Abstract
Developmental pathways such as Notch play a pivotal role in tissue specific stem cell self-renewal as well as in tumor development. However, the role of Notch signaling in breast cancer stem cells (CSC) remains to be determined. We utilized a lentiviral Notch reporter system to identify a subset of cells with a higher Notch activity (Notch+) or reduced activity (Notch-) in multiple breast cancer cell lines. Using in vitro and mouse xenotransplantation assays we investigated the role of Notch pathway in breast CSC regulation. Breast cancer cells with increased Notch activity displayed increased sphere formation as well as expression of breast CSC markers. Interestingly Notch+ cells displayed higher Notch4 expression in both basal and luminal breast cancer cell lines. Moreover, Notch+ cells demonstrated tumor initiation capacity at serial dilutions in mouse xenografts while Notch- cells failed to generate tumors. Gamma-secretase inhibitor (GSI), a Notch blocker but not a chemotherapeutic agent effectively targets these Notch+ cells in vitro and in mouse xenografts. Furthermore, elevated Notch4 and Hey1 expression in primary patient samples correlated with poor patient survival. Our studies reveal molecular mechanism for the role of Notch mediated regulation of breast CSCs and provide a compelling rationale for CSC targeted therapeutics.
Introduction
An increasing body of evidence suggests that a variety of cancers including those of the breast may be driven by a component of tumor initiating cells that retain stem cell properties. Consistent with the cancer stem cell model, the subset of tumor initiating cells is able to generate tumors that recapitulate the phenotypic heterogeneity of the initial tumor. Recent studies, utilizing in situ “lineage tracing”, have demonstrated that the CSC originate from tissue specific stem cells in mouse models of skin, gut and brain cancers (1-3). These studies provide further evidence that self-renewing tissue-specific stem cells may initiate and maintain tumors, mediate metastasis contributing to treatment resistance and relapse. Developmental pathways such as Notch are known to regulate self-renewal of embryonic and tissue specific stem cells (4, 5). In addition, aberrant Notch signaling is associated with several human diseases including malignant transformation of the mammary gland (6). In line with these findings, transactivation of the Notch4 (formerly known as Int-3 gene) as a consequence of insertional mutagenesis by the mouse mammary tumor virus results in malignant transformation of mouse mammary gland (6). These and in vitro studies lend strong support for a role for Notch in the regulation of CSCs.
Notch maintains pluripotent hematopoietic stem cells (HSC) by inhibiting differentiation (7). Consistent with these findings, the Notch pathway is linked to ER-negative human breast tumors with a basal phenotype (8-11) suggesting a restriction in differentiation. Furthermore, we previously provided evidence that Notch induces self-renewal in mammary stem/progenitor cells (5). It has also been reported that Notch induces luminal differentiation of mouse mammary stem cells (12).
In order to investigate the role of Notch in breast CSCs, we utilized a Notch reporter system that allowed us to identification of cells with Notch activity. Cells with Notch activity is isolated by flow cytometry and examined for self-renewal capacity utilizing in vitro CSC assays and stem cell markers such as ALDH. In addition, we determined the tumor initiating capacity of Notch-positive cells in mouse xenografts. Furthermore we provide evidence that GSI effectively targets Notch-positive cells and reduces tumor growth in mouse xenografts. Together these studies reveal a direct role for the Notch pathway in the regulation of breast CSCs and suggest that Notch-targeted therapeutics may be an attractive CSC-specific approach in treatment of breast cancer. In addition, our studies provide molecular mechanism for our previous clinical studies where we demonstrated the clinical efficacy of GSI in combination with docetaxel a chemotherapeutic agent in breast cancer patients.
Materials and Methods
Cell lines and reagents
The breast cancer cell lines MDA-MB 231, MDA-MB 436, ZR-75-1, MCF7, ZR-75-30 and T47D were obtained from American Type Culture Collection (ATCC) more than 5 years ago and maintained in culture conditions as recommended by ATCC. The Sum59 cell line was kindly provided by Dr. Stephen P. Ethier. All cell lines were authenticated by the STR DNA profiling by University of Michigan DNA sequencing core in 2011, prior to our studies presented in this manuscript.
The GSI MRK-003 was kindly provided by Merck&Co., Inc., and stocks at 10 mmol/L in dimethyl sulfoxide were used in in vitro studies. Docetaxel was purchased from Sanofi-Aventis. Results of in vitro experiments are presented as mean ± standard deviation or mean ± standard error representation of 3 independent experiments. Student t-test was used to compare continuous variables. Median time to tumor formation was analyzed using Log rank test and Kaplan Meier method. SPSS version 13 was used for statistical analysis. P-value of less than 0.05 was considered statistically significant.
Notch reporter lentiviral construct
A pGreenFire1-Notch plasmid that expressed destabilized copGFP reporter and firefly luciferase under the control of four Notch response elements and a minimal CMV promoter was purchased from System Biosciences. Lentiviral production was performed by the University of Michigan Vector Core facility. Infection efficiency of cells with lentivirus was optimized using the positive Lentivirus expressing GFP under the control of CMV promoter and the MOI has been determined to be 10. The Notch agonist Delta-Serrate-Lag (DSL) peptide was synthetized by the University of Michigan Peptide Core facility as previously described (5).
