Abstract
Ribosomal subunit association is a key checkpoint in translation initiation, but its structural dynamics are poorly understood. Here, we used a recently developed mixing-spraying, time-resolved, cryogenic electron microscopy (cryo-EM) method to study ribosomal subunit association in the sub-second time range. We have improved this method and increased the cryo-EM data yield by tenfold. Pre-equilibrium states of the association reaction were captured by reacting the mixture of ribosomal subunits for 60 ms and 140 ms. We also identified three distinct ribosome conformations in the associated ribosomes. The observed proportions of these conformations are the same in these two time points, suggesting that ribosomes equilibrate among the three conformations within less than 60 ms upon formation. Our results demonstrate that the mixing-spraying method can capture multiple states of macromolecules during a sub-second reaction. Other fast processes, such as translation initiation, decoding and ribosome recycling, are amenable to study with this method.
Keywords: time-resolved, cryo-EM, mixing-spraying, ribosome subunit association, structural dynamics
INTRODUCTION
Many biological processes occur on the millisecond scale. Example processes include tRNA selection on the ribosome during protein synthesis (Blanchard et al., 2004) (reviewed in (Johansson et al., 2008; Rodnina et al., 2005)), acetylcholine receptor desensitization (Dilger and Brett, 1990; Matsubara et al., 1992), and subunit rotation of F0F1-ATP synthase (Diez et al., 2004). Visualizing the biological molecules engaged in these processes under physiologically relevant conditions can greatly help us understand the underlying molecular mechanisms. We used time-resolved cryogenic electron microscopy (cryo-EM) (Lu et al., 2009) to achieve this goal.
Cryo-EM is able to reveal the structures of biological macromolecules in vitro under close to physiological conditions, and free from the intermolecular packing interactions that are inherent to X-ray crystallographic approaches. In cryo-EM, the solution containing the molecules is applied to a grid, then excess liquid is removed by controlled blotting, and the grid is quickly plunged into the cryogen (i.e., liquid ethane at liquid nitrogen temperature) (Lepault et al., 1983). As a result, the molecules are embedded in a thin (~1000 Å) layer of vitreous ice. Since the fast freezing of the biological specimen takes only a fraction of a millisecond (Cyrklaff et al., 1990), cryo-EM is able, in principle, to capture a pre-equilibrium reaction system as it evolves over a short period of time (e.g., during the span of a second).
The reason why time resolution in the millisecond range is not achievable with the conventional method is that the time for application of the specimen to the grid and the blotting alone takes at least a second. Fast time-resolved cryo-EM techniques (in the range of milliseconds), first developed by Berriman and Unwin (Berriman and Unwin, 1994), overcome this limitation of conventional methods by spraying one reagent directly onto a grid that has been covered with another reagent, and plunging the grid into the cryogen within a short, controlled time. Relatively slow biological processes (over minutes or hours), on the other hand, can be easily studied by conventional EM methods. For example, Mulder and coworkers studied assembly of the small ribosomal subunit by negative staining EM, with time points collected in a range of 1 min to 120 min (Mulder et al., 2010). Likewise, Fischer and coworkers studied reverse translocation of the ribosome by cryo-EM in which specimens with various reaction times from 1 min to 20 min were prepared by the conventional blotting-plunging method (Fischer et al., 2010).
The purpose of the current study was to explore the performance of a novel method of time-resolved cryo-EM (Lu et al., 2009), in which two components are mixed in a microfluidic device, allowed to react for a defined period of time, and then sprayed onto the EM grid as the latter is being plunged into the cryogen (Figure 1). With this device, reactions within the time frame of a second can be studied at time resolutions reaching 10 ms. The advantage of this mixing-spraying method over the method of Berriman and Unwin (Berriman and Unwin, 1994) is its capability to study a reaction involving two macromolecules that are completely mixed in solution. The spraying-freezing method of Berriman and Unwin relies on fast diffusion of one of the reagents after spraying to encounter the other reagents already on the grid for starting the reaction. In contrast, the mixing-spraying method that we use in this work allows the reagents to first be completely mixed and the reaction to proceed in a microfluidic channel, where the mixture of reagents is freely drifting and diffusing.
Figure 1.

Setup of the time-resolved cryo-EM apparatus. (a) Schematic view of the mixing-spraying device. The EM grid moves perpendicular to the paper. (b) Photograph of the spray of droplets, illuminated by a green laser at the point just before the grid passes through the spray mist in the direction perpendicular to the paper. (c) Photograph of the mixing-spraying chip situated inside the environmental chamber. See also Figure S3.
We have applied the mixing-spraying method to study a relatively fast process, the association of the small (30S) and large (50S) ribosomal subunits as they form the 70S ribosome of E. coli. This process is a simplified version of the biologically relevant translation initiation process, as it omits mRNA, initiator aminoacyl-tRNA, and all three initiation factors. When the two ribosomal subunits associate, they form several connections, called intersubunit bridges (Frank et al., 1995; Schuwirth et al., 2005; Valle et al., 2003b). The bridges are important for ribosome dynamics and function. For example, some of the bridges break and reform during ribosomal intersubunit rotation, an essential process in the translocation of the mRNA and tRNAs (Valle et al., 2003b). The subunit association process has been characterized by previous biochemical kinetic studies (Antoun et al., 2004; Hennelly et al., 2005; Nguyenle et al., 2006; Wishnia et al., 1975), one of which proposed that the subunits associate via structural intermediates (Hennelly et al., 2005). Nonetheless, the structures of the ribosome that form in the process remain poorly understood.
The kinetics of subunit association has been studied using ensemble chemical protection assays yielding two conflicting interpretations (Hennelly et al., 2005; Nguyenle et al., 2006). Hennelly and coworkers proposed that the subunit association is a multi-step process (Hennelly et al., 2005): Initially, core bridges B2c and B2a form, followed rapidly by formation of bridge B5, after which the peripheral bridges, such as B7a, are gradually established. According to their model, there should be intermediate conformations of the ribosome in which only subsets of the bridges have formed in the early phase of the reaction. However, Nguyenle and coworkers (Nguyenle et al., 2006) challenged Hennelly and coworkers’ interpretation. They asserted that all the eight bridges they studied, including bridges B5 and B7a, exhibit the same rates of formation, implying that subunit association is a simple one-step reaction. It should be noted that both studies are in the time range of between 25 ms – 50 ms and several seconds. With the capability of capturing time-dependent structural changes and resolving multiple structures of a molecule in one sample, our time-resolved cryo-EM studies of the subunit association reaction can contribute to a resolution of this controversy.
