Abstract
Impaired spatial memory characterizes many mouse models for Alzheimer's disease, but we understand little about how this trait arises. Here we use a transgenic model of amyloidosis to examine the relationship between behavioral performance in tests of spatial navigation and the function of hippocampal place cells. We find that APP mice require considerably more training than controls to reach the same level of performance in a water maze task, and recall the trained location less well 24 hours later. At a single cell level, place fields from control mice become more stable and spatially restricted with repeated exposure to a new environment, while those in APP mice improve less over time, ultimately producing a spatial code of lower resolution, accuracy, and reliability than controls. The limited refinement of place fields in APP mice likely contributes to their delayed water maze acquisition, and provides evidence for circuit dysfunction underlying cognitive impairment.
Keywords: spatial tuning, hippocampal place cell, cognitive decline, CA1, amyloid
Introduction
Many patients with Alzheimer's disease (AD) state that one of the earliest signs of forthcoming dementia was finding themselves lost in a familiar location (Gazova et al., 2012; Lithfous et al., 2013). The initial loss of spatial orientation is usually temporary, but as the disease progresses, can become a persistent symptom with significant repercussions for both patients and caregivers (Carson, 2012; Gitlin et al., 2012). Not surprisingly, some of the same brain regions damaged by Alzheimer's are responsible for spatial orientation. Studies of spatial navigation in rodents have identified a complex network of specialized cells within the hippocampus that respond directly to the animal's physical location in space (Moser et al., 2008; O'Keefe and Dostrovsky, 1971). The coordinated activity of thousands of such place cells provides a neural map of the rodent's surroundings (O'Keefe and Nadel, 1978). The accuracy of this map, and the animal's ability to navigate within this environment, both depend on the function of individual cells at multiple levels within this network.
There are two features of place cells that may link them to functional decline in Alzheimer's disease. First and most simply, they are located in the hippocampus, an area that is particularly prone to degeneration. Second, place cells have properties that make them capable of encoding the sort of episodic memories that are typically lost in the initial stages of Alzheimer's disease (Rolls, 2010; Smith and Mizumori, 2006). Despite their name, place cells encode more than just location. Recent studies have shown that most place cells also encode time, context, and emotional valence, making them uniquely suited to support multifaceted autobiographical memories (Eichenbaum, 2013). Further, encoding of both space and time is facilitated by the innate firing sequences displayed by cells in this area. Trajectories through space are mapped by existing chains of neural activity appropriated to encode the inherently sequential nature of an animal's movement through the environment (Foster and Knierim, 2012). The coordinated chains of neural firing that encode location and time in rodents may be very similar to the pattern of activity used by humans to encode autobiographical experiences that unfold sequentially in both time and space. Late in AD, hippocampal neurodegeneration might cause overt loss of elements in the sequence, but even early damage to this region could affect the precision of their activity. Either form of degeneration could degrade both the encoding and recall of episodic memory.
We hypothesize that the decline in spatial abilities seen in Alzheimer's patients and transgenic mouse models for the disease is caused by progressive dysfunction of hippocampal place cells in response to amyloid accumulation and subsequent neuronal damage. Here we use a mouse model of Alzheimer's amyloidosis to examine the relationship between cognitive function assessed by spatial navigation and the underlying activity of hippocampal neurons responsible for encoding spatial information. By interrogating the process by which amyloid-bearing animals acquire spatial information both at the level of behavioral performance and of single cell activity, we gain valuable insight into the nature of cognitive impairment in our model for AD, and by cautious extension, to the disease itself.
Methods
Transgenic mice
We studied tetracycline-responsive APP transgenic mice expressing a chimeric mouse APP with a humanized Aβ domain encoding the Swedish and Indiana mutations (MMRRC #34845 (Jankowsky et al., 2005)). APP transgenic mice were mated to CaMKIIα-TTA line B mice expressing the tetracycline transactivator (TTA) under control of the calcium-calmodulin kinase type IIα (CaMKIIα) promoter (Jackson Laboratories #3010 (Mayford et al., 1996)) to create bigenic APP/TTA offspring. Single transgenic (TTA only) and non-transgenic (NTG) cagemates were used as controls. Each line had been independently backcrossed to C57BL/6J for >20 generations before being intercrossed to generate stud males that were bred with C57BL/6J females to generate animals for study. All mice used in this study were raised on doxycycline (dox) to suppress transgene expression during postnatal development. We have previously shown this strategy to ameliorate locomotor hyperactivity and normalize body weight of double transgenic animals (Rodgers et al., 2012), permitting reliable cognitive testing. Offspring were started on dox 1-3 days after birth by placing nursing mothers on medicated chow, formulated to 50 mg/kg dox in Purina 5001 chow (F5903, BioServe, Frenchtown, NJ). At weaning, mice were maintained on dox until 6 weeks of age, then returned to regular chow to initiate transgene expression for the remainder of the experiment. Animals were housed on a 13:11 day: night cycle and were used for study between 10 and 14 months of age. All behavioral and electrophysiological studies were done during daylight hours of the light cycle. Animal experiments described here were reviewed and approved by the Baylor College of Medicine Institutional Care and Use Committee.
Behavioral assays
Behavioral testing began after 9.5 months of APP expression (11 months of age) and included open field, Morris water maze (MWM), and radial arm water maze (RAWM). Mice were handled for 3 days prior to the start of behavioral testing, which began with an open field test to assess locomotor activity followed by MWM to assess spatial reference memory and RAWM to assess working memory.
Open field assessment
Animals were placed individually into white acrylic open-top boxes (46 × 46 × 38 cm) in a room lit by indirect white light. Locomotor activity was recorded for 30 min and analyzed using the ANY-maze Video Tracking System (Stoelting Co., Wood Dale, IL).
Morris water maze
Testing was conducted in a circular white acrylic tank measuring 58 cm high and 122 cm in diameter. The water level was 20 cm from the top of the tank and made opaque by adding non-toxic white paint. Water temperature was maintained between 21-23° C. The room was lit with indirect white light and trials were recorded and tracked using ANY-maze Video Tracking System.
Prior to the start of acquisition training, mice received one day of training in a straight swim channel to acclimate them to the water and check for motor deficits. Mice received 4 trials with a 10 min inter-trial interval (ITI) in a channel constructed of white acrylic and measuring 107 × 56 ×14 cm that was placed in the center of pool. Visible cues were removed from the room during straight swim shaping trials. Mice were allowed 60 s to reach a submerged platform on the opposite side of the channel. Mice that failed to reach the platform were guided to the location by the experimenter. Mice were allowed to stay on the platform for 10 s before being removed from the water, dried, and returned to their home cage under an infrared heat lamp between trials.
Acquisition training in the MWM consisted of 4 trials/day with a 15 min ITI. Each training session ended with a short-term memory probe. A square platform (10 × 10 cm) covered in nylon mesh for traction was located 1 cm below the surface in the NE quadrant of the maze, half way between the side and the center of the pool. Mice were placed in the maze facing the wall at each of 4 cardinal start locations and allowed 60 s to locate the hidden platform. As with straight swim, animals that failed to locate the platform in the allotted time were gently guided there by the experimenter. Mice were allowed to stay on the platform for 15 s before being returned to their home cage between trials. Following the 4 daily training trials, the platform was removed from the maze for an immediate probe trial. Animals were placed in the tank half way between the cardinal points (SW, NW, SE, NE) and allowed 45 s to navigate the maze. Proximity to target, percent time, and percent path spent in each quadrant were calculated along with the number of times the animal crossed the platform location compared to the other three potential platform locations (in the SW, NW, and SE quadrants). Mice were trained until they met two performance criteria during the probe trial: first, ≥ 35% of their total swim path had to be located in the trained quadrant, and second, ≥ 40% of their platform crossings had to be over the trained location compared to the three other equivalent sites within the pool.
When mice reached criterion, they were retired from further MWM training, but given a final long-term memory probe 24 hr after reaching criterion. Mice were trained for a maximum of 10 days. After all mice either reached criterion or completed 10 days of training, each was given one additional ‘refresher’ day of training with probe test to ensure equivalent performance between groups prior to starting RAWM training.
Radial arm water maze
The day following MWM refresher training, mice received 1 day of RAWM training consisting of 8 trials with a 15 min ITI. The RAWM was created by installing clear Plexiglas triangular inserts into the existing water maze pool (41.25 cm on each side × 50 cm high), which resulted in 6 open arms joined at the center. Each arm measured 20 cm wide × 34 cm long, and the water was maintained at a depth of 38 cm. The platform was located 3 cm from the end of one arm and submerged 1 cm below the water's surface. Mice were placed into a different arm at the beginning of each trial, with the order of starting positions pseudo-randomly selected prior to training such that no trial began in an arm adjacent to the previous start position. Mice were allowed 60 s to navigate the maze. If a mouse failed to locate the platform in the allotted time, it was gently guided to there by the experimenter and allowed to remain on the platform for 15 s before being returned to its home cage between trials. Latency to locate the platform, swim path length, and number of working memory errors (re-entries into a previously visited arm) were calculated for each trial. Trial 1 scores were excluded from analysis due to the inflated error rate as animals learned the new procedure. Data from trials 2-8 was averaged to provide a performance index for overall RAWM learning. After the final training trial, animals were tested with a short-term probe trial in which the platform was removed and animals were allowed 45 s to navigate the maze. Percent time and path spent in the target arm were calculated and compared among treatment groups.
