Abstract
Antibody polyreactivity can be an obstacle to translating a candidate antibody into a clinical product. Standard tests such as antibody binding to cardiolipin, HEp-2 cells, or nuclear antigens provide measures of polyreactivity, but its causes and the means to resolve are often unclear. Here we present a method for eliminating antibody polyreactivity through the computational design and genetic addition of N-linked glycosylation near known sites of polyreactivity. We used the HIV-1-neutralizing antibody, VRC07, as a test case, since efforts to increase VRC07 potency at three spatially distinct sites resulted in enhanced polyreactivity. The addition of N-linked glycans proximal to the polyreactivity-enhancing mutations at each of the spatially distinct sites resulted in reduced antibody polyreactivity as measured by (i) anti-cardiolipin ELISA, (ii) Luminex AtheNA Multi-Lyte ANA binding, and (iii) HEp-2 cell staining. The reduced polyreactivity trended with increased antibody concentration over time in mice, but not with improved overall protein stability as measured by differential scanning calorimetry. Moreover, glycan proximity to the site of polyreactivity appeared to be a critical factor. The results provide evidence that antibody polyreactivity can result from local, rather than global, features of an antibody and that addition of N-linked glycosylation can be an effective approach to reducing antibody polyreactivity.
Keywords: polyreactivity, antibody engineering, bioinformatics, broadly neutralizing antibody, glycan engineering
Introduction
In recent years, therapeutic antibodies have been successfully used to treat various diseases including rheumatoid arthritis, cancer, and infections of the lower respiratory tract.1–5 A substantial fraction of antibody development often focuses on improving the properties of existing antibodies such as their affinity, neutralization potency, and effector functions, while reducing their polyreactivity.6–11 Antibody polyreactivity,12,13 the ability of an antibody to recognize multiple structurally unrelated antigens, may occur with “natural” antibodies isolated from sera14–16 and may also arise as a byproduct of the antibody engineering process.11 Polyreactivity may not only interfere with normal cellular functions,17,18 but may also reduce antibody half-life,19 thus potentially signaling safety issues or increasing required levels and/or frequency of dosage.20 When polyreactivity originates from alterations generated during the process of antibody engineering, the specific sites on the antibody responsible for the polyreactivity may be localized.21 We hypothesized that one potential means to reduce polyreactivity could involve the attachment of a large molecule, such as an N-linked glycan, proximal to the polyreactive site on the protein, to mask this site from interacting with autoantigens (Fig. 1). We chose N-linked glycosylation since it is genetically encoded and has been used in other aspects of antibody engineering, such as the improvement of neutralization potency,22 solubility,22,23 and stability,24,25 as well as the conjugation of small molecule drugs to antibodies.26
Figure 1.
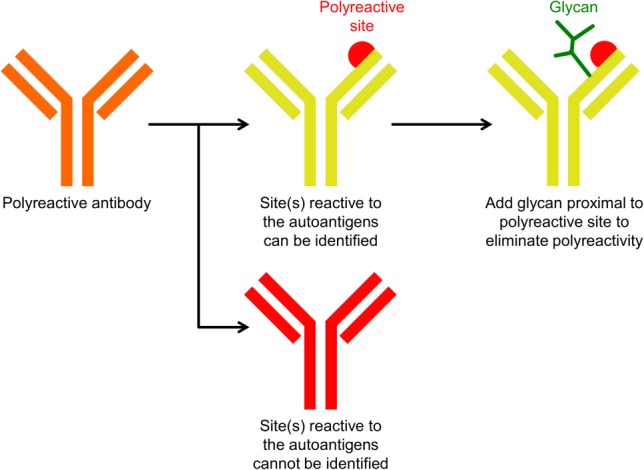
Schematic overview of the method to reduce polyreactivity by introduction of proximal N-linked glycan. Antibody reactivity to autoantigens may be caused by amino acid residue(s) in a localized region (showed as red semi-sphere). The addition of a glycan (colored green) proximal to the polyreactive-enhancing residues can potentially mask the interaction between the localized region and autoantigens.
Here we present proof-of-concept that the polyreactivity of an antibody can be eliminated through addition of an N-linked glycan proximal to a polyreactive site. We tested three different polyreactive variants of the anti-HIV-1 antibody VRC07,11 a more potent somatic variant of antibody VRC01,27,28 closely related to antibody NIH45-46.29 We applied a computational algorithm to identify residues capable of accepting N-linked glycan and measured binding of the resultant glycosylated VRC07 variants to cardiolipin, HEp-2 epithelial cells, and a panel of autoantigens. We performed pharmacokinetics studies in mice, tested proximity requirements of masking glycan, and used differential scanning calorimetry (DSC) measurements to assess the effect of glycan addition on overall protein stability. The methods described here can be integrated into processes of antibody engineering to improve therapeutic antibodies of interest.
Results
Engineering N-linked glycans on polyreactive VRC07 derivatives
In this study, we sought to reduce the polyreactivity of three different polyreactive VR07 derivatives: VRC07-av1, VRC07-av2, and VRC07-av3 (Table1). These derivatives were developed as part of an effort to improve the neutralization potency of VRC07.11 The advantage of using these variants as a test case was that the specific mutations which led to polyreactivity were known (Supporting Information Fig. S1 and Tables S1 and S2). For VRC07-av1, the hydrophobic mutation Gly54Trp appeared to be the cause of polyreactivity, as other heavy chain mutations in this variant, Ile30Gln and Ser58Asn, were not polyreactive, and polyreactivity has been observed with other variant at residue 54 (Supporting Information Fig. S1 and Tables S1 and S2).8,11 For VRC07-av2, polyreactivity resulted from four alanine mutations at the N terminus of the light chain. For VRC07-av3, alteration of the heavy-light chain interface, with an Ala43Arg mutation of the light chain, induced polyreactivity. We used a computational protocol, which included an energetic filter and an N-linked glycan occupancy prediction algorithm,30 to identify positions where N-linked glycans were likely to be successfully added. One or two positions proximal to heavy chain Trp54, to light chain N terminus, and to light chain Arg43 (for VRC07-av1, VRC07-av2, and VRC07-av3, respectively) were then selected for the introduction of an N-glycosylation sequon (Table1 and Fig. 2). In total, five glycan mutants were produced. SDS-PAGE analysis with and without PNGase F digestion (an amidase that cleaves between the protein-proximal N-acetylglucosamine and asparagine moieties of high mannose, hybrid, and complex oligosaccharides on N-linked glycoproteins) demonstrated designed N-linked glycans to be successfully added with all five glycan constructs (Fig. 3). Glycan composition analysis using combination of ultra performance liquid chromatography (UPLC) and exoglycosidase digestion showed VRC07-av1-g1, VRC07-av1-g2, VRC07-av2-g1, and VRC07-av2-g2 to have similar N-glycan profiles, comprising of mixtures of complex type N-glycans with core fucosylation and sialylated structures (Supporting Information Figs. S2 and S3). VRC07-av3-g1 had a distinct glycan profile which appears to correspond to predominantly high mannose N-glycans.
