Abstract
HOX genes are highly expressed in many acute myeloid leukemia (AML) samples, but the patterns of expression and associated regulatory mechanisms are not clearly understood. We analyzed RNA sequencing data from 179 primary AML samples and normal hematopoietic cells to understand the range of expression patterns in normal versus leukemic cells. HOX expression in AML was restricted to specific genes in the HOXA or HOXB loci, and was highly correlated with recurrent cytogenetic abnormalities. However, the majority of samples expressed a canonical set of HOXA and HOXB genes that was nearly identical to the expression signature of normal hematopoietic stem/progenitor cells (HSPCs). Transcriptional profiles at the HOX loci were similar between normal cells and AML samples, and involved bidirectional transcription at the center of each gene cluster. Epigenetic analysis of a subset of AML samples also identified common regions of chromatin accessibility in AML samples and normal CD34+ cells that displayed differences in methylation depending on HOX expression patterns. These data provide an integrated epigenetic view of the HOX gene loci in primary AML samples, and suggest that HOX expression in most AML samples represents a normal stem cell program that is controlled by epigenetic mechanisms at specific regulatory elements.
Introduction
HOX gene expression is a common feature of acute myeloid leukemia (AML), and is thought to reflect “dysregulation” of HOX pathways that lead to abnormal self-renewal and the development of leukemia. Initial studies of HOX gene expression in human hematopoietic cells showed that expression is largely restricted to hematopoietic stem/progenitor cells (1–4), which are uniquely capable of long-term self-renewal. In addition, functional studies in mice demonstrated that expression of specific HOXA and HOXB genes can lead to expansion of long-term repopulating hematopoietic stem cells and a myeloproliferative phenotype (5–9). Mice lacking specific Hox genes also showed deficits in the repopulating ability of hematopoietic cells in competitive transplantation experiments (10–13), although these phenotypes have been variable across studies (14).
In AML patient samples, HOX gene expression is most closely associated with translocations involving MLL, which characteristically show high expression of HOXA genes (15). HOXA9 in particular has been shown to be a “target” of MLL fusion oncoproteins (16–18), and is required for the survival and proliferation of MLL-positive leukemic cells (19,20). Other AML mutations have also been associated with either the presence or absence of HOX gene expression. MLL partial tandem duplications (PTDs) and PICALM-MLLT10 gene fusions have been associated with high levels of HOXA gene expression (21–23), and NPMc mutations are associated with expression of both HOXA and HOXB cluster genes in human AML samples (24,25), and in mice expressing this mutation (26). In contrast, AMLs with the PML-RARA and RUNX1-RUNX1T1 gene fusions (27,28) and mutations in CEBPA (29) have been associated with low or absent HOX gene expression.
Although AML-associated HOX expression phenotypes are often described as “aberrant”, the specific expression patterns reported in the literature are variable and involve multiple genes from either the HOXA or HOXB gene cluster (or both) (30,31). Most studies have relied on targeted gene expression measurements of only selected HOX genes, or they have focused on AMLs with canonical somatic mutations and/or cytogenetic abnormalities. In addition, although some studies have shown that HOX genes are expressed in both AML samples and normal hematopoietic cells (25), the precise patterns of expression in normal versus malignant hematopoietic cells remains unclear. As a result, a comprehensive view of HOX gene expression patterns in AML samples—and their relationships to normal hematopoietic cells—has not yet been established. In this study, we carried out an integrated analysis of HOX gene expression using RNA-sequencing data from 179 primary AML samples that have been previously characterized by either whole-genome or whole-exome sequencing. We compared the HOX expression phenotypes in these AMLs to data from normal bone marrow cells to study the HOX regulatory programs in normal and malignant hematopoiesis. Finally, we performed high-resolution bisulfite sequencing and chromatin accessibility profiling of selected AML samples to identify changes in DNA methylation and chromatin structure at cis-acting regulatory elements within the HOX loci that may be involved in HOX gene regulation in AML. These analyses provide an integrated, unbiased view of HOX gene expression, and have identified potential regulatory elements that may be important for HOX gene expression in normal hematopoietic development, and in AML cells.
Materials and Methods
Clinical and genomic data from primary AML samples
All primary AML samples used in this study were from unfractionated bone marrow cells obtained at the time of diagnosis at the Washington University School of Medicine, using protocols approved by the Human Research Protection Office following informed consent in accordance with the Declaration of Helsinki. Clinical annotation, processing, and analysis of these samples has been described previously (32).
Normal hematopoietic cell populations
Hematopoietic cells from bone marrow aspirates were obtained from three volunteer donors using protocols approved by the Human Research Protection Office following informed consent. Aspirates were processed via ammonium-chloride-potassium red cell lysis, washed, and prepared for fluorescence-activated cell sorting (FACS). A portion of each sample was used to purify promyelocytes (CD14−, CD15+, CD16low/−) (33), monocytes (CD14+), neutrophils (CD14−, CD15+, CD16+) (33), T-cells (CD33−, CD3+) and B-cells (CD33−, CD19+), with the remaining cells enriched for CD34+ cells using bead-based enrichment (MACS human CD34 MicroBead kit, Miltenyi Biotec). CD34-enriched samples were then further purified for CD34+ cells via FACS. The following antibodies were used for FACS: CD34-PE (PE-pool, Beckman Coulter, PN IM1459U), CD14-APC (BD, M5E2), CD15-FITC (BD, clone HI98), CD16-PE (BD, 3G8), CD33-APC (eBioscience, clone WM-53), CD3-V450 (eBioscience, clone OKT3), and CD19-PE (BD, clone HIB19). All samples were cryopreserved in Trizol LS (Life Technologies) for subsequent RNA extraction.
RNA-sequencing, processing, and analysis
Poly-A RNA-sequencing of normal bone marrow cells was performed using established methods (32). Additional RNA-seq data from normal CD34+ cells were obtained from GSE48173. All RNA-seq data were processed from the raw sequencing reads using the tophat (version 2.0) and cufflinks (version 2.2) following the recommended protocols for unstranded libraries (34). FPKM gene expression values were generated simultaneously across all samples (with the “cuffnorm” command) using the Gencode version 18 annotation. Clustering analysis was performed in R using Ward’s method (Murtagh and Legendre 2013).