Initial insights into the structural dynamics of subunit association have come from a recent time-resolved cryo-EM study by Shaikh and coworkers (Shaikh et al., 2014), which favors the interpretation of Hennelly and coworkers (Hennelly et al., 2005). The authors concluded that bridges B2c, B4, B5, and B6 were missing in the 70S reconstruction from time-resolved cryo-EM data collected at both 9.4 ms and 43 ms. They proposed a kinetic model of subunit association according to which the conserved central bridges (e.g., B2a, B3, and B7b) form first, followed by the formation of nonconserved peripheral bridges (B1a, B1b, and B8), and lastly, by the stabilization of the peripheral end of helix 44. A limitation of this study was the small size of single-particle datasets, resulting in low-resolution reconstructions. In the current work, we have studied this process more extensively, sampling different time points, increasing the resolution by improving the data collection efficiency by a factor of ten, and taking advantage of a new particle classification program (see Results – Strategy for classification).
In this work, we first improved the mixing-spraying method of Lu and coworkers (Lu et al., 2009) by designing an environmental chamber (Figure 1c) and optimizing the yield of droplets on the EM grid. We also improved the data collection efficiency by using automated data collection with the Leginon software (Suloway et al., 2005), and employed 3D classification and reconstruction of single particles by the Bayesian approach as implemented in the RELION computer program (Scheres, 2011). By mixing the two subunits and reacting for 60 ms and 140 ms, we were able to capture the subunit association reaction in two stages of the reaction before equilibrium and detected three distinct conformations of the 70S ribosome. Our results show that all bridges are formed within a 60 ms time frame in the nonrotated form of the ribosome.
Thus, this work demonstrates the capability of the mixing-spraying method to capture the states of macromolecules in a reaction within a sub-second time frame.
RESULTS and DISCUSSION
Determination of reaction time window
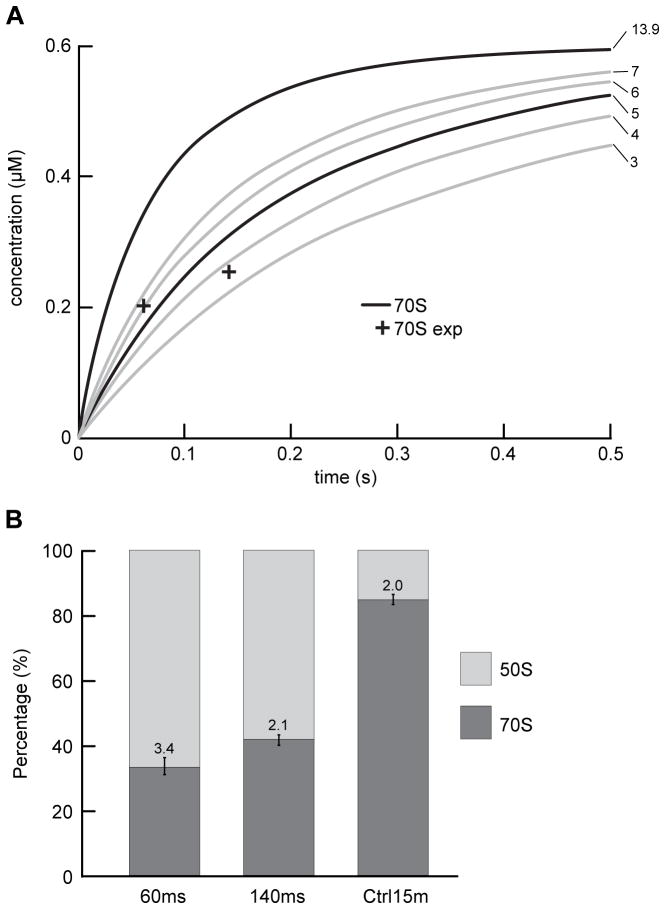
To choose the proper reaction time for studying ribosome subunit association, we first performed a kinetic simulation of the association reaction 30S + 50S <=> 70S. We assumed an association rate constant of ka = 13.9 μM−1 s−1 based on previous light-scattering assays (Hennelly et al., 2005), and a negligible dissociation rate constant (Wishnia et al., 1975). We chose the concentration of subunits to be 1.2 μM for 30S subunits and 0.6 μM for 50S subunits (after mixing). The choice was made to balance particle density on the grid (favoring higher concentration for more particles per micrograph and thereby higher data collection efficiency) against sample consumption rate (favoring lower concentration). The simulation results (Figure 2a, ka = 13.9 μM−1 s−1) indicated that, at the concentration we had chosen, over 90% of the 50S subunits form 70S ribosome within 200 ms. To capture the reaction in a pre-equilibrium system, and also have sufficient data yield for formed 70S particles, we chose the total reaction times of 60 ms and 140 ms (see Supplemental Experimental Procedures – Breakdown of reaction time) for our first and second time points in the time-resolved cryo-EM experiment.
Figure 2.
The proportions of 70S ribosomes from various data sets, and a comparison between the observed proportions and the kinetic simulations of the subunit association reaction. (a) Comparison between the experimentally observed proportions of 70S ribosomes (cross, “70S exp”) and the kinetic simulations of the subunit association reaction (curves). The numbers next to the individual curves indicate the values of association rate constants (ka, unit: μM−1 s−1) used in each of the kinetic simulations. The ka was set to 3, 4, 5, 6, 7 μM−1 s−1 to estimate the value that is most compatible with the experimental results, and ka was set to 13.9 μM−1 s−1 based on previous light scattering assays (Hennelly et al., 2005). (b) Proportions of 70S ribosomes and 50S subunits in the 60 ms, 140 ms, and Ctrl 15 min datasets. The errors in the computational classification were estimated from four independent restarts of RELION 3D classification. See also Figure S1, S2.
We also showed that the purified ribosome subunits are pure and active in peptide synthesis assays (Figure S1, S2, Supplemental Experimental Procedures – Purity and activity of ribosome subunits).