Table 1.
Construct Names and Mutations of VRC07 Variants Evaluated in this Study
| Heavy chain mutations | Light chain mutations | |
|---|---|---|
| VRC07-av1 | G54W-I30Q-S58N | – |
| VRC07-av1-g1 | G54W-I30Q-S58N-M73N-S75T | – |
| VRC07-av1-g2 | G54W-I30Q-S58N-R71N-M73T | – |
| VRC07-av2 | – | AAAA N terminus (E1A-I2A-V3A-L4A) |
| VRC07-av2-g1 | – | AAAA N terminus-R24N |
| VRC07-av2-g2 | – | AAAA N terminus-S26N-Y28T |
| VRC07-av3 | – | A43R |
| VRC07-av3-g1 | – | A43R-R45N-V47T |
Figure 2.
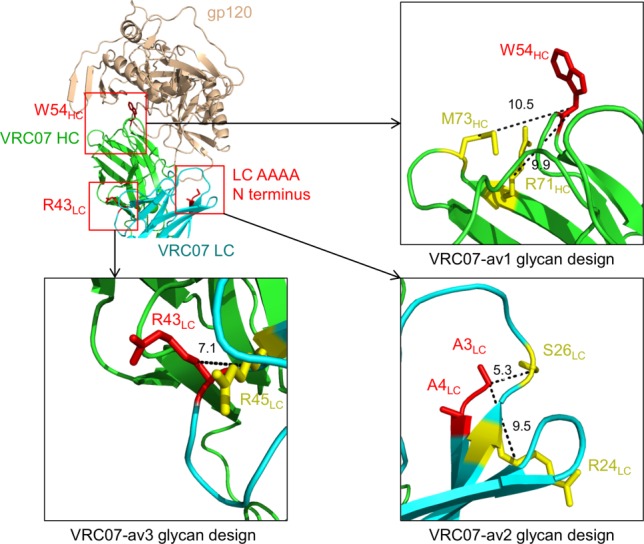
Design of N-linked glycan mutants. Top-left: polyreactive-enhancing residues (colored in red) for each of the three VRC07 variants (W54HC for VRC01-av1, Light chain AAAA N terminus for VRC07-av2, and R43LC for VRC07-av3). Top-right: residues positions to introduce N-linked glycans for VRC07-av1. Bottom left: residues positions to introduce N-linked glycans for VRC07-av3. Bottom right: residues positions to introduce N-linked glycans for VRC07-av2. The polyreactive-enhancing residues and the positions where the N-linked glycans were designed are colored red and yellow, respectively. The distances between the Cβ atoms of polyreactive-enhancing residues (W54HC, R43LC, and A3LC, respectively) and the designed glycans are labeled (in Å).
Figure 3.
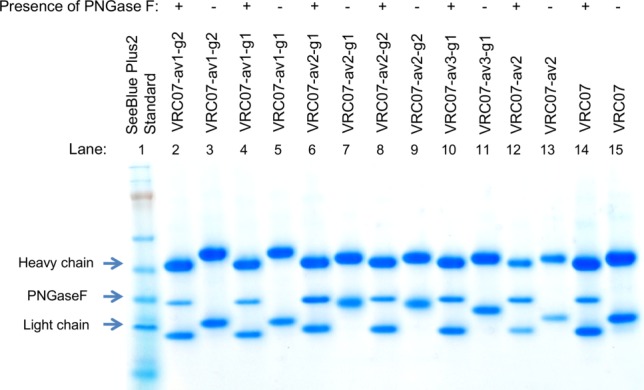
SDS-PAGE analysis of PNGase F treated and untreated glycan constructs. In all PNGase F treated lanes there is a reduction of molecular weight for both the heavy and the light chain due to the presence of the intrinsic N-linked glycans at the constant region of the heavy chain and variable region (residue 72) of the light chain. Deglycosylation of VRC07-av1-g1 and VRC07-av1-g2 (lanes 2,4) resulted in larger reduction of molecular weight for the heavy chain as compared to the other constructs, suggesting that glycans were added to the heavy chain Fab of VRC07-av1-g1 and VRC07-av1-g2, respectively. Deglycosylation of VRC07-av2-g1, VRC07-av2-g2, and VRC07-av3 (lanes 6, 8, 10) resulted in larger reduction of molecular weight as compared to VRC07-av2 and VRC07, suggesting that an additional glycan was added to the light chain of these three constructs.
Addition of N-linked glycosylation substantially reduced polyreactivity
To evaluate the effect of N-linked glycan addition, we used three different assays to evaluate antibody polyreactivity: (1) Anti-cardiolipin ELISA, (2) Luminex AtheNA Multi-Lyte ANA (Wampole Laboratories), which tests antibody reactivity against self antigens (SSA/Ro, SS-B/La, Sm, ribonucleoprotein (RNP), Jo-1, double-stranded DNA, centromere B, and histone), and (3) HEp-2 cell staining. At an antibody concentration of 100 µg/ml, all five N-linked glycan variants displayed substantially lower cardiolipin binding activity (60–90%) compared to the parental polyreactive variants (Fig. 4 and Supporting Information Table S1). In the Luminex AtheNA Multi-Lyte ANA tests, VRC07-av1 exhibited polyreactivity against two self antigens (SSA and Jo-1), VRC07-av2 against three (SSA, Jo-1, and Histone), and VRC07-av3 against three (SSA, Jo-1, and Histone), while all five glycan constructs had no substantial reactivity against any of the tested self-antigens (Fig. 5 and Supporting Information Table S2). For all three polyreactive VRC07 variants, visible fluorescence was observed with HEp-2 cell staining (Fig. 6 and Supporting Information Fig. S4), whereas four of the five glycan mutants showed no substantial fluorescence; in the single case where fluorescence was observed (VRC07-av1-g1), the intensity of HEp-2 cell staining was substantially lowered than for the parental VRC07-av1.