Surgery and behavioral procedures for in vivo multi-tetrode recording
Animals were fixed in a stereotaxic frame and anesthetized with 0.5-2% isoflurane. Ten anchor screws were inserted into the skull, and then a craniotomy was made at −2.0 mm posterior and 1.5 mm lateral from bregma to insert the drive cannula and position the tetrodes just above the cortex. The drive was affixed to the skull with dental cement, and the skin closed around the mount. Ketoprofen analgesic was delivered s.c. prior to recovery from anesthesia and for 3 days after surgery. Animals were given a week to recover before being placed on food restriction to maintain animals at 85% of their projected ad libitum body weight. Food restriction facilitated behavioral training in return for a food reward of diluted condensed milk. Animals were trained for approximately 3 weeks to run laps forward and back along a ∼2 m long rectangular track while the tetrodes were slowly advanced into the CA1 layer of the hippocampus. Recording in the now-familiar environment began when the animals could reliably run at least 10 laps in each direction and multiple stable hippocampal neurons could be isolated as single-units. Daily recording consisted of two sessions on the track of about 15 minutes each, separated by a 20 minute rest session in a raised ‘hammock’ to restrict activity. Once recordings in the familiar room were complete, the animal was introduced to a new linear track ∼1 m long located in a different room. Animals were recorded for 3 days in the novel environment using the same schedule as for the familiar track.
Electrophysiological data acquisition and processing
During the recording sessions, the hyperdrive was connected to a counterbalanced cable that enabled the animal to run freely on the track. Details of recording procedures and data analysis have been described previously (Cheng and Ji, 2013; Ji and Wilson, 2007). Tetrode recordings were acquired using a DigitaLynx system (Neuralynx, Bozeman, MT). Extracellular voltage signals from 4 channels in each tetrode were digitized at 32 kHz and band-pass filtered between 600 Hz and 9 kHz. Spikes were counted when the voltage signal from any of the channels surpassed a trigger threshold of 50–70 μV. Broadband local field potentials (LFPs) of 0.1 Hz–1 kHz were sampled at 2 kHz. The animal's position during recording was monitored by a digital camera tracking two color diodes connected to the tetrode drive. Video data was sampled at 33 Hz with a resolution of ∼0.2 cm. Raw data were stored for off-line analysis. For each animal, we analyzed one day's data from the familiar room (selecting the day with the highest number of recorded neurons) and all three days' data in the novel room. Single neurons were manually isolated from multiunit data using xclust (Matthew Wilson, MIT). Only active putative pyramidal neurons with a mean firing rate between 0.5 and 7 Hz on one trajectory of a track were included for further analysis. We applied the same set of criteria to each cell twice, once for each direction. We also eliminated spikes fired at the track ends. Results are presented either as median and range [25%, 75%] or as mean ± SEM.
Rate curves
Rate curves describe the relationship between the animal's position on the track and a neuron's average firing rate at that position. Each direction on the track is considered a one-dimensional (1D) trajectory, and can be linearized and divided into 2 cm bins. The rate curve of each neuron was computed from the average firing rate at each bin. The rate xi at the ith bin was calculated as the number of spikes fired divided by the time spent in bin i (ti) across all laps run in each trajectory of each session that day. Rate curves for individual sessions were thus calculated for one trajectory during a single session.
Spatial information (SI)
Spatial information (SI) is a measure of how unambiguously the cell's firing reflects the position of the animal. SI was calculated according to the following equation (Skaggs et al., 1993; Skaggs et al., 1996):
where i was the bin number, was the probability of occupancy of bin i, xi was the mean firing rate in bin i, was the overall mean firing rate on each trajectory, and N was the number of bins. For every neuron active on each trajectory, SI was calculated where xi and pi were computed as the number of spikes and the occupancy time during all laps of a trajectory.
Session stability
We computed “session stability” to measure how similar a cell's firing rate curve was in the first session to its rate curve in the second session. For average trajectory firing rates at the i-th bin of xi in session one and yi in session two, the session stability is computed as
Where x ¯ and σx are the mean and standard deviation of [x1, x2, …, xN] and y ¯ and σy are the mean and standard deviation of [y1, y2, …, yN].
Place field analysis
A place field is the track location at which a given neuron is responsive, and its size determines the spatial resolution contributed by that cell. Tentative place fields were defined from the peak(s) in the rate curve for each cell active on a trajectory. A threshold of 10% peak rate was used to define the boundaries of the place field. Neighboring fields separated by less than 4 cm were combined into a single field. Only those fields which had a peak rate > 1 Hz, a mean rate > 0.5 Hz and field length > 10 cm were included in the final analysis.
Statistics
Statistical comparisons of multi-tetrode data were done by Kruskal Wallis test followed by Wilcoxon rank-sum post-hoc comparison using the MATLAB MindCracker package (Daoyun Ji, Baylor College of Medicine). Statistical comparisons of tetrode data between novel and familiar environments was done by two-way ANOVA, followed by Wilcoxon rank-sum post-hoc comparison. Statistical comparisons of behavioral data were done by one-way ANOVA except where indicated for two-way ANOVA, and both were followed by Bonferroni post-hoc comparisons using IBM SPSS Statistics 12.0 or Prism 6.0 analysis software. Statistical values listed in the text are for main effect ANOVA, or where indicated, for post-hoc comparisons. Full statistical details for all significant comparisons are shown in Table 1.
Table 1. Comparative statistics.
| Open Field | ||||||||
|---|---|---|---|---|---|---|---|---|
| Test | Measure | Effect | df | F value | Significance | vs. NTG | vs. TTA | |
| Distance Traveled | One-way ANOVA | Distance Traveled | Genotype | 2, 28 | 4.49 | <0.05 | ns | * |
| % Time in Center | One-way ANOVA | % time | Genotype | 2, 28 | 0.32 | = 0.73 | ns | ns |
| % Path in Center | One-way ANOVA | % path | Genotype | 2, 28 | 2.80 | = 0.08 | ns | ns |
| Test | Measure | Effect | df | F value | Significance | vs. NTG | vs. TTA | |
| RM ANOVA | Path length | Day | 3, 81 | 2.08 | < 0.001 | --- | --- | |
| Training | Genotype | 2, 28 | 8.59 | <0.01 | * D2 & D4 | * D2 & D4 | ||
| Interaction | 6, 81 | 0.17 | = 0.98 | ns | ns | |||
| RM ANOVA | Proximity | Day | 3, 81 | 31.89 | < 0.0001 | |||
| STM Probe | Genotype | 2, 28 | 8.11 | <0.01 | *** D3 *** D4 | **D3 *** D4 | ||
| Interaction | 6, 81 | 3.86 | <0.01 | --- | --- | |||
| Days to Criteria | One-way ANOVA | Number if Days | Genotype | 2, 28 | 9.31 | < 0.001 | ** | *** |
| LTM Probe | One-way ANOVA | % Time in Target | Genotype | 2, 25 | 5.30 | <0.05 | ** | * |
| LTM Probe | One-way ANOVA | % Path in Target | Genotype | 2, 25 | 3.47 | <0.05 | p = 0.06 | p = 0.07 |
| Retraining | One-way ANOVA | Path length | Genotype | 2, 28 | 1.91 | = 0.12 | ns | ns |
| Retraining Probe | One-way ANOVA | Proximity | Genotype | 2, 28 | 2.78 | = 0.17 | ns | ns |
| RAWM | ||||||||
| Test | Measure | Effect | df | F value | Significance | vs. NTG | vs. TTA | |
| RM ANOVA | WM Errors | Trial | 6. 150 | 4, 80 | < 0.001 | --- | --- | |
| Training | Genotype | 2, 28 | 9.23 | < 0.001 | * Trials 3-8 | * Trials 3-5 & 8 | ||
| Interaction | 12, 150 | 1.44 | = 0.15 | ns | ns | |||
| STM Probe | One-way ANOVA | % Path in Target | Genotype | 2.25 | 5.21 | < 0.001 | ** | * |
| Novel Day 3 Place Cell Data | ||||||||
| Test | df | X value | Significance | vs NTG | vs TTA | |||
| Spatial Information | Kruskal-Wallis | 3, 389 | 15.58 | < 0.001 | * | *** | ||
| Place Field Length | Kruskal-Wallis | 3, 316 | 10.21 | <0.01 | * | * | ||
| Familiar Day Place Cell Data | ||||||||
| Test | df | X value | Significance | vs NTG | vs TTA | |||
| Spatial Information | Kruskal-Wallis | 3, 823 | 72.73 | < 0.001 | **** | **** | ||
| Session Stability | Kruskal-Wallis | 3, 551 | 56.63 | < 0.001 | **** | *** | ||
| Peak Firing Rate | Kruskal-Wallis | 3, 1434 | 37.23 | < 0.0001 | **** | ** | ||
| Novel vs Familiar Place Cell Data | ||||||||
| Test | Effect | df | F value | Significance | vs NTG | vs TTA | ||
| Spatial Information | Two-way ANOVA | Day | 1, 1170 | 575.9 | < 0.001 | |||
| Genotype | 2, 1170 | 18.12 | < 0.0001 | *** | *** | |||
| Interaction | 2, 1170 | 17.67 | < 0.0001 | --- | --- | |||
| Session Stability | Two-way ANOVA | Day | 1, 967 | 38.69 | < 0.001 | |||
| Genotype | 2, 967 | 7.461 | < 0.001 | **** | * | |||
| Interaction | 2, 967 | 6.508 | <0.01 | --- | --- | |||
| Peak Firing Rate | Two-way ANOVA | Day | 1, 1211 | 26.7 | < 0.0001 | |||
| Genotype | 2, 1211 | 11.43 | < 0.0001 | **** | *** | |||
| Interaction | 2, 1211 | 4.986 | <0.01 | --- | --- | |||
Histology
After recording was completed, animals were sacrificed with an overdose of pentobarbital and the final position of the tetrodes was marked by electrolytic lesion (30 μA, 6–10 s). Brains were immersion fixed in 4% paraformaldehyde. Coronal 35 μm sections were stained with cresyl violet to locate the electrode tracks and confirm tetrode positions. Additional sections were stained with silver and thioflavine-S to verify amyloid load in APP/TTA animals.
