SUMMARY
Bacterial biofilms in the colon alter the host tissue microenvironment. A role for biofilms in colon cancer metabolism has been suggested but to date has not been evaluated. Using metabolomics, we investigated the metabolic influence that microbial biofilms have on colon tissues and the related occurrence of cancer. Patient-matched colon cancers and histologically normal tissues, with or without biofilms, were examined. We show the upregulation of polyamine metabolites in tissues from cancer hosts with significant enhancement of N1, N12-diacetylspermine in both biofilm positive cancer and normal tissues. Antibiotic treatment, which cleared biofilms, decreased N1, N12-diacetylspermine levels to those seen in biofilm negative tissues, indicating that host cancer and bacterial biofilm structures contribute to the polyamine metabolite pool. These results show that colonic mucosal biofilms alter the cancer metabolome, to produce a regulator of cellular proliferation and colon cancer growth potentially affecting cancer development and progression.
INTRODUCTION
Colon cancer has numerous risk factors (Botteri et al., 2008; Gay et al., 2012; Moskal et al., 2007), but recent studies highlight a potential role for the colon microbiota in colon cancer development (Sears and Garrett, 2014) . The colon microbiota can form structures, termed biofilms, that line the mucosal surface and indicate disruption of the normal colon mucous barrier (Probert and Gibson, 2002; Shah and Swiatlo, 2008). Biofilms have been associated with non-malignant pathologies such as inflammatory bowel disease (Swidsinski et al., 2005) but have not been analyzed in the setting of human colon cancer. We have recently demonstrated that biofilms are associated with human colon cancer and linked to cancer location, with virtually all right-sided colon adenomas and cancers possessing biofilms, while left-sided cancers are infrequently biofilm positive (Dejea et al., 2014). Importantly, histologically normal colon mucosa, collected from the surgical resection margin, was also biofilm positive or negative demonstrating 100% concordance with their paired cancer. While differences in clinical prognosis and genetic characteristics between right- and left-sided colon cancers have been documented (Benedix et al., 2010; Meguid et al., 2008) the role of specific microbes in different regions of the colon has not yet been investigated. We thus hypothesized that biofilm structure might affect cancer biology by modulating the metabolome, yielding metabolites that enhance cancer growth. We approached our hypothesis by investigating the colon tissue metabolome using four independent metabolomic platforms permitting the examination of: (i) metabolic changes in cancer tissues with and without biofilms, (ii) the spatial organization of metabolite distributions, and (iii) the unbiased assimilation of stable isotope labelled metabolites into metabolic pathways. Global (untargeted) and targeted analyses were performed with liquid chromatography quadrupole time-of-flight mass spectrometry (LC-QTOF-MS) and liquid chromatography triple quadrupole mass spectrometry (LC-QqQ-MS), respectively, whereas nanostructure-initiator mass spectrometry (NIMS) imaging was utilized to identify the spatial distribution of metabolites. Ultimately, we determined the metabolic changes that biofilms have on colon tissue, the origin of the metabolites and their potential biological roles in the host and microbiota.