Optimization of data yield and quality
Low data yield is a major challenge for time-resolved cryo-EM by the mixing-spraying method, because only a small fraction of the EM grid is covered with ice that is thin enough for collecting high-quality images. We were able to improve the data yield by optimizing the protocols for preparing time-resolved cryo-EM specimens and EM data collection. Specifically, we first optimized the size of the droplets and their spreading, to increase the area on the EM grid that is suitable for collecting data. We optimized the pressure of the nitrogen gas to yield a dense distribution of small droplets from the spray (Figure 1b). We also plasma-cleaned the EM grids immediately before use to ensure that the surface of the carbon-coated grid is hydrophilic, so that the droplets can spread thinly on the grid. As a result, the majority of the droplets on the EM grid had a diameter smaller than 50 μm and spread thinly on the EM grids, with a thickness suitable for collecting high-magnification images (Figure S3a). Second, the high yield of suitable droplets enabled us to use the program Leginon (Suloway et al., 2005) for data collection.
The quality of time-resolved cryo-EM high-magnification images is comparable to that of images collected from a control experiment performed by incubating the subunits for 15 min, then preparing the cryo-EM grids using the conventional blotting-plunging method (referred to hereafter as the Ctrl 15 min sample). The time-resolved cryo-EM images are affected negligibly by the ramping effect due to uneven ice thickness, which produces uneven brightness of the images (Figure S3c). As a result, for the 140 ms dataset, we collected 2,586 high-quality micrographs out of a total of 8,797 micrographs from 8 time-resolved cryo-EM grids in a total of about 120 hours, and selected 85,880 particles (50S subunits or 70S ribosomes), using the automatic particle selection program Autopicker (Langlois et al., 2014). Similarly, we collected 816 high-quality micrographs for the 60 ms dataset.
Strategy for classification
At every stage in its time course, a pre-equilibrium system is intrinsically heterogeneous, i.e., it contains molecules differing in composition and conformation, so there is a need for computational classification to disentangle the different subpopulations, as an essential prerequisite of time-resolved methods of visualization. In order to determine the changes in composition (quantified as 50S subunits versus 70S ribosomes, with 30S subunits in stoichiometric abundance) over time, we pooled the time-resolved 60 ms, time-resolved 140 ms, and Ctrl 15 min datasets into one single dataset (Figure S4a). The purpose of pooling is to ensure that all the data are classified using the same criterion, so that the proportions of subpopulations in the three time points are strictly comparable.
After classification, we tracked the particles corresponding to the associated 70S ribosomes and the 50S subunits back to the individual experiments, and calculated the proportion of the total number of 50S subunits that are found in associated 70S ribosomes (referred to hereafter as the proportion of 70S ribosomes) in each dataset. This proportion is zero at time point 0 and should reach 1 once all 50S subunits are consumed in forming 70S ribosomes.
Time course of the subunit association reaction
To confirm that the time-resolved cryo-EM method can capture the subunit association reaction in the pre-equilibrium states, we quantified the proportion of 70S ribosomes in the total 50S-containing particles (i.e., 50S subunits and 70S ribosomes) in each dataset. The observed proportions of 70S ribosomes are 33% at 60 ms, 42% at 140 ms, and 85% at 15 min (Ctrl 15 min) (Figure 2b, Table S1). The errors, in the range of 2% – 4%, are estimated by comparing the results from four runs of RELION classification, and represent the errors in reproducing the proportion of 70S ribosomes with random seeds (i.e., the random assignment of particles into several classes at the start of each classification). Single-tailed t-tests show that the increase in the proportion of 70S ribosomes as a function of time is significant (p = 3×10−3 for 60 ms vs. 140 ms data, p = 5×10−8 for 140 ms vs. 15 min data, using 0.05 as the level of significance). In comparing the time-resolved datasets with the Ctrl 15 min dataset, however, we must note that the Ctrl 15 min sample was diluted ~20× with buffer after the prolonged 15 min incubation time and prior to grid preparation using the blotting-plunging method. This dilution will shift the 30S + 50S <=> 70S equilibrium toward the free 30S and 50S subunits, lowering the equilibrium concentration of the 70S ribosomes. Nonetheless, assuming that all of the subunits are competent to associate into 70S ribosomes and that the Ctrl 15 min sample has reached equilibrium, we can use the proportion of 70S ribosomes in the Ctrl 15 min dataset to estimate the equilibrium dissociation constant (KD) of the subunit association reaction. Performing this calculation yields a KD of 6×10−9 M, which is within the same order of magnitude as that estimated previously (KD of ~1×10−9 M – 4×10−9 M at 3 mM Mg2+ – 4 mM Mg2+, with KD presumably leveling off at higher Mg2+ concentrations than 4 mM) (Wishnia et al., 1975). Using this estimated KD and the initial concentrations of 30S and 50S subunits in the 60 ms and 140 ms time-resolved experiments, we could then estimate that, had the time-resolved experiments reached equilibrium, we would expect 99% of the 50S subunits to have associated into 70S ribosomes (see Supplemental Experimental Procedures – Estimation of the dissociation constant and the equilibrium concentration of 70S ribosome). Thus, the fact that we observe only 33% and 42% of the 50S subunits in the 60 ms and 140 ms datasets, respectively, to have associated into 70S ribosomes demonstrates that the time-resolved 60 ms and 140 ms samples indeed represent pre-equilibrium states of the subunit association reaction.
Our observed proportions of 70S ribosomes at the three reaction time points follow the same trend with, but are lower than, the values expected from the kinetic simulation (Figure 2a), which are 58% at 60 ms, 83% at 140 ms, and 100% at 15 min. The proportions are more compatible with a ka value between 4 μM−1 s−1 and 5 μM−1 s−1 than with the previously reported ka of 13.9 μM−1 s−1 (Hennelly et al., 2005) (Figure 2a), suggesting an approximately three-fold lower value in the apparent association rate constant estimated from our time-resolved cryo-EM experiments relative to that measured by light scattering assays (Hennelly et al., 2005).
