Antibiotic-resistant bacteria are a global health issue, which necessitate the development of new effective therapeutics. Proline-rich antimicrobial peptides (PrAMPs), which include oncocins, are an extensively studied class of AMPs counteracting bacterial infection at submicromolar concentrations. Oncocins enter and kill bacteria by inhibiting certain targets, and not through membrane lysis. Although recently reported to bind DnaK and the bacterial ribosome, their mode of inhibition remains elusive. Here we report the crystal structure of the oncocin derivative, onc112, bound to the Thermus thermophilus 70S ribosome. Strikingly, this 19-residue-long proline-rich peptide manifests the features of several known classes of ribosome inhibitors by simultaneously blocking the peptidyl transferase center and the peptide exit tunnel of the ribosome. This high-resolution structure thus reveals the mechanism by which oncocins inhibit protein synthesis, providing an opportunity for structure-based design of new generation therapeutics.
Throughout the course of evolution, the innate immune system in plants and animals has developed efficient ways to counter infections. One of the defense lines is through the synthesis of antimicrobial peptides that kill bacterial and fungal pathogens 1–3. These AMPs are an effective weapon against the ability of microorganisms to develop resistance 4. Most AMPs are small peptides of 15–70 amino acid residues generated from posttranslational processing of larger precursors, whose synthesis is induced through the Toll and Toll-like receptors 4,5. Although AMPs are extremely diverse, they can be categorized into several groups based on their structures or sequences as α-helical, cysteine-rich, glycine-rich and proline-rich peptides 1.
The PrAMPs, expressed in mammals and insects, have attracted particular attention due to their wide distribution and unique mechanism of killing bacteria without cell membrane disruption 6. PrAMPs oncocin, apidaecin, drosocin and pyrrhocoricin interact with the substrate-binding domain of the chaperone DnaK with dissociation constants in the micromolar range, resulting in protein misfolding and aggregation, and subsequent bacterial death 7–11. This mechanism is bacteria specific, because PrAMPs are only slightly toxic towards mammalian cells due to their inability to penetrate the mammalian cell membrane 12. Structural studies of the bacterial DnaK complexes with PrAMPs revealed two binding modes for PrAMPs to the conventional binding cleft of DnaK 13,14, indicating that DnaK is rather promiscuous in peptide binding.
Recent biochemical data demonstrated that an apidaecin derivative Api88 and its truncated mutants are equally efficient in entering E. coli cell and binding to DnaK. However, only the full-length Api88 is functional 15. This, combined with the susceptibility of a DnaK null mutant to PrAMPs, suggest that DnaK is not the main target for this group of peptides 16. Cross-linking, cell-free translation and binding assays show that apidaecin derivatives, api88 and api137, and oncocin derivatives, onc72 and onc112, inhibit protein synthesis by binding the 70S ribosome with dissociation constants in the nanomolar range 16. Remarkably, onc112 binds to the 70S ribosome approximately 50-fold stronger than it does to DnaK 16, making the ribosome the preferred target.
To gain insights into the mode of its interaction, we have determined the 2.9 Å resolution crystal structure of onc112 bound to the Thermus thermophilus 70S ribosome in complex with mRNA and a tRNAfMet in the P site (Online Methods and Table 1). Remarkably, unlike most known antibiotics, a single onc112 molecule interacts with not just one, but with three adjacent functional sites of the ribosome. Its N-terminus binds near the peptidyl transferase center (PTC) of the 50S subunit, where it interferes with the A-site tRNA and the peptidyl-tRNA in the P site. The rest of onc112 binds inside the peptide exit tunnel of the 50S subunit and blocks it completely (Fig. 1). Our study provides insights into the mechanism by which PrAMPs inhibit translation and builds a foundation for further structure-based design of a new generation of antimicrobial therapeutics, which may be more effective against the development of bacterial drug resistance. In addition, the visualization of a peptide in the 50S subunit exit tunnel at high-resolution sheds light on the function of the tunnel during translation.
Table 1.