RESULTS
Untargeted metabolomic reveals increased polyamine metabolites in colon cancers
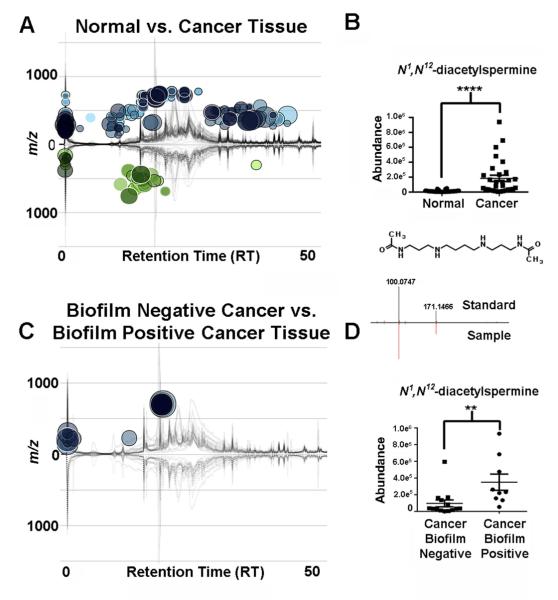
Two patient groups of samples were obtained from Johns Hopkins University (JHU) School of Medicine, Baltimore, MD, USA and Karolinska University Hospital, Stockholm, Sweden. Using an untargeted metabolomics analysis, 304 differentially regulated features were observed (JHU samples) (Figure 1A) when comparing the colon cancer tissues to their paired normal tissues (n=30 patients). The feature with one of the largest fold changes of 9.4-fold (p=3.7e−9, m/z=287.240) was identified as N1, N12-diacetylspermine (Figure 1B). Other polyamine metabolites were also upregulated in the colon cancer tissue samples 3.7-fold, N1-acetylspermidine (m/z=188.172, p=1.9e−8) and 3.6-fold, N1-acetylspermine (m/z=245.235, p=3.5e−8). Further analysis revealed that a number of features could be classified as phospholipids and fatty acids (Table S1). Analysis of the Karolinska University Hospital samples (n=30 patients) validated the upregulation of N1, N12-diacetylspermine 8.1-fold (p=2.9e−3), N1-acetylspermidine (p=5.6e−4, 3.2-fold) and N1-acetylspermine (p=4.6e−3, 2.2-fold). Many of the other metabolites seen in the JHU samples (phospholipids, carnitines, fatty acids and hypoxanthine) could not be observed in this sample set (Table S1). These metabolites have previously been identified with many other diseases as well as colon cancer, and have low specificity to one disease (Gonzalez-Dominguez et al., 2014; Sampey et al., 2012). However, the observation of increased polyamine metabolites in colon cancer tissues from both patient populations shows the potential specificity of these metabolites, which are further validated here. The presence of the acetylated polyamines, phospholipids and fatty acids suggest the involvement of acetyl-CoA and upregulation of fatty acid oxidation. However untargeted analysis by hydrophilic liquid interaction chromatography (HILIC) in positive and negative mode, showed no presence of acetyl-CoA or malonyl-CoA or further metabolic dysregulation in the colon tissues.
Figure 1. Untargeted metabolomics.
(A) Cloud plot showing dysregulated features between normal tissues and patient-matched colon cancer tissues (n=30, two-tailed Wilcoxon test). Total ion chromatograms for each sample can be seen on the plot. (B) Relative abundance of N1, N12-diacetylspermine (two-tailed Wilcoxon test, ****p<0.0001). (C) Comparison of biofilm negative cancers, n=11 (lower part of plot) to biofilm positive cancers n=8 (upper part of plot) (two-tailed Mann-Whitney test). (D) Relative abundance of N1, N12-diacetylspermine (two-tailed Mann-Whitney test) **p<0.01. Tandem MS spectrum of N1, N12-diacetylspermine in samples and standards is shown. See also Table S1 and Figure S1.
Stratification by biofilm status reveals an upregulation of N1, N12-diacetylspermine
We next stratified our untargeted metabolomic analysis to determine if colon tissues with biofilms differed in their metabolic features from those colon tissues lacking biofilms. Only 28 dysregulated metabolites were seen (Figure 1C). In the biofilm positive cancer tissues the upregulation of N1, N12-diacetylspermine 3.8-fold (p=2.5e−3) (Figure 1D), N1-acetylspermidine (p=4.1e−2, 1.7-fold) and N1-acetylspermine (p=2.0e−2, 2.0-fold) was observed. Of note, our sample set contained two biofilm positive cancer samples located on the left side of the colon which had the highest concentration of N1, N12-diacetylspermine out of all the cancer samples examined, suggesting that this metabolite is not simply upregulated based upon location of the cancer in the colon but rather specifically related to the presence of biofilm. The other metabolites that were significantly changed included undecanoic acid, dodecanoic acid, capryloylglycine, and isobutrylcarnitine.
