of published regulators controlling the epidermal progenitor, early and late differentiation gene sets, ranked by the number of genes regulated.
(F) Multi-dimensional GSEA conducted on published epidermal regulator gene sets against the three differentiation signatures (-log10 enrichment p value; maximum p < 0.05), with progenitor to late differentiation gene regulation ordered from top to bottom (see also Figure S4).
Comparison to Other Epidermal Regulators
To place MAF:MAFB in the context of known epidermal gene regulators, we next compared MAF:MAFB target genes to 42 published target gene sets. After p63, KLF4, and ZNF750, MAF:MAFB regulates the fourth largest number of genes within progenitor and differentiation gene sets (Figures 3E and 3F). Multi-dimensional GSEA was then performed to categorize known epidermal factors as regulators of specific gene signatures throughout differentiation. For factor knockdowns or knockouts, we used the downregulated gene set, and for overexpressed factors, we utilized the upregulated gene set as a proxy for gene activation. Using human sub-confluent keratinocyte and calcium-differentiated keratinocyte gene sets (Sen et al., 2010) as benchmarks, we found enrichment for the proliferating keratinocyte gene set in our progenitor gene set, whereas the calcium-differentiated gene set was enriched for our early and late differentiation gene sets (Figure 3F). Additionally, we found enrichment for known progenitor-maintenance factors DNMT1 and ANCR in the progenitor signature, whereas factors such as KLF4 and GRHL3 were enriched for the early and late differentiation signatures. The reverse GSEA, where upregulated gene sets with loss of factors and downregulated gene sets of overexpressed factors were used as a proxy for repressed genes, provided similarly congruent findings (Figure S4C). Consistent with the number of genes they regulate, MAF:MAFB’s enrichment score for the late differentiation signature was similar to that of KLF4, p63, and ZNF750, known essential regulators of epidermal differentiation.
LncRNAs Are Upstream Regulators of MAF:MAFB
Given the induction of MAF:MAFB during epidermal differentiation, we next searched for upstream regulators of these two TFs. The compendium of published gene sets was used to identify candidates by determining whether or not MAF and MAFB expression changed in these gene sets. This approach predicted a network of MAF:MAFB regulators in which MAF:MAFB is regulated by two specific lncRNAs (Figure S4D). ANCR, the anti-differentiation lncRNA, was predicted to act as an MAF: MAFB repressor, and TINCR, the terminal differentiation-induced lncRNA, was predicted to act as an MAF:MAFB activator. Further supporting a MAF:MAFB-inducing role for TINCR, we found that the TINCR gene set was the most strongly enriched known regulator gene set in the MAF:MAFB gene set (Figure 4A). Consistent with the multi-dimensional GSEA, we also observed enrichment in MAF:MAFB for the ZNF750, KLF4, and p63 gene sets, suggesting cooperative regulation of genetic programs by these TFs with MAF:MAFB. Furthermore, we observed significant overlap of the MAF:MAFB gene set with genes controlled by both TINCR and ANCR (Figure 4B). To verify that TINCR and ANCR are upstream of MAF:MAFB, we analyzed expression of MAF and MAFB in TINCR- and ANCR-depleted cells and epidermal tissue. Consistent with the prediction that TINCR is a positive regulator of MAF:MAFB, we observed that TINCR depletion in tissue reduced levels of MAF:MAFB mRNA (Figure 4C). In contrast, ANCR depletion in progenitor keratinocyte populations highly upregulated MAF: MAFB mRNA (Figure 4D). Notably, we did not observe significant effects on TINCR or ANCR expression by MAF:MAFB (Figure S5A), placing MAF:MAFB downstream of these lncRNAs. Thus, TINCR and ANCR lncRNAs control expression of epidermal MAF:MAFB.
Figure 4. LncRNAs Are Upstream Regulators of MAF:MAFB.
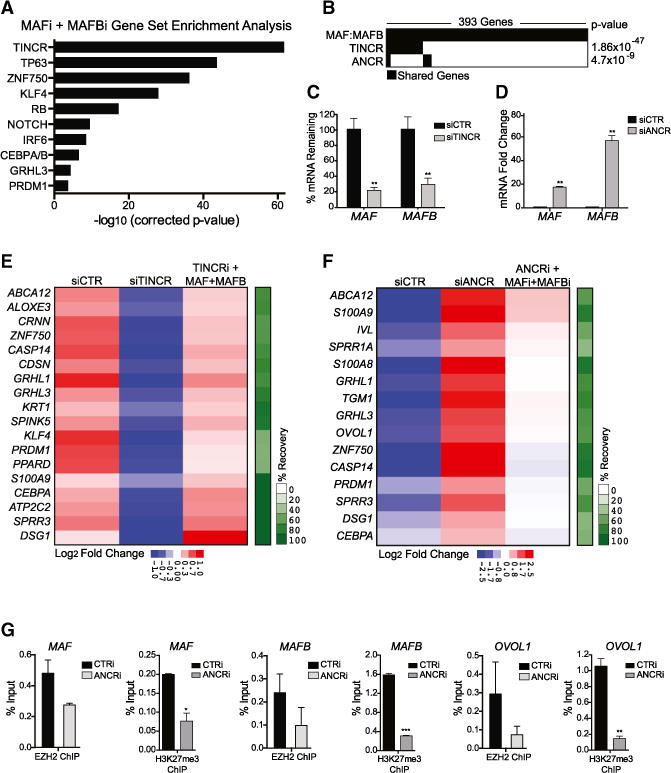
(A) GSEA of genes downregulated in MAF:MAFB-depleted tissue overlaps with gene sets of known epidermal differentiation regulators.