Results
Spatial learning is significantly slowed in amyloid-bearing mice
We studied a mouse model in which amyloid pathology is recreated through overexpression of APP encoding two AD-associated familial mutations (APPswe/ind; (Jankowsky et al., 2005). Transgenic APP expression in this model is dependent on the tetracycline transactivator (TTA) that is limited to forebrain neurons by the CaMKIIα promoter (Mayford et al., 1996). Using this system, we can avoid any confounding effects of APP overexpression on brain development by delaying transgene onset with doxycycline until the animals reach adulthood (Rodgers et al., 2012). As a consequence of this transgenic system, our studies required inclusion of an additional control group expressing TTA alone to ensure that any differences we observed between bigenic APP/TTA mice and non-transgenic (NTG) siblings were due to APP and amyloid rather than the transactivator required to drive APP expression.
Many transgenic models encoding familial mutations in the amyloid precursor protein (APP) display spatial memory impairments akin to the cognitive symptoms of AD (Dodart et al., 2002; Morgan, 2003). Few studies, however, have examined the possibility that delayed acquisition may be distinct from impaired recall, and that each process may be differently affected by underlying deficits in spatial encoding. We therefore designed a variation of the Morris water maze that allowed us to distinguish spatial learning from long-term memory. The task was based on training animals for as long as needed to reach a pre-specified level of performance before examining long-term recall. This train-to-criteria protocol yielded several added metrics of spatial learning to complement average path length per day, including the number of training days required to reach criteria performance, the percentage of animals at criteria performance each day, and the accuracy of immediate recall for the trained location at the end of each session.
Before proceeding with cognitive testing, we first ensured that our mice had no locomotor abnormalities that might confound behavioral analyses. Open field testing showed that APP/TTA mice ran slightly farther during the 30 min trial than TTA single-transgenic controls, but were not significantly different from NTG (post hoc vs. TTA p < 0.05; Table 1; n = 9-12 per genotype, 4 female and 6-8 male mice per group,). We found no difference between genotypes in percent path or time spent in the center 1/3 of the field, indicating that the APP/TTA mice showed no sign of heightened anxiety (p > 0.05).
We next assessed motor abnormalities in a straight swim channel that served the added purpose of acclimating them to water. All three groups performed similarly and we saw no specific motor deficits in any of the animals, so we proceeded with water maze testing the following day. All mice received 4 daily training trials per day immediately followed by a probe trial to assess short-term memory (STM). Mice underwent successive days of training until they reached two criteria of performance during their end-of-day probe test (see Materials and Methods for details). Once a mouse reached criteria it was retired from further training, but underwent one final probe trial 24 h later to assess long-term memory.
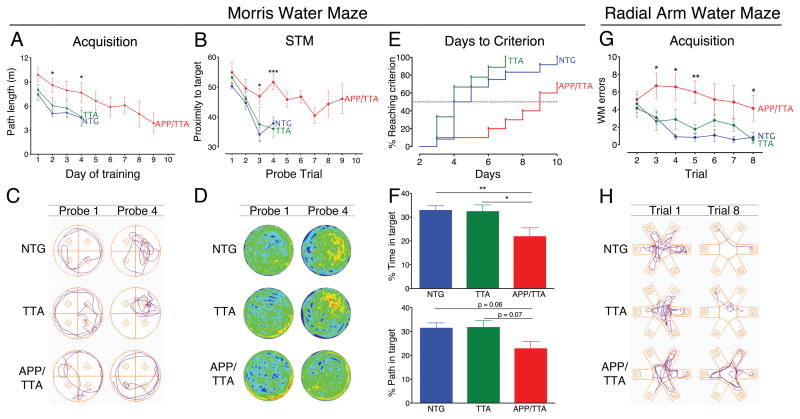
All three genotypes improved their navigation as training progressed. Path length required to reach the hidden platform decreased over successive days of training for all groups (main effect of day, p < 0.001), however, APP/TTA mice improved more slowly than controls, swimming significantly farther to reach the platform on days 2 and 4 (post-hoc p < 0.05 vs. NTG and TTA; Figure 1A). APP/TTA mice also showed poorer performance in probe trials at the end of each training session. While the accuracy of short-term memory improved over successive testing days for all three genotypes (main effect of day, p < 0.0001), APP/TTA mice again improved more slowly than control mice (p < 0.05; Figure 1B). By days 3 and 4 of testing, APP/TTA mice had a higher proximity score - meaning they swam farther from the target - than either control group during the end-of day probe trials (probe 3, post hoc p < 0.001 for NTG and 0.01 for TTA; probe 4, p < 0.001 for both NTG and TTA; (Gallagher et al., 1993)). The APP/TTA learning impairment was most evident by looking at traces of the swim path taken by individual animals from each genotype. During the first probe trial on day 1 of training, mice from all three genotypes circled the pool in an undirected fashion. By probe 4, the control animals swam in a path that focused on the platform location, while the APP/TTA mouse was still swimming in circles (Figure 1C). The same pattern emerged when data from the entire group was averaged and displayed as heat maps of target proximity (Figure 1D). Given these results, it is not surprising that APP/TTA mice required more days of training than control mice to reach criteria performance (post-hoc p < 0.01 vs. NTG and 0.001 vs. TTA, Figure 1E). Half of the NTG and TTA mice reached criteria performance within 4 days of training, and all passed this threshold within 10 days. In contrast, it took 9 days for half of the APP/TTA mice to reach criteria performance, and 30% never met criteria at all. Collectively, these results indicate that APP/TTA mice acquire spatial information less efficiently than controls, with some animals failing to demonstrate any proficiency even after 10 days of training.
Figure 1. APP/TTA mice learn spatial tasks less efficiently and effectively than age-matched controls.
A, Average path length required to reach the hidden platform across testing days in MWM. APP/TTA mice performed significantly worse than NTG or TTA controls on days 2 and 4 of testing. B, Average proximity to the target during short term memory (STM) probe trials conducted after the final training trial each day demonstrate a memory deficit in APP/TTA mice on days 3 and 4 of testing. C, Representative track plots showing swim paths of NTG, TTA, and APP/TTA littermates on the first and fourth MWM probe trials. Mice spent nearly equal time in each quadrant at the beginning of training (probe 1). By probe trial 4, NTG and TTA mice swam nearer the target (located in the northeast quadrant) than the APP/TTA mouse. D, Heat plots showing average target proximity for each genotype on probe trials 1 and 4. NTG and TTA mice spend much of their swim path concentrated around the platform (denoted by red/orange shading) compared with APP/TTA mice who show little preference for the trained location. E, Proportion of each genotype reaching criteria on consecutive days of testing. APP/TTA mice require significantly more days to reach criteria than NTG and TTA controls. F, Percentage of time spent in the target quadrant during the LTM probe is significantly lower for APP/TTA mice than for NTG or TTA controls (upper panel). The percentage of total path length spent in the target quadrant during LTM testing shows the same trend as percentage time, but falls just short of statistical significance. G, Comparison of RAWM errors made by each genotype highlights the slow rate of acquisition in APP/TTA mice. APP/TTA mice made significantly more errors on trials 3, 4, and 5 than NTG or TTA mice. I, Representative track plots from NTG, TTA, and APP/TTA littermates on the first and last RAWM training trials. All genotypes perform similarly during trial 1. By the end of training, control mice locate the hidden platform without error, while the APP/TTA mouse remains unable to navigate effectively.* p< 0.05, ** p < 0.01, *** p< 0.001. See Table 1 for complete statitsical comparisons.
Not only did APP/TTA mice take longer to learn the trained location than controls, they also recalled the platform location less well later. Twenty-four hours after each animal passed criteria, they were given one final probe trial to examine long-term recall. The APP/TTA mice spent significantly less time in the trained quadrant than either control group (post hoc p < 0.01 for NTG and 0.05 for TTA, Figure 1F, top panel). The difference between groups showed a similar trend when performance was measured as percent of path in the trained quadrant (post hoc, p = 0.06 for NTG and 0.07 for TTA, Figure 1F, bottom panel), however, the effect was not detected by all measures. Specifically, the performance of APP/TTA mice was no different than controls when measured as proximity to the target location (p = 0.08). Taken together, these analyses suggest that APP/TTA mice show some impairment in long-term spatial memory at this stage, but it is not yet severe enough to affect every aspect of their performance.