Nanostring nCounter gene expression analysis
Orthogonal measurements of HOX expression in AML samples and normal CD34+ cells were obtained using a custom Nanostring nCounter codeset targeting 54 genes, including all HOXA-HOXD genes and additional controls for expression normalization and quality control. 100 ng of total RNA was used for each assay, and all experiments were performed in duplicate. Expression values were obtained via normalization to the control genes using the NanoStringNorm package in R (35).
Microarray gene expression analysis
Microarray data from the Affymetrix U133+2 array platform were downloaded from the GEO repository, and included datasets from AML samples (GSE10358), normal bone hematopoietic cells (GSE12662), and purified hematopoietic progenitors (GSE24006). Processed data for GSE10358 were used directly from the GEO repository, but all other data were reprocessed from raw images. Signal intensities and gene expression values for these samples were processed together with the RMA processing algorithm using the oligo package in R (36). Clustering analysis was performed in R as above.
Bisulfite sequencing and analysis
Bisulfite sequencing was performed using either whole-genome bisulfite-converted sequencing libraries generated with the Epigenome library preparation kit, or with the Agilent SureSelect Methyl-Seq kit (Agilent, Santa Clara, CA). Indexed sequencing was performed on Illumina HiSeq 2000 instruments and reads were mapped with BSMap using default parameters (37). Methylation values for the HOX gene clusters were obtained using the Bis-SNP program with default parameters (38). Differential methylation analysis was performed on pooled methylation data using a chi-squared test of methylated vs. unmethylated counts for each AML type, and required a bonferroni-corrected p-value of 0.05 and minimum difference between any pooled dataset of 0.5 for significance. Smoothed methylation values were generated for visualization using the BSSeq R package (39).
Chromatin accessibility profiling (ATAC-seq)
Transposase-mediated chromatin accessibility profiling was performed using the Nextera library preparation kit as described in (40) using 50,000 viable cells per sample. Nextera libraries were size-fractionated into small (<300 bp) and large (300–800 bp) libraries and sequenced on separate lanes of Illumina 2500 instruments (two libraries per lane). Reads were mapped with the bwa mem program and resulting data were merged and filtered to retain unique, properly-paired reads. Peaks were identified using homer (41) and coverage profiles were generated using bamCoverage (42).
Data availability
RNA-seq expression data from AML samples are available from the TCGA portal (https://tcga-data.nci.nih.gov/tcga/). Raw RNA-seq data from normal bone marrow cells, as well as ATAC-seq and bisulfite sequencing data from AML samples, will be deposited in dbGaP (http://www.ncbi.nlm.nih.gov/gap). Processed ATAC-seq and methylation datasets from the HOX loci are available from the authors upon request.
Results
HOX gene expression patterns in AML samples are associated with recurrent AML mutations and a canonical pattern found in normal hematopoietic progenitors
To understand the range of HOX expression patterns in AML and their relationship to recurrent AML mutations and normal hematopoietic HOX expression, we studied poly-A selected RNA-sequencing data from 179 well-characterized AML samples (32) and 29 samples from normal bone marrow cells at different stages of development (from (43), and this study). We first compared gene expression levels defined by RNA-seq to microarray expression data, and also to data from a custom Nanostring assay targeting all HOX genes. All platforms demonstrated variable levels of expression of the HOXA and HOXB cluster genes, with HOXA3-HOXA10 and HOXB2-HOXB6 showing the greatest expression, and no detectable expression of HOXC and HOXD genes (Figures S1A–C). HOXA and HOXB gene expression was generally concordant across the expression platforms, although RNA-seq showed better agreement with orthogonal expression measurements from the Nanostring platform than array-based expression values, perhaps due to the design of the array probes that target the HOX genes (Figures S1D, S1E).
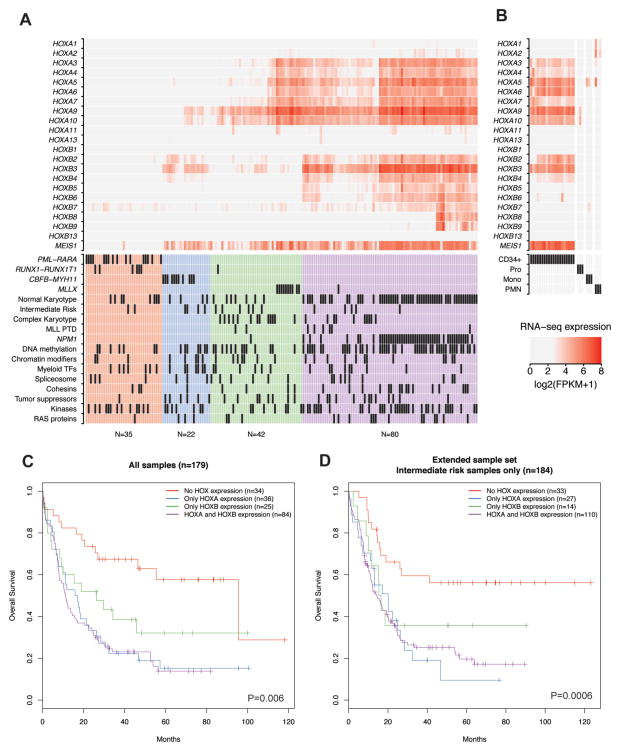
We next explored HOXA and HOXB expression patterns in the AML samples and their relationship to somatic mutations identified by whole genome and exome sequencing. Figure 1A shows unsupervised hierarchical clustering of RNA-seq gene expression for all HOXA and HOXB genes (and the HOX cofactor MEIS1) from all 179 AML samples, along with cytogenetic classification and mutational status of recurrently mutated genes organized into functional categories (see Table S1). This analysis resulted in distinct HOX expression groups that were characterized by the presence or absence of expression of genes in either the HOXA or HOXB gene clusters (or both), which were associated with recurrent chromosomal rearrangements and certain somatic mutations (Table 1). Although MEIS1 was frequently expressed in samples with HOX gene expression, the overall correlation between expression levels of MEIS1 and HOX genes was relatively low (Figure S1F). The first HOX expression group had essentially undetectable HOX expression at both gene clusters (Figure 1A, red; mean expression across both HOXA and HOXB genes: 0.25 FPKM; range: 0–6.8 FPKM), and was comprised of ~20% of the AML samples, including nearly all samples with the RUNX1-RUNX1T1 and PML-RARA gene fusions (Figure S2A). A second group had primarily HOXA expression (Figure 1A, green), and included all of the samples with MLL translocations (Figure S2A); this is consistent with previous studies of HOX expression in AML, and the established role of HOXA genes in MLL leukemias. AMLs with other mutations in the MLL gene, including partial tandem duplications (PTDs, n=8), were also associated with HOX expression, although these cases tended to show expression of both HOXA and HOXB genes (Figure S2B). AMLs with inv(16), resulting in the CBFB-MYH11 gene fusion, defined a third group with low to moderate expression of HOXB genes (Figure 1A, blue), and little or no HOXA gene expression (Figure S2A). The last group demonstrated moderate to high expression of a canonical subset of genes in both the HOXA and HOXB gene clusters (Figure 1A, purple), including the HOXA3-HOXA10 and HOXB2-HOXB6 genes, although in some cases the 5′ HOXB genes HOXB8 and HOXB9 were also expressed. The samples in this group were comprised of primarily normal karyotype AMLs, and included nearly all of the samples with the NPMc mutation (Figure S2C), although AMLs with wild type NPM1 were also found in this set. Statistical analysis of mutations and mutation categories with respect to these HOX expression phenotypes demonstrated that NPMc mutations, mutations in genes involved in DNA methylation (specifically DNMT3A mutations), and recurrent gene fusions were significantly associated with one or another of the HOX expression patterns after correction for multiple comparisons (Table 1 and Table S2).