Biofilm positive cancer tissues were compared to their paired normal tissues and revealed an upregulation of N1, N12-diacetylspermine 62.2-fold (p=1.6e−2), N1-acetylspermidine (p=1.6e−2, 6.5-fold), N1-acetylspermine (p=1.6e−2, 5.8-fold) and spermidine (p=2.7e−2, 2.3-fold) in the cancer tissues (Figure S1A). Biofilm negative paired colon cancer and normal tissues were also compared and showed that N1, N12-diacetylspermine, N1-acetylspermidine, N1-acetylspermine and spermidine were upregulated 7.2-fold (p=2.0−3), 3.0-fold (p=2.9e−3), 3.1-fold (p=1.2e−3) and 1.4-fold (p=1.3e−2), respectively, in cancer tissues (Figure S1B). These results demonstrate upregulation of acetylated polyamine production within colon cancer tissues that is further enhanced in biofilm positive colon cancer tissues, most markedly for N1, N12-diacetylspermine.
Targeted validation confirms polyamine metabolite changes
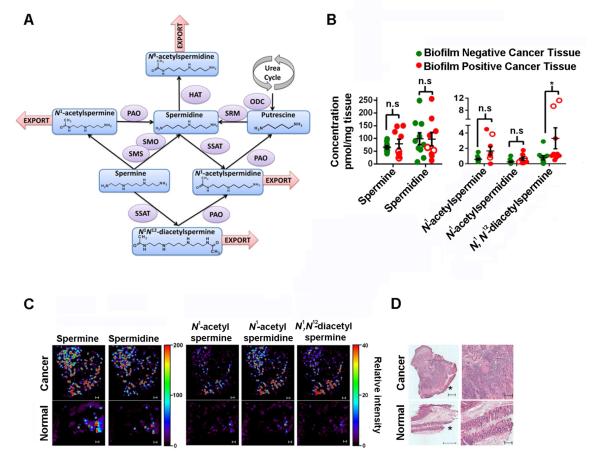
The polyamine metabolites were analyzed by QqQ-MS selected reaction monitoring (Figure 2A) which confirmed that these metabolites were significantly upregulated in cancer tissues, regardless of biofilm status, compared to their paired normal colon tissues (Figures S2A and S2B). Comparison of biofilm negative to biofilm positive cancer tissues, revealed significant upregulation of N1, N12-diacetylspermine only in the biofilm positive tissues (p=1.8e−2) (Figure 2B). Importantly, the normal tissues which were concordantly biofilm negative or positive with their paired cancer tissues revealed an upregulation of N1, N12-diacetylspermine and N1-acetylspermine (p=2.3e−2 and p=4.1e−2 respectively) in the biofilm positive normal colon tissues compared to biofilm negative normal colon tissues obtained from colon cancer hosts (Figure S2C).
Figure 2. Biofilm effects on metabolites in colon tissues.
(A) Scheme of polyamine metabolism. Polyamine oxidase (PAO), spermidine/spermine N1-acetyltransferase (SSAT), ornithine decarboxylase (ODC), histone acetyltransferase (HAT), spermidine synthase (SRM), spermine oxidase (SMO), spermine synthase (SMS). (B) Targeted metabolomics, concentrations of metabolites in cancers with (n=9) or without (n=10) biofilms (two-tailed Mann-Whitney test). Empty symbols indicate left-sided biofilm positive samples. Error bars are SEM, *p<0.05, n.s = not significant. (C) Nanostructure-initiator mass spectrometry imaging on biofilm positive normal and cancer tissue. Scale =100 μm. (D) Hematoxylin and eosin staining, *mucosal edge. Scale = 500 μm left, 200 μm right column. See also Figure S2.
In situ imaging reveals metabolite spatial specificity
To further validate the LC/MS experiments and explore spatial specificity within and between the normal and cancer tissues we used NIMS imaging (Northen et al., 2007). Figures 2C and 2D correlate normal and cancer tissues with polyamine metabolite levels providing evidence that polyamine metabolite concentrations (relative intensity) are higher in the cancer than in normal tissue in situ. The enhanced detection of the acetylated metabolites at the mucosal edge of the cancer tissue further suggested that the microbial biofilm could be contributing to the signal detected by NIMS.