The difference in the apparent ka values may be attributable to two possible sources: (1) Existence of a fraction of purified ribosomal subunits that cannot form 70S ribosomes following the kinetics that was characterized by the light scattering assays in the time range of tens of seconds (Hennelly et al., 2005), but will eventually form 70S ribosomes after a prolonged incubation time of 15 min; (2) The notable differences in the experimental conditions (e.g. buffer composition, temperature), when comparing the light scattering assays (Hennelly et al., 2005) with our time-resolved cryo-EM experiments. Specifically, the previous light scattering assays used a Tris-based buffer system at 20°C (Hennelly et al., 2005), while our time-resolved experiments used a HEPES-M12 buffer system at 24–26°C (see Supplemental Experimental Procedures – Buffers, Environmental chamber). Indeed, several research groups have reported different ribosomal subunit association rate constants ranging from 1 μM−1 s−1 all the way to 18 μM−1 s−1, using E. coli ribosome subunits in the absence of mRNA and tRNA under a variety of experimental conditions (Antoun et al., 2004; Goerisch et al., 1976; Hennelly et al., 2005; Wishnia et al., 1975). In view of the differences in experimental conditions and the large range of reported values for ka, the second source of the discrepancy is likely to be more important.
Conformational differences of 70S ribosomes
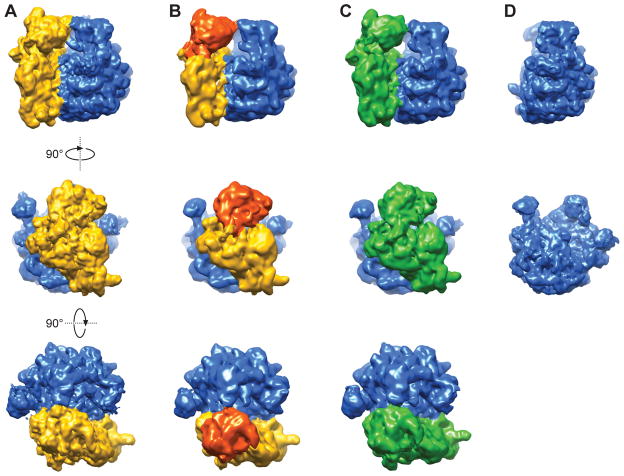
Interestingly, we were able to identify three classes of 70S ribosomes, in addition to a single class of 50S subunit, from the classification (Figure 3). The nonrotated 70S (NR) and rotated 70S (RT) conformations of ribosomes have been observed in previous cryo-EM studies (e.g. (Agrawal et al., 1999; Frank and Agrawal, 2000; Valle et al., 2003b)). The 30S subunit in the 70S RT conformation has undergone an ~8° counterclockwise rotation compared to the 70S NR conformation, as viewed from the 30S subunit side (Figure 3c, side view). The third class of 70S ribosomes is termed “nonrotated 70S with 30S head swivel” (NRS 70S), because the orientation of the 30S subunit body relative to the 50S subunit is identical to that in the NR 70S conformation, while the 30S subunit head domain is swiveled (~4°) counterclockwise when viewed from the top of the ribosome, compared to its position in the NR 70S (Figure 3b, top view).
Figure 3.
Cryo-EM maps of 70S in three conformations and 50S from the experiments. (a) Non-rotated 70S ribosome (NR). (b) Non-rotated 70S ribosome with 30S head swivel (NRS). (c) Rotated 70S ribosome (RT). (d) 50S ribosome subunit. See also Figure S4.
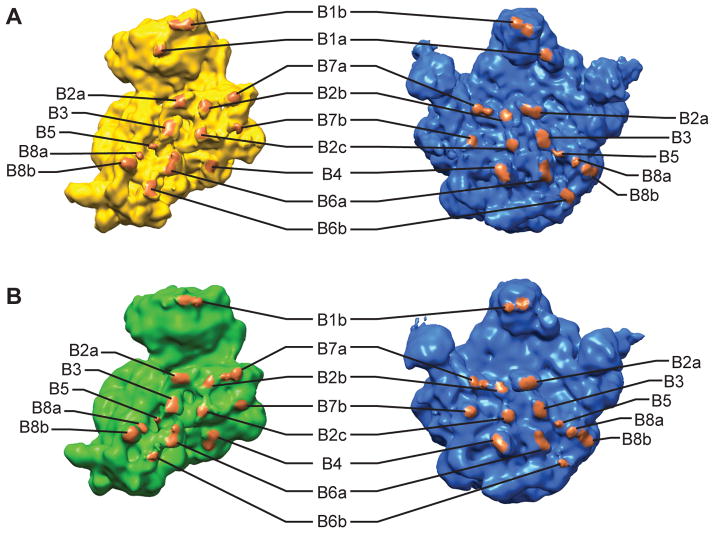
All of the intersubunit bridges are present in the NR 70S, and most of the bridges are present in the NRS 70S and RT 70S (Figure 4, Table 1). To verify that all of the bridges have formed in NR 70S as early as 60 ms, we performed 3D reconstruction on the 70S particles corresponding to the NR 70S conformation from only the time-resolved 60 ms dataset (5,499 particles). This reconstruction shows clear density for helix 44 in the 30S subunit, as well as bridges B2a, B3, B5, B6a and B6b along helix 44 (Figure S5), indicating that all of the intersubunit bridges are formed in NR 70S by the time 60 ms is reached. The NRS 70S ribosome lacks bridge B1a and has weak density of B1b compared to NR 70S. On the other hand, the RT 70S ribosome lacks bridges B1a and has weak density for B6b. The bridge B1b in the RT 70S ribosome appears rearranged compared with the NR 70S ribosome. Bridge B6b at the distal end of helix 44 of 30S subunit is likely disrupted and rearranged during the intersubunit rotation in the RT 70S ribosome.
Figure 4.
Intersubunit bridges in the 70S ribosomes. (a) Segmentation of the 70S NR map, showing the bridges that are present in 70S NR map. (b) Segmentation of the 70S RT map, showing the bridges that are present in 70S RT map. See also Figure S5 and Table 1.
Table 1.
Presence of intersubunit bridges in different conformations of 70S ribosome. Related to Figure 4.
| Bridge a | NR | NRS | RT |
|---|---|---|---|
| B1a | + | − | − |
| B1b | + | + | + |
| B2a | + | + | + |
| B2b | + | + | + |
| B2c | + | + | + |
| B3 | + | + | + |
| B4 | + | + | + |
| B5 | + | + | + |
| B6a | + | + | + |
| B6b | + | + | − b |
| B7a | + | + | + |
| B7b | + | + | + |
| B8a | + | + | + |
| B8b | + | + | + |
Bridge assignment is based on (Dunkle et al., 2011; Gabashvili et al., 2000; Schuwirth et al., 2005; Yusupov et al., 2001).