Figure 4.
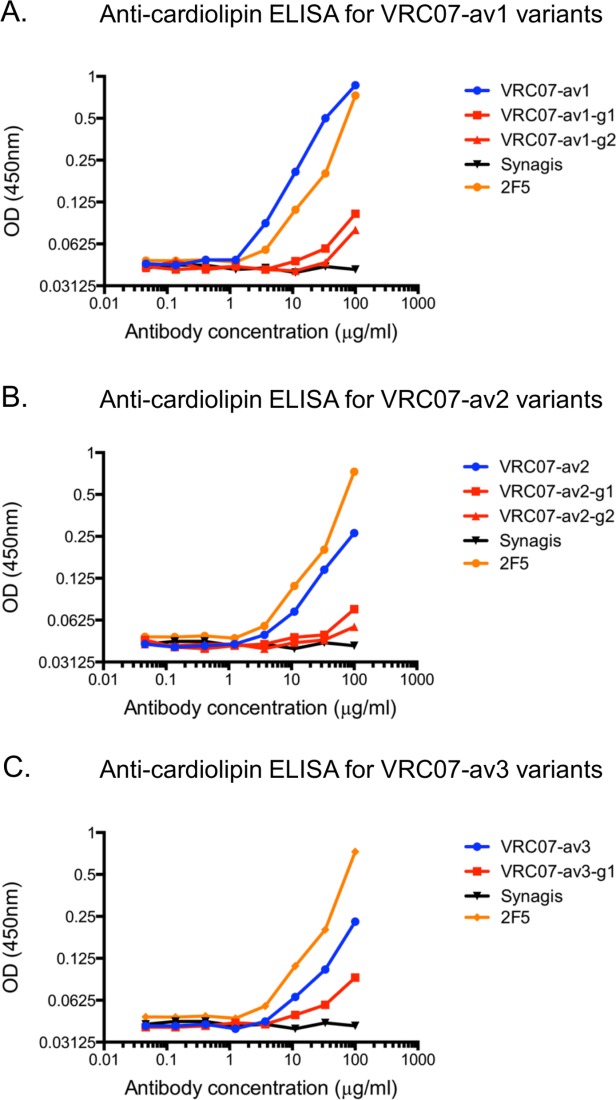
Cardiolipin reactivity of VRC07 variants. Shown is the cardiolipin ELISA results at different antibody concentrations for (A) VRC07-av1 and its glycan mutants (B) VRC07-av2 and its glycan mutants, and (C) VRC07-av3 and its glycan mutant. Synagis (Palivizumab), an anti-RSV monoclonal antibody, was used as the negative control. 2F5, a polyreactive HIV-1 antibody, was used as the positive control. In all three cases, the glycan variants had a decrease of approximately 60% to 90% in cardiolipin reactivity at the highest antibody concentration (100 µg/ml).
Figure 5.
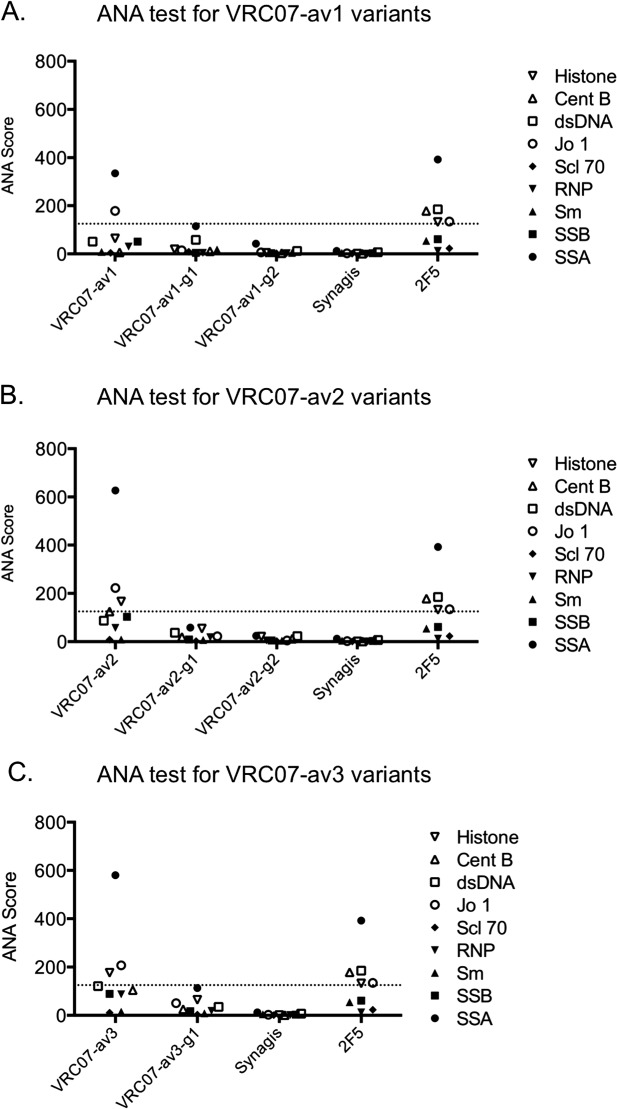
Luminex AtheNA Multi-Lyte ANA test results for each VRC07 variant. Shown is the Athena ANA results for (A) VRC07-av1 and its glycan mutants (B) VRC07-av2 and its glycan mutants, and (C) VRC07-av3 and its glycan mutant. The horizontal dash line indicates an ANA score cutoff of 125. Synagis (Palivizumab), an anti-RSV monoclonal antibody, was used as the negative control. 2F5, a polyreactive HIV-1 antibody, was used as the positive control. In all three cases, the parental VRC07 variants were reactive to a number of autoantigens, while for the glycan variants no positive reactivity was observed against any autoantigen.