Figure 1.
A) Patterns of HOX gene expression in primary AML samples. HOXA and HOXB gene expression values from RNA-seq data (log2 (FPKM+1)) are shown for 179 primary AML samples following unsupervised hierarchical clustering of the samples based solely on the expression of the genes shown, which identified four patterns defined by the presence or absence of HOXA and/or HOXB genes. The mutation status and cytogenetic category for each AML is shown in the table below the heatmap (32). B) HOXA and HOXB gene expression in normal hematopoietic cells at different developmental stages from RNA-seq data, including CD34+ progenitors (n=20), promyelocytes (Pro, n=3), monocytes (Mono, n=3), and neutrophils (PMN, n=3). C) Kaplan-Meier analysis of overall patient survival stratified by the HOX expression groups identified in panel A (n=179, P=0.006 across all groups; performed using Cox proportional hazard regression). D) Survival analysis of 184 AML patients with intermediate risk cytogenetic status based on HOX expression phenotype from microarray expression (data obtained from (48); see Figure S5; P=0.0006, Cox proportional hazard regression).
Table 1.
Association of clinical variables and somatic mutation categories* with HOX expression patterns in primary AML samples.
| Variable | No HOX expression | HOXA expression | HOXB expression | HOXA and HOXB expression | Adjusted P-value# |
|---|---|---|---|---|---|
| (n=35) | (n=42) | (n=22) | (n=80) | ||
| White blood cell count; x 109/l, mean (range) | 29 (0.4–224) | 23 (0.8–115) | 37 (1–172) | 46 (0.6–297) | 0.04 |
| Bone marrow blasts; %, mean (range) | 69 (34–100) | 70 (30–100) | 59 (33–90) | 71 (32–100) | 0.05 |
| Monocytic differentiation; n, (%) | 1 (1.8) | 16 (28.6) | 11 (19.6) | 28 (50.0) | 0.03 |
| CD34 marker positive; n, (%) | 20 (57.1) | 33 (78.6) | 20 (9.1) | 39 (48.8) | 0.01 |
| Cytogenetic categories; n (%) | 1.00E-10 | ||||
| Normal karyotype | 8 (10.6) | 7 (9.3) | 4 (5.3) | 56 (74.7) | |
| Complex cytogenetics | 0 | 13 (56.5) | 0 | 10 (43.4) | |
| PML-RARA | 16 (100) | 0 | 0 | 0 | |
| RUNX1-RUNX1T1 | 6 (85.7) | 1 (14.3) | 0 | 0 | |
| CBFB-MYH11 | 0 | 0 | 11 (100) | 0 | |
| MLL-X fusions | 0 | 10 (90.9) | 1 (9.1) | 0 | |
| Somatic mutation categories; n, (%) | |||||
| NPMc | 0 | 1 (2.1) | 0 | 47 (97.9) | 0.005 |
| DNA methylation | 7 (9.6) | 13 (17.8) | 7 (9.6) | 46 (63.0) | 0.005 |
| Tumor suppressors | 1 (3.6) | 8 (28.6) | 6 (21.4) | 13 (46.4) | N.S. |
| Kinases | 13 (20.6) | 8 (12.7) | 7 (11.1) | 35 (55.6) | N.S. |
| Myeloid transcription factors | 7 (24.1) | 6 (20.7) | 7 (24.1) | 9 (31.0) | N.S. |
| Chromatin modifiers | 3 (11.1) | 7 (25.9) | 5 (18.5) | 12 (44.4) | N.S. |
| Cohesins | 3 (12.5) | 4 (16.7) | 1 (4.2) | 16 (66.7) | N.S. |
| Spliceosome | 4 (18.2) | 6 (27.3) | 3 (13.6) | 9 (40.9) | N.S. |
| RAS pathway | 1 (5) | 3 (15) | 3 (15) | 13 (65.0) | N.S. |
See Table S1 for the specific genes included in each mutation category.
Significance testing performed via ANOVA or chi-squared test, followed by bonferroni correction for multiple comparisons.
N.S = not significant.
We next compared HOXA and HOXB expression in AML to RNA-seq-based HOX expression from normal hematopoietic progenitors (CD34+; n=20) and myeloid cells at various stages of development, including promyelocytes (n=3), monocytes (n=3), and neutrophils (n=3). As expected, HOX genes were expressed in CD34+ cells (Figure 1B). Expression was substantially lower in promyelocytes, and nearly absent in more mature myeloid cells (see Figure S3A and S3B), consistent with previous studies of developmental HOX expression during hematopoiesis (1–3,44). Expression in CD34+ cells was also restricted to the HOXA and HOXB genes, with no expression of HOXC and HOXD genes (Figure S4A). Remarkably, this pattern was highly similar to the common HOXA/HOXB signature observed in the majority of the normal karyotype AML samples. The presence of this normal stem cell signature in AML was supported by array-based HOX expression values, and analysis of additional microarray expression data from progenitor cell populations within the CD34+ compartment (45) suggested that this HOX expression pattern is enriched in CD34+/CD38− hematopoietic stem/progenitors and multipotent progenitors (Figure S4B). This observation suggests that HOX gene expression in many AML samples may not be dysregulated per se, but rather reflects a normal stem/progenitor cell pattern that is “captured” in AML cells.