Bridge B6b has weak density in the 70S RT conformation.
Our observation that the associated 70S ribosomes exist in three conformations suggests that intersubunit rotation and 30S head swiveling represent intrinsic motions of associated 70S ribosomes that lack any bound mRNA and/or tRNAs. In an attempt to elucidate the order in which ribosomal conformational changes take place during subunit association, we compared the proportion of 70S ribosomes in different conformations as a function of the reaction time.
Quantifying the percentages of ribosome in different conformations
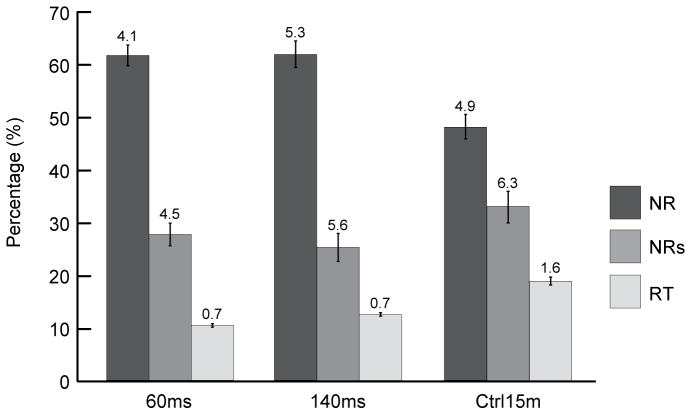
We quantified the proportion of 70S ribosomes in each conformation in the total 70S ribosomes for individual datasets (Figure 5, Table S2). In comparing the proportions of the three conformations in the 60 ms and 140 ms data sets, we find that they are quite similar, with approximately 60% NR 70S, 30% NRS 70S and 10% RT 70S. Single-tailed t-tests yield p = 0.5 for NR 70S, p = 0.2 for NRS 70S, and p = 2×10−3 for RT 70S conformations. In contrast, comparing the proportions in the 140 ms with those in the Ctrl 15 min data set, we find that they appear to differ for all three conformations, with NR 70S decreasing and the other two conformations increasing with time. Single-tailed t-tests yield p = 5×10−3 for NR 70S, p = 6×10−3 for NRS 70S, and p = 2×10−4 for RT 70S conformations. However, we note that the 3D classification process may be less accurate in distinguishing among the three 70S ribosome conformations than in distinguishing between 70S ribosomes and 50S subunits, a fact that is reflected in the larger errors for the former relative to the latter (Figure 2b, 5). This difference in accuracy is most likely attributable to the nature of the differences among the reconstructions – the 70S ribosome and 50S subunit have a massive, easily detectable compositional difference, whereas the various 70S ribosome conformations present more subtle conformational differences that are primarily confined to the peripheral region of the ribosome. Based on this limitation, we regard the change in the proportion of RT 70S from 60 ms to 140 ms as within margins of error, in particular because the proportion is rather small (~10%).
Figure 5.
Proportions of 70S ribosomes in different conformations from the various datasets. Three conformations of 70S ribosomes were observed: rotated (RT), nonrotated with 30S head swivel (NRS), and nonrotated (NR). The errors in the computational classification were estimated from four independent restarts of RELION 3D classification. See also Tables S2.
In addition to the potential inaccuracies associated with distinguishing among the three 70S ribosome conformations, the differences in proportions of 70S conformations between the time-resolved and 15 min datasets may be attributable to the different grid preparation procedures for the time-resolved experiments relative to the Ctrl 15 min experiment (see Experimental Procedures). Factors that may affect the kinetics of the transitions among the 70S conformations include the interactions of ribosomes with the carbon support film and/or the air-water interface, as suggested in previous studies (Lu et al., 2009), and the ~20× dilution of the ribosome sample required for the blotting-plunging method. The potential effects of different grid preparation procedures on these possibly grid-specific transition kinetics warrant further study. Regardless, based on the similarity of the proportions of 70S ribosome conformations at 60 ms and 140 ms, we propose that the conformational changes in the 70S ribosome (i.e., intersubunit rotation and 30S head swivel) are likely to have reached equilibrium before the first time point in our experiment (60 ms). Future mixing-spraying time-resolved cryo-EM experiments collected at earlier time points will be needed to test this hypothesis and determine the relative timing with which these conformational changes occur during the formation of 70S ribosomes from 30S and 50S subunits.
Model of the structural dynamics of ribosome subunit association
Our study unveils multiple conformations of the vacant (i.e., lacking bound mRNA and tRNAs) 70S ribosome during the early phase (i.e., pre-equilibrium) of ribosome subunit association and the proportions of ribosomes in these conformations. Based on these structural and quantitative observations, we propose a structural dynamic model of subunit association: (1) Upon initial subunit association, the 70S ribosomes can rapidly interconvert among the NR 70S, NRS 70S, and RT 70S conformations and reach equilibrium among these three conformations earlier than the first time point we studied (60 ms). (2) The 30S head is able to swivel in the nonrotated 70S ribosome conformation, which is indicated by the observation that the 30S subunit body domains in the NR 70S and NRS 70S maps overlap, but their 30S subunit head domains exhibit different conformations distinguishable at the current resolution (9 Å – 12 Å). However, we did not observe different extents of 30S head swiveling in the rotated 70S ribosome. The fact that we had only 11,000 particles corresponding to RT 70S may have limited our ability to distinguish more subtle differences, such as 30S head swiveling, in the RT 70S. Comparison of our observations on the vacant ribosomes with the previous cryo-EM and crystallography studies in the presence of mRNA and tRNAs (Agirrezabala et al., 2012; Ratje et al., 2010; Valle et al., 2003b; Zhou et al., 2013) highlights the importance of mRNA and tRNAs in defining and modulating the dynamics of the 30S head. (3) The rotated conformation of 70S ribosome, RT 70S, is likely less energetically stable than the nonrotated conformations, which is indicated by the low proportion of RT 70S in our studies (10% – 20% at each reaction time). Future studies are needed to answer the questions how the buffer conditions, as well as the presence of mRNA, tRNAs and initiation factors during translation initiation, affect the distribution of the intermediate conformations of the ribosome and the dynamics of interconversions among these intermediates.