N1, N12-diacetylspermine is potentially acetylated by bacterial enzymes and is an end-product of polyamine metabolism
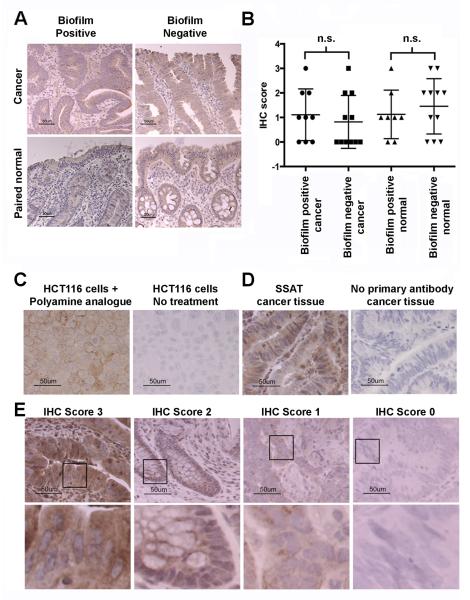
Increased acetylated polyamine metabolite synthesis may originate from the host tissue and indicate cellular proliferation; Ki67 staining showed a significant increase in crypt epithelial cell proliferation in normal tissues that were biofilm positive compared to biofilm negative (Dejea et al., 2014). Increases in acetylated polyamine metabolites could also arise from increased spermidine/spermine N1-acetyltransferase (SSAT) expression; an increase in SSAT could occur as a stress response to bacterial infection and biofilm formation (Gerner et al., 1993). In the human host, SSAT is required for acetylation of spermine to generate N1, N12-diacetylspermine; thus, mucosal SSAT expression was examined by immunohistochemical (IHC) staining and quantification (Figure 3). No significant difference was detected in epithelial cell SSAT between tissues with and without a biofilm (Figure 3B), indicating that the upregulation in biofilm-covered tissues is not due to increased mucosal SSAT acetylation of polyamines by the host. Indeed as polyamine metabolism is universal among bacterial species and essential for bacterial survival, alternative bacterial SSAT acetyltransferases may contribute (e.g., Bacillus subtilis (Woolridge et al., 1999) and we hypothesize polyamine metabolites may be produced by cooperatively within biofilm positive bacterial communities.
Figure 3. Immunohistochemical analysis (IHC) of spermidine/spermine N1-acetyltransferase (SSAT).
(A) SSAT IHC of cancer and paired normal tissues from patients with and without a biofilm (scale bar 50 μm). (B) Scoring of epithelial SSAT IHC in cancers and paired normal tissues with (n=9) and without (n=11) a biofilm. (C) HCT116 cells stimulated with N1,N11-bis(ethyl) norspermine with positive SSAT staining compared with unstimulated HCT116 cells lacking SSAT. (D) SSAT staining of cancer tissue with and without primary antibody. IHC 0-3 scoring system (E) with representative cytoplasmic staining intensities. Selected inserts are displayed in the bottom panel. All images were captured at 400x (scale bars 50 μm).
To further ascertain the biological effects of N1, N12-diacetylspermine, colon cancer cell lines (HT-29) were dosed with [U-14N]- and [U-15N]-N1, N12-diacetylspermine, to assess its metabolic fate by global isotope metabolomics. We observed no reversible conversion of N1, N12-diacetylspermine to its precursors. Instead we observed the [U-14N]- N1, N12-diacetylspermine and [U-15N]- N1, N12-diacetylspermine ions still present and unconverted in the cells 24 h after dosing. This shows that the labeled metabolites entered the cells but were not further metabolized, and are most likely a metabolic end product.
Correlations between N1, N12-diacetylspermine and taxonomic composition were observed
The correlation between the taxonomic composition of biofilm positive samples and acetylated polyamines was also investigated. A positive association with some Clostridia groups was seen, Sporobacter (ρ = 0.754, p=0.0018), Peptostreptococcaceae (ρ = 0.691, p=0.0062) and Veillonellaceae (ρ = 0.668, p=0.0091), indicating a possible contribution to N1, N12-diacetylspermine production in biofilm positive tumors. A negative association was seen with the order Bacteroidales from the Bacteroidetes class (ρ = 0.733, p=0.0028).