(B–D) (B) Overlap of MAFi:MAFBi, TINCRi, and ANCRi gene sets in keratinocytes. Quantification of MAF and MAFB mRNA levels in (C) TINCR-depleted tissue and (D) ANCR-depleted progenitor keratinocytes. Mean ± SEM; n = 2 biological replicates; **p < 0.01.
(E) Enforced expression of MAF:MAFB in TINCR-depleted tissue partially rescues the differentiation defect observed with TINCR knockdown; heatmap shows average fold change by qRT-PCR (n = 2; p < 0.05, t test) and percentage recovery of differentiation genes in green bar at right.
(F) Concurrent knockdown of MAF and MAFB partially rescues the ANCRi phenotype of premature differentiation gene induction; heatmap shows average fold by qRT-PCR (n = 2; p < 0.05, t test) and percentage recovery of differentiation genes.
(G) ChIP-qPCR showing binding of EZH2 at ANCR-regulated genes in CTRi- and ANCRi-treated progenitor keratinocytes and the H3K27me3-repressive histone mark at these loci. Mean ± SEM; n = 2; *p < 0.05, **p < 0.01, ***p < 0.001 (see also Figure S5).
We next explored if MAF:MAFB represents downstream mediators of these two lncRNAs in controlling differentiation. First, we examined if MAF:MAFB could rescue the loss of differentiation seen with TINCR depletion. Restoring expression of MAF:MAFB in TINCR-depleted tissue recovered expression of TINCR differentiation targets (Figures 4E, S5B, and S5C), indicating that MAF: MAFB can partially rescue the differentiation defects produced by TINCR loss. Likewise, we attempted to rescue the premature progenitor differentiation gene induction that occurs with ANCR loss by preventing the MAF:MAFB upregulation that occurs with ANCR depletion in undifferentiated keratinocytes (Figures S5D and S5E). Partial reduction in differentiation gene induction was observed in MAF-, MAFB-, and ANCR-depleted progenitors, indicating that ANCR progenitor maintenance also, at least in part, is sustained by the repression of MAF:MAFB (Figure 4F).
TINCR exerts a portion of its effects via direct mRNA binding and recruitment to STAU1 to regulate mRNA stability (Kretz et al., 2013). Analysis of published data (Kretz et al., 2013) demonstrated that TINCR binds both MAF and MAFB mRNA (Figure S5F). Furthermore, MAF mRNA stability decreased in the absence of TINCR with modest but significant effects on MAFB mRNA (Figures S5G and S5H). Correspondingly, protein levels of MAF:MAFB were decreased in the absence of TINCR (Figure S5I). The mechanism by which ANCR suppresses differentiation gene induction in epidermis has not yet been established; however, a recent study in osteoblast differentiation showed that ANCR is required for recruitment of the EZH2 Polycomb protein to repress gene transcription (Zhu and Xu, 2013). Chromatin immunoprecipitation (ChIP)-qPCR of EZH2 and its accompanying modified histone, H3K27me3, was therefore performed to assess EZH2 recruitment to ANCR-repressed differentiation genes. ANCR depletion decreased EZH2 recruitment to MAF and MAFB and to another differentiation gene, OVOL1, and yielded a significant decrease in the repressive H3K27me3 mark (Figure 4G). Taken together, these results point to ANCR and TINCR lncRNAs as upstream regulators of MAF:MAFB, whereby ANCR represses MAF:MAFB in association with EZH2 gene targeting to prevent premature MAF:MAFB-driven progenitor differentiation, and whereby TINCR potentiates differentiation gene expression through enhancement of MAF:MAFB mRNA stability.
Genome-wide MAF:MAFB Occupancy in Differentiation
After observing that ANCR and TINCR lncRNAs control MAF: MAFB expression, we next looked for direct downstream effectors of MAF:MAFB-driven differentiation. To do this, we performed ChIP sequencing (ChIP-seq) analysis on MAF and MAFB in differentiated keratinocytes, and identified 2,403 peaks associated with MAF and 5,286 peaks associated with MAFB (Figure 5A). MAF:MAFB-binding peaks were found throughout the genome and overlapped to a significant degree (Figure 5A). Consistent with this co-localization, we observed physical proximity of these proteins in the nuclei of differentiating epidermal layers by proximity ligation assay (PLA), as well as by co-immunoprecipitation of endogenous MAF and MAFB (Figures S6A–S6C). To find additionally proximally binding TFs, we performed feature overlapper for chromosomal interval subsets (FOCIS) analysis (Webster et al., 2014) for binding site enrichment of other TFs, and identified the p53/p63 motif as highly enriched adjacent to MAF:MAFB-binding regions (Figure 5B; Table S4). Binding sites for p63, an epidermis-specifying TF that controls more genes within the epidermal gene sets above than any other studied TF (Figure 3E), were highly correlated with sites of MAF:MAFB binding, implying a potential cooperative interaction by MAF:MAFB with p63 (Figure S6D). Interestingly, overexpression of p63 resulted in MAF and MAFB gene induction, suggesting an upstream regulatory role in addition to cooperation (Figure S6E). Evidence for direct MAF:MAFB-p63 association, however, could not be detected by PLA, suggesting proximal binding at target genes may not involve direct physical interactions between MAF:MAFB and p63.