HOX gene expression patterns correlate with patient outcomes
HOX gene expression patterns have been shown to be associated with patient outcomes in some studies (27,46,47). We therefore compared patient survival across the HOX expression phenotypes identified by our clustering analysis. Indeed, overall patient survival was significantly longer in the groups with no HOX expression or HOXB expression alone, compared to those with HOXA or with the HOXA/HOXB signatures (Figure 1C). Since these expression patterns were closely correlated with recurrent cytogenetic abnormalities that have well-established relationships to patient outcome, we conducted a separate survival analysis on 184 intermediate risk AML samples from this and a previously published study (48). HOX expression phenotypes were obtained from unsupervised clustering of microarray-based HOX gene expression values from all samples included in this study (Figure S5), and the overall patient survival was compared between samples with normal karyotype or intermediate risk cytogenetic abnormalities and different HOX expression groups. Importantly, this analysis also showed that HOX expression phenotypes were significantly associated with overall survival, with no HOX expression having longer survival compared to samples that express HOXA alone, or both HOXA and HOXB genes (Figure 1D). Since the AML samples in this set did not have favorable risk cytogenetic findings, the lack of HOX expression in AML appears to predict good survival regardless of the cytogenetic risk status.
Transcriptional activity at the HOXA and HOXB gene clusters occurs in a canonical fashion that is shared between AML samples and normal hematopoietic progenitors
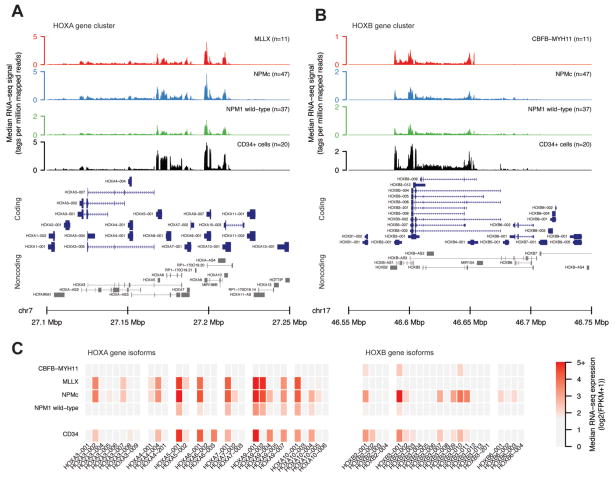
The coordinated expression of specific “sets” of genes in the HOXA or HOXB clusters in both AML and normal hematopoietic progenitors suggests that these genes may be coordinately regulated in both normal and malignant hematopoietic cells. Indeed, we observed expression of the same HOXA and HOXB genes across all AMLs in which a specific HOX cluster was expressed (i.e. the same HOXA genes tend to be highly expressed in AMLs with MLL translocations vs. those with the HSPC-like signature; Figure 1A), as well as normal progenitors (although the expression levels of some genes were significantly different among AML samples with different HOX expression patterns, and between AML samples and normal CD34+ cells, see Figures S3A and S3B). To determine whether these characteristic expression patterns extended beyond summarized RNA-seq expression values (i.e., gene-based FPKM values), we compared the raw RNA-seq signals and isoform usage across the HOXA and HOXB loci in normal CD34+ cells, and in AMLs with different HOX expression phenotypes and mutation profiles. Figure 2A shows the median RNA-seq signal (normalized read depth) at the HOXA locus for normal CD34+ cells, AMLs with MLL translocations, and AMLs with the HOXA/HOXB HSPC-like expression signature (with and without NPMc mutations). Samples in each group displayed a similar HOXA coverage profile, with the highest transcriptional activity localized to the central region within the locus (HOXA5-HOXA10). Likewise, the RNA-seq coverage signal at the HOXB locus was greatest within a region between HOXB2-HOXB6, and was similar between CD34+ cells and AMLs with the HSPC-like expression pattern, regardless of the presence of the NPMc mutation (Figure 2B). Inv(16)/CBFB-MYH11 AMLs, which showed low level HOXB expression, demonstrated a highly similar transcriptional profile, but with lower coverage depth. Transcriptional activity at both loci was detected on both strands, with expression levels of HOX antisense transcripts that were highly correlated with coding gene expression at both HOXA and HOXB gene clusters (Figure S6A), and which demonstrated similar mutation-specific expression patterns (Figure S6B). Analysis of transcript isoform usage at the HOXA and HOXB loci also showed that the dominant isoforms of the highly expressed HOXA and HOXB genes were shared across all AML types and normal CD34+ cells (Figure 2C). These findings imply that transcriptional activity at the HOXA and HOXB loci occurs in a canonical, bidirectional fashion in normal hematopoietic progenitors, and in a subset of AML samples.
Figure 2.
Transcriptional profiles of the HOXA and HOXB gene clusters from different AML subtypes. A) and B) show the median normalized RNA-seq read depth from the HOXA and HOXB loci, respectively, from AMLs with MLL translocations (MLLX, n=11), Normal Karyotype and NPMc mutations (n=47), Normal Karyotype without NPMc mutations (n=33), and normal CD34+ cell (n=20). Protein-coding genes (in blue) and noncoding transcripts (in gray) are shown below each plot. Note difference in scale of the Y axis across the sample types, indicating different expression levels despite the similarity in transcriptional patterns. C) Median expression (in FPKM) of transcript isoforms for expressed genes in the HOXA and HOXB clusters for AMLs in the indicated mutation or cytogenetic category and CD34+ cells.