Data collection and refinement statistics
| Tth70S complex | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 210.1, 450.7, 623.5 |
| (°) | 90, 90, 90 |
| Resolution (Å) | 50–2.9 (2.90–2.98) |
| Rmerge | 16.9 (138.1) |
| I / I | 8.62 (1.20) |
| Completeness (%) | 99.7% (99.9%) |
| Redundancy | 4.6 (4.7) |
| Refinement | |
| Resolution (Å) | 50.0 - 2.9 |
| No. reflections | 1,271,909 |
| Rwork / Rfree | 20.3/24.8 |
| No. atoms | |
| RNA | 191,321 |
| Protein | 90,537 |
| Ions | 2,366 |
| Waters | 3,990 |
| B-factor (average) | 59.85 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.717 |
Single crystal dataset
Values in parentheses are for highest-resolution shell.
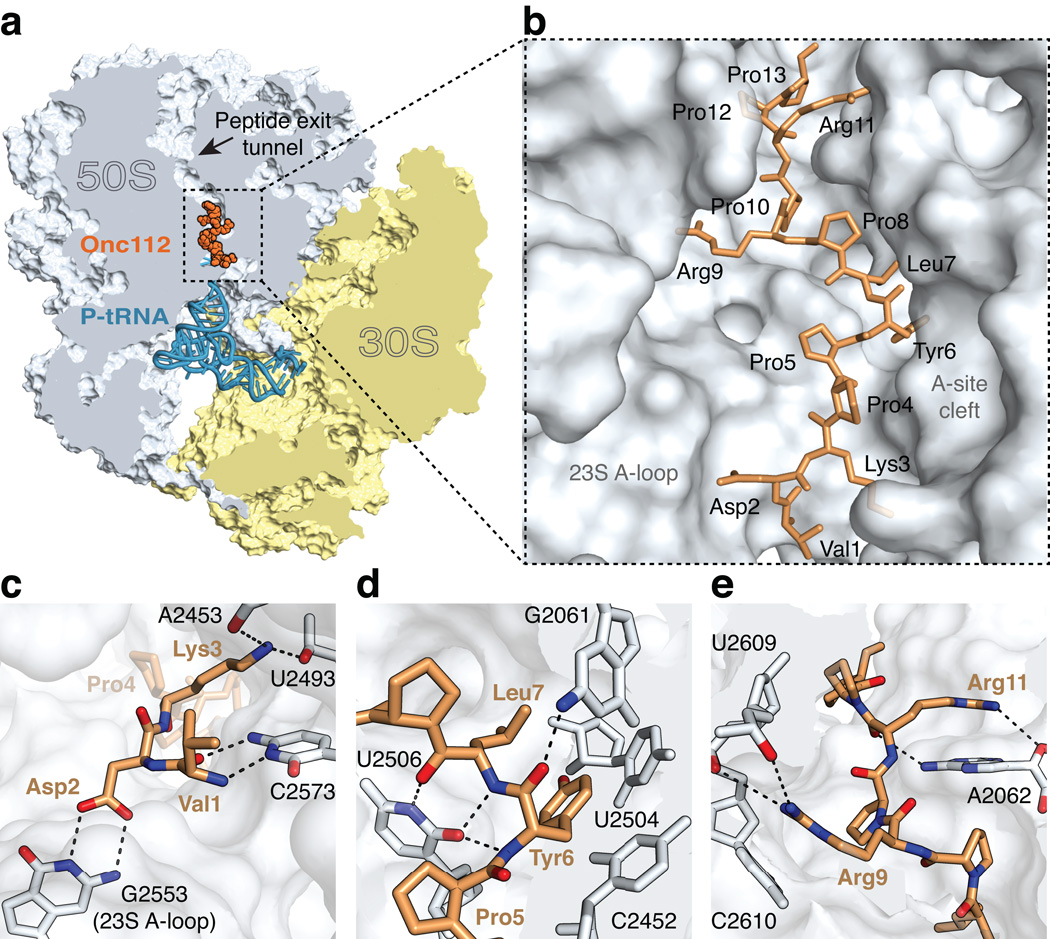
Figure 1. The structure of onc112 bound to the ribosome.