All of the mice that reached criteria performance in the MWM were advanced to testing in the radial arm water maze (RAWM; n = 12 out of 12 NTG, 9/9 TTA, and 7/11 APP/TTA). This test used the same room and spatial cues as the MWM, but with a new maze configuration and platform location. The task thus examined cognitive flexibility for the new demands of a reconfigured maze. We first gave each of the mice refresher training in the MWM to ensure that the performance of all genotypes was rebalanced in RAWM testing. There were no differences between groups in either the mean path length during re-training (p = 0.12) or proximity to target during the probe trial for immediate recall (p = 0.17). We therefore moved forward with RAWM training after reconfiguring the pool into the shape of an asterisk. In this maze, performance is assessed by the efficiency of the swim path and an error is counted when the animal re-enters an arm it has already explored during that trial. Using this measure, we found that the performance of all genotypes improved with training (main effect of trial, p < 0.001), however, the APP/TTA mice again learned the task more slowly than controls. APP/TTA mice made more errors than controls overall (main effect of genotype, p < 0.001) with significant pairwise differences on trials 3-8 (post hoc p < 0.05 vs. TTA on trials 3-5 and 8 and vs. NTG on trials 3-8, Figure 1G). The APP/TTA learning impairment was most evident by looking at traces of the swim path taken by representative animals from each genotype (Figure 1H). On the first trial mice from all three groups made multiple reentry errors while learning the task. By the eighth trial, the NTG and TTA mice swam in a nearly direct line to the escape platform, while the APP/TTA mouse continued to swim in an undirected manner and indeed never reached the target. After the eighth training trial, all mice underwent a probe test during which APP/TTA mice spent a significantly smaller fraction of their swim path in the target arm than NTG mice (post hoc p < 0.01 for NTG and p = 0.05 for TTA). Despite having spent as much and often more time in the test room as controls, APP/TTA mice were less able to learn the new task.
APP/TTA mice display normal place fields on initial exposure to a novel environment
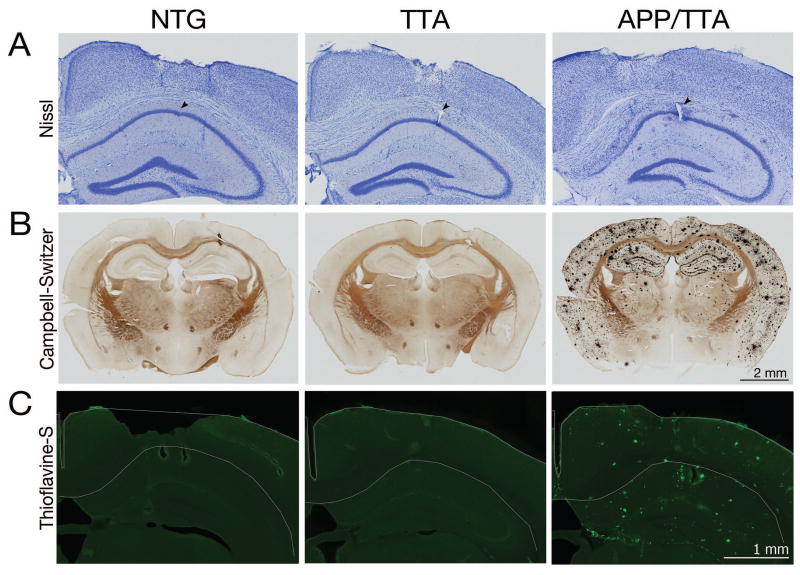
We were curious to understand whether spatial learning impairments in the APP/TTA mice resulted from hippocampal dysfunction at the level of spatial mapping or if the deficit was in their ability to use that information to navigate the environment. Restated, were the APP/TTA mice working from an imprecise neural map, or was their map fine but their ability to use it impaired? We therefore tested the spatial response properties of hippocampal CA1 neurons, where pyramidal cells display precise positional information that collectively generates a neural map of the surrounding environment. Because of the inherent challenge of performing electrophysiological recordings in a pool, we instead examined place cell properties while animals were trained to run laps on a dry linear track in return for food rewards. By artificially constraining their trajectory, track running provides the added advantage of forcing the animal to pass through all points of the ‘environment’ so that the activity of individual cells can be evaluated over repeated trials. Animals from all three genotypes were implanted with multi-tetrode microdrives to isolate the firing of individual CA1 neurons while animals navigated a linear track, two sessions per day, 15 minutes per session, completing 10-40 laps per session. Tetrode position within dorsal CA1, along with the extent of pathology in APP/TTA mice at the time of recording, is shown in Figure 2.
Figure 2. Tetrodes were positioned in the CA1 pyramidal cell layer to record electrical activity during spatial navigation.
A, Nissl stained coronal brain sections show tetrodes targeting the dorsal hippocampal CA1 pyramidal layer. Arrows denote small electrolytic lesions induced to aid in locating final tetrode position once recording was complete. NTG, left; TTA, middle; APP/TTA, right. Adjacent silver (B) and thioflavine-S stained (C) sections demonstrate the degree of amyloid pathology present in APP/TTA animals at the time of recording. Note the prevalence of amyloid plaques throughout the hippocampus and cortex in APP/TTA mice.
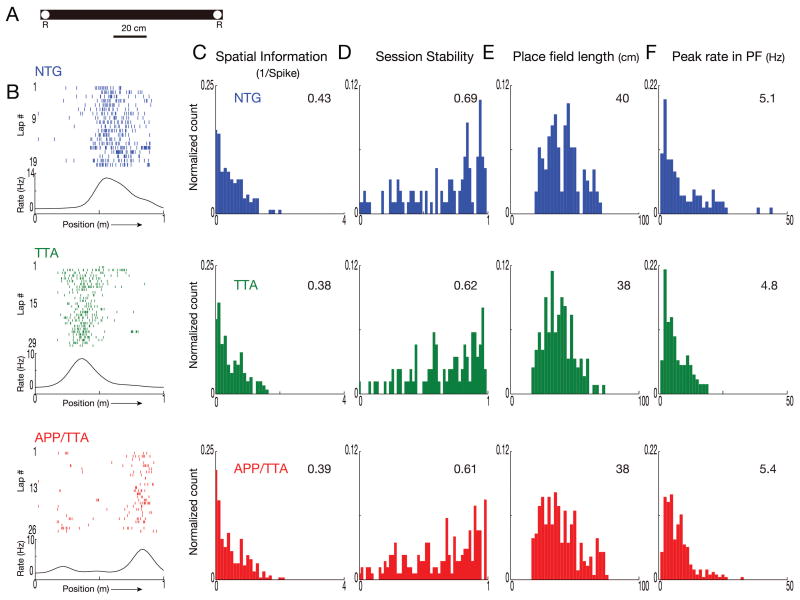
Because we were interested to see how CA1 place cell properties emerge in our mice, we first trained the animals to run along a well-worn linear track in exchange for food rewards, and then recorded their activity from the first day they were moved to another, novel track in an unfamiliar room (Figure 3A). Prior to data analysis, we confirmed that run speed in each maze was similar among genotypes (p > 0.05, Table 2). We recorded a total of 354 CA1 neurons in the novel environment, yielding 63 putative pyramidal neurons from NTG mice, 55 from TTA mice, and 101 from APP/TTA mice (n = 4-5 males and 1-2 females per genotype). In each group, the majority of putative pyramidal neurons were also spatially responsive (NTG: 57%, TTA: 63%, APP/TTA: 65%; Table 2) The median firing rates did not differ significantly between genotypes, nor did the animals' running speed (p > 0.05; see Table 3 for detailed information). As expected for a novel environment, raster plots of firing events during each lap reveal that spikes were not well confined to specific locations, with many isolated spikes scattered along the trajectory (Figure 3B). Rate curves reflecting each neuron's average firing rate across all laps as a function of the animal's position along the track show only broad peaks that correspond to their as-yet still poorly-defined place fields, with little qualitative difference between genotypes.
Figure 3. Place cells in APP/TTA and control mice behave similarly on initial exposure to a novel environment.
A. Diagram of the linear track used for training in a novel environment. R: location of food rewards. B. Lap by lap raster plots and rate curves for firing of a representative neuron from each genotype show poor spatial clustering during their first day on the novel track. Each tick represents one spike, each lap is shown in stacked rows. Firing rates were averaged across all laps and plotted as rate curves shown beneath each raster plot. Direction of the animal's movement is indicated by an arrow along the x axis. With just one day's training to refine spatial responses, rate curves appear broad and low, with poor specificity. C-F. Distribution of spatial information (C), session stability (D), place field length (E), and peak firing rates in place field (F) for each genotype. Histograms are normalized by the total number of samples per analysis, calculated by trajectory for each neuron. Median values for the population are shown in the top right corner of each graph. At this early point in their training, none of the measured properties differed significantly between genotypes.