Figure 5. Genome-wide MAF:MAFB Binding in Differentiated Keratinocytes.
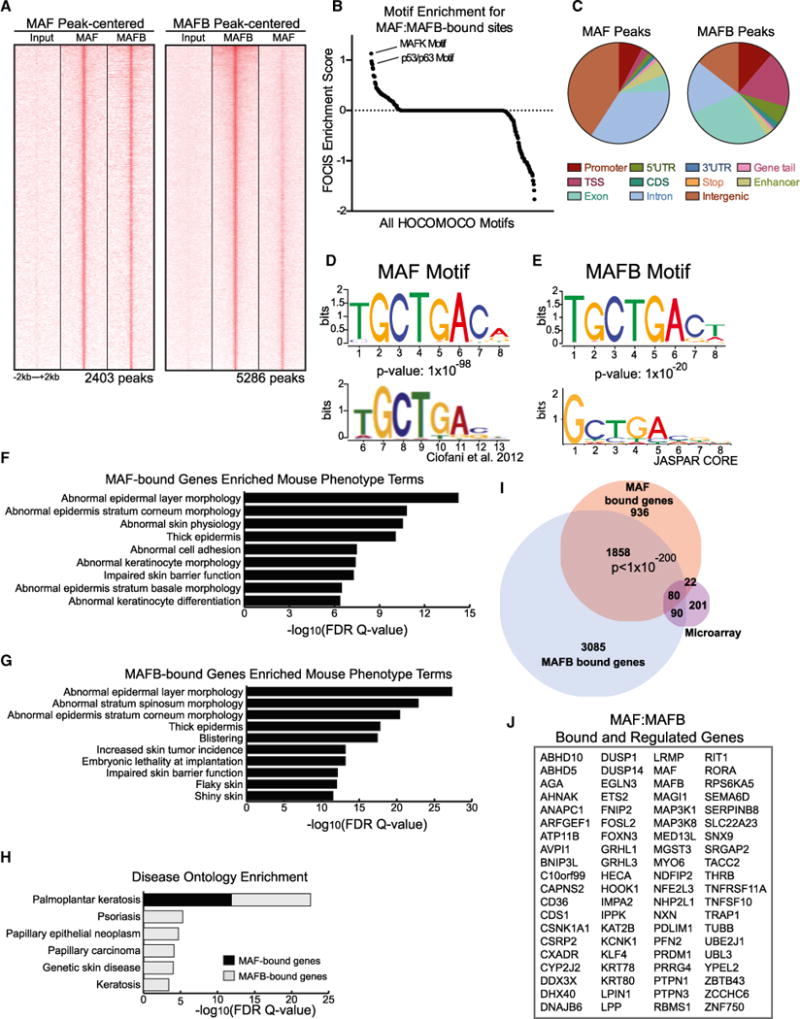
(A) Heatmap of MAF-centered ChIP-seq peaks aligned with input and MAFB ChIP-seq peaks (left) and MAFB-centered ChIP-seq peaks with input and MAF ChIP-seq peaks (right).
(B) FOCIS analysis was used to identify motifs predicted to co-occur within MAF and MAFB peaks; noted are two of the top hits.
(C–G) (C) Distribution of MAF peaks (left) and MAFB peaks (right) across the genome. Position-weight matrix of motifs discovered with ChIP-seq peaks for MAF with published (D) MAF motif from Ciofani et al. and MAFB-discovered motif with JASPAR (E) MAFB motif. Enriched mouse phenotype terms for genes associated with (F) MAF ChIP-seq peaks and (G) MAFB ChIP-seq peaks.
(H) Disease ontology enrichment observed for genes associated with MAF and MAFB peaks.
(I) Venn diagram showing overlap of MAFi:MAFBi microarray genes with MAF-bound and MAFB-bound genes.
(J) Eighty genes whose expression changes with knockdown of both MAF and MAFB and that are associated with both MAF and MAFB ChIP-seq peaks (see also Figure S6).