Antibiotic treatment removed biofilms and decreased N1, N12-diacetylspermine levels
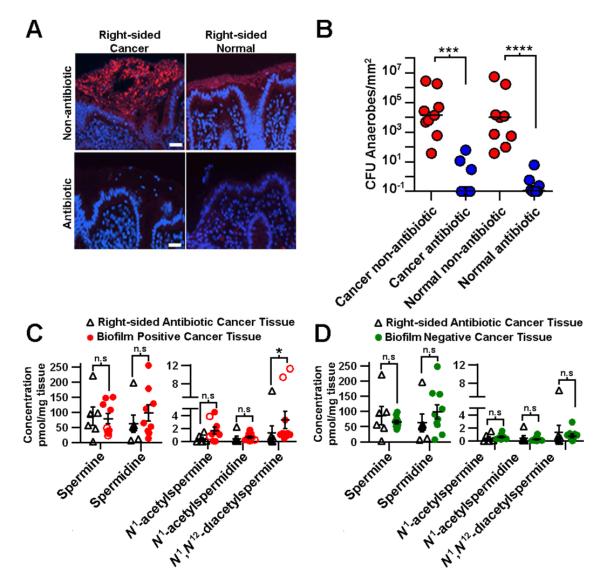
To further delineate the microbial versus host source of the upregulated polyamine metabolites identified, we collected tissues from nine colon cancer patients treated with oral antibiotics 24 hours prior to surgery. FISH analyses of both right and left colon tissues from these antibiotic-treated patients revealed no biofilms (Figure 4A), and microbial culture revealed little to no anaerobic or aerobic microbial growth on nutrient rich agar, suggesting that oral antibiotic treatment is effective at lowering the cultivatable microbial load (Figure 4B). In the context of the finding that essentially all right-sided cancers display biofilms while 88% of left-sided cancers are biofilm negative, the right and left cancer samples were compared to each other, revealing no significantly increased metabolites (Figure S3A). However, comparison of all antibiotic-treated cancers to their paired normal tissue by targeted metabolomics revealed an increase in N1-acetylspermine (p=1.6e−2), N1-acetylspermidine (p=1.6e−2) and N1, N12-diacetylspermine (p=1.6e−2) in the cancer tissue (Figure S3B). A further comparison of antibiotic-treated right-sided cancers to all biofilm positive cancers (from patients not receiving oral antibiotics prior to surgery) revealed significantly less N1, N12-diacetylspermine (p=3.3e−2) in the antibiotic-treated right-sided cancers (Figure 4C). Moreover, antibiotic-treated right-sided cancers and biofilm negative cancer tissues from patients not treated with oral antibiotics prior to surgery (Figure 4D) were metabolically similar. Collectively the data are consistent with the interpretation that both host cells and microbial biofilms contribute to upregulation of polyamine metabolites in colon cancer.
Figure 4. Antibiotic-treated cancers.
(A) Fluorescent in situ hybridization of all bacteria (red). Right-sided normal and paired cancer tissue from non-antibiotic and antibiotic-treated patients, DAPI counterstain (scale 10 μm). (B) Microbial culture data from antibiotic-treated (n=8) (blue) and non-antibiotic-treated patients (n=8) (red). Absolute concentrations of metabolites in (C) right-sided cancers from antibiotic-treated patients (n=6) and cancers with biofilms (n=9) from non-antibiotic-treated patients. (D) Right-sided cancers from antibiotic-treated patients (n=6) and cancers without biofilms (n=10) from non-antibiotic-treated patients. Statistics; two-tailed Mann-Whitney test, error bars are SEM, *p<0.05, ***p<0.001, **p<0.0001, n.s = not significant. See also Figure S3.
Assessment of colonoscopy control tissues revealed no acetylated polyamines
To test the specificity of the polyamine metabolite changes to the cancer host and the acetylated metabolites to biofilm positive tissues, we examined biopsies from ascending (right) and descending (left) colon undergoing routine screening colonoscopy. No acetylated polyamines were observed by targeted metabolomics in the left- and right-sided normal colonoscopy biopsies from healthy individuals (Figure S3C). A comparison of the normal colon biopsies to biofilm positive normal tissues, revealed upregulation of spermine and N1-acetylspermidine in cancer host tissues (p=5.0e−2, p=7.0e−3) (Figure S3D). Spermine was also increased in the biofilm-negative normal tissues from cancer hosts compared to the normal colonoscopy biopsies from healthy individuals (p=2.0e−2) (Figure S3E). These results are consistent with cellular metabolic changes occurring in histologically normal colon tissues of the cancer-bearing host due to far-reaching field effects covering a large proportion of the colon (Dakubo et al., 2007).