Epigenetic profiling identifies potential regulatory regions in the HOX gene loci of AML cells and normal hematopoietic progenitors
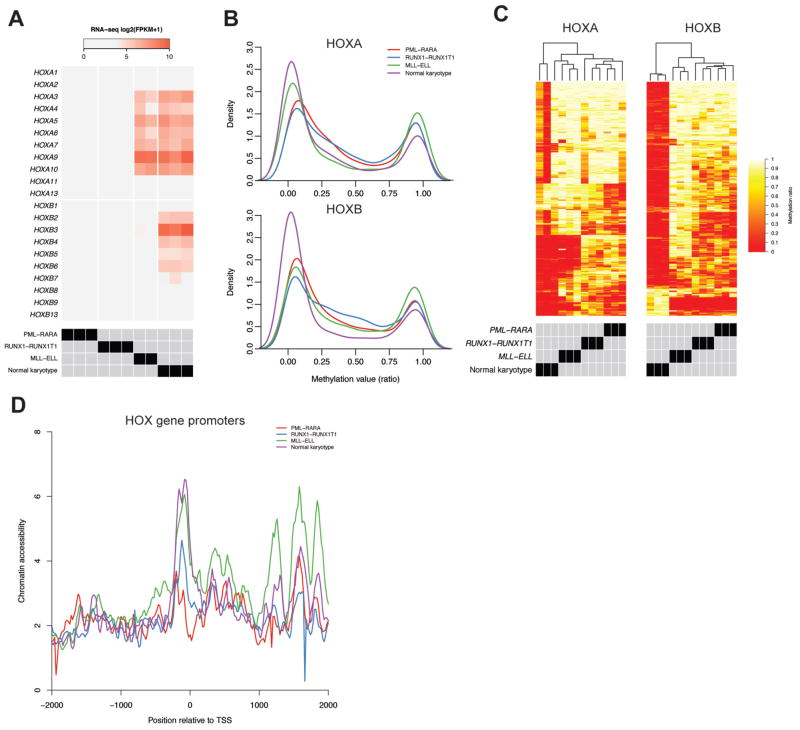
The close association between HOX expression phenotypes and recurrent PML-RARA, RUNX1-RUNX1T1 and MLL gene fusions in AML suggests that they may have direct effects on gene regulation at the HOX loci. These fusion products are thought to function via direct interactions with target gene loci, and/or recruitment of transcription factors and/or epigenetic regulators (17,49,50). We therefore performed whole-genome and targeted bisulfite sequencing and transposase-mediated chromatin accessibility profiling (ATAC-seq) on AML samples with these mutations to identify potential regulatory elements within or near the HOX gene loci that may be the targets for such interactions. We selected four sets of AML samples with characteristic HOX expression patterns, including MLL translocations (t(11;19)/MLL-ELL; n=2), normal karyotype AMLs with NPMc mutations that exhibit the HSPC-like HOX pattern (n=3), and AMLs with RUNX1-RUNX1T1 (n=3) or PML-RARA (n=3) gene fusions (Figure 3A); similar data were also obtained for normal CD34+ cells from previously published datasets (51).
Figure 3.
Epigenetic analysis of the HOX loci in primary AML samples with characteristic HOX expression patterns. A) RNA-seq expression of the HOX genes from the samples used for epigenetic analysis, including PML-RARA, RUNX1-RUNX1T1, MLL-ELL, and normal karyotype AMLs with the HSPC-like HOX expression pattern and NPMc mutations (n=3 each). B) Methylation distributions from bisulfite sequencing of the HOX loci in primary AML samples, showing skewing towards less methylation in samples with HOXA expression (MLL-ELL and normal karyotype) and HOXB expression (normal karyotype only). C) Clustering of methylation values identified from differential methylation analysis of CpGs at the HOXA and HOXB locus between each AML set. D) Aggregate chromatin accessibility profiles at HOX gene promoters from each AML type. Each curve shows the mean normalized ATAC-seq signal across all HOXA and HOXB gene promoters from the indicated AML types (n=3 each), which demonstrates that AMLs with HOX expression (MLL-ELL and normal karyotype samples) have more open chromatin compared to those without HOX expression.
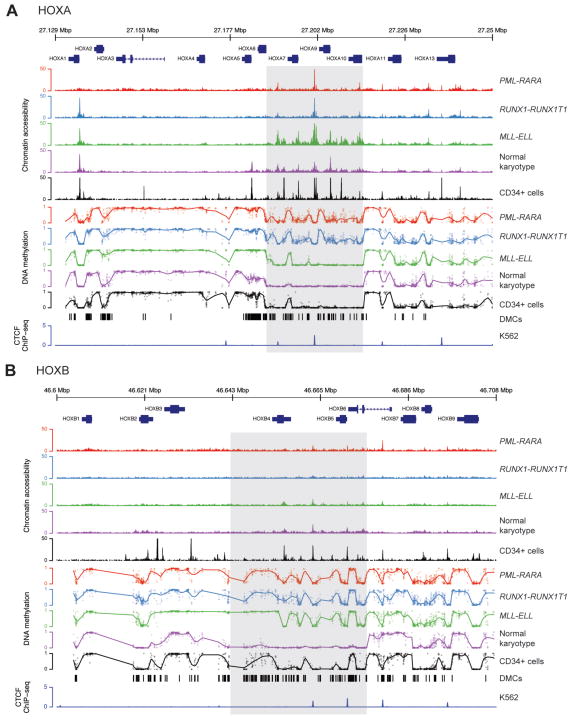
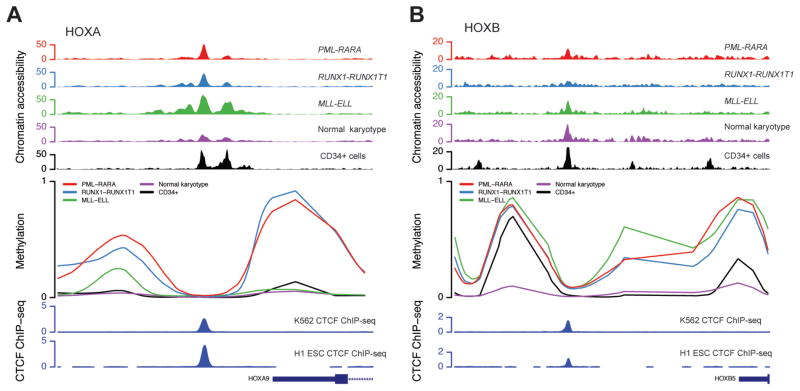
Bisulfite sequencing of the AML samples resulted in high-confidence methylation values for 2,893 and 2,419 CpG dinucleotides at the HOXA and HOXB loci, respectively, which were relatively hypomethylated in the samples with HOX expression (Figure 3B). Differential methylation analysis identified 427 (14.7%) and 458 (18.9%) differentially methylated CpGs among the four AML sets at the HOXA and HOXB loci, respectively, which clearly distinguished each group of AML samples upon hierarchical clustering (Figures 3C). Chromatin accessibility also correlated with HOX expression, and was increased in the MLL-ELL and normal karyotype samples at HOXA gene promoters (Figure 3D); genome-wide ATAC-seq signals were similar across all the samples (data not shown), suggesting that these findings were not due to differences in assay performance across this set of samples. Analysis of the methylation profiles and chromatin accessibility at the HOXA and HOXB loci demonstrated remarkable similarity across all of the samples, but also identified regions with differences that correlated with HOX expression. This was most apparent in a 38 kbp region within the HOXA locus, where the normal karyotype, HSPC-like, MLL-ELL AMLs, and normal CD34+ cells were largely unmethylated; they also contained regions of accessible chromatin at gene promoters and intergenic loci between the HOXA5 and HOXA10 genes (Figure 4A, shaded region). Similar findings were observed in a ~50 kbp region at the HOXB locus that was largely unmethylated, and that contained more accessible chromatin in both the normal karyotype AML cases with HSPC-like HOX expression, and CD34+ cells (Figure 4B, shaded region). We also observed similar methylation patterns at the same loci in array-based methylation data from an extended set of AML samples with these mutations (see Figures S7A and S7B), implying that these differences are not limited to this small set of samples. These data suggest that epigenetic regulatory activity is concentrated within distinct regions in both HOX loci.