The model presented in the previous paragraph is consistent with observations from single-molecule fluorescence resonance energy transfer (FRET) studies demonstrating that the vacant 70S ribosomes strongly prefer to occupy the nonrotated state and rarely undergo transient excursions to the rotated state (Cornish et al., 2008; Cornish et al., 2009; Ning et al., 2014). Indeed, our conclusion that 70S ribosomes rapidly equilibrate between nonrotated and rotated conformations is strongly supported by the fact that the proportion of rotated 70S at 60 ms reaction time in our study (11%) mirrors that in the fully equilibrated smFRET experiments (15% ~ 21%) (Cornish et al., 2008; Cornish et al., 2009; Ning et al., 2014). Combining our time-resolved cryo-EM observations and smFRET observations, it is likely that the 70S ribosomes that initially associate in the rotated conformation will rapidly convert to the lower-energy, more-stable nonrotated conformation (either NR or NRS) within tens of milliseconds. In contrast, the 70S ribosomes that initially associate in the nonrotated conformation will remain in the nonrotated conformation and rarely convert to the rotated conformation.
Our model is also in good agreement with that proposed from ensemble kinetic studies by Noller group (Nguyenle et al., 2006), and provides more microscopic details since we were able to resolve multiple conformations. We did not observe that the intersubunit bridges form in a stepwise manner, as proposed by Hennelly and coworkers (Hennelly et al., 2005). Instead, the 70S NR conformation has all the bridges formed, and the missing bridges in NRS and RT conformations are readily explained by the large-scale changes in the subunit-subunit interactions accompanying intersubunit rotation and/or 30S head swiveling.
Our observations and the proposed model exhibit several differences compared with the results of a previous time-resolved cryo-EM study of subunit association (Shaikh et al., 2014), despite the fact that both studies used the same buffer composition, similar ribosomal subunit purification procedures, and the same mixing-spraying chip design (for our 60 ms experiment and their 43 ms experiment). The only difference in the experimental setup is the addition of an environmental chamber in our experiments and, consequently, our plunging time (18 ms) is longer than theirs (5 ms) (see Supplemental Information for additional comparison).
While Shaikh and coworkers concluded that bridges B2c, B4, B5, and B6 form later than 43 ms, our 60 ms dataset yielded an NR 70S ribosome containing all the bridges, including these four. One possible reason for the difference between our observations and theirs may be the different activities of the ribosome samples. A second possible reason may be the difference in plunging time. A drastic climate change happens to the ribosome sample during the plunging, from room-temperature, humid air to near-liquid-nitrogen-temperature, dry air, and the effects of such climate change on the kinetics of 70S ribosome formation from 30S and 50S subunits are not well understood. However, a third possible reason is the difference in the resolution of the maps, with 23–33Å in their study versus 9–12Å in ours (Figure S4b–S4d). The higher resolution of our maps stems from our improved data yield, and allows us to detect the intersubunit bridges with more confidence. Future time-resolved cryo-EM studies of ribosome subunit association containing additional time points at sufficiently high resolution will be needed to resolve this discrepancy.
To sum up, in this study we used the mixing-spraying method of time-resolved cryo-EM to study ribosomal subunit association within tens of millisecond time scale, and were able to capture pre-equilibrium states of this association reaction. In addition, we revealed three conformations of the 70S ribosome formed in the early phase (i.e., pre-equilibrium) of the association reaction. Quantification of the proportions of 70S ribosomes in these conformations suggested that, upon formation, the 70S ribosomes rapidly equilibrates among these conformations on a timescale faster than 60 ms. The observed large-scale conformational changes of the vacant ribosome (i.e., in the absence of tRNAs and mRNA), namely the rotation of the 30S subunit and the swiveling of the 30S subunit head, likely represent spontaneous thermal motions of the 70S ribosome (Frank and Gonzalez, 2010).
In the future, it will be interesting to study translation initiation using this method of time-resolved cryo-EM, to shed light on the role of mRNA, initiator tRNA and initiation factors on the structural dynamics of the subunit association reaction. The mixing-spraying method can also be applied to a wide variety of reactions involving ribosomes, including aminoacyl-tRNA selection, translocation, termination, and ribosome recycling.
EXPERIMENTAL PROCEDURES
Mixing-spraying device
This device, developed by Lu and coworkers (Lu et al., 2009), is a small silicon chip containing nano-fabricated channels, which combines the functionalities of mixing, reacting, and spraying (Figure 1). The two solutions are injected into the chip by syringes, and mixed passively by turbulence (within 0.5ms at total flow rate of 6 μL/s). The mixture reacts for a certain time while flowing through the channel, then it is made into spray at the outlet nozzle where it meets the compressed nitrogen gas. In this work we used chips with 38 ms and 107 ms mean reaction time.
Environmental chamber
We designed an environmental chamber to monitor and maintain the temperature and humidity during the time the time-resolved cryo-EM grid is sprayed and plunged (Figure 1c). The environmental chamber is connected to a water tank, which is equipped with a sonicator and an air pump to generate humidified air. During the experiment, the ambient conditions can be maintained at 24–26 °C and 80%–90% relative humidity (equivalent to 0.016 – 0.018 gram of water per gram of dry air).
Preparation of time-resolved cryo-EM grids
The ribosome subunit association reaction was performed using the mixing-spraying device as previously described (Lu et al., 2009), with some alterations. Specifically, Quantifoil R2/2 300 mesh Cu EM grids were carbon-coated, then glow-discharged within 3 h before grid preparation, using Gatan Solarus 950 with H2 at flow rate 6.4 Standard Cubic Centimeters per Minute (sccm) and O2 at flow rate 27.5 sccm, at 25 W for 25 sec. Equal volume of 1.2 μM 30S and 0.6 μM 50S (final concentration after mixing) were injected into the mixing-spraying device each at flow rate of 3 μL/s. Compressed nitrogen gas was kept at 3.4×105 Pa and humidified by passing through two consecutive water tanks. The computer-controlled plunging device was purchased from Dr. Howard White (Eastern Virginia Medical School, VA).