Mammosphere Assay
Dissociated single MCF7-Notch and MCF7-mCMV cells were plated on six-well ultra-low cluster plates (Corning Inc.) at a density of 1×105 cells/ml and grown for seven–ten days. Subsequent cultures after dissociation of primary spheres were plated on new plates at a density of 1×104 cells/ml. Mammosphere cultures were grown in a serum-free mammary epithelium basal medium as previously described (13)
In vivo tumorigenicity and MRK-003 treatments
Drug solutions were prepared fresh before each dose using a dounce homogenizer. MRK-003 (Merck Research Laboratories) was synthesized according to standard medicinal chemistry procedures. For the in vitro experiments, stocks were prepared at 10 mM in DMSO and dilutions were made directly before use. For the in vivo experiments, MRK-003 was dosed as a suspension in 0.5% methylcellulose at a 10 ml/kg dosing volume, made fresh daily. Before administering each dose the compound was thoroughly mixed to distribute suspension evenly. The dosing schedule for MRK-003 was 75 mg/kg by oral gavage, once daily using intermediate schedule (3 days on, 4 days off- four cycles), which was well tolerated.
Taxotere was dosed as a solution in 0.9% normal saline at a 10 mg/kg solution, intravenously, once per week.
All mice were housed in the AAALAC-accredited specific pathogen-free rodent facilities at the University of Michigan. Mice were housed in sterilized, ventilated racks and supplied with commercial chow and sterile water both previously autoclaved. All experimentation involving live mice were conducted in accordance with standard operating procedures approved by the University Committee on the Use and Care of Animals at the University of Michigan.Athymic 4- to 6-week-old NOD/SCID mice were used. Single cells were flow cytometry sorted sorted based on GFP expression and implanted into mammary fat pads. Mice were monitored via bioluminescence imaging over the course of 10 weeks utilizing the in vivo bioluminescence Caliper IVIS imaging systems. For reimplantation studies, tumors were removed, chopped, and processed with collagenase for 1 to 2 hours at 37C. Cells were then washed with PBS, trypsinized, and passed through a 40-mm filter. The single cells obtained were labeled with H-2Kd antibody and 40,6-diamidino-2-phenylindole (DAPI) and then sorted with flow cytometry. Alive human cells were reinoculated subcutaneously to determine reimplantation capacity. For in vivo treatment studies, mice were randomly assigned to different groups when the tumors were palpable. Tumors were measured weekly using a caliper and tumor size was calculated using the following formula: tumor volume = (length x width2)/2.
RNA extraction and real-time RT-PCR
Total RNA was extracted using RNeasy Mini kit (QIAGEN) and 500ng of RNA was used for making cDNA using Reverse Transcription System (Promega). Each cDNA was analyzed in triplicate using real-time quantitative reverse transcription–PCR (qRT-PCR) assays in an ABI PRISM 7900HT sequence detection system with 384-well block module and automation accessory (Applied Biosystems). The information of the PCR primers and fluorogenic probes used are available on the Applied Biosystems website (GAPDH: Hs00266705, Notch1: Hs01062014_m1, Notch2: Hs01050702_m1, Notch3: Hs01128541_m1, Notch4: Hs00965889_m1). The relative expression mRNA level was normalized against the internal control GAPDH gene (ΔCt = Ct (target gene) – Ct (GAPDH)). The relative fold change was measured by 2-ΔΔCt formula compared to SUM159-Notch- control cells.
In vivo experiments
Mouse tumor xenografts experiments are presented with means and standard errors of the mean. Experiments with multiple tumors per mouse had standard errors of the mean calculated using clustering methods. Tumor growth experiments were analyzed using linear mixed models to account for the repeated measures. Correlation structures were dependent upon the experimental design. Luciferase experiments were assumed to have a first order autoregressive correlation structure. Pairwise comparisons were made within the models at cross-sections and the Bonferroni multiple comparisons adjustment was used for each experiment. Analyses were completed using SAS 9.3 (SAS Institute, Cary, NC) with alpha of 0.05 determining statistical significance as determined by a statistician (SD).
Immunofluorescene
Notch+ and Notch- FACS sorted cells were cyto-spun onto slides for immunofluorescence analysis. Slides were labeled with anti-human ALDH1A1 (ab51028, Abcam, Cambridge, MA) or anti-human Hey1 (ab22614, Abcam, Cambridge, MA) followed by species-specific alexafluor 555 nmconjugated secondary antibody (Invitrogen). Nuclei were labeled with DAPI (Sigma-Aldrich, St Louis, MO) and slides were mounted with mounting media for fluorescence (Vector Laboratories Inc., Burlingame, CA). Images were captured with a Zeiss LSM 510 laser scanning confocal microscope.