Figure 4.
Integrated epigenomic profiles of the HOXA and HOXB gene clusters in primary AML samples and normal CD34+ cells. Panels A and B show DNA methylation and transposase-mediated chromatin accessibility (ATAC-seq) profiles from primary AML samples at the HOXA and HOXB loci, respectively. Methylation values reflect the coverage-weighted mean methylation ratio across all samples in each group. Chromatin accessibility values represent the median number of tags per 1 kbp per million mapped tags across each AML set (n=3 each). The bottom tracks show differentially methylated CpGs (DMCs) at each locus, and the ChIP-seq signal for CTCF from the K562 cell line from the ENCODE consortium (UT Austin) (52), respectively.
Although there were characteristic epigenetic differences at these loci that correlated with HOX expression, samples with the same expression patterns showed very similar methylation and chromatin accessibility profiles. For example, AMLs with PML-RARA and RUNX1-RUNX1T1 had the same patterns of hypermethylation. Further, regions with accessible chromatin in the MLL-ELL and normal karyotype AMLs were largely shared, or overlapped with DNAse hypersensitive sites found in normal CD34+ cells. Interestingly, several non-promoter-associated regions with accessible chromatin were shared across all AML samples (regardless of HOX expression) and normal CD34+ cells, suggesting that these may be common regulatory elements involved in the activation or repression of HOX gene expression in both normal and AML cells. Regions at both the HOXA and HOXB loci showed remarkable similarity in the ATAC-seq signals in all of the AML samples, but differed dramatically in their methylation states (Figure 5A, 5B). Further analysis of these regions revealed that they occurred near binding sites for the epigenetic regulator CTCF in a variety of cell types, including embryonic stem cells and the K562 cell line (52). These findings suggest that epigenetic regulation of HOX gene expression in both normal and malignant hematopoietic cells may be orchestrated by CTCF (and perhaps other factors) at defined regulatory elements within the HOX loci.
Figure 5.
Differential methylation at common chromatin-accessible regions within the HOXA (panel A) and HOXB (panel B) gene clusters. Each panel shows chromatin accessibility and methylation at chromatin-accessible sites present in all AML samples and normal CD34+ cells, with common chromatin accessibility patterns and hypermethylation in samples without expression of nearby HOX genes. These loci occur near previously identified CTCF binding events in H1 hESCs and the K562 erythroleukemia cell line from the ENCODE consortium (UT Austin), (52).
Discussion
Understanding the regulatory mechanisms controlling self-renewal pathways in hematopoietic cells could provide insights into the biology of normal hematopoietic stem cells, as well as the events that initiate and sustain leukemia. HOX transcription factors have been proposed to be master regulators of self-renewal in HSPCs, and are expressed in the majority of AML samples, which has lead to the hypothesis that dysregulation of these transcription factors promotes abnormal self-renewal in AML. Indeed, previous studies have described the remarkable variability in HOX expression in primary AML samples, as well as mutations and clinical outcomes that are associated with the presence or absence of HOX expression in AML (25,27,46).
In this study, we conducted an integrated epigenetic analysis of the HOX loci in primary AML samples to understand HOX expression patterns and epigenetic regulation in AML. We found that HOX expression phenotypes in AML fall into four broad categories, including little to no expression, expression of either the HOXA or HOXB cluster genes alone, or a canonical pattern with expression of genes from both clusters that was nearly identical to the expression signature in normal HSPCs. These patterns were tightly linked to AML-subtype-defining mutations and were associated with patient survival, which is consistent with previous studies associating HOX expression and stem cell signatures with patient outcomes (27,45,53). In addition, we found that the transcriptional profiles, isoform usage, and noncoding transcript expression were similar across all AML samples (regardless of mutation status or subtype) and normal CD34+ cells, suggesting that each locus is regulated in a canonical fashion as a single unit, and not at the level of individual genes. Epigenetic analysis also identified distinct regions in each gene cluster that display the hallmarks of regulatory elements, which appear to be coordinating HOX expression in both normal and leukemic hematopoietic cells.