(a) Overview of onc112 (brown) and the P-site tRNAfMet (blue) bound to the 70S ribosome. The 50S and 30S subunits are shown in light blue and yellow, respectively. Portions of the ribosome are omitted for clarity. (b) Close-up view of the positioning of onc112 in the peptide exit tunnel. For reference, landmark features of the ribosome peptidyl transferase center (PTC) are indicated. (c–e) Interactions of onc112 with the ribosome. Putative hydrogen bonds between oxygen (red) and nitrogen (blue) atoms are shown as black dashes. (c) Interactions of the N-terminus of onc112 with the 23S rRNA. (d) Interactions of onc112 within the PTC. The middle part of onc112 occupies the A-site cleft in the PTC. Residues Leu7 and Tyr6 form a three-layer stack with the nucleotide base of C2452 (2463). Additional stabilisation of onc112 is provided by interactions with nucleotides U2506 (2517) and G2061 (2082). (e) Interactions of the C-terminus of onc112 with the peptide exit tunnel. Arg9 and Arg11 form a stacking interaction with the nucleotide bases of C2610 (2621) and A2062 (2083), respectively.
PyMOL 33 was used to generate all figures.
RESULTS
We determined the structure of the 70S ribosome complex with onc112 by molecular replacement using a high-resolution model of the 70S ribosome with its ligands removed 17 as a search model (Online Methods). Clear unbiased electron density for the mRNA, P-site tRNAfMet and onc112 inside the peptide tunnel of the 50S subunit with its N-terminus near the acceptor stem of the tRNAfMet appeared after the difference Fourier map was calculated using initially phased diffraction data (Fig. 1a, b and Supplementary Fig. 1). Throughout the text, the E. coli nucleotide numbering is used with the T. thermophilus numbers provided within brackets.
Interactions of the onc112 N-terminus with the ribosome
The universally conserved PTC of the ribosome, which is made exclusively of RNA, catalyzes peptide bond formation between the incoming aminoacyl-tRNA and the peptidyl-tRNA, both of which must be properly aligned in the A and P sites, respectively 17. While most antibiotics target only certain parts of the PTC or the peptide exit tunnel, onc112 overlaps with all of those binding sites located in the 50S exit tunnel (Figs. 1a, b, 2c). Surprisingly, the orientation of onc112 inside the peptide tunnel is opposite to that of a natural nascent peptide chain for which incoming amino acids are added at its C-terminus in the PTC. The N-terminus of onc112 (residues 1 to 5) follows the path of the CCA-end of the acceptor stem and of the aminoacyl moiety of an A-site tRNA (Fig. 2a and Supplementary Fig. 2a). The first three residues of onc112 form multiple interactions with the 23S ribosomal RNA (rRNA). For instance, the main chain peptide backbone of Val1 and the side chain of Asp2 form hydrogen bonding (H-bonding) interactions with nucleotides C2573 (2584) and G2553 (2564), respectively (Fig. 1c and Supplementary Fig. 3a). The latter nucleotide is part of the A-loop of the 23S rRNA and normally forms a Watson-Crick base pair with C75 of the A-site tRNA 17. This interaction is partially replaced by the γ-carboxyl group of the Asp2 side chain that superposes with the Watson-Crick edge of nucleotide C75 of an accommodated tRNA in the A site (Supplementary Fig. 3b) 17. The side chain of Lys3 is fully extended and forms non-specific electrostatic interactions with 23S rRNA (Fig. 1c). These interactions will ensure that the binding of onc112 to the ribosome and of the tRNA to the A site of the 50S subunit are mutually exclusive. Moreover, the binding of onc112 reorients nucleotide U2585 (2596) in a way that it would collide with the amino acid moiety attached to the peptidyl-tRNA in the P site (Supplementary Fig. 2b). By interfering with the binding of the CCA-end of the aminoacyl-tRNA in the PTC and indirectly with the peptidyl-tRNA via the displacement of nucleotide U2585 (2596), onc112 presumably blocks the peptidyl transferase function of the ribosome, thereby accounting for its antimicrobial properties.