MAF:MAFB, however, also bound some regions independently of each other. For example, a portion of MAF peaks was present in intergenic regions, whereas MAFB was more equally distributed between genic and intergenic regions (Figure 5C). Consistent with these findings, MAF peaks were slightly more enriched for enhancer marks than MAFB peaks, as measured by enrichment for H3K4me1 marks acquired from ENCODE normal human epidermal keratinocytes data (Rada-Iglesias et al., 2011; Bonn et al., 2012; Figures S6F and S6G). HOMER motif discovery software analysis of MAF peaks identified a motif that resembled both the canonical MAF motif and a recently published MAF motif (Figure 5D, top; Ciofani et al., 2012). Likewise, the motif identified for MAFB was similar to that of MAF, and resembled and slightly expanded the JASPAR MAFB motif (Figure 5E).
We next performed Genomic Regions Enrichment of Annotations Tool (GREAT) analysis on MAF and MAFB peaks to identify genes potentially associated with these peaks. Interestingly, for both MAF and MAFB, we identified genes that were enriched for skin-relevant mouse phenotype terms, including abnormal epidermal layer morphology and keratinocyte differentiation (Figures 5F and 5G). Furthermore, disease ontology enrichment for MAF:MAFB gene-associated peaks revealed enrichment for human skin diseases characterized by abnormal epidermal differentiation, including psoriasis and hyperkeratotic genodermatoses (Figure 5H). Candidate direct MAF:MAFB targets were identified by intersecting genes bound by MAF:MAFB with those regulated by MAF:MAFB in tissue (Figure 5I). We observed a significant overlap between MAF:MAFB-bound genes seen by ChIP-seq and MAF:MAFB-controlled genes seen by profiling, with 102/2,896 genes identified as putative direct targets of MAF (p value = 3.78 × 10−22) and 170/5,113 genes as putative direct targets of MAFB (p value = 2.07 × 10−40). Additionally, we found a significant overlap in the MAF and MAFB peaks with a p value < 1 × 10−200, and identified 80 putative direct targets of both MAF and MAFB (Figure 5J), a number of which are known epidermal differentiation-promoting TFs.
MAF:MAFB Regulates Epidermal TFs
The 80 MAF:MAFB-bound and -regulated genes are putative direct targets of both MAF and MAFB. These genes were enriched for GO terms related to the regulation of transcription and gene expression (Figure S7A), suggesting that MAF: MAFB-driven differentiation may involve impacts on downstream TFs. Consistent with this, the expression of a panel of known epidermal differentiation-inducing TFs seen altered in MAF:MAFB microarray analysis was confirmed by qPCR to be reduced with the depletion of MAF:MAFB in epidermal tissue (Figure S7B), confirming that MAF:MAFB is required for full induction of other transcriptional drivers of epidermal differentiation. To study how MAF:MAFB regulates its differentiation-promoting TF targets, we selected the four best characterized of these TFs for further analysis. GRHL3, KLF4, ZNF750, and PRDM1 were re-verified to each be reduced by MAF:MAFB loss in tissue and to each display proximal regions of MAF:MAFB binding (Figures 6A, 6B, and S7C); weak binding of MAF was observed proximal to the KLF4 gene body, however, MAF also bound a site 95 kb away and controlled KLF4 expression, suggesting it may also act as a more distal regulatory element in this context. Binding peaks were verified by ChIP-qPCR and sequential ChIP-qPCR (re-ChIP) of first MAF then MAFB, as well as in the opposite direction, namely first MAFB then MAF (Figures 6C, 6D, S7D, and S7E), confirming that both MAF and MAFB bound these genes. GRHL3, KLF4, and ZNF750 are each also known p63 target genes. Consistent with enriched co-localization of p63 with peaks of MAF:MAFB binding noted above, re-ChIP of MAF then p63 or MAFB then p63 identified binding of both proteins at the promoters of KLF4, ZNF750, and GRHL3 (Figure S7F), suggesting that MAF:MAFB acts cooperatively with p63 to induce these TFs during epidermal differentiation.
Figure 6. MAF and MAFB Control Differentiation-Inducing Epidermal TFs.
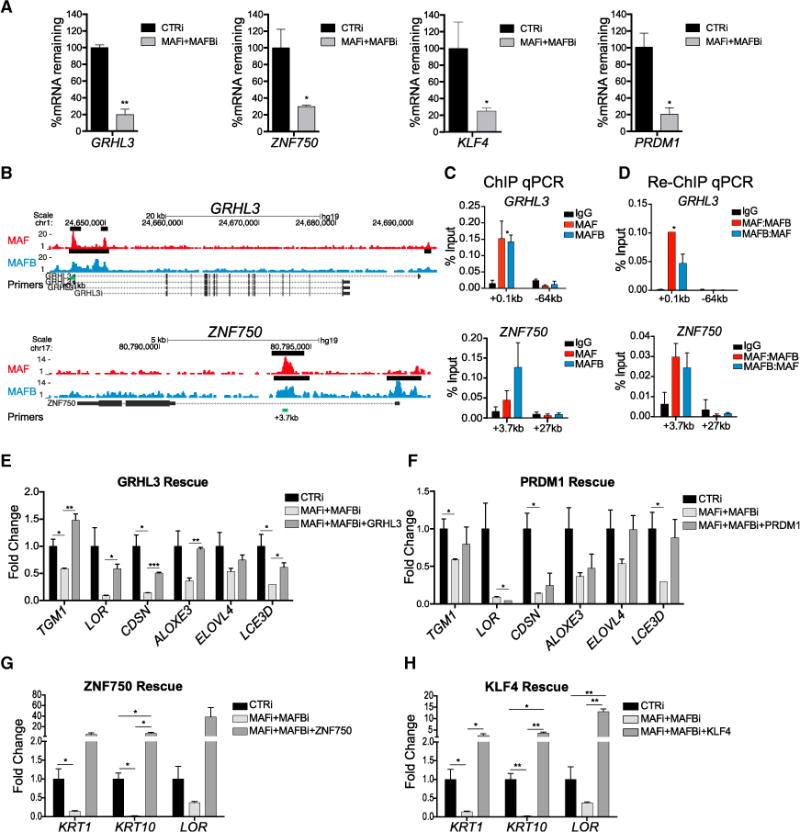
(A) mRNA levels of epidermal TFs in MAF:MAFB-depleted epidermal tissue.