Although HOX expression in AML is typically described as dysregulated, the similarity between normal and leukemic HOX expression patterns we observed suggests that most HOX expression in AML in fact reflects a normal stem cell state that is “captured” in the transformed cells. With this view, non-HSPC-like expression patterns (e.g., no expression, or the expression of genes from only one cluster) truly represent the dysregulated state in AML. The absence of HOX gene expression could potentially occur because of direct suppression of the HOX gene loci by oncogenic fusions, such as PML-RARA and RUNX1-RUNX1T1, or it may result from the inactivation of a common pathway required for maintenance of HOX expression. Alternatively, AMLs with non-HSPC-like expression patterns may simply originate from a more differentiated hematopoietic progenitor—one that has already downregulated one or both of the HOX gene clusters as part of normal development. If this is the case, and if AML cells truly require self-renewal as part of their defining biology, then the AMLs with no HOX expression may have their self-renewal signals provided by novel mechanisms. Indeed, both PML-RARA and RUNX1-RUNX1T1 are known to “reprogram” myeloid progenitors to have a self renewal phenotype (54,55). We suspect that the other AML cases with no HOX expression (and neither of these fusions) may have alternative self-renewal pathways that have not yet been recognized. Regardless, the fact that AMLs with the same HOX expression profiles have similar patient outcomes suggests that they indeed share some fundamental biological property that correlates with responses to current, established therapies.
In addition to the striking differences in expression patterns at the HOX loci, the levels of HOX expression were also somewhat variable among the AML samples. This could be due to true differences in expression levels, or to more technical issues, such as the fraction of cells in samples that were part of the malignant clone. Some genes did show statistically different expression levels among AML types, but the range of expression values overlapped, arguing against systematic differences in transcript abundance across AMLs with different mutations and HOX expression phenotypes (the notable exception being AML samples with inv(16), which all showed low level expression of only the HOXB genes). The range of HOX expression values in normal CD34+ cells was similar to that observed in AML samples with the stem cell pattern, supporting the notion that the HOX genes are regulated in similar fashion in normal and malignant HSPCs. It is also interesting to note that the HOX expression patterns we observed always involved multiple genes. This suggests that functional studies based on overexpression of a single gene should be interpreted with caution, since this approach may not accurately model HOX transcription factors in normal or leukemic cells, where multiple HOX genes are always expressed together.
The findings presented here also provide evidence for distinct regulatory regions within the HOXA and HOXB gene clusters that may be acting as “locus control regions” in hematopoietic cells. These regions contain open chromatin in AML samples and normal cells, regardless of HOX expression, and appear to be at the center of epigenetic differences associated with HOX expression, including DNA methylation and chromatin accessibility. It is not yet clear whether these regions are also targets for certain transcription factors or oncogenic mutations, although we observed canonical epigenetic patterns that appear to be associated with only the presence or absence of HOX expression (rather than with specific mutations). These candidate regulatory regions are located within or near CTCF binding sites, which may be organizing higher order chromatin structures that are involved in locus regulation. Experimental evidence from other systems has shown that chromatin structure at the HOX loci is dynamic throughout development and cellular differentiation (56–58); similar regulatory pathways may be involved with AML. However, additional experiments will be necessary to understand whether direct interactions with oncogenic gene fusions (or other mutant genes) play a direct role in guiding such processes, or whether additional factors (or the HOX transcription factors themselves) are involved in regulating HOX gene expression. These studies may identify “upstream” factors involved in regulating HOX expression in normal cells and in AML, and may point to novel approaches for disrupting HOX gene expression in AML for therapeutic purposes.
Supplementary Material
Acknowledgments
This work was supported by grants to D.H.S. (CDP-1402, P50 CA171963-01 (D. Link, P.I.)), and T.J.L. from the National Cancer Institute (P01CA101937 and R01CA162086). We thank the staff of The Genome Institute at Washington University for technical assistance. Technical assistance was also provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core, the High Speed Cell Sorting Core, and the Molecular and Genomic Analysis Core at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, which are all supported by the National Cancer Institute Cancer Center Support Grant P30CA91842. We also thank David Russler-Germain for critical reading of the manuscript.
Footnotes
The authors have no conflicts of interest relating to the work described in this manuscript.
Supplementary information is available at Leukemia’s website.
References
- 1.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proceedings of the National Academy of Sciences. 1994 Dec 6;91(25):12223–7. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Experimental Hematology. 2002 Jan;30(1):49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 3.Giampaolo A, Sterpetti P, Bulgarini D, Samoggia P, Pelosi E, Valtieri M, et al. Key functional role and lineage-specific expression of selected HOXB genes in purified hematopoietic progenitor differentiation. Blood. 1994 Dec 1;84(11):3637–47. [PubMed] [Google Scholar]
- 4.Magli MC, Barba P, Celetti A, De Vita G, Cillo C, Boncinelli E. Coordinate regulation of HOX genes in human hematopoietic cells. Proceedings of the National Academy of Sciences. 1991 Jul 15;88(14):6348–52. doi: 10.1073/pnas.88.14.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Molecular and Cellular Biology. 2001 Jan;21(1):224–34. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Molecular and Cellular Biology. 1997 Jan;17(1):495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, et al. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes & Development. 1995 Jul 15;9(14):1753–65. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 8.Antonchuk J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Experimental Hematology. 2001 Sep;29(9):1125–34. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- 9.Bach C, Buhl S, Mueller D, Garcia-Cuellar M-P, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010 Apr 8;115(14):2910–8. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence HJ, Christensen J, Fong S, Hu Y-L, Weissman I, Sauvageau G, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005 Dec 1;106(12):3988–94. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brun ACM, Björnsson JM, Magnusson M, Larsson N, Leveén P, Ehinger M, et al. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood. 2004 Jun 1;103(11):4126–33. doi: 10.1182/blood-2003-10-3557. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997 Mar 15;89(6):1922–30. [PubMed] [Google Scholar]