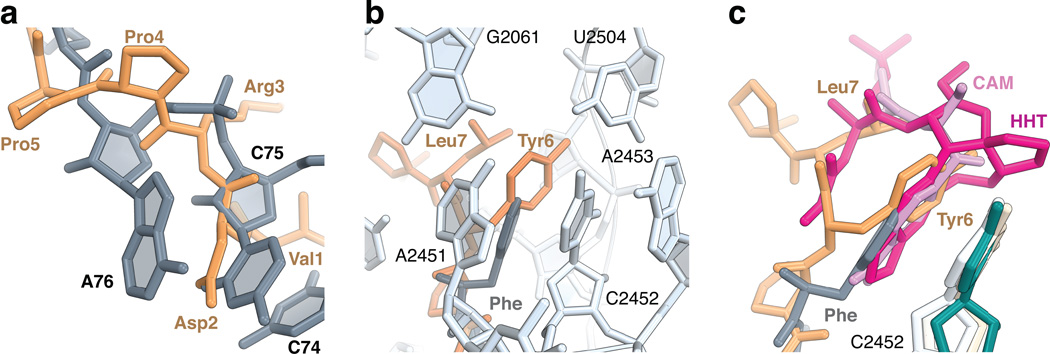
Figure 2. Peptide binding to the A-site cleft in the peptidyl transferase center.
(a) Residues 1 to 5 of onc112 (brown) follow the path of the CCA-end of an accommodated tRNA in the A site (grey) (PDB accession 1VY4) 17. (b) The aromatic ring of Tyr6 of onc112 (brown) forms a stacking interaction with the nucleotide base of C2452 (2463) of the 23S rRNA in a manner that is analogous to the phenylalanine residue attached to a Phe-tRNAPhe bound in the A site (grey) (PDB accession 1VY4) 17. Nucleotides of the 23S rRNA (light blue) forming the A-site cleft are labelled. (c) Structures of antibiotics chloramphenicol (CAM, magenta; 23S rRNA, ivory) (PDB accession 4V7W) 18 and homoharringtonine (HHT, pink; 26S rRNA, darkcyan) (PDB accession 4U4Q) 20 bound in the A-site cleft. Nucleotide C2452 (2463) of the 23S rRNA in the complex of onc112 with the ribosome is light blue.
Interactions of onc112 within the PTC
The middle part of onc112 (residues 6 to 8) occupies the A-site cleft in the PTC, which forms one side of the peptide exit tunnel wall where the aminoacyl moiety of an A-site tRNA would normally bind (Fig. 1d). The Tyr6 residue, located right after a consecutive pair of prolines, is rotationally confined such that its phenyl ring forms a stacking interaction with the nucleotide base of C2452 (2463) in a manner that is analogous to the phenyl moiety of a Phe-tRNAPhe bound in the A site 17 (Fig 2b), while its hydroxyl group forms a H-bond with a water molecule (Supplementary Fig. 4b). This binding mode is also similar to that of the antibiotics chloramphenicol (CAM) and homoharringtonine (HHT) 18–21 (Fig 2c). Like onc112, CAM also displays broad-spectrum activity in bacteria, but does not inhibit eukaryotic translation. However, in contrast to the structure of the CAM complex, nucleotide U2585 (2596) in this onc112-70S complex is rotated by about 55° and the base of A2062 (2083) is rotated by about 45° to accommodate Pro5 and Pro8 of the peptide, respectively (Fig. 3a, b). In addition, the interactions of onc112 with the A-site cleft in the PTC are further stabilized by the Leu7 side chain, capping a three-layer stacked unit, and by the N3 and O2 of U2506 (2517) that interact with the main chain of residues Tyr6 and Leu7 (Fig. 1d and Supplementary Fig. 4a). The high conservation of this region of the ribosome suggests that onc112 will likely inhibit eukaryotic protein synthesis, provided it passes through the cell membrane.