Flow Cytometry
Cells were harvested in HBSS with 2% FBS, and GFP expression was measured on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ), and data were analyzed using ModFit software (Verity Software House, Topsham, ME).
Results
Lentiviral Notch reporter system identifies a subset of breast cancer cells with increased Notch activity and CSC characteristics
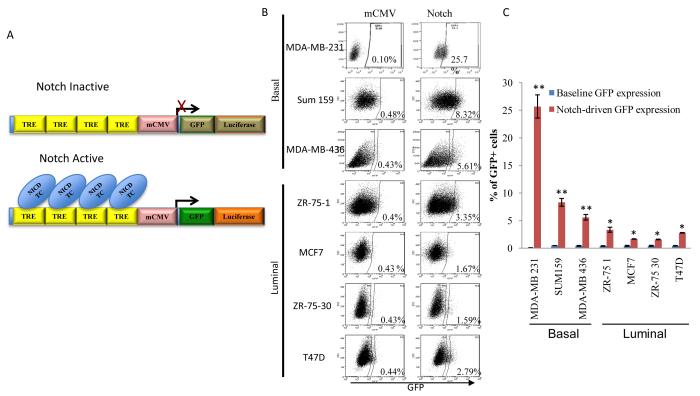
Infection of multiple breast cancer cell lines with a lentiviral Notch reporter construct reveals a subset of cells with increased Notch activity (Notch+ cells) as determined by mCMV-driven GFP expression under the control of a Notch specific transcriptional response element (Figure 1A, B). Breast cancer cell lines, MDA-MB-231, Sum159 and MDA-MB-436 representing the basal-like or claudin-low subtype contained a higher Notch+ cell population compared to cell lines, ZR-75-1, MCF7, ZR-75-30 and T47D which represent the luminal subtype (Figure 1B, C). The developmental Notch pathway has been shown to be essential for normal mammary stem cell self-renewal and lineage-specific differentiation as demonstrated by mammopshere formation and differentiation assays in vitro (5). In order to determine whether the CSC marker, ALDH1 expression overlaps with the Notch activity (14), Notch-reporter expressing cells were cyto-spun and stained with ALDH1 antibody. ALDH1 expression was primarily found in GFP expressing Notch+ cells (Figure 2A) suggesting a higher Notch activity in breast CSCs. There was also non-statistically significant overlap between Notch activity and CD44+CD24- phenotype (Figure 1B). In agreement with CSC properties, we demonstrated that the Notch+ cells possessed a higher sphere forming capacity as compared to Notch- cells (Figure 2C). Further analyses of these cells revealed that Notch+ cells had reduced levels of ER expression compared to the Notch- cells (Figure 2D). To determine whether these Notch+ cells are responsive to Notch stimulation, the Notch agonist peptide, Delta-Serrate-Lag (DSL) was used to activate the Notch pathway leading to increased GFP expression and expansion of Notch+ cell population (Figure 2E). This was further confirmed with dose dependent activation of Notch reporter upon stimulation of cells with recombinant DLL4 (Figure 2F). Endothelial cells were reported to have higher Notch ligands particularly DLL4 expression (15). Consistent with these reports, there was a time dependent increase in the Notch activity when Notch reporter expressing MCF7 cells co-cultured with endothelial HUVEC cells (Figure 2G).
Figure 1. Identification of Notch activity in multiple breast cancer cell lines is determined by the Notch reporter system.
Breast cancer cell lines infected with the lentiviral Notch reporter that expresses GFP and Luciferase under the minimal CMV promoter driven by Notch transcription response element (GTGGGAACGGCATTGTAGCG). (TRE: transcription response element, NICD Notch intracellular domain, TC: transcription complex). (A) A schematic illustration of the lentiviral constructs used in studies presented here (B, C) Basal breast cancer cell lines, MDA-MB-231, Sum159, MDA-MB-436 display a higher Notch activity compared to luminal breast cancer cell lines, ZR-75-1, MCF7, ZR-75-30, T47D. Means ± SD (n=3),*p≤0.05, ** p≤0.005.
Figure 2. Notch reporter positive (Notch+) cells display CSC features.
(A) FACS sorted Notch+ MCF7 cells showed a higher ALDH1 expression compared to Notch- cells. (B) Notch activity and CD44+CD24- phenotype in MCF7 cells are not correlated with statistical significance. (C) A higher sphere forming capacity was observed in Notch+ cells compared to the Notch- cells. (D) Notch+ MCF7 cells show reduced ER expression compared to the Notch- cells (E, F) Notch agonist Delta-Serrate-Lag (DSL) peptide or recombinant DLL4 ligand induces Notch reporter activity in MCF7 cells as demonstrated by GFP expression. (G) Notch activity is increased when MCF7 cells co-cultured with the human endothelial HUVEC cells. (Means ± SD (n=3),*p≤0.05, ** p≤0.005.