Figure 6.
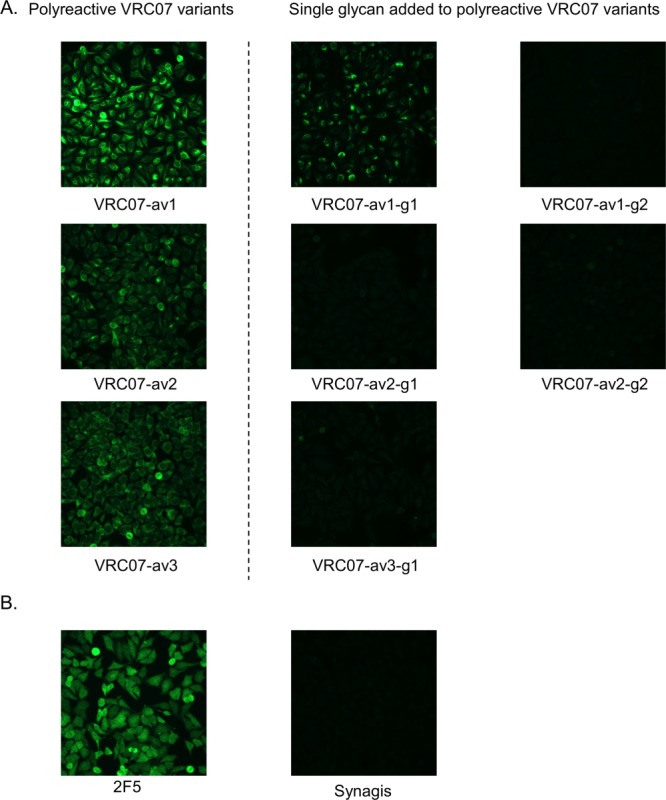
HEp-2 cell staining results at antibody concentration of 50 μg/ml. (A) Results for VRC07 variants, and (B) results for control antibodies. Synagis, an anti-RSV monoclonal antibody, was used as the negative control. 2F5, a polyreactive HIV-1 antibody, was used as the positive control. The addition of glycan reduced the fluorescent signal in all test cases, indicating lower reactivity toward HEp-2 cells.
Addition of N-linked glycosylation had variable effects on antibody neutralization potency
To measure the effect of the addition of N-linked glycan on HIV-1-neutralization potency, both parental polyreactive variants and N-linked glycan variants were evaluated on a panel of 20 HIV-1 strains (Supporting Information Table S3). For VRC07-av1, the neutralization potency of the N-linked glycan variants was substantially reduced (approximately 60- and 150-fold for VRC07-av1-g1 and VRC07-av2-g2, respectively), while for VRC07-av2 and VRC07-av3, the addition of N-linked glycan induced a modest change in neutralization potency (approximately 2.5- and 9.5-fold for VRC07-av2-g1 and VRC07-av2-g2, respectively, from VRC07-av2; approximately 2.5-fold for VRC07-av3-g1 from VRC07-av3). In general, the effect of the N-glycan addition on antibody neutralization trended with its proximity to the paratope.
Addition of N-linked glycan generally increased antibody concentration over time
To assess whether the reduction of polyreactivity in vitro via addition of N-linked glycan translated to increased antibody concentration over time in vivo, antibody VRC07, three polyreactive variants, and four glycan mutants were injected into mice (five animals in each group), and antibody concentration at different time points was determined (Fig. 7; Supporting Information Fig S5 and Table S4); note that in this experiment, we were unable to evaluate VRC07-av1-g2 as its binding toward the RSC3 probe,27 which was used to measure serum-antibody concentration, was too low to be accurately quantified (Supporting Information Fig. S6). VRC07-av1 and VRC07-av2 showed significantly lower average area under the serum-antibody concentration-time curve (AUC) as compared to wild type VRC07 (49.5%, P < 0.01 for VRC07-av1; 47.5%, P < 0.005 for VRC07-av2). VRC07-av2-g1 showed significantly higher AUC (52.2%, P < 0.05) relative to VRC07-av2, while VRC07-av1-g1 and VRC07-av2-g2 also showed modestly higher average AUC compared to VRC07-av1 (9.0%) and VRC07-av2 (32.0%), respectively. VRC07-av3 also showed modest decrease (16.2%) in terms of average AUC as compared to the wild type. Unexpectedly, VRC07-av3-g1 showed significant decrease in terms of average AUC as compared to VRC07-av3 (55.0%, P < 0.0001). Thus, addition of polyreactivity-inducing mutations reduced AUC relative to the parent VRC07 in three out of three cases (av1-av3), while the addition of masking glycan increased AUC, in three out of four cases, with VRC07-av3-g1 being the sole outlier.
Figure 7.
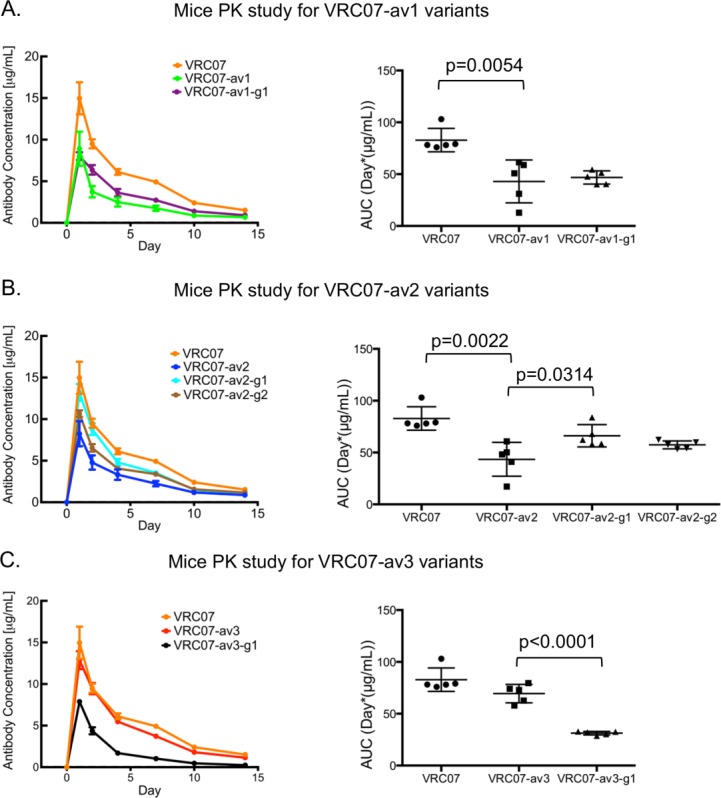
Pharmacokinetics (PK) studies for VRC07 variants in mice. Shown are the average sera antibody concentration at different time points (left) and the AUCs for each individual animal (right) for (A) VRC07, VRC07-av1, and VRC07-av1-g1 (B) VRC07, VRC07-av2, VRC07-av2-g1, and VRC07-av2-g2, and (C) VRC07, VRC07-av3, and VRC07-av3-g1.