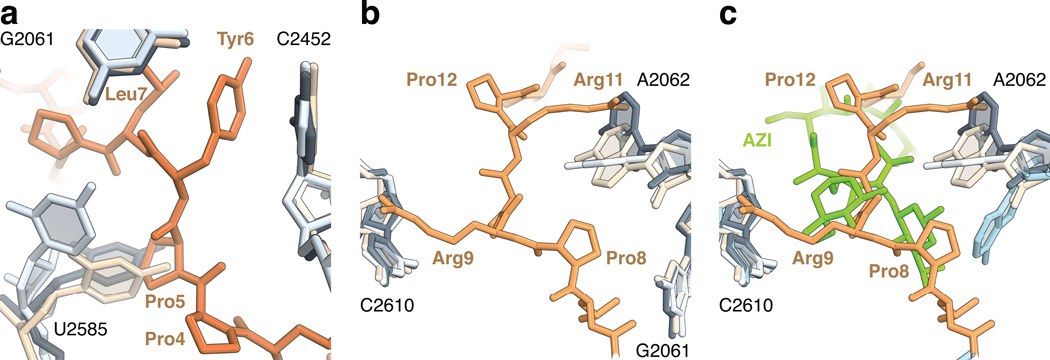
Figure 3. Conformational changes in the peptide exit tunnel.
(a) In the complex of onc112 with the 70S ribosome, the universally conserved nucleotide U2585 (2596) (light blue) changes its position to accommodate Pro5 (70S-CAM complex, ivory, PDB accession 4V7W 18; 70S complex with A- and P-site tRNAs, dark grey, PDB accession 1VY4 17). (b) In the upper chamber of the peptide exit tunnel, the binding of onc112 fixes the conformation of the nucleotide base of A2062 (2083) (light blue) such that it forms a stacking interaction with Arg11 and accommodates Pro8. (c) Same as in panel b, but showing the macrolide antibiotic azithromycin (AZI, green) and A2062 (blue) from the structure of the 70S-AZI complex (PDB accession 4V7Y) 18.
Interactions of onc112 in the exit tunnel
In the upper chamber of the peptide exit tunnel, the nucleotide base of A2062 (2083) adopts a conformation allowing the side chain of Arg11 to form a favorable stacking interaction with its nucleotide base (Figs. 1e, 3b). A similar interaction is also observed between Arg9 and C2610 (2621) (Figs. 1e, 3b). These extended arginine side chains span about 16 Å across the ribosome peptide exit tunnel, thereby completely plugging the upper chamber (Figs. 1e, 3b). Interestingly, nucleotide A2062 (2083) adopts different conformations when interacting with erythromycin on the stalled ribosome 22,23. Its mutation, as well as mutation of C2610 (2621), substantially reduce nascent peptide- and antibiotic-dependent ribosome stalling 24,25. The location of onc112 overlaps with the macrolide antibiotics erythromycin and azithromycin 18 (Fig. 3c), explaining the antimicrobial effects of onc112, which occupies the binding sites of several classes of antibiotics. Our data explain previous observation that alanine substitutions of Lys3, Tyr6, Leu7 and Arg11 severely reduce the antimicrobial activity of oncocin 26 and decrease its binding affinity to the 70S ribosome by more than 30-fold 16. The side chains of these residues are all involved in tight interactions with the 23S rRNA and their mutation will consequently affect the binding of oncocin to the ribosome. We do not see electron density for the last six amino acid residues of onc112, which suggests they are flexible and likely not essential for the binding to the ribosome.
DISCUSSION
The crystal structure of the proline-rich peptide onc112 bound to the 70S ribosome highlights the network of extensive interactions between the peptide and the 23S rRNA. This peptide forms a 34 Å-long plug that blocks access to the A and P sites, the peptidyl transferase center and the peptide exit tunnel of the 50S ribosomal subunit. The path of the peptide overlaps with the binding sites of multiple classes of antibiotics, suggesting that it inhibits the bacterial ribosome through a concerted mode of action. This would likely prevent the transition to the elongation phase of protein synthesis and result in the accumulation of 70S ribosomal particles. The structure presented here sheds light on the mechanism that has been evolved by eukaryotic organisms to defeat bacterial resistance to the ribosome targeting compounds. Such peptides, which simultaneously occupy multiple drug binding sites, limit the probability of the appearance of resistance mutations. Our study builds a platform for structure-based design of next generation of improved antimicrobial agents.