(B) MAF (top, red) and MAFB (bottom, blue) ChIP-seq tracks at epidermal TF genes, GRHL3 and ZNF750; black bars denote ChIP-seq peaks called by MACS and green bars denote sequences for ChIP-qPCR.
(C) ChIP-qPCR of enriched MAF and MAFB peaks at GRHL3 and ZNF750.
(D) Re-ChIP-qPCR of enriched MAF and MAFB peaks showing occupancy of both MAF and MAFB on TF gene loci.
(E–H) Overexpression of (E) GRHL3, (F) PRDM1, (G) ZNF750, or (H) KLF4 partially rescues impaired differentiation observed in MAFi:MAFBi tissue. Mean ± SEM; n = 2 biological replicates; *p < 0.05, **p < 0.01, ***p < 0.001 (see also Figure S7).
To determine if MAF:MAFB-bound and -regulated TFs are sufficient to rescue differentiation gene expression in the setting of MAF:MAFB loss, we individually and separately restored the expression of GRHL3, ZNF750, KLF4, or PRDM1 in MAF: MAFB-depleted organotypic epidermal tissue. Each TF was able to recover a subset of impaired differentiation gene expression elicited by MAF:MAFB loss (Figures 6E–6H and S7G–S7L). Intriguingly, KLF4 and ZNF750, consistent with recent work demonstrating their cooperative action in this setting (Boxer et al., 2014; Sen et al., 2012), were able to rescue similar subsets of differentiation genes, whereas GRHL3 and PRDM1 were able to rescue a distinctly different subset (Figures 6G and 6H versus Figures 6E and 6F). These findings suggest that MAF:MAFB controls the expression of epidermal differentiation-inducing TFs that, in turn, act downstream of MAF:MAFB to activate discrete gene subsets in the epidermal differentiation program.
DISCUSSION
Here we characterized specific gene sets enriched during the regeneration of differentiated human epidermis to identify a new role for MAF:MAFB in epidermal homeostasis and to place MAF:MAFB in context among previously known epidermal regulators, including upstream lncRNAs and downstream effector TFs (Figure 7). The genetic circuitry underlying progenitor differentiation in somatic tissues is beginning to unfold as more functionally active regulators are identified. To fully understand the specific genetic programs controlled by individual factors, it is helpful to characterize the landscape of genomic expression occurring during the regeneration of differentiated tissue. In this study, we assessed mRNA expression changes during the regeneration of differentiated epidermis, starting from a progenitor cell population into a fully stratified tissue, every 24 hr over 7 days, by which point the full suite of terminal differentiation genes are induced. Previous studies focused on either short-term (up to 2 days) in vitro analysis or long-term (weeks) whole-skin transcriptional events (Janich et al., 2013; Klingenberg et al., 2010), potentially overlooking important transcriptional changes that were observed here between days 2 and 7. Analysis of our expression data identified distinct expression correlations between differentiation genes and up to 100 predicted gene regulators, 64 of which are as of yet uncharacterized in epidermis.
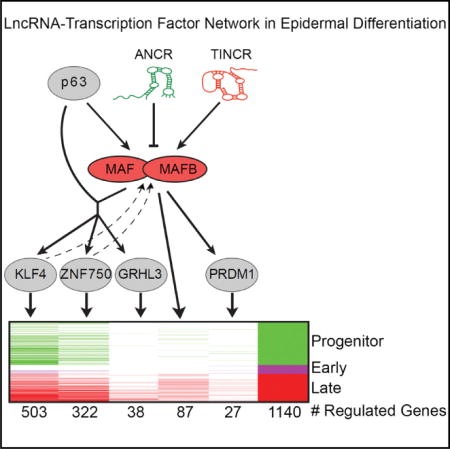
Figure 7. LncRNA:TF Network Regulating Epidermal Differentiation.