Table 2. Single unit statistics.
| Putative place cells | Total cells | Proportion of total cells with place fields | Number of place fields per cell/trajectory | |
|---|---|---|---|---|
| Novel day 1 | ||||
| NTG | 63 | 111 | 0.57 | 0.43 |
| TTA | 55 | 88 | 0.63 | 0.54 |
| APP/TTA | 101 | 155 | 0.65 | 0.55 |
| Novel day 2 | ||||
| NTG | 41 | 67 | 0.61 | 0.58 |
| TTA | 47 | 88 | 0.53 | 0.47 |
| APP/TTA | 91 | 137 | 0.66 | 0.58 |
| Novel day 3 | ||||
| NTG | 27 | 46 | 0.59 | 0.62 |
| TTA | 64 | 103 | 0.62 | 0.45 |
| APP/TTA | 73 | 126 | 0.58 | 0.52 |
| Familiar | ||||
| NTG | 103 | 151 | 0.68 | 1.29 |
| TTA | 63 | 92 | 0.68 | 1.11 |
| APP/TTA | 137 | 205 | 0.67 | 1.25 |
Table 3. Place cell statistics.
| Average firing rate | Average run speed/trajec. | Average no. of laps | Trajectory SI | Session stability | Field length | Peak rate | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Range | Median | Range | Median | Range | Median | Range | Median | Range | Median | Range | Median | Range | |
| Novel day 1 | ||||||||||||||
| NTG | 1.7 | 1.06, 3.61 | 24 | 17.85, 29.42 | 21 | 19, 32 | 0.43 | 0.15, 0.77 | 0.69 | 0.26, 0.87 | 40 | 32, 48 | 5.1 | 2.80, 11.92 |
| TTA | 1.7 | 1.12, 3.08 | 19 | 17.03, 23.33 | 22 | 22, 29 | 0.38 | 0.14, 0.81 | 0.62 | 0.27, 0.86 | 38 | 28, 45 | 4.8 | 2.73, 8.14 |
| APP/TTA | 1.8 | 0.88, 3.57 | 21 | 14.92, 28.65 | 21 | 21, 31 | 0.39 | 0.12, 0.84 | 0.61 | 0.26, 0.87 | 38 | 28,50 | 5.4 | 3.66, 8.31 |
| Novel day 2 | ||||||||||||||
| NTG | 1.9 | 0.87,3.31 | 23 | 19.97, 31.38 | 23 | 17, 36 | 0.50 | 0.19, 0.91 | 0.78 | 0.42, 0.95 | 35 | 28, 43 | 5.3 | 3.20, 9.20 |
| TTA | 2.0 | 1.02, 3.59 | 23 | 19.15, 27.04 | 28 | 22, 32.5 | 0.44 | 0.16, 0.99 | 0.78 | 0.59, 0.93 | 33 | 28, 43 | 7.1 | 3.50, 10.65 |
| APP/TTA | 2.1 | 1.04, 3.92 | 26 | 21.15, 35.44 | 33 | 28.5, 33.5 | 0.37 | 0.16, 0.80 | 0.71 | 0.33, 0.90 | 38 | 28, 49 | 5.5 | 3.69, 9.80 |
| Novel day 3 | ||||||||||||||
| NTG | 2.0 | 1.10,3.88 | 23 | 19.48, 25.67 | 30 | 24, 34 | 0.48 | 0.31, 0.83 | 0.75 | 0.19, 0.93 | 33 | 25, 43 | 5.6 | 3.39, 10.50 |
| TTA | 1.8 | 0.99, 3.29 | 26 | 21.07, 27.85 | 29 | 26,41 | 0.54 | 0.24, 1.09 | 0.72 | 0.41, 0.91 | 33 | 25, 42 | 5.7 | 3.37, 10.50 |
| APP/TTA | 1.8 | 0.89, 3.44 | 29 | 20.75, 35.07 | 32 | 27.5, 34 | 0.34** | 0.09, 0.75 | 0.78 | 0.43, 0.91 | 38* | 30, 50 | 6.4 | 3.53, 8.83 |
| Familiar | ||||||||||||||
| NTG | 1.6 | 0.93,2.63 | 20 | 16.15, 22.58 | 17 | 13, 19 | 1.7 | 1.05, 2.38 | 0.87 | 0.68, 0.96 | 24 | 18, 32 | 7.2 | 3.68, 14.59 |
| TTA | 1.3 | 0.83,2.10 | 12 | 9.70, 24.25 | 11* | 10, 15.5 | 1.6 | 1.02, 2.17 | 0.83 | 0.50, 0.94 | 24 | 18, 32 | 6.6 | 3.23, 12.19 |
| APP/TTA | 1.5 | 0.90, 2.73 | 19 | 15.64, 22.55 | 15 | 12, 18 | 1.0*** | 0.23, 1.64 | 0.65*** | 0.29, 0.86 | 28** | 19, 35 | 5 3*** | 2.94, 9.29 |
These average rate curves for each neuron were used to examine the overall location specificity of each place cell in the form of spatial information (SI). SI provides a measure of how much information the firing of a place cell provides about the animal's position in space, measured in units of ‘bits’ of information per spike. During the first day animals were exposed to the novel track, place cells from all three groups displayed similar SI (p > 0.05; Figure 3C). We also found no difference between groups in the stability of firing patterns from the first to the second recording session (p > 0.05; Figure 3D). In addition, place field length – the average distance over which each place cell responded – was indistinguishable between genotypes (p > 0.05; Figure 3E). Finally, there was no difference between genotypes in peak firing rates of the place cells within their respective place fields (p > 0.05; Figure 3F). In essence, all three genotypes displayed equally poor place cell resolution during their first day's exposure to a new environment.
Experience-dependent scaling and stability of place fields is impaired in APP/TTA mice
Having found that place cell properties in the APP/TTA mice started out identical to controls, we next examined how their responses evolved with continued training over the next several days. Mice were returned to the novel track for two additional days, two sessions per day. We recorded 41 NTG, 47 TTA and 91 APP/TTA putative pyramidal neurons on day 2 and 27 NTG, 64 TTA and 73 APP/TTA neurons on day 3. Both raster plots of lap-by-lap spike firing and average rate curves showed the place fields of control mice becoming more refined as training progressed (Figure 4A). Nonetheless, the parity between genotypes that we saw on day 1 continued on day 2, with no significant differences detected in SI, stability between sessions, place field length, or peak firing rate in field (all p > 0.05, Figure 4B-E). However, by the third day of training, significant differences between the genotypes began to emerge. By day 3, cells from APP/TTA mice had lower SI values than those of either NTG or TTA, indicating that their place cells provided less information about the animal's location than cells in control mice (post hoc p < 0.01 vs. NTG and 0.005 vs. TTA; Figure 4B). Consistent with the difference in SI, place fields in APP/TTA mice were larger on day 3 than either control group (post hoc p < 0.05 vs. NTG and 0.01 vs. TTA; Figure 4D). Neither session stability nor peak firing rate differed between genotypes by day 3. These findings suggest that place cell properties evolve with experience in control mice, but either fail to do so, or do so much more slowly, in APP/TTA mice. Indeed, there is a striking similarity between the poor refinement of place cells during repeated exposure to a novel environment and the limited improvement of spatial navigation by APP/TTA mice in the water maze.
Figure 4. Genotype differences slowly emerge as place cells mature through additional training.
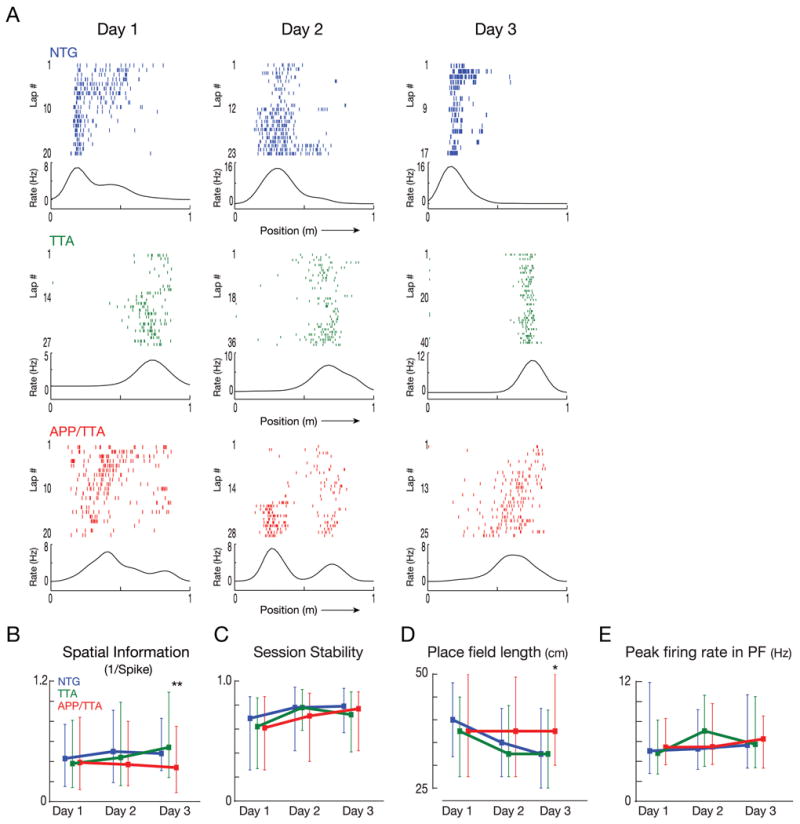
A. Lap by lap raster plots of representative neurons recorded from each genotype across subsequent days of training on the novel track. Although the same cell cannot be reliably identified from one day to the next, the graphs show data recorded from the same tetrode in each mouse. B-E. Comparison of median spatial information (B), session stability (C), place field length (D) and peak firing rates (E) from each genotype across days. Error bars represent the 25% and 75% range. By day 3, cells from APP/TTA mice carried less spatial information and were active in larger place fields than NTG or TTA animals. * p < 0.05, ** p < 0.01. See Table 1 for complete statistical comparisons.