Collection of time-resolved cryo-EM data using the Leginon program
The cryo-EM data were collected, as previously described (Grassucci et al., 2008), in low-dose mode on an FEI (Hillsboro, Oregon) Tecnai F20 TEM at 200-kV extraction voltage with the automated image collection program Leginon (Suloway et al., 2005). Micrographs were recorded on a Gatan (Warrendale, PA) UltraScan 4000 CCD camera with effective CCD magnification of 66,813× and pixel size of 2.25 Å on the object scale. For the time-resolved cryo-EM specimens, only the holes close to the edge of an ice blot are suitable for collecting high-magnification images.
3D classification by using RELION
In the classification of single particles to identify 70S ribosome in different conformations, the datasets of time-resolved 60 ms, time-resolved 140 ms, and Ctrl 15 min were pooled together for classification and 3D reconstructions, so that all the individual datasets were classified using the same criteria, and therefore the classification results (i.e., the relative proportion of each conformation) among the datasets were comparable. The 50S and 70S particles were then traced back to each experiment, to quantify the proportion of 70S in 50S-containing particles, and the proportion of each 70S conformation in the total 70S particles. We used the RELION program (Scheres, 2012) in a stepwise hierarchical classification to discard bad particles identified by the automatic particle-picking program, to separate the 50S subunit from the 70S ribosome, and to sort out the various conformations of the 70S ribosome assisted by visual examination of the reconstructions (Figure S4a). Specifically, the reference volume for the initial alignment of the total set of putative particles was chosen to be a 50S subunit density map (the cryo-EM map of empty 70S ribosome (Valle et al., 2003a) with 30S subunit computationally removed) low-pass filtered to 60 Å. We used the particle data in un-decimated form in the 3D classification, as well as in the final steps of 3D reconstruction and refinement.
In step (1), the datasets of time-resolved 60 ms (after discarding bad particles, 38,316 particles), time-resolved 140 ms (82,574 particles), and Ctrl 15 min (55,369 particles) were combined to form the total dataset (176,259 particles) (Figure S4a). In step (2), the total dataset was separated into 10 classes using RELION 3D classification. The classification results were analyzed using a novel quantitative analysis method (Chen et al., 2014; Shen et al., 2014), and the reconstructions from the different classes were visually examined by using the UCSF Chimera program (Pettersen et al., 2004). The classes yielding a reconstruction of 70S ribosome (94,860 particles) were kept for step (3) of the classification. The classes yielding a reconstruction of 50S subunit (81,399 particles) were kept for refinement in step (4). In step (3), the classes yielding 70S reconstructions (94,860 particles) were classified into 8 classes. The classes resulting in bad reconstructions, likely due to remaining unrecognizable particles, were discarded, keeping 75,254 particles of 70S ribosomes. The remaining classes were visually examined by using UCSF Chimera and regrouped based on the conformation of their resulting reconstructions: non-rotated 70S (NR) (39,678 particles), non-rotated 70S with 30S-head swivel (NRS) (24,447 particles), and rotated 70S (RT) (11,129 particles). In step (4), each class of 70S ribosomes or 50S subunits was refined using RELION 3D auto-refinement (Scheres, 2012) with data in un-decimated form. In step (5), the particles were traced back to each experiment, to quantify the proportion of 70S and the proportion of 70S in each conformation. Steps (2) to (5) were repeated three more times to estimate the error of the 3D classification process.
Resolution measurement
We assessed the resolutions of the reconstructions of 70S ribosomes in their different conformations and the 50S subunit from step (4) using the gold-standard criterion (Scheres and Chen, 2012). For each of the classes, we first performed RELION 3D auto-refinement separately on two randomly split half datasets, following the gold-standard protocol, yielding two unfiltered volumes. Then we multiplied these two unfiltered volumes with a soft Gaussian mask having 0.5 fall-off at 5 pixels (11.2 Å) outside the ribosome, to eliminate peripheral noise. We then calculated the Fourier Shell Correlation (FSC) between the two masked volumes (Figure S4b–S4d), and used the FSC = 0.143 criterion to determine the gold-standard resolution as 9.7 Å (70S NR), 11.1 Å (70S NRS), 11.6 Å (70S RT), 9.5 Å (50S).
Identification of intersubunit bridges in the cryo-EM maps of the 70S ribosome
First, for comparison with each cryo-EM map of the 70S ribosome in this study, a simulated density map of the 70S ribosome was generated from crystallographic structures (PDB ID: 2AVY, 2AW4, the non-rotated 70S ribosome, for comparison with both 70S NR and 70S NRS; PDB ID: 2AW7, 2AWB, the rotated 70S ribosome, for comparison with 70S RT) and low-pass filtered to 10 Å resolution. Second, to bring the simulated density map and our cryo-EM map of the 70S ribosome into register, the 50S portions of the two simulated maps were fitted into the corresponding 50S subunit density map in this study using the Fit in Map tool in USCF Chimera (Pettersen et al., 2004), the rationale being that the 50S subunit is larger and more stable than the 30S subunit. Also, the surface regions around ribosomal proteins L4, L15 and L21, which have pronounced features, were visually inspected to check the alignment of the map from fitting results. Third, the threshold of the simulated density map was adjusted to a level such that the contour of the simulated map just enclosed all the atoms in the crystallographic structure, using default values in Chimera of Van der Waals radii for atoms of C, O, N, P, and S. Specifically, the threshold level was adjusted such that the surface regions around ribosomal proteins L4, L15 and L21 were enclosed within the contour. Fourth, the cryo-EM map was segmented into 30S and 50S subunit density maps. The segmentation reveals the boundaries of the subunits, and hence facilitates locating the bridging contacts on the intersubunit interface. The locations of bridges identified in cryo-EM (Gabashvili et al., 2000) and crystallographic structures (Dunkle et al., 2011; Schuwirth et al., 2005; Yusupov et al., 2001) were marked in our segmented density maps using the Volume Tracer tool in chimera, and connectivity of densities linking 30S and 50S subunits was inspected at each point.
Supplementary Material
Highlights.
Used time-resolved cryo-EM to image sample after 60 ms and 140 ms reaction times.
Captured pre-equilibrium states of the ribosome subunit association reaction.
Visualized, and quantified the occupancies of, three ribosome conformations.