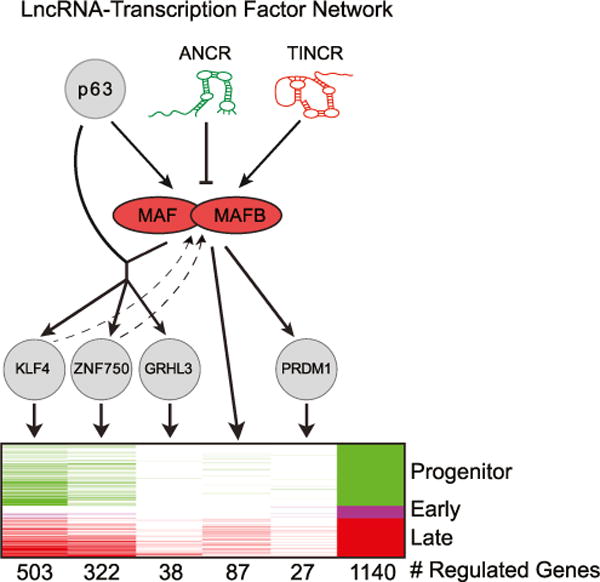
Proposed model of a lncRNA-MAF:MAFB-TF network regulating epidermal differentiation.
Among these new candidate regulators are MAF and MAFB TFs, which we observed to be necessary and sufficient for the regulation of a subset of the epidermal progenitor differentiation program. Notably, MAF and MAFB expression has been detected in the inner root sheath of the hair follicle, with c-maf−/− and mafb−/− mice displaying irregular cuticle patterning of whiskers, suggesting their function is not limited to the epidermis of the skin (Miyai et al., 2010). Although our work has focused on interfollicular epidermal homeostasis, this study suggests that MAF:MAFB contributes to morphogenesis of additional skin appendages, which can be addressed by conditional double-gene ablation in mice, since maf−/− mafb−/− mice are embryonic lethal (Aziz et al., 2009). Pro-differentiation functions have been attributed previously to both MAF and MAFB in other settings, notably the hematopoietic system. In these settings, however, they largely regulate cell cycle. Identified MAF:MAFB differentiation roles in other tissues include pancreas, bone, and, most recently, sensory neurons (Blank and Andrews, 1997; Artner et al., 2007; Sarrazin et al., 2009; Smink et al., 2009; Rutz et al., 2011; Wende et al., 2012). Another study also demonstrated that both MAF and MAFB are required for macrophage differentiation by preventing self-renewal (Aziz et al., 2009). Likewise, we have observed that MAF:MAFB impairs self-renewal of keratinocyte progenitors. However, our study suggests that MAF:MAFB is not only suppressing proliferation, but also promoting epidermal differentiation by activating essential differentiation TFs.
To help integrate the newly observed actions of MAF:MAFB in epidermal differentiation with existing knowledge of epidermal gene regulation, we incorporated gene sets from published epidermal regulators and placed them into the context of our progenitor differentiation gene sets. Such analysis permitted a view of the individual regulators that control specific aspects of the differentiation program. Although upstream regulation of MAFB by C/EBPd has been reported in skin, and recapitulated in our network analysis (Figure S4D), downstream targets of MAFB have not been fully characterized (Borrelli et al., 2010). Integration of MAF:MAFB ChIP-seq data with the spectrum of genes whose expression they control facilitated identification of MAF:MAFB downstream targets. Among these are TFs, including GRHL3, ZNF750, KLF4, and PRDM1, each of which plays an essential and non-redundant role in epidermal differentiation (Yu et al., 2006; Ting et al., 2005; Peyrard-Janvid et al., 2014; Birnbaum et al., 2006; Sen et al., 2012; Segre et al., 1999; Magnúsdóttir et al., 2007). MAF:MAFB genomic binding displayed significant, although not exclusive, colocalization with p63, the dominant epidermal TF identified to date, with indispensable roles in development, self-renewal, and differentiation of the tissue (Truong et al., 2006; Senoo et al., 2007; Mills et al., 1999) that also is mutated in a spectrum of human genetic skin disease (van Bokhoven and McKeon, 2002). p63 also influences expression of a number of MAF:MAFB target genes, in addition to MAF:MAFB themselves, including ZNF750 (Truong et al., 2006; Zarnegar et al., 2012), suggesting that MAF:MAFB represents an essential cooperative gene regulator for a subset of p63 targets.
The integrative approach allowed for prediction of upstream regulators of MAF and MAFB, revealing an unexpectedly prominent role for specific lncRNAs in this process. While not previously characterized as important regulators of any differentiation TFs in stratified epithelial tissues, such as epidermis, lncRNA regulation has been demonstrated for TFs important in other processes. For example, HOTAIR regulates expression of the HOXD TF locus during development by targeting chromatin-modifying complexes (Rinn et al., 2007; Wang et al., 2011), and multiple lncRNAs regulate Snai1 in hematopoiesis by mechanisms that are still being defined (Ørom et al., 2010). In epidermis, ANCR and TINCR lncRNAs, which act in opposing roles to either maintain progenitor status or to promote differentiation, respectively, appear to converge on MAF and MAFB by either repressing them in progenitors or by stimulating them in differentiation. In this setting, it appears that ANCR repression of MAF:MAFB expression may occur, at least in part, by facilitating binding of EZH2-repressive complexes to differentiation genes in progenitor keratinocytes. In the case of TINCR, it appears that TINCR upregulation of MAF:MAFB also may occur, at least in part, via the major known mechanism of TINCR action, specifically mRNA binding and stabilization. Taken together, these findings identify a regulatory network with features that include ANCR and TINCR lncRNA control of MAF:MAFB induction, with MAF:MAFB in turn inducing expression of genetically non-redundant TFs, including GRHL3, ZNF750, KLF4, and PRDM1, to engage terminal epidermal differentiation.