We next wanted to examine whether place cell properties might diverge further between genotypes once the animals became completely familiar with the track and its environment through additional weeks of training. This experiment can be easily conducted over the period it takes to slowly lower the tetrodes into the hippocampus when the animals are simultaneously being trained to run laps on a track in exchange for food rewards. Obviously, because the tetrodes are not yet in place when the animals are first introduced to this room, they cannot be used to examine place cell properties following initial exposure, but it can be used to examine place cell properties on a well-trained track once the electrodes are positioned. To ensure that the two tracks were perceived as being different, the familiar track used for initial training and tetrode positioning had a configuration distinct from that of the novel track and was located in a different room (Figure 5A). We recorded a total of 448 CA1 neurons, from which we identified 103 putative pyramidal neurons from NTG mice, 63 from TTA, and 137 from APP/TTA, of which 67-68% were spatially response in all three genotypes (Table 2). As before, all three genotypes had similar median firing rates and similar running speeds (Table 3; p > 0.05), however, the diminished spatial responsiveness of APP/TTA cells observed by day 3 on the novel track was exacerbated with additional training. Raster plots and rate curves of activity in individual neurons from control mice show close alignment between sequential laps, with tightly restricted spiking. In contrast, the firing of place cells in APP/TTA mice was much less precise (Figure 5B). Raster plots revealed chatter along much of the track, producing rate curves with low, broad peaks that spanned large stretches of distance. The spatial information provided by place cell activity was significantly lower in APP/TTA mice than in controls (post hoc p < 0.005 vs. NTG and TTA, Figure 5C). Additionally, the stability of place fields from one session to another was significantly lower in APP/TTA mice than in NTG and TTA controls, suggesting that their ability to reactivate neural firing on return to the same environment is degraded (post hoc p < 0.005 vs. NTG and TTA, Figure 5D). Place cells from APP/TTA mice also had larger place fields (post hoc p < 0.01 vs. NTG and TTA, Figure 5E) and lower peak firing rates within those fields than either control group (post hoc p < 0.005 vs. NTG and TTA, Figure 5E). Collectively, these data show that spatial representations of familiar environments in the APP/TTA mice are significantly less accurate than in healthy controls.
Figure 5. Familiar environments are much less accurately and reliably represented in APP/TTA mice than controls.
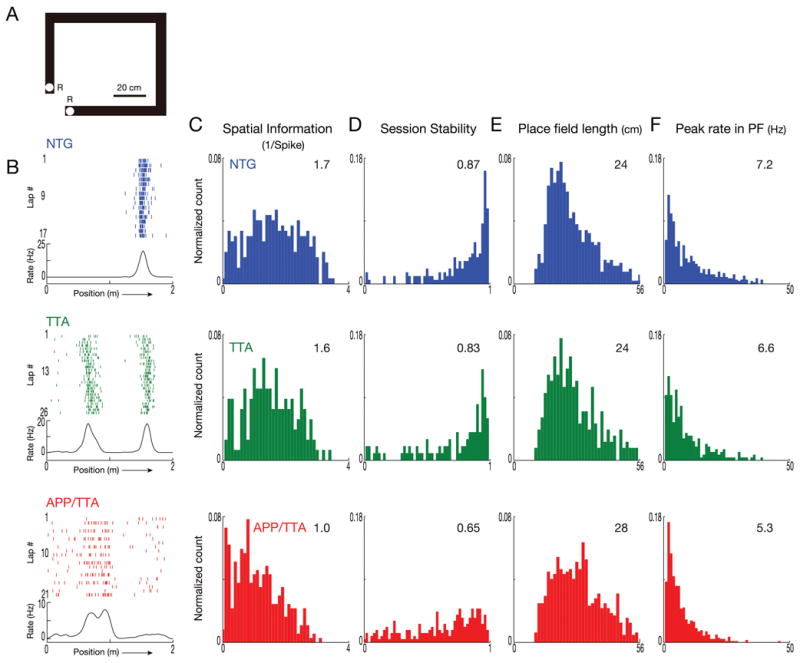
The emerging difference in place cell function noted after 3 days in a novel environment becomes considerably more pronounced after several weeks in a familiar setting. A. Diagram of the linear track used for recording in a familiar environment. Animals were trained on this track every day for 2-3 weeks during tetrode positioning. R: location of food rewards. B. Lap by lap raster plots and rate curves for firing of a representative neuron from each genotype show well-developed place fields in NTG and TTA mice, but poor alignment of activity with position in the APP/TTA animal. C-F. Distribution of spatial information (C), session stability (D), place field length (E), and peak firing rates in place field (F) for each genotype. Median values for the population are shown in the top right corner of each graph. After repeated exposure to the now-familiar track, place cell function in APP/TTA mice was significantly poorer than controls by every measure evaluated (SI, stability, and peak firing rate, p < 0.005; field length p < 0.01). See Table 1 for complete statistical comparisons.
The inferior evolution of spatial representations in APP/TTA mice becomes even more apparent when group data for novel day 1 and familiar responses are plotted side by side. Although spatial information provided by place cell firing increased in all three genotypes between novel and familiar sessions (p < 0.0001), the APP/TTA mice improved less than either control group (post hoc p < 0.005 vs. NTG and TTA; Figure 6A). Similarly, session-to-session stability increased for all three genotypes, but to a considerably lesser extent in APP/TTA mice than in NTG or TTA controls (post hoc p < 0.005 vs. NTG and TTA; Figure 6B). Since the shape and length of the two tracks differed, place field length between conditions could not be directly compared. However, firing rates within the place fields could, and again revealed that place cells in APP/TTA mice did not mature to the same degree as controls. Place cells in NTG and TTA mice fired more rapidly within their place fields as the track became familiar, but the rate of firing in APP/TTA mice remained flat (post hoc p < 0.005 vs. NTG and TTA; Figure 6C). Although spatial encoding began with equal imprecision, improvements in spatial specificity, firing rate, and stability through repeated exposure was substantially more limited in APP/TTA mice than in either control group. Ultimately, APP/TTA mice were working from a map of lower resolution and reliability than was available to either NTG or TTA animals.
Figure 6. Experience-dependent scaling and stability of place fields is impaired in APP/TTA mice.
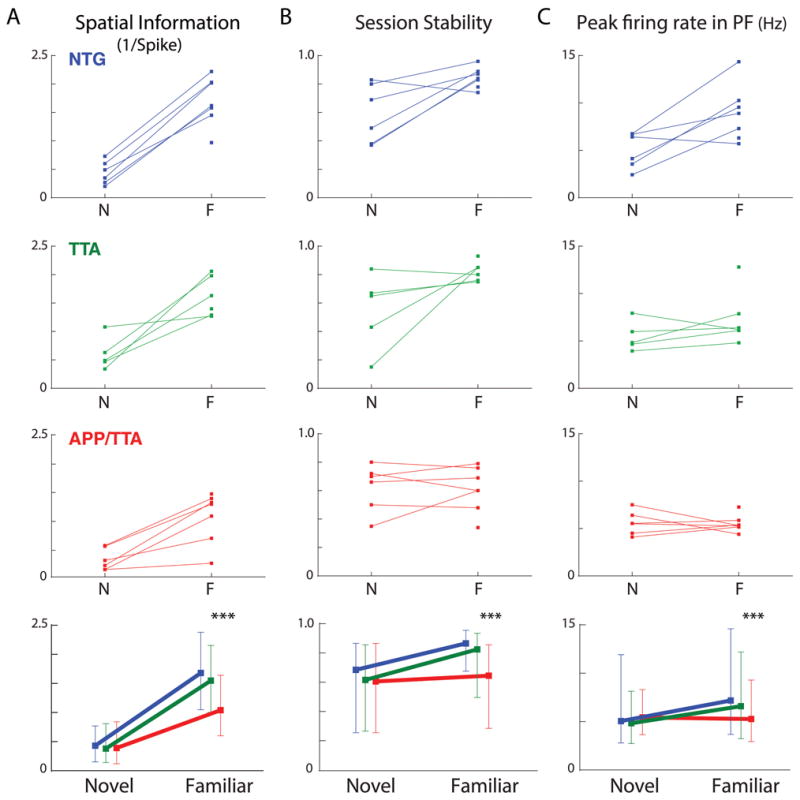
Comparison of spatial information (A), session stability (B), and peak firing rates (C) from the first day in a novel environment and following several weeks of training in a familiar setting exposes the widening gap between controls and APP/TTA animals. Top panels show the median values for individual animals across environments. Data from one mouse in each group was not available on the novel track. Bottom panels show median values for each genotype across environments. Vertical bars represent 25% and 75% range values. The interaction between genotype and time was significant for all three comparisons. *** p < 0.005 for both NTG and TTA. See Table 1 for complete statistical comparisons.
Discussion
We have investigated the neural basis of impaired spatial navigation in a mouse model of Alzheimer's amyloidosis. We show that APP/TTA mice required significantly more training to learn the position of a hidden platform in tests of spatial navigation, and then recall this location less accurately than control mice 24 hours later. These behavioral deficits arise in part from functional alterations in the hippocampal place cells responsible for encoding spatial information. Although APP/TTA mice form initial place fields as quickly and efficiently as controls, the scaling and stability of place cell properties with repeated exposure to the same environment is impaired. Our data provide a clear example of how behavioral deficits are inexorably linked with abnormal neural circuit function in a mouse model of Alzheimer's disease.