At 60 ms all intersubunit bridges are fully formed in the associated 70S ribosome.
Acknowledgments
We thank R. Grassucci for assistance with cryo-EM data collection, Dr. H. White for help with installing the plunging device and with the design of the environmental chamber, K. M. Headley for assistance in the initial stage of methodology improvement, and M. Thomas-Baum for assistance in the preparation of the figures. This work was supported by the Howard Hughes Medical Institute and the National Institute of Health Grants R01 GM55440 and GM29169 to J.F., and by the National Institute of Health Grant R01GM084288 to R.L.G.
Footnotes
AUTHOR CONTRIBUTIONS
B.C., R.L.G., and J.F. designed the experiments. Z.L., D.B., and T.-M. L. provided the mixing-spraying chips and training to B.C. on the time-resolved cryo-EM method. B.C. performed kinetic simulation and biochemical experiments, and improved the time-resolved cryo-EM method. B.C., S.K., M.S., and B.S. collected time-resolved cryo-EM data. B.C., M.S., and B.S. analyzed the initial cryo-EM datasets. S.K. and B.C. analyzed the final cryo-EM datasets. B.C., S.K., R.L.G., and J.F. wrote the manuscript. All authors approved the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes. The electron microscopy (EM) maps have been deposited in the EMBL-European Bioinformatics Institute EM Data Bank under accession codes EMD-2976, EMD-2977 and EMD-2978 (70S NR, NRS, RT maps, respectively).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Tenson T, Ehrenberg M. Ribosome formation from subunits studied by stopped-flow and rayleigh light scattering. Biol Proced Online. 2004;6:35–54. doi: 10.1251/bpo71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman J, Unwin N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy. 1994;56:241–252. doi: 10.1016/0304-3991(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Chen B, Shen B, Frank J. Particle migration analysis in iterative classification of cryo-EM single-particle data. J Struct Biol. 2014;188:267–273. doi: 10.1016/j.jsb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M, Adrian M, Dubochet J. Evaporation during preparation of unsupported thin vitrified aqueous layers for cryo-electron microscopy. J Electron Microsc Tech. 1990;16:351–355. doi: 10.1002/jemt.1060160407. [DOI] [PubMed] [Google Scholar]
- Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CA, et al. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- Dilger JP, Brett RS. Direct measurement of the concentration- and time-dependent open probability of the nicotinic acetylcholine receptor channel. Biophys J. 1990;57:723–731. doi: 10.1016/S0006-3495(90)82593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JHD. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Gonzalez JRL. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Verschoor A, Li Y, Zhu J, Lata RK, Radermacher M, Penczek P, Grassucci R, Agrawal RK, Srivastava S. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem Cell Biol. 1995;73:757–765. doi: 10.1139/o95-084. [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CMT, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Goerisch H, Goss DJ, Parkhurst LJ. Kinetics of ribosome dissociation and subunit association studied in a light-scattering stopped-flow apparatus. Biochemistry. 1976;15:5743–5753. doi: 10.1021/bi00671a010. [DOI] [PubMed] [Google Scholar]
- Grassucci RA, Taylor D, Frank J. Visualization of macromolecular complexes using cryo-electron microscopy with FEI Tecnai transmission electron microscopes. Nature protocols. 2008;3:330–339. doi: 10.1038/nprot.2007.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennelly SP, Antoun A, Ehrenberg M, Gualerzi CO, Knight W, Lodmell JS, Hill WE. A time-resolved investigation of ribosomal subunit association. J Mol Biol. 2005;346:1243–1258. doi: 10.1016/j.jmb.2004.12.054. [DOI] [PubMed] [Google Scholar]
- Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Curr Opin Microbiol. 2008;11:141–147. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Langlois R, Pallesen J, Ash JT, Nam Ho D, Rubinstein JL, Frank J. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J Struct Biol. 2014;186:1–7. doi: 10.1016/j.jsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepault J, Booy FP, Dubochet J. Electron microscopy of frozen biological suspensions. J Microsc. 1983;129:89–102. doi: 10.1111/j.1365-2818.1983.tb04163.x. [DOI] [PubMed] [Google Scholar]
- Lu Z, Shaikh TR, Barnard D, Meng X, Mohamed H, Yassin A, Mannella CA, Agrawal RK, Lu T-M, Wagenknecht T. Monolithic microfluidic mixing-spraying devices for time-resolved cryo-electron microscopy. J Struct Biol. 2009;168:388–395. doi: 10.1016/j.jsb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara N, Billington AP, Hess GP. How fast does an acetylcholine receptor channel open? Laser-pulse photolysis of an inactive precursor of carbamoylcholine in the microsecond time region with BC3H1 cells. Biochemistry. 1992;31:5507–5514. doi: 10.1021/bi00139a012. [DOI] [PubMed] [Google Scholar]
- Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyenle T, Laurberg M, Brenowitz M, Noller HF. Following the dynamics of changes in solvent accessibility of 16 S and 23 S rRNA during ribosomal subunit association using synchrotron-generated hydroxyl radicals. J Mol Biol. 2006;359:1235–1248. doi: 10.1016/j.jmb.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Ning W, Fei J, Gonzalez RL., Jr The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc Natl Acad Sci. 2014;111:12073–12078. doi: 10.1073/pnas.1401864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2011;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 Å Resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Yassin AS, Lu Z, Barnard D, Meng X, Lu T-M, Wagenknecht T, Agrawal RK. Initial bridges between two ribosomal subunits are formed within 9.4 milliseconds, as studied by time-resolved cryo-EM. Proc Natl Acad Sci. 2014;111:9822–9827. doi: 10.1073/pnas.1406744111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Chen B, Liao H, Frank J. Quantitative analysis in iterative classification schemes for cryo-EM applications. In: Herman GT, Frank J, editors. Computational Methods for Three-Dimensional Microscopy Reconstruction. Birkhauser; Basel: 2014. [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Mol Biol. 2003a;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wishnia A, Boussert A, Graffe M, Dessen P, Grunberg-Manago M. Kinetics of the reversible association of ribosomal subunits: Stopped-flow studies of the rate law and of the effect of Mg2+ J Mol Biol. 1975;93:499–515. doi: 10.1016/0022-2836(75)90242-9. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G–ribosome complexes trapped in intermediate states of translocation. Science. 2013;340 doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.