N-glycan proximity and polyreactivity masking
To examine whether reductions in polyreactivity through N-glycan addition were due to local versus general masking, we designed 10 VRC07-glycan variants in which the masking N-linked glycan was positioned further from the polyreactivity-inducing mutations than with the initial five variants (Supporting Information Table S5). Cβ distances between these N-glycan variants and the polyreactive-inducing mutations were greater than 20 Å, while Cβ distances for the initial five N-glycan variants were between 5.3 and 10.5 Å. Based on the anti-cardiolipin ELISA assay (Supporting Information Fig. S7 and Table S6), the reduction in VRC07–av1 binding to cardiolipin was substantially less than all three control glycan mutants as compared to VRC01-av1-g1 and VRC01-av1-g2, for VRC07-av2 the reduction of reactivity was substantially smaller for two of the three control glycan mutants (VRC07-av2-c1 and VRC07-av2-c3) as compared to VRC01-av2-g1 and VRC01-av2-g2, and for VRC07-av3 the reduction of reactivity was substantially smaller for two of the four control glycan mutants as compared to VRC07-av3-g1. All control glycan mutants showed low reactivity toward autoantigens in Luminex AtheNA Multi-Lyte ANA tests, except for VRC07-av2-c3 which showed reactivity toward SSB (Supporting Information Fig. S8 and Table S7). As for the Hep-2 cell staining experiments (Supporting Information Fig. S9), two control glycan mutants showed strong fluorescence, and one showed minor fluorescence; for VRC07-av2 all three control glycan mutants showed minimal to no visible fluorescence; for VRC07-av3 one control glycan mutant (VRC07-av3-c1) showed minor fluorescence, while the other three control glycan mutants showed minimal to no visible fluorescence (Supporting Information Fig. S9). Taken together, these results suggested that the addition of N-linked glycan over 20 Å from the polyreactivity-enhancing mutations can reduce polyreactivity (as observed with three of the ten >20 Å variants), but this reduction is less substantial and less frequent than the reduction observed with more proximal N-glycans.
Relationship between overall protein stability and reduction in polyreactivity via addition of N-linked glycan
N-linked glycan can affect protein stability, and protein stability may in turn affect polyreactivity. To investigate whether altered protein stability might account for altered polyreactivity, we evaluated the thermostability of VRC07 variants using DSC (Supporting Information Fig. S10 and Table S8). For VRC07-av1 and VRC07-av3, the addition of polyreactivity-enhancing mutations did not alter thermostability compared to the parent VRC07, and addition of polyreactivity-masking glycans to these variants did not improve thermostability (in most cases a decrease of transition temperature Tm1 was observed), suggesting that the polyreactivity of VRC07-av1 and VRC07-av3 was not a result of reduced thermostability. For VRC07-av2, there was a 4 °C decrease of Tm1 as compared to VRC07. However, among the three variants (VRC07-av2-g1, VRC07-av2-g2, and VRC07-av2-c2) which were deemed not polyreactive based on all three assays, two of them (VRC07-av2-g1 and VRC07-av2-c2) did not have better thermostability as compared to VRC07-av2, suggesting that for these two cases the reduction of polyreactivity by N-linked glycan addition was not a result of improved thermostability. Overall, these results indicate that the reduction of polyreactivity by addition of N-linked glycan that we observed was likely not a result of improved overall protein stability.
Discussion
In addition to the polyreactivity that arises through antibody engineering such as with the polyreactive variants of antibody VRC07 described here, antibody polyreactivity can arise naturally. Polyreactive antibodies are generally removed from the B-cell repertoire through clonal deletion;31 a complementary mechanism, however, was recently described for the polyreactive Hy10 antibody, which involved N-glycan masking of the polyreactive site.32 Thus, the approach described in this article for masking polyreactivity through the introduction of a proximal N-linked glycan appears to have precedent in natural processes of antibody development.31,32 Overall, our results suggest that the glycan-masking approach can be used as part of the antibody engineering process to improve the safety and half-life of lead antibody candidates. In addition to the N-linked glycan approach described here, other methods that introduce large molecules at specific sites of the protein, such as site-specific PEGylation,33 may also have utility in reducing antibody polyreactivity.
The impact of N-linked glycan on neutralization potency of the VRC07 variants appeared to correlate with the position of the added glycan relative to the paratope. Two of the five glycan variants, VRC07-av1-g1 and VRC07-av1-g2 showed substantially impaired neutralization compared to the parental VRC07-av1 construct; this was likely due to the central position of the polyreactive-inducing Trp54 residue in the middle of the antibody paratope [Fig. 2], with the addition of a proximal N-linked glycan likely sterically interfering with antibody-antigen binding or removing critical functional residues directly (e.g., Arg7134). Three other glycan variants mostly retained their parental neutralization potency, and these were positioned near polyreactive-enhancing residues at the rim of the paratope (VRC07-av2) or outside of the paratope altogether (VRC07-av3). We note that the benefits of reducing polyreactivity might outweigh modest reductions in antibody potency; alternatively, other sites of N-linked glycan—positioned further from the paratope—might better retain function.