Initial place fields emerge within minutes after animals are placed into a novel open space (Wilson and McNaughton, 1993). During those first few minutes, silent cells may suddenly become active while active cells may change or fine-tune their firing locations (Cheng and Frank, 2008; Frank et al., 2004; Leutgeb et al., 2004). Our data indicate that these short-term place field dynamics are normal in APP/TTA mice on their first day in the novel environment. Indeed, it was not until the third day of training on the novel track that differences began to emerge. Even so, the small rise in spatial information and the small decrease in field size shown by control mice still only hinted at the imminent deficits in APP/TTA mice. Although APP/TTA mice were capable of forming appropriate initial place fields, they were less able to achieve the smaller-scale, longer-term refinements that hone place cell properties as a new environment becomes familiar. Place fields from control mice had significantly higher spatial information, greater stability, and higher peak firing rates in the familiar environment than in the novel setting. Refinement of these properties was considerably less pronounced in the APP/TTA mice. After weeks of training on the familiar track, place fields in APP/TTA mice had less spatial information, poorer stability, and lower peak rates than similarly trained control animals. Ultimately this failure meant that APP/TTA mice were left to navigate their environment using a neural map that was both less accurate and less reliable than that available to their healthy siblings.
Our study is the first to examine the formation and focusing of hippocampal place fields in an APP transgenic model. By examining place cells in both novel and familiar environments, we identified a unique deficit in the convergence of firing rate and spatial specificity that emerges with experience. Previous study had shown that place fields in aged Tg2576 mice were less precise than controls in a familiar environment, but did not explore whether this was due to disability in their initial mapping of a new environment, or to a deficit in experience-dependent refinement of the map (Cacucci et al., 2008). Our data suggests that place field impairments were due to poor stabilization and spatial focusing on repeated exposure rather than initial formation.
The inferior place field scaling and stability of APP/TTA mice likely contributes to their poor acquisition and recall of the hidden platform location in Morris water maze. While the performance of both APP/TTA and control mice improved with additional training, it took APP/TTA mice a median of nine days to reach criteria performance while it took control mice only four. Their delayed acquisition closely parallels the slow improvement of place fields observed in APP/TTA mice. Because the same place cells are re-engaged when the animal revisits a location they have explored before, their activity also encodes a memory of the location (Kentros, 2006; Thompson and Best, 1990; Ziv et al., 2013). The encoded memory can be compromised by either poor specificity, producing a low-resolution cognitive map for the space, or poor stability, producing a weaker memory code. On the first day in a novel environment, the resolution and strength of the newly formed memory codes were similar across genotypes, and consistent with this, all three genotypes showed roughly equivalent performance on the first day of water maze training. As training progressed, place field precision and stability increased in control mice, but improved much less in APP/TTA mice. The resulting lower-resolution cognitive map may render APP/TTA unable to precisely locate the target location during water maze training. This impairment would be compounded by an unstable memory code rendering APP/TTA animals unable to remember the platform location during probe trials. Our data thus suggest a neural mechanism for delayed MWM acquisition and impaired long-term recall in APP/TTA mice.
APP/TTA mice were also impaired in the RAWM task, which in our study examined the ability to use familiar cues for navigation in addition to its primary measure of working memory for locations previously visited during each trial. While control mice improved quickly over successive training trials, the APP/TTA mice continued to make re-entry errors across trials indicative of a working memory deficit. Consistent with their impaired recall in MWM, APP/TTA mice also performed more poorly than controls during the final RAWM probe test, indicative of a deficit in spatial reference memory. As with MWM, incomplete place cell stabilization and scaling may also underlie inferior performance in RAWM. Because the RAWM was done in the same room as MWM, the animals had several weeks to become familiar with the surrounding environment and spatial cues. The poorer resolution and stability of cognitive maps for familiar environments in APP/TTA mice may have left them less able to remember both the last places they visited and the target location. Alternatively, by modifying the environment with acrylic inserts from an open space to a radial maze, the space may have been remapped as a novel environment to form a new memory code (Colgin et al., 2008). The degree of change required for the hippocampus to ‘recognize’ a known location as new is relatively subtle in healthy animals (Jeffery, 2011). Although our experiments do not address this possibility, we predict that APP/TTA mice may display a different threshold for remapping than control mice. A case could be made for a shift in either direction to explain the RAWM deficits we observed. If the APP/TTA mice remap too readily, they would be unable to draw on the existing cognitive map available to control mice for navigation in the new maze. Conversely, if APP/TTA mice are more resistant to remapping, they might be less flexible in learning the new configuration and escape location. Future experiments to distinguish between these possibilities may provide additional insight into the neural basis for cognitive dysfunction in APP models.
How might amyloid accumulation and the resulting pathological response interfere with place cell function? On one hand, it is easy to envision how physical disruption of neural circuits throughout the brain might limit the spatial responsiveness of hippocampal neurons. Amyloid plaques are associated with synaptic depletion, neurite distortion and separation, and in some mouse models, overt neuronal loss (Brendza et al., 2005; Dong et al., 2007; Koffie et al., 2009; Oakley et al., 2006; Spires-Jones and Knafo, 2012; Tsai et al., 2004). These alterations might impair signaling between the primary sensory cortices and the hippocampus, or might desynchronize the arrival of information from different sensory domains (Stern et al., 2004). Even before reaching the hippocampus the processing of sensory information may be disrupted. For example, calcium imaging in amyloid-bearing APP23 mice revealed that orientation tuning of neurons in the visual cortex became progressively degraded as plaque burden worsened (Grienberger et al., 2012). Given the magnitude of physical damage to sensory and limbic networks that occurs in amyloid-bearing models, it is instead surprising that the place cell deficits we observed were not more severe. Rather, the impairments we found suggest that more subtle deficits in hippocampal synaptic plasticity may be at play. Soluble aggregates of Aβ can depress synaptic transmission and inhibit LTP, and both effects are modulated through the NMDA receptor (Malinow, 2012; Ondrejcak et al., 2009; Venkitaramani et al., 2007). Aβ-dependent alterations in NMDA-R activation may serve as a mechanistic link to the impairments in place cell stabilization and spatial focusing documented here, as past work has shown that experience-dependent alteration of CA1 place fields are also dependent on NMDA-R function (Ekstrom et al., 2001).
One technical advantage we had in the current studies was the ability to withhold APP overexpression in the APP/TTA mice until the brain had matured, thus eliminating the possibility for transgenic APP/Aβ to alter neural development (Rodgers et al., 2012). By avoiding embryonic and early postnatal expression, we ensured that APP/TTA mice ran the same distance as controls in open field and at the same speed on familiar and novel tracks. In contrast, standard APP transgenic models such as Tg2756 mice can be hyperactive (Bardgett et al., 2011; Bedrosian et al., 2011; Rustay et al., 2010), and this phenotype could confound measurement of both place cells and behavior. Running speed can affect place field properties such as peak firing rate, making it an important parameter to control in order to produce fair comparisons between mice of different genotypes. One concern we did have in using the tet-off system was the potential for TTA expression to induce degeneration and alter behavior on certain strain backgrounds (Han et al., 2012). We took two steps to ensure that TTA was not a factor in the current study. First, we studied mice on a congenic B6 background that is resistant to TTA-induced neurodegeneration and behavioral changes (Han et al., 2012). Second, we included age- and gender-matched TTA single-transgenic siblings as controls for all of the current experiments. These measures allow us to be reasonably certain that the behavioral and neurophysiological changes we observe in the APP/TTA mice are correctly attributed to APP overexpression and its associated amyloid pathology.
We have leveraged several decades of work elucidating the complex spatial properties of the hippocampal region as a starting point from which to explore the circuit basis for cognitive decline in models of Alzheimer's disease. In doing so, we have uncovered a unique deficit in the neural encoding of spatial information that may underlie navigational impairments of APP transgenic mice. We must now work forward from this point to understand how downstream areas process this degraded map, and work backward to identify what defect in local circuitry or sensory input restricts experience-dependent honing of place field properties. For nearly two decades, rodents' innate capacity for spatial navigation has allowed us to probe the relationship between pathology and cognitive decline in Alzheimer's models. Now the same ability offers us a means to explore the neural mechanism of that decline.
Acknowledgments
We thank Bryan Song, Anna Gumpel, Carolyn Allen, and Yuanyuan Zhang for animal care and Rich Paylor for advice on behavioral testing.