EXPERIMENTAL PROCEDURES
Organotypic Culture and Skin Grafts
Primary human keratinocytes were isolated from fresh surgically discarded skin, and cultured in Keratinocyte-SFM (Life Technologies 17005-142) and Medium 154 (Life Technologies M-154-500). Organotypic regeneration of human epidermis was performed as previously described (Truong et al., 2006). Biologic replicates were performed in all cases using cells from two or more unrelated donors. Organotypic epidermis was cultured for 4 days then grafted onto 6-week-old female NOD scid gamma mice (Jackson ImmunoResearch Laboratories) (Scholl et al., 2007). Skin grafts were harvested 17 days later. All animal protocols were approved by the Stanford Panel on Laboratory Animal Care (Protocol 9863).
Gene Transfer, RNAi, and CRISPR/Cas9 Knockout
Gene transfer was performed by viral transduction (Sen et al., 2010). For siRNA knockdown, 1 × 106 cells were electroporated with 1 nmol siRNA using Amaxa Human Keratinocyte Nucleofector Kit (Lonza VPD-1002). Human keratinocytes were transduced with lentivirus generated using lentiCRISPRv2 (Sanjana et al., 2014; Shalem et al., 2014) to introduce FLAG-Cas9 and a sgRNA targeting either control, MAF, or MAFB. Post-puromycin selection, a second sgRNA was introduced using pLX-sgRNA (Wang et al., 2014) and cells were selected with blasticidin. Double-gene-targeted cells were screened for MAF and MAFB deletion by western blot.
ChIP and ChIP-Seq Analysis
Human keratinocytes were cross-linked with 1% formaldehyde and chromatin was sonicated to an average fragment length of 150–250 bp. Chromatin was immunoprecipitated overnight at 4°C. Following cross-link reversal, samples were treated with RNaseA and the DNA was purified using a PCR Purification Kit (QIAGEN). Sequencing reads were uniquely aligned to hg19 with Bowtie (Langmead et al., 2009). ChIP signals were normalized to ten million mapped reads with peaks called using MACS (p value < 10−5, FDR < 0.05, fold enrichment > 5) (Zhang et al., 2008). Sequences for 500 MAF and MAFB peaks in each dataset, within ±100 bp of peak summits, were extracted and de novo motif analysis against these sequences was performed using HOMER (Heinz et al., 2010). GREAT was used to identify genes and gene set enrichments associated with peaks (McLean et al., 2010). FOCIS analysis was also performed (Webster et al., 2014).
Other Bioinformatic Analysis
Gene module map was performed (Segal et al., 2004). Data were acquired from GEO, supplemental data, or directly from authors. Gene expression was normalized in every dataset separately using the RMA package in R. The modules and their regulation programs were learned and predicted using the Module Networks algorithm in Genomica (Novershtern et al., 2011; Segal et al., 2003, 2004). A set of modules and their associated regulation programs were automatically inferred by an iterative procedure given expression values of 1,046 candidate regulator genes. One hundred permutations were performed and for each run 20 modules were selected. Regulators were ranked by frequency of prediction as a regulator. Multi-dimensional GSEA was conducted using Genomica with a p value cutoff of 0.01 (hypergeometric test). Gene sets were compared in a bar code graph by identifying shared genes in gene sets, and p value was determined by Fisher’s exact test. Cytoscape software was used to generate network models.
Supplementary Material
Highlights.
Epidermal gene sets in a progenitor differentiation time course are identified
Module mapping identified MAF:MAFB as epidermal differentiation regulators
ANCR and TINCR lncRNAs regulate epidermal MAF:MAFB expression
GRHL3, ZNF750, KLF4, and PRDM1 are MAF:MAFB effectors
Acknowledgments
We thank M. Fuller, A. Oro, H. Chang, G. Crabtree, M. Wernig, K. Wang, A. Bhaduri, X. Bao, A. Rubin, O. Wapinksi, S. Atwood, B. Sun, and A. Ungewickell for pre-review of the manuscript. We thank I. Obrien for technical support and L. Morcom and P. Bernstein for administrative assistance. This work was supported by the U.S. Department of Veterans Affairs Office of Research and Development and the National Institute of Arthritis and Musculoskeletal and Skin Diseases NIH R01 AR45192 (P.A.K), NIH F32 CA1420902, and NIH Diversity Supplement to AR45192 (V.L.-P).
Footnotes
ACCESSION NUMBERS
Profiling data and ChIP-seq data have been deposited with GEO accession code GSE52954.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2015.01.028.