A mouse model of bacterial-mediated colon tumorigenesis showed no correlation between polyamine metabolites and tissues
In a relevant mouse model of bacterial-mediated colon tumorigenesis and polyamine metabolism, colitis and distal colon tumors that are biofilm negative were induced after enterotoxigenic Bacteroides fragilis (ETBF) colonization (Goodwin et al., 2011). We examined normal and tumor tissues from these mice through targeted metabolomics and did not see a correlation between polyamine levels and tissues, supporting the observation that polyamine levels are not perturbed in biofilm negative tissues.
DISCUSSION
The observation in this study is a direct correlation between biofilm formation on colon cancers and the upregulation of N1, N12-diacetylspermine; a polyamine metabolite that may affect the growth of both cancer and its associated biofilm. Importantly, although biofilms are nearly universal in association with right colon cancers, the measurement of upregulated acetylated polyamine metabolites on left biofilm positive cancer and paired normal tissues highlights that biofilm status, not merely the colon region, drives the changes identified in polyamine metabolism.
It is well known that the microbiota and human tissues exhibit a symbiotic relationship, and polyamines and their metabolites are essential for both (Ridaura et al., 2013). Increased polyamine concentrations are associated with eukaryotic proliferation (Gerner and Meyskens, 2004), and microbiota require polyamines for growth, cell-wall synthesis, and biofilm formation (Patel et al., 2006; Shah and Swiatlo, 2008). Biofilm formation, even in the normal colon tissue, was associated with increased colonic epithelial cell proliferation (Dejea et al., 2014), and enhanced polyamine metabolism. Thus, our data indicate biofilms increase polyamine metabolite concentrations in both normal and cancer tissues. The increase of carnitines and fatty acid metabolites in the colon cancer tissues suggests increased inflammation. Indeed an increase in both interleukin 6 (IL-6) and signal transducer and activator of transcription 3 (Stat3) activation were seen in biofilm positive compared to biofilm negative normal tissues from the cancer host (Dejea et al., 2014).
Although the molecular details remain to be identified, we propose a model in which host and bacterial polyamine metabolites act synergistically to promote biofilm formation and cellular proliferation, creating conditions conducive to oncogenic transformation in colonic epithelial cells. Consistent with this hypothesis, studies have shown that ornithine decarboxylase (ODC) and SSAT mRNA expression are affected by microbiota in human cancer cell lines (Linsalata et al., 2010). Helicobacter pylori, for example, can upregulate c-MYC, activating ODC (Bussiere et al., 2005). However, here, SSAT was not increased in host normal or cancer mucosa when biofilms were present, indicating that N1, N12-diacetylspermine is produced in biofilm positive tissues through bacterial acetylation. Therefore, changes in host cell metabolism may provide polyamines to stimulate biofilm formation in colon mucosa. Indeed bacterial transporters for uptake of extracellular polyamines exist (Patel et al., 2006). Collectively, the upregulation of polyamine metabolism can enhance cancer growth, invasion and metastasis (Soda, 2011). Although ideal for further mechanistic studies, a murine model of biofilm positive proximal colon tumors is not available and unlikely to emerge given the reported differences in mucus: bacterial interactions between murine and human hosts(Johansson and Hansson, 2011; Swidsinski et al., 2009).
Treatment of colon cancer models and clinical trials with polyamine-metabolism inhibitors have resulted in ambiguous findings (Babbar and Gerner, 2011), however targeting both polyamine production and biofilm interactions could prove to be a more successful strategy.
EXPERIMENTAL PROCEDURES
Sample collection
Colon cancers and paired histologically normal tissues were collected from patients undergoing surgery at JHU Hospital and Karolinska University Hospital, see Extended Experimental Procedures.
Fluorescent in situ hybridization (FISH) analysis
FISH analysis was carried out as previously described (Dejea et al., 2014) and is provided in Extended Experimental Procedures.