Notch activity correlates with higher Notch4 expression and downstream targets
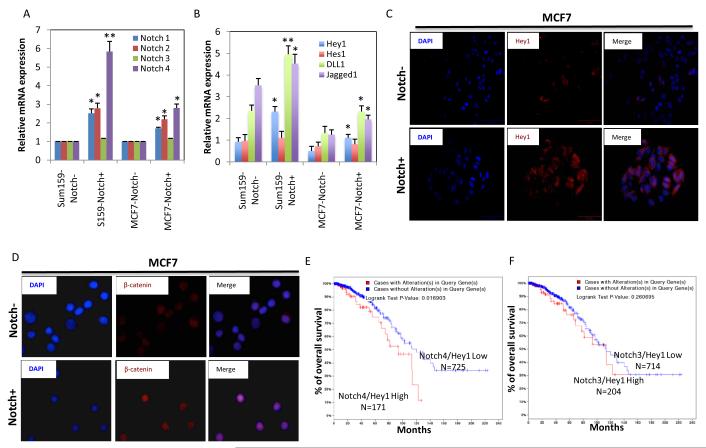
In order to determine what Notch receptor may be responsible for Notch activity, we quantified the expression of Notch1, Notch2, Notch3, Notch4 in Notch+ and Notch- cells. qPCR experiments demonstrated that Notch4 receptor was expressed at 3-5 fold higher levels in the Notch+ cells than the Notch- cells (Figure 3A). We next examined the expression of Notch specific ligands and downstream targets (16) by qPCR and immunofluorescent (IF) experiments. Downstream effectors Hey1 but not Hes1 is significantly higher in Notch+ cells compared to the Notch- cells (Figure 3B,C). Expression of Notch ligands DLL1 and Jagged1 were significantly higher in Notch+ cells as compared to the Notch - cells (Figure 2B). We reasoned whether higher Notch4 expression was correlated with these ligands and downstream targets as well as with patient survival. We utilized the cancer genome atlas (TCGA) data set (1061 patient samples) submitted to free cBioPortal website (17) and demonstrated that there is indeed a positive correlation between the Notch4 mRNA expression and DLL1, DLL4, Jagged1 and Hey1 mRNA expressions (Supplementary Figure 1A,B). It has been reported that Notch is required for Wnt mediated malignant transformation of mammary epithelial cells (18). Moreover our previous studies demonstrated that Wnt/β-catenin pathway is selectively activated in breast cancer stem cells (19) To demonstrate a possible link between Notch and Wnt/β-catenin pathways, FACS sorted Notch+ and Notch- cells were cytospun on slides and stained for β-catenin. Notch+ cells displayed a higher β-catenin nuclear localization compared to the Notch- cells (Figure 3D) suggesting a link between these two pathways. Furthermore, elevated Notch4-Hey1 mRNA expressions but not Notch3-Hey1 correlated with poor patient survival as demonstrated by using the cBioPortal (Figure 3 E,F). These results strongly suggest that Notch4 receptor play a major role in Notch activity and correlates with expression of ligands and downstream effectors and appears to have a clinical relevance in patient samples.
Figure 3. Notch reporter positive cells express higher Notch1, Notch2 and Notch4 receptors and downstream effectors compared to the negative cells.
(A, B) qPCR analyses show increased expressions of Notch1, 2 and 4, Notch ligands Jagged 1 and DLL1 and downstream effector Hey1 in Notch+ cells compared to Notch- cell populations. (C) Notch+ cells compared to the Notch- cells display higher Hey1 expression and (D) increased nuclear β-catenin localization as assessed by IF. (E, F) Analyses of TCGA breast cancer data set (cBioPortal) revealed that elevated Notch4/Hey1 expressions but bot Notch2/Hey1 expressions correlated with poor patient survival. Means ± SD (n=3),*p≤0.05, ** p≤0.005.
Single Notch+ cells, but not Notch- cells has the capacity to repopulate initial heterogeneity
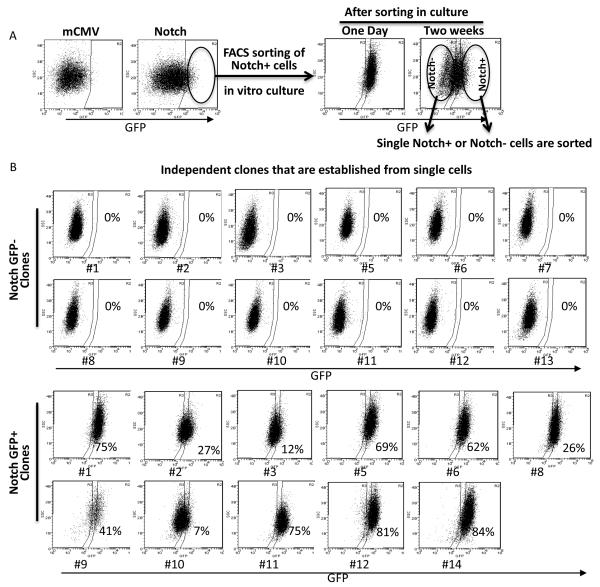
Like their normal counterparts, the hallmark of CSCs has been defined as self-renewal and lineage differentiation (20). To provide further evidence that Notch+ cells possess self-renewing and lineage differentiation capacity, we established multiple independent clones from Notch+ and Notch- cells. We first ensured that all the cells had the Notch reporter construct by presorting Notch+ cells in bulk. These Notch+ cells generated both Notch+ and Notch- cell populations in two weeks (Figure 4A). However, FACS sorting in bulk is associated with 5-10% impurity (21) and does not eliminate the possibility of contamination of Notch- cells within the Notch+ cell population. Thus we generated multiple independent clones from single FACS sorted Notch+ or Notch- cells from Sum159, MCF7 and T47D and analyzed their lineage differentiation capacity. Here we demonstrate that independent Notch+ or Notch- clones from Sum159 cells (Figure 4B). All of the Notch- clones (n=12) remained Notch- over the course of twelve weeks in culture. In contrast, Notch+ clones (n=11) were able to generate both Notch+ and Notch- cell populations suggesting a lineage differentiation capacity (Figure 4B). We obtained similar results in single Notch+ cell generated clones of MCF7 and T47D cell lines (data not shown).