All three polyreactive VRC07 variants showed lower AUC as compared to the parent VRC07, and three of the four polyreactivity-reducing glycan additions showed increased AUC, suggesting that increased polyreactivity—as evaluated by the assays used in this study—does generally translate into reduced AUC. For one glycan addition, however, we did observed a substantial decrease in AUC indicating the factors governing AUC to not be completely predictive. These results confirm the potential of glycan masking as a means of improving the pharmacokinetics profile of a target antibody as well as the importance of experimentally validating the AUC of glycan-masked antibodies to ensure the engineered glycans to not lead to unfavorable immunogenicity or other issues.
We observed that the reduction in polyreactivity by N-glycan masking was more pronounced when the masking glycans were positioned closer to the polyreactive-inducing mutations and that the reduction of polyreactivity via N-glycan addition was not a result of increased overall protein stability. This suggests a steric mechanism of N-glycan masking. We did observe, however, 3 of 10 glycan variants, in which glycans were added over 20 Å from the site of the polyreactive-inducing mutation, to have reduced polyreactive. In this context we note that N-glycan additions can be over 2 kDa in mass and over 20 Å in size, and that further investigation is likely needed to define the long-distance effect of N-glycan additions.
As part of the proof-of-principle for N-linked glycan reduction of polyreactivity, we provide a computational protocol for the identification of suitable positions on a protein surface to introduce N-linked glycan. The protocol first filters residue positions where asparagine mutation and threonine/serine mutation at two residues downstream will likely disrupt the overall fold of the protein through side chain modeling and energetic evaluation. Then NGlycPred, a Random Forest classifier35 that predicts N-linked glycan occupancy based on structural and sequence information, was used to evaluate the glycan occupancy of the remaining candidate residues. For the five glycan mutants we designed, the overall fold was not disrupted by the insertion of an N-X-T/S sequon, and an N-linked glycan could be successfully attached to each antibody. In addition to antibodies, the procedures described here could be used more generally to modulate proteins of interest, by site-specific addition of N-linked glycosylation, which—in addition to reducing polyreactivity and improving pharmacokinetics—could also be used to positively modulate other properties such as solubility or immunogenicity.
Materials and Methods
Computational design of glycan mutants
We designed glycan variants for three different VRC07 variants: VRC07-av1, VRC07-av2, and VRC07-av3 (Table1). The residue positions of the VRC07 Fab that were compatible with introducing an N-linked glycan were first determined using an in-house computational protocol (Supporting Information Fig. S11). Briefly, starting from a input protein structure (here we used a crystal structure of VRC07- I30QHC-G54WHC -S58NHC 11), the protocol first identified residue positions i for further consideration only if: (1) the residue type of i was not a proline, a cysteine that formed a disulfide bridge, or a glycine that did not have general Ramachandran torsional angles (determined by PROCHECK36); (2) the residue type of i + 1 was not a proline; and (3) the residue type of i + 2 was not a proline, a cysteine that formed a disulfide bridge, or a glycine that did not have general Ramachandran torsional angles. Next, for each selected position i, homology models of the input protein with either N-X-T or N-X-S substitutions (mutations of position i to asparagine and position i + 2 to threonine or serine) were generated using SCAP.37,38 Entries for which the side chain energies of the mutated residues evaluated by SCAP were greater than 10 kcal/mol were filtered. Finally, for each of the remaining entries, the NGlycPred score30 (which predicts the glycan occupancy of the N-linked glycosylation sequon using structural and sequence information) for the asparagine position was evaluated, and the entries with a NGlycPred score of greater than 0.9 were considered compatible for introducing an N-linked glycan. For each of the three VRC07 derivatives, we designed one or two single glycan mutants by adding an N-linked glycan to residues that were (a) compatible for introducing an N-linked glycan and (b) were proximal to the residue(s) causing polyreactivity (W54HC for VRC07-av1, light chain AAAA N terminus for VRC07-av2, and R43LC for VRC07-av3), resulting in a total of five glycan mutants (Table1). Additional glycan variants were designed as controls by adding on to the polyreactive variants glycan designed for the other two polyreactive variants (e.g., adding LC S24N, which were designed for VRC07-av2, to VRC07-av1), resulting in a total of 10 control antibodies (Supporting Information Table S5).
Expression and purification of antibodies
Antibodies were expressed by transient co-transfection of heavy and light chain plasmids into HEK293F cells (Invitrogen) in suspension at 37 °C for 4–5 days. The cell supernatants were passed over Protein A agarose (Pierce), and bound antibodies were washed with PBS and eluted with IgG elution buffer into 1/10th volume of 1M Tris-HCl pH 8.0.
PNGaseF digestion analysis
Three microgram of antibody was digested with 1.5 µl of PNGaseF (500,000 units/ml, NEB) in a total reaction volume of 15 µl. 1.5 µl of 10% NP 40 was added to each reaction. The digest reactions were incubated at 37 °C for 1 h. Digested samples were analyzed on NuPAGE Bis-Tris Mini Gels 4–12% (Invitrogen) in MES running buffer. Protein marker was SeeBlue Plus2 Pre-stained Protein Standard (Invitrogen).
Glycan Characterization with UPLC
100 μg of IgG sample was incubated with 10 µg of IdeS protease (Ulrich von Pawel-Rammingen et al., EMBO, 2002) at 37 °C overnight. Fab and Fc domains were separated by SDS PAGE. The gel bands corresponding to intact Fab domains (100 kDa) were cut out and sequentially washed with Acetonitrile and MilliQ Water and incubated overnight at 37 °C with the enzyme Peptide N Glycosidase F (New England biolabs, UK). Glycans released were eluted in MilliQ water and dried down in a centrifugal evaporator. Dry glycans were labeled with 2-aminobenzoic acid following supplier's protocol (LudgerTag™ 2AA glycan labeling kit, Ludger, UK) and purified with LudgerClean™ T1 Glycan Cleanup Cartridges T1 cartridges. Eluted glycans were then analyzed by UPLC using a BEH Amide column on a Waters Acuity UPLC system (Waters, UK) employing a gradient of solvent A (1.25 mM ammonium formate, pH 4.4) and solvent B (acetonitrile). The sample was solvated in 50% MilliQ water, 50% solvent B and loaded onto the column pre-equilibrated with 35% A. The gradient was increased from 35 to 46% A over 22 min at 0.5 ml/min and was further increased from 46 to 50% B over 2 min at 0.5 ml/min. The gradient was then set at 100% A for 2 min at 0.5 ml/min. Fluorescence was detected using an excitation wavelength of 360 nm and a detection wavelength of 425 nm. Assignment of glycan structures was further clarified by sequential digestion of free glycans with a panel of exoglycosidase enzymes.