ONLINE METHODS
mRNA, tRNA, and peptide synthesis
The mRNA with a Shine-Dalgarno sequence and an initiation codon in the P site was synthesized by integrated DNA technologies with the sequence, 5′ GGC AAG GAG GUA AAA AUG UUC UAA 3′. The fMet-tRNAfMet was prepared as previously described 27. The oncocin derivative, peptide onc112, with the sequence VDKPPYLPRPRPPRrIYNr-NH2 16 was chemically synthesized by GenScript USA Inc. (Piscataway, NJ), where 'r' stands for D-arginine.
Complex Formation and Crystallisation
The Thermus thermopilus 70S ribosomes were purified, crystallized, and cryo-protected as previously described 28. Essentially, 4 µM ribosomes were incubated with 8 µM mRNA and fMet-tRNAfMet in 50 mM KCl, 5 mM Hepes-KOH (pH 7.6), 10 mM NH4Cl, 10 mM Mg acetate and 6 mM β-mercaptoethanol at 55 °C for 6 minutes. The complex was further incubated at room temperature for 10–15 minutes in the presence of 50 µM onc112. Crystals were grown in sitting drop trays in which 3 µl of ribosome complex was mixed with 3.5–4.5 µl reservoir solution containing 0.1 M Tris-HCl pH 7.6, 2.9% (w/v) PEG 20K, 9% (v/v) MPD, 0.175 M L-Arginine and 0.5 mM β-mercaptoethanol, and incubated at 19 °C. The crystals were transferred stepwise to a cryo-protectant solution containing 10 mM NH4Cl, 0.1 M Tris-HCl pH 7.6, 10 mM Mg acetate, 50 mM KCl, 6 mM β-mercaptoethanol, 2.9% (w/v) PEG20K and 40% (v/v) MPD, with the last stabilization step also containing 50 µM onc112. The crystals were left to equilibrate for about 24 hours at 19 °C and were frozen at 80 °K in a N2 stream before plunging in liquid nitrogen.
Data collection, model building and structure refinement
All data collection was carried out at 100 °K. X-ray diffraction data were collected using beamline 24ID-C at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) using 0.3° oscillations. We used the XDS program package 29 to integrate and scale collected data. All crystals belong to the orthorhombic P212121 space group and exhibit similar cell dimensions as previously reported for the wild-type T. thermophilus 70S crystals (Table 1).
We used PHASER from the CCP4 program suite 30 to determine initial solution for the structure by molecular replacement. The search model was generated from the previously published structure of the T. thermophilus 70S ribosome 17 (PDB accession 1VY4) with all its ligands removed. The refinement with two 70S ribosomes in the asymmetric unit was performed by rigid body refinement, and then by five cycles of position and B-factor refinement using PHENIX 31. After initial refinement, unambiguous difference electron density for onc112, mRNA, P-site tRNAfMet became clearly visible in the Fo – Fc difference Fourier map. The onc112 peptide, mRNA and the tRNAfMet in the P site were built into the Fo – Fc electron density map using Coot 32 and the model was further refined using PHENIX 31. The electron density for onc112 allowed us to build unambiguously the first thirteen residues. The electron density for the last six residues was not seen indicating their conformational flexibility. The absence of electron density for the fMet moiety of the tRNAfMet is likely due to the hydrolysis of the aminoacyl group from the tRNA or its flexibility. The final statistics of refinement are provided in Table 1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank P. Moore for valuable discussions, suggestions and critical reading of the manuscript. We thank R. Grodzicki for preparing the fMet-tRNAfMet. We also thank the staffs of the Advanced Photon Source beamline 24-ID for help with data collection and the Center for Structural Biology facility at Yale University for computational support. This work was supported by US National Institutes of Health grant GM022778 to T.A.S.
Footnotes
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4Z8C.