Figure 4. Notch- cells fails to generate Notch+ cells in vitro.
(A) Notch reporter lentivirus infected Sum159 cells were presorted by FACS and Notch+ cells were kept in culture for two weeks during which Notch+ cells generated Notch+ as well as Notch- cells. (B) Using the presorted Notch+ cells, single Notch+ or Notch- cells were FACS sorted and independent clones were grown. We analyzed these independent clones that are generated from single Notch- (n=13) or Notch+ (n=12) cells. None of the clones of single Notch- cells was able to generate the Notch+ cell population while the clones generated from single Notch+ cells were able to generate both Notch+ as well as Notch- cell populations. Means ± SD (n=3),*p≤0.05, ** p≤0.005.
Notch+ cells from multiple breast cancer cell lines possess self-renewing capacity and generate tumors in mouse xenografts
The in vivo self-renewal capacity of CSCs is determined by the generation of tumors in a serial transplantation assays (20). Although the Notch pathway has been shown to play a crucial role in mammary gland development by regulating self-renewal and lineage-specific differentiation (5, 12), its direct role in breast CSC self-renewal remains unknown. In order to determine whether the Notch pathway is involved in breast CSC self-renewal in vivo, we evaluated the ability of Notch+ or Notch- cell populations to form tumors in a serial mouse transplantation assays. The Notch reporter system drives the expression of GFP and luciferase genes under the Notch responsive element, and thus the luciferase expression is utilized to monitor Notch activity and tumor growth in live animals. As outlined in Supplementary Figure 2, primary Sum159 and MCF7 tumors that express Notch reporter were dissociated, Notch+ or Notch- cells were FACS sorted and implanted into secondary mice to evaluate the ability of Notch+ or Notch- cells to form tumors. Notch+ Sum159 (50K) and MCF7 (100K) cells formed significantly larger tumors with shorter latency periods compared to Notch- cells (Figure 5A and C, Supplementary Figure 2). While Notch- tumors were small (and precluded the FACS analyses), both Notch+ Sum159 and MCF7 tumors displayed initial phenotypic heterogeneity by generating both Notch+ cells as well as Notch- cells (Figure 5B and D). We next assessed the CSC frequency of Notch+ and Notch- cells in limiting-dilution transplantation assays in tertiary mouse xenografts. Limiting dilutions of Notch+ or Notch- negative cells (from Sum159 or MCF7 cell lines) were implanted and the tumor growths were determined in each group at ten weeks. These tertiary limiting-dilution xenotransplantation assays demonstrated that the CSC frequency was significantly higher in Notch+ cells compared to Notch- cells from both Sum159 or MCF7 cell lines (Table 1 and 2) suggesting a higher Notch activity in the CSC population. Furthermore, Notch+ cells meet the criteria of CSC features by generating tumors in a serial transplantation assays and differentiating into Notch- cell population.
Figure 5. Notch+ cells possess tumor initiating capacity and show lineage differentiation in mouse xenografts.
(A, C) Notch+ Sum159 and MCF7 cells compared to Notch- cells demonstrated a higher tumor forming capacity in mice xenografts. (B, D) FACS analyses of primary tumor cells showed that Notch+ tumors reconstituted the initial heterogeneity in vivo as evidenced by the presence of both Notch+ and Notch- cells (two independent FACS data presented).
Table 1.
Tertiary tumor growth and the CSC frequency of Notch+ and Notch− Sum159 breast cancer cell subpopulations in NOD-SCID mice using the serial transplantation assay
| Cell populations | Cell number |
Tumors/Number of Implantations |
CSC frequency |
|---|---|---|---|
| Sum159-Notch+ | 15000 | 6/6/ | Notch+: 67 |
| Sum159-Notch+ | 1000 | 3/3 | |
| Sum159-Notch+ | 500 | 2/3 | |
| Sum159-Nocth+ | 100 | 1/2 | |
| Sum159-Notch− | 15000 | 2/6 | Notch−: 9848 |
| Sum159-Notch− | 1000 | 0/3 | |
| Sunnl 59-Notch− | 500 | 0/3 | |
| Sunnl 59-Notch− | 100 | 0/3 | |
| p =0.000000199 | |||
Table 2.