Assays to measure polyreactivity
Quanta Lite ACA IgG III ELISA Assay (INOVA Diagnostics, catalog number 708625) was used to test for IgG cardiolipin reactivity per the manufacturer's instructions. Antibodies were tested at dilutions starting at 100 µg/ml and titrated three fold. Here we considered OD (450 nm) values of equal to or greater than three fold of the background ELISA signal as positives.
Reactivity to HIV-1 negative human epithelial (HEp-2) cells was determined by indirect immunofluorescence binding of mAbs to HEp-2 cells (Zeuss Scientific, Branchburg, NJ) as described previously.14 Briefly, 20 μl of antibody at 25 and 50 μg/ml was placed on a predetermined spot on the surface of an ANA HEp-2 kit slide, incubated for 25 min at room temperature, washed, and developed with 20 μl of goat anti-human Ig-FITC at 20 μg/ml (Southern Biotech, Birmingham, AL) for 25 min. Incubations were performed in humid chambers in the dark. Slides were washed and dried; a drop of 33% glycerol was placed on each spot prior to the fixing of coverslips. Images were taken on an Olympus AX70 with SpotFlex FX1520 charge-coupled device (CCD) with a UPlanFL 40× (numerical aperture, 0.75) objective at 25 °C in the FITC channel using SPOT software. All images were acquired for the time specified in the figure legend. Image layout and scaling were performed in Adobe Photoshop without image manipulation.
The Luminex AtheNA Multi-Lyte ANA test (Wampole Laboratories) was used to test for monoclonal antibody reactivity to SSA/Ro, SS-B/La, Sm, RNP, Jo-1, double-stranded DNA, centromere B, and histone and was performed as per the manufacturer's specifications and as previously described.14 Monoclonal antibody concentrations assayed were 50, 25, 12.5, and 6.25 µg/ml. Ten microliter of each concentration were incubated with the luminex fluorescent beads and the test performed per the manufacturer's specifications. Here we considered result outcome of equal to or greater than 125, which is slightly higher than 120 suggested by the manufacturer, as positive.
HIV-1 neutralization
Neutralization was measured using single-round-of-infection HIV-1 Env-pseudoviruses and TZM-bl target cells (Hela cells engineered to express CD4 and CCR5), as described previously.27,39 Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation. The 50% inhibitory concentrations (IC50) were reported as the antibody concentration or serum dilution required to inhibit infection by 50%.
Mice pharmacokinetics study
Three polyreactive VRC07 variants, four designed VRC07 glycan constructs, and VRC07 wild type were evaluated in the mice pharmacokinetics study. Groups of five 88-days-old female CB17SC-F SCID mice (Taconic Biosciences) were intravenously injected with 40 µg/500 µl of purified monoclonal antibodies in PBS on day 0. Blood was collected before injection on day 0, and 1, 2, 4, 7, and 14 days after injection. Plasma antibody levels of VRC07 and its mutants were quantified as described before.40 ELISA plates (96 well) were coated with 2 μg/ml RSC327 in PBS, incubated at 4 °C overnight and blocked with blocking buffer (PBS containing 5% Skim milk, 2% BSA and 0.1% Tween-20) at room temperature for 1 h. Sera were diluted in blocking buffer and added to the plate and incubated at room temperature for 1 h, Horseradish peroxidase (HRP)-conjugated anti-human IgG, Fcγ-specific (Jackson ImmunoResearch Laboratories), was added and incubated at room temperature for 1 h. Tetramethylbenzidine (Sigma) HRP substrate was added to each well, and the absorbance of the yellow color that developed after adding 0.5M H2SO4 was measured at 450 nm. Purified monoclonal antibodies were used as a standard, and the limit of detection was 10 ng/ml. Sera antibody concentration—time AUC values were calculated using the WinNonlin software (Pharsight). Two-tailed paired t-test was performed for AUC between the polyreactive variants and their respective glycan mutants or the wild type.
Differential scanning calorimetry
DSC was performed using VP-DSC instrument (MicroCal). Data were collected in the 40–100°C range at a scanning rate of 1 °C/min. Antibody concentration was 0.5 mg/ml. The resulting thermograms were corrected for the heat capacity of the solvent by subtraction of the corresponding PBS buffer scans. The data were analyzed using Origin 7.0 software (MicroCal).