Microbial Culture
Anaerobic tissue specimens collected in specialized transport media (Anaerobe Systems) were washed twice with 0.016% DTT in saline prior to hand homogenization in saline under anaerobic conditions. Tissue homogenate was diluted (100-106) and plated on pre-reduced non-selective Brucella blood agar (Bru) plates. Plates were stored under anaerobic conditions at 37°C until colony forming unit counts could be obtained (24-72 hours).
Untargeted metabolomics
Samples were analyzed by RPLC and HILIC ESI-QTOFMS as previously described (Ivanisevic et al., 2013). The full dataset is available as a public share on XCMS Online. See Extended Experimental Procedures.
Targeted metabolomics of polyamines
A Scherzo SM-C18 column (Imtakt, Philadelphia, PA) effectively retained and separated the polyamines and polyamine metabolites. Samples were analyzed using an Agilent Technologies series 1200 HPLC connected to an Agilent Technologies 6410 QqQ-MS as described in Extended Experimental Procedures.
NIMS analysis
NIMS substrates were prepared as previously described (Woo et al., 2008) and are detailed in the Extended Experimental Procedures. Hematoxylin and eosin and SSAT immunohistochemical staining. Standard protocols were used, see Extended Experimental Procedures.
Global isotope metabolomics
HT-29 cell lines were dosed with 14N1, 14N12-diacetylspermine or 15N1, 15N12-diacetylspermine for 24 h and extracted in organic solvent for HPLC-ESI-QTOFMS as for the untargeted metabolomics method described above. See Extended Experimental Procedures.
Supplementary Material
Highlights.
Colonic mucosal biofilms alter the cancer metabolome
N1, N12-diacetylspermine was significantly upregulated in tissues with biofilms
Biofilms create conditions conducive to oncogenic transformation in colon cells
Global isotope metabolomics reveals the metabolite fate of N1, N12-diacetylspermine
ACKNOWLEDGMENTS
We thank Katharine Romans, Bert Vogelstein, Kenneth W. Kinzler for providing samples for these studies and Ruchi Badani and Annemarie Boleij for experimental assistance. We would also like to thank Samejima Keijiro from Tokyo Metropolitan Institute of Medical Science for providing 14N and 15N- N1,N12-diacetylspermine.This work was supported by the California Institute of Regenerative Medicine no. TR1-01219; the US National Institutes of Health nos. R01 CA170737, R24 EY017540, P30 MH062261, RC1 HL101034, P01 DA026146, R01 CA151393, R21 CA170492, K087856, P30 DK089502, P30 CA006973, R01 CA051085, R01 CA098454 and T32AI007417; 300-2344 (Alexander and Margaret Stewart Trust, JHU School of Medicine); and U.S. Dept. Energy nos. FG0207ER64325 and DE-AC0205CH11231.
Footnotes
AUTHOR CONTRIBUTIONS
G.S., C.L.S., D.M.P., B.F., R.A.C., G.J.P., supervised the work. C.H.J., C.M.D., D.E., L.T.H., A.F.S., K.C., E.C.W., E.M.H., W.U., L.G., C.L.S., R.A.C., J.W., performed the experiments and data analysis. C.H.J., C.L.S., C.M.D., wrote the manuscript. All authors read and revised the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Babbar N, Gerner EW. Targeting polyamines and inflammation for cancer prevention. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2011;188:49–64. doi: 10.1007/978-3-642-10858-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedix F, Kube R, Meyer F, Schmidt U, Gastinger I, Lippert H. Comparison of 17,641 patients with right- and left-sided colon cancer: differences in epidemiology, perioperative course, histology, and survival. Diseases of the colon and rectum. 2010;53:57–64. doi: 10.1007/DCR.0b013e3181c703a4. [DOI] [PubMed] [Google Scholar]
- Botteri E, Iodice S, Bagnardi V, Raimondi S, Lowenfels AB, Maisonneuve P. Smoking and Colorectal Cancer A Meta-analysis. Jama-J Am Med Assoc. 2008;300:2765–2778. doi: 10.1001/jama.2008.839. [DOI] [PubMed] [Google Scholar]
- Bussiere FI, Chaturvedi R, Cheng Y, Gobert AP, Asim M, Blumberg DR, Xu H, Kim PY, Hacker A, Casero RA, Jr., et al. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. The Journal of biological chemistry. 2005;280:2409–2412. doi: 10.1074/jbc.C400498200. [DOI] [PubMed] [Google Scholar]
- Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer cell international. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. PNAS. 2014 doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay LJ, Mitrou PN, Keen J, Bowman R, Naguib A, Cooke J, Kuhnle GG, Burns PA, Luben R, Lentjes M, et al. Dietary, lifestyle and clinicopathological factors associated with APC mutations and promoter methylation in colorectal cancers from the EPIC-Norfolk study. J Pathol. 2012;228:405–415. doi: 10.1002/path.4085. [DOI] [PubMed] [Google Scholar]
- Gerner EW, Kurtts TA, Fuller DJ, Casero RA., Jr. Stress induction of the spermidine/spermine N1-acetyltransferase by a post-transcriptional mechanism in mammalian cells. The Biochemical journal. 1993;294(Pt 2):491–495. doi: 10.1042/bj2940491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr. Polyamines and cancer: old molecules, new understanding. Nature reviews Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Dominguez R, Garcia A, Garcia-Barrera T, Barbas C, Gomez-Ariza JL. Metabolomic profiling of serum in the progression of Alzheimer’s disease by capillary electrophoresis-mass spectrometry. Electrophoresis. 2014;35:3321–3330. doi: 10.1002/elps.201400196. [DOI] [PubMed] [Google Scholar]
- Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisevic J, Zhu ZJ, Plate L, Tautenhahn R, Chen S, O’Brien PJ, Johnson CH, Marletta MA, Patti GJ, Siuzdak G. Toward ’omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Analytical chemistry. 2013;85:6876–6884. doi: 10.1021/ac401140h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Hansson GC. Keeping Bacteria at a Distance. Science. 2011;334:182–183. doi: 10.1126/science.1213909. [DOI] [PubMed] [Google Scholar]
- Linsalata M, Cavallini A, Messa C, Orlando A, Refolo MG, Russo F. Lactobacillus rhamnosus GG influences polyamine metabolism in HGC-27 gastric cancer cell line: a strategy toward nutritional approach to chemoprevention of gastric cance. Current pharmaceutical design. 2010;16:847–853. doi: 10.2174/138161210790883598. [DOI] [PubMed] [Google Scholar]
- Meguid RA, Slidell MB, Wolfgang CL, Chang DC, Ahuja N. Is there a difference in survival between right- versus left-sided colon cancers? Ann Surg Oncol. 2008;15:2388–2394. doi: 10.1245/s10434-008-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskal A, Norat T, Ferrari P, Riboli E. Alcohol intake and colorectal cancer risk: A dose-response meta-analysis of published cohort studies. Int J Cancer. 2007;120:664–671. doi: 10.1002/ijc.22299. [DOI] [PubMed] [Google Scholar]
- Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. Polyamines are essential for the formation of plague biofilm. Journal of bacteriology. 2006;188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert HM, Gibson GR. Bacterial biofilms in the human gastrointestinal tract. Current issues in intestinal microbiology. 2002;3:23–27. [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampey BP, Freemerman AJ, Zhang J, Kuan PF, Galanko JA, O’Connell TM, Ilkayeva OR, Muehlbauer MJ, Stevens RD, Newgard CB, et al. Metabolomic Profiling Reveals Mitochondrial-Derived Lipid Biomarkers That Drive Obesity-Associated Inflammation. Plos One. 2012:7. doi: 10.1371/journal.pone.0038812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell host & microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 2008;68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- Soda K. The mechanisms by which polyamines accelerate tumor spread. J Exp Clin Canc Res. 2011:30. doi: 10.1186/1756-9966-30-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn’s disease and ulcerative colitis - an overview. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2009;60(Suppl 6):61–71. [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. Journal of clinical microbiology. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HK, Northen TR, Yanes O, Siuzdak G. Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis. Nature protocols. 2008;3:1341–1349. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- Woolridge DP, Martinez JD, Stringer DE, Gerner EW. Characterization of a novel spermidine/spermine acetyltransferase, BltD, from Bacillus subtilis. The Biochemical journal. 1999;340(Pt 3):753–758. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.