Tertiary tumor growth and the CSC frequency of Notch+ and Notch− MCF7 breast cancer cell subpopulations in NOD-SCID mice using the serial transplantation assay
| Cell Populations |
Cell number | Tu mors/ Number of Implantations |
CSC frequency |
|---|---|---|---|
| MCF7-Notch+ | 100000 | 3/3 | Notch+ : 16554 |
| MCF7-Notch+ | 60000 | 3/3 | |
| MCF7-Notch+ | 30000 | 3/3 | |
| MCF7-Notch+ | 10000 | 3/3 | |
| MCF7-Notch− | 100000 | 3/3 | Notch−: 62385 |
| MCF7-Notch− | 60000 | 3/3 | |
| MCF7-Notch− | 30000 | 2/3 | |
| MCF7-Notch− | 10000 | 0/3 | |
| p =0.00758 | |||
As demonstrated in Table 1, Sum159 Notch+ cells display a higher CSC frequency (1 in 67) compared to the Notch- cell population. Thus, we investigated the ability of single Notch+ cells to generate tumors by implanting single Notch+ or single Notch- cells into the fat pads of NOD-SCID mice. As expected from limiting-dilution transplantation experiments, single Notch- cell implantations failed to generate tumors in mice (Supplementary Figure 3A-left fat pads). In contrast, one out of ten single Notch+ cell implantations generated tumors as demonstrated by bioluminescence imaging starting four weeks post-injection (Supplementary Figure 3A-right fat pads). We evaluated this single Notch+ cell-generated tumor to determine the ability of single cell reconstituting the initial phenotypic heterogeneity. FACS analyses revealed that a single Notch+ cell was able to generate both Notch+ and Notch- cell populations as demonstrated by GFP expression (Supplementary Figure 3B).
Targeting Notch with GSI inhibits Notch activity and reduces the tumor-initiating capacity of breast cancer cells
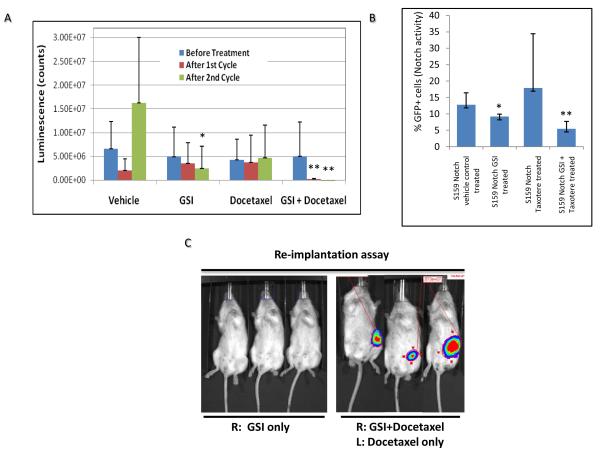
Recent preclinical and clinical studies suggest that the efficacy of GSI in breast cancer patients may be due to its effect on the breast CSCs (22-24). Our studies provide evidence that the Notch-reporter system with luciferase expression may be an attractive tool to monitor Notch activity in growing tumors in live animals. Therefore, we utilized GSI to determine whether the inhibition of Notch would efficiently target breast CSCs and inhibit tumor growth. We implanted the Notch-reporter expressing Sum159 cells into the fat pads of NOD-SCID mice and treated them with two cycles of GSI alone or in combination with a chemotherapeutic agent (Docetaxel) at three weeks post-implantation (Supplementary Figure 4). We measured the tumor size over the course of the study and determined not surprisingly that GSI treatment did not have much effect on primary tumor size. However, Notch activity was significantly diminished after the second cycle of GSI alone or GSI plus Docetaxel treatment as measured by luciferase imaging in live animals (Figure 6A). Furthermore, analyses of these tumors by flow cytometry also confirmed that Notch-driven GFP expression was significantly reduced in GSI treated tumors (Figure 6B). Therefore, we reasoned that inhibition of Notch activity by GSI in primary tumors may target CSCs and if so these tumors should have reduced tumor-initiating capacity in secondary re-implantations at limiting dilutions. Thus the primary tumors from mice that were either untreated or treated with Docetaxel or GSI alone or in combination were re-implanted at indicated dilutions into the fat pads of secondary mice. Following ten weeks of monitoring, we determined that the GSI alone or GSI in combination with Docexatel treated tumors not only displayed significantly lower tumor initiating capacity, but also had reduced CSC frequency (1 in 504 in GSI alone, 1 in 1602 in GSI+Docetaxel compared to 1 in 23 in Control and 1 in 88 in Docetaxel treated tumors) (Figure 6C and Table 3).