- 13.Björnsson JM, Larsson N, Brun ACM, Magnusson M, Andersson E, Lundström P, et al. Reduced proliferative capacity of hematopoietic stem cells deficient in Hoxb3 and Hoxb4. Molecular and Cellular Biology. 2003 Jun;23(11):3872–83. doi: 10.1128/MCB.23.11.3872-3883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bijl J, Thompson A, Ramirez-Solis R, Krosl J, Grier DG, Lawrence HJ, et al. Analysis of HSC activity and compensatory Hox gene expression profile in Hoxb cluster mutant fetal liver cells. Blood. 2006 Jul 1;108(1):116–22. doi: 10.1182/blood-2005-06-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004 Nov 23;14(22):2063–9. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010 Jun 15;17(6):609–21. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-Rearranged Leukemia Is Dependent on Aberrant H3K79 Methylation by DOT1L. Cancer Cell. 2011 Jul;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Cancer Research. 24. Vol. 65. American Association for Cancer Research; 2005. Dec 15, Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications; pp. 11367–74. [DOI] [PubMed] [Google Scholar]
- 19.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009 Mar 12;113(11):2375–85. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, et al. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc Natl Acad Sci USA National Acad Sciences. 2011 May 10;108(19):7956–61. doi: 10.1073/pnas.1103154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soulier J, Clappier E, Cayuela J-M, Regnault A, García-Peydró M, Dombret H, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005 Jul 1;106(1):274–86. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 22.Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006 Oct;116(10):2707–16. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, et al. CALM-AF10+ TALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005 Nov;19(11):1948–57. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 24.Mullighan CG, Kennedy A, Zhou X, Radtke I, Phillips LA, Shurtleff SA, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007 Sep;21(9):2000–9. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 25.Alcalay M. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005 Apr 12;106(3):899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 26.Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011 Mar 27;43(5):470–5. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreeff M, Ruvolo V, Gadgil S, Zeng C, Coombes K, Chen W, et al. HOX expression patterns identify a common signature for favorable AML. Leukemia. 2008 Jul 31;22(11):2041–7. doi: 10.1038/leu.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson A, Quinn MF, Grimwade D, O’Neill CM, Ahmed MR, Grimes S, et al. Global down-regulation of HOX gene expression in PML-RARalpha + acute promyelocytic leukemia identified by small-array real-time PCR. Blood. 2003 Feb 15;101(4):1558–65. doi: 10.1182/blood.V101.4.1558. [DOI] [PubMed] [Google Scholar]
- 29.Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol American Society of Clinical Oncology. 2010 Feb 1;28(4):570–7. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
- 30.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007 Oct 15;26(47):6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 31.De Braekeleer E, Douet-Guilbert N, Basinko A, Le Bris M-J, Morel F, De Braekeleer M. Hox gene dysregulation in acute myeloid leukemia. Future Oncol. 2014 Feb;10(3):475–95. doi: 10.2217/fon.13.195. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May 30;368(22):2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elghetany MT, Ge Y, Patel J, Martinez J, Uhrova H. Flow cytometric study of neutrophilic granulopoiesis in normal bone marrow using an expanded panel of antibodies: Correlation with morphologic assessments. J Clin Lab Anal. 2004;18(1):36–41. doi: 10.1002/jcla.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012 Mar 1;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waggott D, Chu K, Yin S, Wouters BG, Liu F-F, Boutros PC. Bioinformatics. 11. Vol. 28. Oxford University Press; 2012. Jun 1, NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data; pp. 1546–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho BS, Irizarry RA. Bioinformatics. 19. Vol. 26. Oxford University Press; 2010. Oct 1, A framework for oligonucleotide microarray preprocessing; pp. 2363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xi Y, Li W. BMC Bioinformatics. 1. Vol. 10. BioMed Central Ltd; 2009. BSMAP: whole genome bisulfite sequence MAPping program; p. 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biology. 2012;13(7):R61. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen KD, Langmead B, Irizarry RA. Genome Biology. 10. Vol. 13. BioMed Central Ltd; 2012. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions; p. R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013 Dec;10(12):1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010 May;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. Nucleic Acids Research. Web Server issue. Vol. 42. Oxford University Press; 2014. Jul, deep Tools: a flexible platform for exploring deep-sequencing data; pp. W187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macrae T, Sargeant T, Lemieux S, Hébert J, Deneault E, Sauvageau G. RNA-Seq reveals spliceosome and proteasome genes as most consistent transcripts in human cancer cells. In: Sobol RW, editor. PLoS ONE. 9. Vol. 8. 2013. p. e72884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Identification of homeobox genes expressed in human haemopoietic progenitor cells. 1994 Jul 8;144(2):213–9. doi: 10.1016/0378-1119(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 45.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010 Dec 22;304(24):2706–15. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roche J, Zeng C, Barón A, Gadgil S, Gemmill RM, Tigaud I, et al. Hox expression in AML identifies a distinct subset of patients with intermediate cytogenetics. Leukemia. 2004 Apr 15;18(6):1059–63. doi: 10.1038/sj.leu.2403366. [DOI] [PubMed] [Google Scholar]
- 47.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002 Feb;16(2):186–95. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 48.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008 May 1;111(9):4797–808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun X-J, Wang Z, Wang L, Jiang Y, Kost N, Soong TD, et al. A stable transcription factor complex nucleated by oligomeric AML1–ETO controls leukaemogenesis. Nature. 2013 Jun 30;500(7460):93–7. doi: 10.1038/nature12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002 Feb 8;295(5557):1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 51.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012 Sep 5;489(7414):83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 5;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature Medicine. 2011 Sep;17(9):1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 54.Tonks A, Pearn L, Tonks AJ, Pearce L, Hoy T, Phillips S, et al. The AML1-ETO fusion gene promotes extensive self-renewal of human primary erythroid cells. Blood. 2003 Jan 15;101(2):624–32. doi: 10.1182/blood-2002-06-1732. [DOI] [PubMed] [Google Scholar]
- 55.Wojiski S, Guibal FC, Kindler T, Lee BH, Jesneck JL, Fabian A, et al. PML-RARalpha initiates leukemia by conferring properties of self-renewal to committed promyelocytic progenitors. Leukemia. 2009 Aug;23(8):1462–71. doi: 10.1038/leu.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011 Oct 14;334(6053):222–5. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 57.Rousseau M, Crutchley JL, Miura H, Suderman M, Blanchette M, Dostie J. Nucleic Acids Research. 3. Vol. 42. Oxford University Press; 2014. Feb, Hox in motion: tracking HoxA cluster conformation during differentiation; pp. 1524–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007 Oct 11;449(7163):731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq expression data from AML samples are available from the TCGA portal (https://tcga-data.nci.nih.gov/tcga/). Raw RNA-seq data from normal bone marrow cells, as well as ATAC-seq and bisulfite sequencing data from AML samples, will be deposited in dbGaP (http://www.ncbi.nlm.nih.gov/gap). Processed ATAC-seq and methylation datasets from the HOX loci are available from the authors upon request.