References
- Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet b cell maturation. Proc Natl Acad Sci USA. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–871. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12:193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum RY, Zvulunov A, Hallel-Halevy D, Cagnano E, Finer G, Ofir R, Geiger D, Silberstein E, Feferman Y, Birk OS. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat Genet. 2006;38:749–751. doi: 10.1038/ng1813. [DOI] [PubMed] [Google Scholar]
- Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Borrelli S, Fanoni D, Dolfini D, Alotto D, Ravo M, Grober OM, Weisz A, Castagnoli C, Berti E, Vigano MA, Mantovani R. C/EBPd gene targets in human keratinocytes. PLoS ONE. 2010;5:e13789. doi: 10.1371/journal.pone.0013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eychène A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008;8:683–693. doi: 10.1038/nrc2460. [DOI] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedge SP, Kumar A, Kurschner C, Shapiro LH. c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol Cell Biol. 1998;18:2729–2737. doi: 10.1128/mcb.18.5.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cisregulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, Andersen B. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Toufighi K, Solanas G, Luis NM, Minkwitz S, Serrano L, Lehner B, Benitah SA. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell. 2013;13:745–753. doi: 10.1016/j.stem.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg JM, McFarland KL, Friedman AJ, Boyce ST, Aronow BJ, Supp DM. Engineered human skin substitutes undergo large-scale genomic reprogramming and normal skin-like maturation after transplantation to athymic mice. J Invest Dermatol. 2010;130:587–601. doi: 10.1038/jid.2009.295. [DOI] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GXY, Chow J, Kim GE, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci USA. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Ferreira LMR, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet β cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Miyai M, Tanaka YG, Kamitani A, Hamada M, Takahashi S, Kataoka K. c-Maf and MafB transcription factors are differentially expressed in Huxley’s and Henle’s layers of the inner root sheath of the hair follicle and regulate cuticle formation. J Dermatol Sci. 2010;57:178–182. doi: 10.1016/j.jdermsci.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci USA. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmány G, Donati G, Uribe-Lewis S, Pavlidis P, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Nakashima T, Takeda S, Isogai M, Hamada M, Kimura A, Kodama T, Yamaguchi A, Owen MJ, Takahashi S, Takayanagi H. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest. 2010a;120:3455–3465. doi: 10.1172/JCI42528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Nakashima T, Hayashi M, Fukunaga T, Kato S, Kodama T, Takahashi S, Calame K, Takayanagi H. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc Natl Acad Sci USA. 2010b;107:3117–3122. doi: 10.1073/pnas.0912779107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama R, Nguyen BC, Talora C, Ogawa E, Tommasi di Vignano A, Lioumi M, Chiorino G, Tagami H, Woo M, Dotto GP. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1-caspase 3 regulatory mechanism. Dev Cell. 2004;6:551–562. doi: 10.1016/s1534-5807(04)00098-x. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. 2014;94:23–32. doi: 10.1016/j.ajhg.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Noubade R, Eidenschenk C, Ota N, Zeng W, Zheng Y, Hackney J, Ding J, Singh H, Ouyang W. Transcription factor c-Maf mediates the TGF-β-dependent suppression of IL-22 production in T(H)17 cells. Nat Immunol. 2011;12:1238–1245. doi: 10.1038/ni.2134. [DOI] [PubMed] [Google Scholar]
- Sadl V, Jin F, Yu J, Cui S, Holmyard D, Quaggin S, Barsh G, Cordes S. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev Biol. 2002;249:16–29. doi: 10.1006/dbio.2002.0751. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L, Kelly Modis L, Kastner P, Chan S, Duprez E, et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138:300–313. doi: 10.1016/j.cell.2009.04.057. [DOI] [PubMed] [Google Scholar]
- Scholl FA, Dumesic PA, Barragan DI, Harada K, Bissonauth V, Charron J, Khavari PA. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 2007;12:615–629. doi: 10.1016/j.devcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieweke MH, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- Smink JJ, Bégay V, Schoenmaker T, Sterneck E, de Vries TJ, Leutz A. Transcription factor C/EBPbeta isoform ratio regulates osteoclastogenesis through MafB. EMBO J. 2009;28:1769–1781. doi: 10.1038/emboj.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med. 2002;8:133–139. doi: 10.1016/s1471-4914(01)02260-2. [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DE, Barajas B, Bussat RT, Yan KJ, Neela PH, Flockhart RJ, Kovalski J, Zehnder A, Khavari PA. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. 2014;24:751–760. doi: 10.1101/gr.166231.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H, Lechner SG, Cheret C, Bourane S, Kolanczyk ME, Pattyn A, Reuter K, Munier FL, Carroll P, Lewin GR, Birchmeier C. The transcription factor c-Maf controls touch receptor development and function. Science. 2012;335:1373–1376. doi: 10.1126/science.1214314. [DOI] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, Andersen B. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Webster DE, Lopez-Pajares V, Vander Stoep, Hunt B, Qu K, Yan KJ, Berk DR, Sen GL, Khavari PA. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. Am J Hum Genet. 2012;91:435–443. doi: 10.1016/j.ajhg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