Figure 6. Gamma-secretase inhibitor (GSI) reduces the Notch activity and targets breast CSCs in mouse xenografts.
Mice were implanted with Notch reporter expressing Sum159 cells and treated with two cycles of GSI or Docetaxel alone or in combination starting at three weeks post implantation (Supplementary Figure 2). (A) Bioluminescent imaging of mice during the course of treatment showed that the Notch activity was significantly reduced in GSI alone or GSI plus Docetaxel treated animals. (B) Reduced Notch activity in tumors was also confirmed by reduced GFP expression in GSI treated animals. (C) Secondary re-implantation of residual tumors from GSI treated mice revealed a reduced tumor initiating capacity. Means ± SD (n=3),*p≤0.05, ** p≤0.005.
Table 3.
Residual tumor growth following indicated treatments and the CSC frequency estimated in re-implantation assays in NOD-SCID mice
| Treatment | Number of Cells | Tumors/ Implantations |
CSC Frequency |
|---|---|---|---|
| Control | 1000 | 3/3 | 23 |
| Control | 500 | 3/3 | |
| Control | 100 | 3/3 | |
| GSI | 1000 | 2/3 | 504 |
| GSI | 500 | 0/3 | |
| GSI | 100 | 1/3 | |
| Docetaxel | 1000 | 3/3 | 88 |
| Docetaxel | 500 | 2/3 | |
| Docetaxel | 100 | 2/3 | |
| GSI+Docetaxel | 1000 | 0/3 | 1602 |
| GSI+Docetaxel | 500 | 0/3 | |
| GSI+Docetaxel | 100 | 0/3 |
| Group 1 | Group 2 | P value |
| Control | Docetaxel | 0.219 |
| Control | GSI | 0.000231 |
| Control | GSI+Docetaxel | 1.21E-06 |
| Docetaxel | GSI | 0.00543 |
| Docetaxel | GSI+Docetaxel | 4.25E-05 |
| GSI | GSI+Docetaxel | 0.0813 |
Discussion
Despite recent advances in biology and improved diagnostics, intrinsic and acquired resistance to endocrine, trastuzumab or other chemotherapeutic agents, remains one of the most significant challenges for treatment of metastatic breast cancer (25, 26). Clinical implications of the cancer stem cell concept and the role of these cells in mediating the tumorigenesis suggest that these cells might provide a promising therapeutic target (27, 28). Furthermore, accumulating evidence now from a number of solid tumors suggest that CSCs are more resistant to chemotherapy and radiation therapy as compared to bulk tumor cells (28-32). The Notch pathway has been implicated in mediating resistance to chemotherapeutic agents in many human malignancies including breast (22, 33, 34). Utilizing a novel Notch reporter system, we investigated its direct role in tumor initiation and therapeutic resistance in in vitro assays and in mouse xenografts models. In agreement with previous loss of function studies (35), we demonstrated that breast cancer cells with higher Notch activity indeed display stem cell features and expression of CSC markers (14). As previously reported (36), we found higher Notch activity in basal-like breast cancer cell lines compared to luminal breast cancer cell lines. Interestingly Notch4 receptor expressed 3-5 fold higher in Notch+ cells compared Notch- cells suggesting a major role on Notch pathway.
The notch reporter system has been shown to be a useful tool to monitor Notch activity in developing organs in live animals as well as in lung tumor xenografts (37, 38). Thus using this reporter system, we determined that Notch+ cells had significantly higher tumor initiating capacity compared to Notch- cells in serial transplantation assays. Primary and secondary tumors generated by Notch+ cells had the in vivo lineage differentiation capacity since they generated both Notch+ and Notch- cells. Limiting-dilution transplantation assays demonstrated a significantly higher CSC frequency within the Notch+ cell population (1 in 67) compared to the Notch- cell population (1 in 9848). Together these findings provide strong evidence that Notch activity substantially enriches breast CSCs in vivo and thus may have therapeutic utility. Elevated expressions of Notch4 and Hey1 significantly correlated with poor patient survival among breast cancer patients further supporting the clinical relevance of Notch pathway.
In early phase clinical trials, targeting Notch in breast cancer patients with GSI provided a clinical benefit which may be explained by its CSC specific activity (22). To provide a more direct molecular role for Notch in mouse xenografts, we treated mice bearing the Notch reporter expressing Sum159 cells and monitored Notch activity in growing tumors in live animals. Two cycles of GSI treatment reduced the Notch activity and reduced the CSC population. This activity was further enhanced when combined with Docetaxel since there was no tumor growth in secondary tumor implantations.
In summary, our studies provide direct evidence for a role of the Notch pathway in CSC self-renewal and tumor initiation in in vitro assays and in live animals by utilizing a novel Notch reporter system. Together with previous reports, it further emphasizes the importance of CSC-targeted therapeutics in cancer treatments.