Acknowledgments
The authors thank the Structural Biology Section, Structural Bioinformatics Core, Humoral Immunology Section, and Humoral Immunology Core at the NIH Vaccine Research Center for helpful discussions or comments on the manuscript, and Josh Eudailey for expert technical assistance. They also thank Marlon Dillon for assisting with the animal experiments.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- Mejias A, Ramilo O. Review of palivizumab in the prophylaxis of respiratory syncytial virus (RSV) in high-risk infants. Biol Targets Ther. 2008;2:433–439. doi: 10.2147/btt.s3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol. 2010;10:301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- Leavy O. Therapeutic antibodies: past, present and future. Nat Rev Immunol. 2010;10:297. doi: 10.1038/nri2763. [DOI] [PubMed] [Google Scholar]
- Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10:317–327. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- Baker MP, Jones TD. Identification and removal of immunogenicity in therapeutic proteins. Curr Opin Drug Discov Dev. 2007;10:219–227. [PubMed] [Google Scholar]
- Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Tsunoda H, Kuramochi T, Sampei Z, Ishii S, Hattori K. Engineering the variable region of therapeutic IgG antibodies. MAbs. 2011;3:243–252. doi: 10.4161/mabs.3.3.15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derer S, Kellner C, Berger S, Valerius T, Peipp M. Fc engineering: design, expression, and functional characterization of antibody variants with improved effector function. Methods Mol Biol. 2012;907:519–536. doi: 10.1007/978-1-61779-974-7_30. [DOI] [PubMed] [Google Scholar]
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, Shi W, Yang ZY, Doria-Rose NA, McKee K, O'Dell S, Schmidt SD, Chuang GY, Druz A, Soto C, Yang Y, Zhang B, Zhou T, Todd JP, Lloyd KE, Eudailey J, Roberts KE, Donald BR, Bailer RT, Ledgerwood J, Program NCS, Mullikin JC, Shapiro L, Koup RA, Graham BS, Nason MC, Connors M, Haynes BF, Rao SS, Roederer M, Kwong PD, Mascola JR, Nabel GJ. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix-Desmazes S, Kaveri SV, Mouthon L, Ayouba A, Malanchere E, Coutinho A, Kazatchkine MD. Self-reactive antibodies (natural autoantibodies) in healthy individuals. J Immunol Methods. 1998;216:117–137. doi: 10.1016/s0022-1759(98)00074-x. [DOI] [PubMed] [Google Scholar]
- Notkins AL. Polyreactivity of antibody molecules. Trends Immunol. 2004;25:174–179. doi: 10.1016/j.it.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Wiehe K, Grimm SK, Lynch R, Yang G, Kozink DM, Perrin F, Cooper AJ, Hwang KK, Chen X, Liu M, McKee K, Parks RJ, Eudailey J, Wang M, Clowse M, Criscione-Schreiber LG, Moody MA, Ackerman ME, Boyd SD, Gao F, Kelsoe G, Verkoczy L, Tomaras GD, Liao HX, Kepler TB, Montefiori DC, Mascola JR, Haynes BF. An autoreactive antibody from an SLE/HIV-1 individual broadly neutralizes HIV-1. J Clin Invest. 2014;124:1835–1843. doi: 10.1172/JCI73441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Tzioufas AG, Notkins AL. Properties and function of polyreactive antibodies and polyreactive antigen-binding B cells. J Autoimmun. 2007;29:219–228. doi: 10.1016/j.jaut.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcheray F, Fraser JW, Gao B, McColl A, DeVito J, Dargon I, Helou Y, Wong W, Girouard TC, Saidman SL, Colvin RB, Palmisano A, Maggiore U, Vaglio A, Smith RN, Zorn E. Polyreactive antibodies developing amidst humoral rejection of human kidney grafts bind apoptotic cells and activate complement. Am J Transplant. 2013;13:2590–2600. doi: 10.1111/ajt.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigounas G, Harindranath N, Donadel G, Notkins AL. Half-life of polyreactive antibodies. J Clin Immunol. 1994;14:134–140. doi: 10.1007/BF01541346. [DOI] [PubMed] [Google Scholar]
- Thomson AH. Introduction to clinical pharmacokinetics. Paed Perinatal Drug Ther. 2000;4:3–11. [Google Scholar]
- Martin T, Crouzier R, Weber JC, Kipps TJ, Pasquali JL. Structure-function studies on a polyreactive (natural) autoantibody. Polyreactivity is dependent on somatically generated sequences in the third complementarity-determining region of the antibody heavy chain. J Immunol. 1994;152:5988–5996. [PubMed] [Google Scholar]
- Song R, Oren DA, Franco D, Seaman MS, Ho DD. Strategic addition of an N-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat Biotechnol. 2013;31:1047–1052. doi: 10.1038/nbt.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Luo J, O'Neil KT, Kang J, Lacy ER, Canziani G, Baker A, Huang M, Tang QM, Raju TS, Jacobs SA, Teplyakov A, Gilliland GL, Feng Y. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng Des Sel. 2010;23:643–651. doi: 10.1093/protein/gzq037. [DOI] [PubMed] [Google Scholar]
- Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci USA. 2009;106:3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. MAbs. 2011;3:568–576. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Ramakrishnan B, Li J, Wang Y, Feng Y, Prabakaran P, Colantonio S, Dyba M, Qasba P, Dimitrov D. Site-specific antibody-drug conjugation through an engineered glycotransferase and a chemically reactive sugar. MAbs. 2014;6:1190–1200. doi: 10.4161/mabs.29889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Boyington JC, Joyce MG, Zhu J, Nabel GJ, Kwong PD, Georgiev I. Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics. 2012;28:2249–2255. doi: 10.1093/bioinformatics/bts426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Verkoczy L, Kelsoe G. Redemption of autoreactive B cells. Proc Natl Acad Sci USA. 2014;111:9022–9023. doi: 10.1073/pnas.1407877111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri Z, Schofield P, Horikawa K, Spierings E, Kipling D, Randall KL, Langley D, Roome B, Vazquez-Lombardi R, Rouet R, Hermes J, Chan TD, Brink R, Dunn-Walters DK, Christ D, Goodnow CC. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc Natl Acad Sci USA. 2014;111:E2567–E2575. doi: 10.1073/pnas.1406974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan LQ, Wang HB, Lai J, Xu YC, Zhang C, Chen SQ. Site-specific PEGylation of a mutated-cysteine residue and its effect on tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) Biomaterials. 2013;34:9115–9123. doi: 10.1016/j.biomaterials.2013.08.020. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Machine Learn. 2001;45:5–32. Random forests. [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thorton JM. PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Xiang Z, Honig B. Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol. 2001;311:421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou T, Yang ZY, Svehla K, O'Dell S, Louder MK, Xu L, Mascola JR, Burton DR, Hoxie JA, Doms RW, Kwong PD, Nabel GJ. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol. 2009;83:5077–5086. doi: 10.1128/JVI.02600-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SY, Pegu A, Rudicell RS, Yang ZY, Joyce MG, Chen X, Wang K, Bao S, Kraemer TD, Rath T, Zeng M, Schmidt SD, Todd JP, Penzak SR, Saunders KO, Nason MC, Haase AT, Rao SS, Blumberg RS, Mascola JR, Nabel GJ. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514:642–645. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