AUTHOR CONTRIBUTIONS
R.N.R., I.B.L. and M.G.G. designed and performed experiments, analyzed data and wrote the manuscript; T.A.S. analyzed data, wrote the manuscript and oversaw the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98:5807–5822. doi: 10.1007/s00253-014-5792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Souza Candido E, et al. The use of versatile plant antimicrobial peptides in agribusiness and human health. Peptides. 2014;55:65–78. doi: 10.1016/j.peptides.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Li W, et al. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids. 2014;46:2287–2294. doi: 10.1007/s00726-014-1820-1. [DOI] [PubMed] [Google Scholar]
- 4.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 5.Brogden KA, Ackermann M, McCray PB, Jr, Tack BF. Antimicrobial peptides in animals and their role in host defences. Int J Antimicrob Agents. 2003;22:465–478. doi: 10.1016/s0924-8579(03)00180-8. [DOI] [PubMed] [Google Scholar]
- 6.Otvos L., Jr Antibacterial peptides isolated from insects. J Pept Sci. 2000;6:497–511. doi: 10.1002/1099-1387(200010)6:10<497::AID-PSC277>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Kragol G, et al. Identification of crucial residues for the antibacterial activity of the proline-rich peptide, pyrrhocoricin. Eur J Biochem. 2002;269:4226–4237. doi: 10.1046/j.1432-1033.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- 8.Otvos L, Jr, et al. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;39:14150–14159. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]
- 9.Otvos L., Jr Antibacterial peptides and proteins with multiple cellular targets. J Pept Sci. 2005;11:697–706. doi: 10.1002/psc.698. [DOI] [PubMed] [Google Scholar]
- 10.Kragol G, et al. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry. 2001;40:3016–3026. doi: 10.1021/bi002656a. [DOI] [PubMed] [Google Scholar]
- 11.Knappe D, et al. Oncocin (VDKPPYLPRPRPPRRIYNR-NH2): a novel antibacterial peptide optimized against gram-negative human pathogens. J Med Chem. 2010;53:5240–5247. doi: 10.1021/jm100378b. [DOI] [PubMed] [Google Scholar]
- 12.Hansen A, Schafer I, Knappe D, Seibel P, Hoffmann R. Intracellular toxicity of proline-rich antimicrobial peptides shuttled into mammalian cells by the cell-penetrating peptide penetratin. Antimicrob Agents Chemother. 2012;56:5194–5201. doi: 10.1128/AAC.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahn M, et al. Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J Mol Biol. 2013;425:2463–2479. doi: 10.1016/j.jmb.2013.03.041. [DOI] [PubMed] [Google Scholar]
- 14.Zahn M, et al. Structural identification of DnaK binding sites within bovine and sheep bactenecin Bac7. Protein Pept Lett. 2014;21:407–412. doi: 10.2174/09298665113206660111. [DOI] [PubMed] [Google Scholar]
- 15.Berthold N, Hoffmann R. Cellular uptake of apidaecin 1b and related analogs in Gram-negative bacteria reveals novel antibacterial mechanism for proline-rich antimicrobial peptides. Protein Pept Lett. 2014;21:391–398. doi: 10.2174/09298665113206660104. [DOI] [PubMed] [Google Scholar]
- 16.Krizsan A, et al. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 s ribosome. Angew Chem Int Ed Engl. 2014;53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 17.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci U S A. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson DN. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci. 2011;1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 20.Garreau de Loubresse N, et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 21.Gurel G, Blaha G, Moore PB, Steitz TA. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol. 2009;389:146–156. doi: 10.1016/j.jmb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arenz S, et al. Drug Sensing by the Ribosome Induces Translational Arrest via Active Site Perturbation. Mol Cell. 2014;56:446–452. doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arenz S, et al. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez-Laslop N, et al. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci U S A. 2011;108:10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Knappe D, et al. Rational design of oncocin derivatives with superior protease stabilities and antibacterial activities based on the high-resolution structure of the oncocin-DnaK complex. Chembiochem. 2011;12:874–876. doi: 10.1002/cbic.201000792. [DOI] [PubMed] [Google Scholar]
- 27.Junemann R, et al. In vivo deuteration of transfer RNAs: overexpression and large-scale purification of deuterated specific tRNAs. Nucleic Acids Res. 1996;24:907–913. doi: 10.1093/nar/24.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 30.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. The PyMOL Molecular Graphics System. 2006 http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.