Supplementary Material
Acknowledgements
This work was supported by the NIH grants: CA101860 and CA129765 to MSW. Research funding and GSI MRK-003 was kindly provided by Merck&Co., Inc.
Financial Support: This work was supported by the NIH grants: CA101860, CA129765 and research funding by Merck&Co., Inc. to MS. Wicha.
Footnotes
Conflict of Interest: MSW has financial holdings and is a scientific adviser for OncoMed Pharmaceuticals, is a scientific adviser for Verastem, Paganini and MedImmune.
References
- 1.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 2.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, Zou J, Ye Z, Hammond H, Chen G, Tokunaga A, et al. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell. 2008;2:461–71. doi: 10.1016/j.stem.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:605–15. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–55. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 7.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–22. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–31. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–35. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CW, Raskett CM, Prudovsky I, Altieri DC. Molecular dependence of estrogen receptor-negative breast cancer on a notch-survivin signaling axis. Cancer Res. 2008;68:5273–81. doi: 10.1158/0008-5472.CAN-07-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CW, Simin K, Liu Q, Plescia J, Guha M, Khan A, et al. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res. 2008;10:R97. doi: 10.1186/bcr2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–41. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;5:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shutter JR, Scully S, Fan W, Richards WG, Kitajewski J, Deblandre GA, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;11:1313–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol. 2007;31:461–6. [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci U S A. 2006;10:3799–804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;6:e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer Stem Cells--Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 21.Savage EC, Vanderheyden AD, Bell AM, Syrbu SI, Jensen CS. Independent diagnostic accuracy of flow cytometry obtained from fine-needle aspirates: a 10-year experience with 451 cases. Am J Clin Pathol. 2011;135:304–9. doi: 10.1309/AJCPHY69XVJGULKO. [DOI] [PubMed] [Google Scholar]
- 22.Schott AF, Landis MD, Dontu G, Griffith KA, Layman RM, Krop I, et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin Cancer Res. 2013;19:1512–24. doi: 10.1158/1078-0432.CCR-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandya K, Meeke K, Clementz AG, Rogowski A, Roberts J, Miele L, et al. Targeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrence. Br J Cancer. 2011;105:796–806. doi: 10.1038/bjc.2011.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grudzien P, Lo S, Albain KS, Robinson P, Rajan P, Strack PR, et al. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010;30:3853–67. [PubMed] [Google Scholar]
- 25.Normanno N, Morabito A, De Luca A, Piccirillo MC, Gallo M, Maiello MR, et al. Target-based therapies in breast cancer: current status and future perspectives. Endocr Relat Cancer. 2009;16:675–702. doi: 10.1677/ERC-08-0208. [DOI] [PubMed] [Google Scholar]
- 26.Coley HM. Mechanisms and strategies to overcome chemotherapy resistance in metastatic breast cancer. Cancer Treat Rev. 2008;34:378–90. doi: 10.1016/j.ctrv.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Wicha MS. Targeting breast cancer stem cells. Breast. 2009 Oct;18(Suppl 3):S56–8. doi: 10.1016/S0960-9776(09)70274-7. [DOI] [PubMed] [Google Scholar]
- 28.Korkaya H, Wicha MS. Selective targeting of cancer stem cells: a new concept in cancer therapeutics. BioDrugs. 2007;21:299–310. doi: 10.2165/00063030-200721050-00002. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu M-F, et al. Therapeutic resistance and tumor-initiation: Molecular pathways involved in breast cancer stem cell self-renewal. J Natl Cancer Inst. 2008;7100:672–9. [Google Scholar]
- 30.Ischenko I, Seeliger H, Schaffer M, Jauch KW, Bruns CJ. Cancer Stem Cells: How can we Target them? Curr Med Chem. 2008;15:3171–84. doi: 10.2174/092986708786848541. [DOI] [PubMed] [Google Scholar]
- 31.Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–90. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kvinlaug BT, Huntly BJ. Targeting cancer stem cells. Expert Opin Ther Targets. 2007;11:915–27. doi: 10.1517/14728222.11.7.915. [DOI] [PubMed] [Google Scholar]
- 33.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–48. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair P, Somasundaram K, Krishna S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J Virol. 2003;77:7106–12. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suman S, Das TP, Damodaran C. Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br J Cancer. 2013;109:2587–96. doi: 10.1038/bjc.2013.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speiser JJ, Ersahin C, Osipo C. The functional role of Notch signaling in triple-negative breast cancer. Vitam Horm. 2013;93:277–306. doi: 10.1016/B978-0-12-416673-8.00013-7. [DOI] [PubMed] [Google Scholar]
- 37.Vilas-Boas F, Fior R, Swedlow JR, Storey KG, Henrique D. A novel reporter of notch signalling indicates regulated and random Notch activation during vertebrate neurogenesis. BMC Biol. 2011;9:58. doi: 10.1186/1741-7007-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan KA, Wang L, Korkaya H, Chen G, Maillard I, Beer DG, et al. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clin Cancer Res. 2013;19:1972–80. doi: 10.1158/1078-0432.CCR-12-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.