Grant sponsor: NIH/OD, Grant number: DP2 OD001734
Grant sponsor: NIH/NIA, Grant number: T32 AG000183
Grant sponsor: The Robert A. and Rene E. Belfer Family Foundation
Grant sponsor: The Bill and Melinda Gates Foundation, Grant number: Gates Millennium Scholarship
Grant sponsor: BCM Alzheimer's Disease and Memory Disorders Center and the George and Cynthia Mitchell Foundation
References
- Bardgett ME, Davis NN, Schultheis PJ, Griffith MS. Ciproxifan, an H3 receptor antagonist, alleviates hyperactivity and cognitive deficits in the APP Tg2576 mouse model of Alzheimer's disease. Neurobiol Learn Mem. 2011;95(1):64–72. doi: 10.1016/j.nlm.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrosian TA, Herring KL, Weil ZM, Nelson RJ. Altered temporal patterns of anxiety in aged and amyloid precursor protein (APP) transgenic mice. Proc Natl Acad Sci U S A. 2011;108(28):11686–91. doi: 10.1073/pnas.1103098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza RP, Bacskai BJ, Cirrito JR, Simmons KA, Skoch JM, Klunk WE, Mathis CA, Bales KR, Paul SM, Hyman BT, et al. Anti-Aβ antibody treatment promotes the rapid recovery of amyloid-associated neuritic dystrophy in PDAPP transgenic mice. J Clin Invest. 2005;115(2):428–33. doi: 10.1172/JCI23269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacucci F, Yi M, Wills TJ, Chapman P, O'Keefe J. Place cell firing correlates with memory deficits and amyloid plaque burden in Tg2576 Alzheimer mouse model. Proc Natl Acad Sci U S A. 2008;105(22):7863–8. doi: 10.1073/pnas.0802908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson VB. Wandering: a common challenging behavior. Caring. 2012;31(1):52–3. [PubMed] [Google Scholar]
- Cheng J, Ji D. Rigid firing sequences undermine spatial memory codes in a neurodegenerative mouse model. Elife. 2013;2:e00647. doi: 10.7554/eLife.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Frank LM. New experiences enhance coordinated neural activity in the hippocampus. Neuron. 2008;57(2):303–13. doi: 10.1016/j.neuron.2007.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgin LL, Moser EI, Moser MB. Understanding memory through hippocampal remapping. Trends Neurosci. 2008;31(9):469–77. doi: 10.1016/j.tins.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Dodart JC, Mathis C, Bales KR, Paul SM. Does my mouse have Alzheimer's disease? Genes Brain Behav. 2002;1(3):142–55. doi: 10.1034/j.1601-183x.2002.10302.x. [DOI] [PubMed] [Google Scholar]
- Dong H, Martin MV, Chambers S, Csernansky JG. Spatial relationship between synapse loss and β-amyloid deposition in Tg2576 mice. J Comp Neurol. 2007;500(2):311–21. doi: 10.1002/cne.21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H. Memory on time. Trends Cogn Sci. 2013;17(2):81–8. doi: 10.1016/j.tics.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom AD, Meltzer J, McNaughton BL, Barnes CA. NMDA receptor antagonism blocks experience-dependent expansion of hippocampal “place fields”. Neuron. 2001;31(4):631–8. doi: 10.1016/s0896-6273(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Foster DJ, Knierim JJ. Sequence learning and the role of the hippocampus in rodent navigation. Curr Opin Neurobiol. 2012;22(2):294–300. doi: 10.1016/j.conb.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LM, Stanley GB, Brown EN. Hippocampal plasticity across multiple days of exposure to novel environments. J Neurosci. 2004;24(35):7681–9. doi: 10.1523/JNEUROSCI.1958-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher M, Burwell R, Burchinal M. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav Neurosci. 1993;107(4):618–26. doi: 10.1037//0735-7044.107.4.618. [DOI] [PubMed] [Google Scholar]
- Gazova I, Vlcek K, Laczo J, Nedelska Z, Hyncicova E, Mokrisova I, Sheardova K, Hort J. Spatial navigation-a unique window into physiological and pathological aging. Front Aging Neurosci. 2012;4:16. doi: 10.3389/fnagi.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin LN, Kales HC, Lyketsos CG. Nonpharmacologic management of behavioral symptoms in dementia. JAMA. 2012;308(19):2020–9. doi: 10.1001/jama.2012.36918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Rochefort NL, Adelsberger H, Henning HA, Hill DN, Reichwald J, Staufenbiel M, Konnerth A. Staged decline of neuronal function in vivo in an animal model of Alzheimer's disease. Nat Commun. 2012;3:774. doi: 10.1038/ncomms1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Allen CC, Buchovecky CM, Yetman MJ, Born HA, Marin MA, Rodgers SP, Song BJ, Lu HC, Justice MJ, et al. Strain background influences neurotoxicity and behavioral abnormalities in mice expressing the tetracycline transactivator. J Neurosci. 2012;32(31):10574–86. doi: 10.1523/JNEUROSCI.0893-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Slunt HH, Gonzales V, Savonenko AV, Wen JC, Jenkins NA, Copeland NG, Younkin LH, Lester HA, Younkin SG, et al. Persistent amyloidosis following suppression of Aβ production in a transgenic model of Alzheimer's disease. PLoS Medicine. 2005;2(12):e355. doi: 10.1371/journal.pmed.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery KJ. Place cells, grid cells, attractors, and remapping. Neural Plast. 2011;2011:182602. doi: 10.1155/2011/182602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D, Wilson MA. Coordinated memory replay in the visual cortex and hippocampus during sleep. Nat Neurosci. 2007;10(1):100–7. doi: 10.1038/nn1825. [DOI] [PubMed] [Google Scholar]
- Kentros C. Hippocampal place cells: the “where” of episodic memory? Hippocampus. 2006;16(9):743–54. doi: 10.1002/hipo.20199. [DOI] [PubMed] [Google Scholar]
- Koffie RM, Meyer-Luehmann M, Hashimoto T, Adams KW, Mielke ML, Garcia-Alloza M, Micheva KD, Smith SJ, Kim ML, Lee VM, et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc Natl Acad Sci U S A. 2009;106(10):4012–7. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Treves A, Moser MB, Moser EI. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science. 2004;305(5688):1295–8. doi: 10.1126/science.1100265. [DOI] [PubMed] [Google Scholar]
- Lithfous S, Dufour A, Despres O. Spatial navigation in normal aging and the prodromal stage of Alzheimer's disease: insights from imaging and behavioral studies. Ageing Res Rev. 2013;12(1):201–13. doi: 10.1016/j.arr.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Malinow R. New developments on the role of NMDA receptors in Alzheimer's disease. Curr Opin Neurobiol. 2012;22(3):559–63. doi: 10.1016/j.conb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274(5293):1678–83. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Morgan D. Learning and memory deficits in APP transgenic mouse models of amyloid deposition. Neurochem Res. 2003;28(7):1029–34. doi: 10.1023/a:1023255106106. [DOI] [PubMed] [Google Scholar]
- Moser EI, Kropff E, Moser MB. Place cells, grid cells, and the brain's spatial representation system. Annu Rev Neurosci. 2008;31:69–89. doi: 10.1146/annurev.neuro.31.061307.090723. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34(1):171–5. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L. The hippocampus as a cognitive map. Oxford University Press; 1978. [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five famlial alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26(40):10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrejcak T, Klyubin I, Hu NW, Barry AE, Cullen WK, Rowan MJ. Alzheimer's Disease Amyloid beta-Protein and Synaptic Function. Neuromolecular Med. 2009 doi: 10.1007/s12017-009-8091-0. [DOI] [PubMed] [Google Scholar]
- Rodgers SP, Born HA, Das P, Jankowsky JL. Transgenic APP expression during postnatal development causes persistent locomotor hyperactivity in the adult. Mol Neurodegener. 2012;7:28. doi: 10.1186/1750-1326-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls ET. A computational theory of episodic memory formation in the hippocampus. Behav Brain Res. 2010;215(2):180–96. doi: 10.1016/j.bbr.2010.03.027. [DOI] [PubMed] [Google Scholar]
- Rustay NR, Cronin EA, Curzon P, Markosyan S, Bitner RS, Ellis TA, Waring JF, Decker MW, Rueter LE, Browman KE. Mice expressing the Swedish APP mutation on a 129 genetic background demonstrate consistent behavioral deficits and pathological markers of Alzheimer's disease. Brain Res. 2010;1311:136–47. doi: 10.1016/j.brainres.2009.11.040. [DOI] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL, Gothard KM, Markus EJ. An information-theoretic approach to deciphering the hippocampal code. In: Hanson SJ, Cowan JD, Giles CI, editors. Advances in neural information processing systems. San Mateo, CA: Morgan Kaufmann; 1993. pp. 1030–1037. [Google Scholar]
- Skaggs WE, McNaughton BL, Wilson MA, Barnes CA. Theta phase precession in hippocampal neuronal populations and the compression of temporal sequences. Hippocampus. 1996;6(2):149–72. doi: 10.1002/(SICI)1098-1063(1996)6:2<149::AID-HIPO6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Smith DM, Mizumori SJ. Hippocampal place cells, context, and episodic memory. Hippocampus. 2006;16(9):716–29. doi: 10.1002/hipo.20208. [DOI] [PubMed] [Google Scholar]
- Spires-Jones T, Knafo S. Spines, plasticity, and cognition in Alzheimer's model mice. Neural Plast. 2012;2012:319836. doi: 10.1155/2012/319836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern EA, Bacskai BJ, Hickey GA, Attenello FJ, Lombardo JA, Hyman BT. Cortical synaptic integration in vivo is disrupted by amyloid-β plaques. J Neurosci. 2004;24(19):4535–40. doi: 10.1523/JNEUROSCI.0462-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LT, Best PJ. Long-term stability of the place-field activity of single units recorded from the dorsal hippocampus of freely behaving rats. Brain Res. 1990;509(2):299–308. doi: 10.1016/0006-8993(90)90555-p. [DOI] [PubMed] [Google Scholar]
- Tsai J, Grutzendler J, Duff K, Gan WB. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat Neurosci. 2004;7(11):1181–3. doi: 10.1038/nn1335. [DOI] [PubMed] [Google Scholar]
- Venkitaramani DV, Chin J, Netzer WJ, Gouras GK, Lesne S, Malinow R, Lombroso PJ. β-amyloid modulation of synaptic transmission and plasticity. J Neurosci. 2007;27(44):11832–7. doi: 10.1523/JNEUROSCI.3478-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Dynamics of the hippocampal ensemble code for space. Science. 1993;261(5124):1055–8. doi: 10.1126/science.8351520. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16(3):264–6. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]