Abstract
The AAA+ ATPase Vps4 disassembles ESCRT-III and is essential for HIV-1 budding and other pathways. Vps4 is a paradigmatic member of a class of hexameric AAA+ ATPases that disassemble protein complexes without degradation. To distinguish between local displacement versus global unfolding mechanisms for complex disassembly, we carried out hydrogen-deuterium exchange during Saccharomyces cerevisiae Vps4 disassembly of of a chimeric Vps24-2 ESCRT-III filament. EX1 exchange behavior shows that Vps4 completely unfolds ESCRT-III substrates on a time scale consistent with the disassembly reaction. The established unfoldase ClpX showed the same pattern, demonstrating a common unfolding mechanism. Vps4 hexamers containing a single cysteine residue in the pore loops were cross-linked to ESCRT-III subunits containing unique cysteine within the folded core domain. These data support a mechanism in which Vps4 disassembles its substrates by completely unfolding them and threading them through the central pore.
Introduction
The Endosomal Sorting Complex Required for Transport (ESCRT) machinery is essential for the budding and release of HIV-1 1, multivesicular body biogenesis 2, cytokinesis 3,4, exosome biogenesis 5, and membrane wound repair 6. The ESCRTs consist of ALIX and ESCRT-0, -I and –II, which bind to ubiquitin and viral and cargo proteins 7; the ESCRT-III proteins that severs membrane necks 8; and the AAA+ ATPase Vps4 9,10. Vps4 is required for the recycling of ESCRT-III from membrane-bound filaments back to the cytosol 11 and probably has additional roles in the ESCRT pathway. As such, the absence or inhibition of Vps4 leads to a complete shutdown of HIV-1 budding and multivesicular body formation.
There are seven ESCRT-III proteins in yeast and twelve in humans. These proteins are monomers or dimers in solution, but carry out their functions as membrane-bound oligomeric assemblies. All ESCRT-III proteins share a common fold, consisting of a long α1-α2 helical hairpin and three shorter helices α3-α5 (12-14). The C-termini of ESCRT-III proteins encode autoinhibitory elements that prevent premature assembly 15-17. ESCRT-III proteins form a variety of assemblies: filaments 18, spirals 19-22, and helical tubes 9,13. These assemblies are do not spontaneously dissociate. Rather, the enzymatic action of Vps4 is required to disassemble them.
Some ESCRT-III subunits contain C-terminal MIT(microtubule interacting and transport)-interacting motifs (MIMs) that bind to Vps4 (23,24). These include the α-helical MIM1 motif of Vps2 (human CHMP2), Did2 (CHMP1), and Ist1, and the extended MIM2 motif of Vps20 (CHMP6) 25,26. Other ESCRT subunits bind to Vps4 with low affinity if at all. Vps4 binds to the MIM1 and MIM2 motifs of ESCRT-III subunits via two sites on its N-terminal MIT domain 23,24,26. In the presence of ATP, wild-type Vps4 exists predominantly as hexamers 27 (although dodecamers predominate when ATP hydrolysis is blocked), and is therefore believed to function as a ring-shaped hexamer. In the absence of ATP, Vps4 exists as monomers and dimers, and available crystal structures of Vps4 reflect these inactive states 27-31. EM reconstructions have been obtained for yeast Vps4 oligomers stabilized by the active site Glu residue mutant E233Q or AMPPNP 30,32,33 and intrepreted in terms of dodecamers or tetradecamers consisting of two rings. None of these reconstructions were at a resolution high enough to place the crystallographic Vps4 monomers. However, the lower ring in one of the reconstructions contains a central pore and is consistent with a hexameric model of Vps4 (34).
This model predicts that conserved Trp and Leu residues line the central pore. These residues are important for HIV-1 budding in vivo 28. Mixed Vps4 hexamers containing a single catalytically active ATPase site and a single MIT domain can disassemble ESCRT-III as long as the putative central pore is intact 35. These data suggest that portions of ESCRT-III subunits enter the pore during disassembly. In one model, ESCRT-III unfolds completely and translocates through the pore, similar to substrates processed by the AAA+ (ATPases associated with diverse cellular activities) unfoldase ClpX 36,37. ClpX degrades proteins in conjunction with the peptidase ClpP, but it also has disassembly functions independent of ClpP 38. In a second model, Vps4 breaks up filaments by locally disrupting contacts in the assembly. This would be similar to the mechanism of SNARE disassembly by NSF, which appears to unwind SNAREs but does not translocate them through the pore 39. In the second model, the region entering the Vps4 pore would likely be limited to parts of the C-terminal region. We set out to address whether Vps4 denatures its substrate globally or whether a more localized perturbation is involved.
We selected the chimeric ESCRT-III subunit Vps24-2 (Fig. 1a) as a model substrate, because its core is a stably folded unit 12, it is forms homooligomeric filaments (Supplementary Fig.1a), and it binds to Vps4 via the MIM1 of Vps2 (18). The Vps24-2 filament is the only homooligomeric ESCRT–III assembly that has been shown to be completely disassembled by Vps4 in vitro. We measured the baseline conformational dynamics of Vps24-2 filaments and its changes on exposure to Vps4, using hydrogen-deuterium exchange and mass spectrometry (HDX-MS). These experiments showed that Vps24-2 is completely unfolded concurrently with its removal from the filament. We went on to show that the unfolding process has close parallels with that of the established unfoldase ClpX. We then showed that unfolding is coupled to translocation through a central pore in the Vps4 hexamer. Pore loop residues were required for disassembly, presumably by directly engaging the substrate and mechanically pulling on it in response to ATP-hydrolysis driven conformational changes of the Vps4 ring. Finally, cross-linking between an engineered cysteine residue in the Vps4 pore and cysteines in Vps24-2 revealed that the core region of ESCRT-III comes into direct contact with the pore, indicating complete threading through the Vps4 hexamer pore.
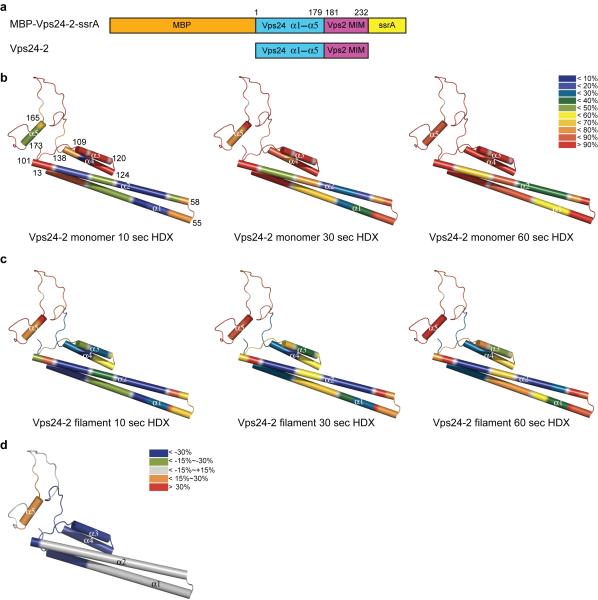
Figure 1. Dynamics of a model ESCRT-III substrate of Vps4.
(a) Schematic diagram of MBP-Vps24-2-ssrA and Vps24-2 constructs used in the study. Residue numbers from Vps24 and Vps2 are shown for the construct boundaries. (b-c) Deuteron incorporation over time for Vps24-2 monomer (b) and Vps24-2 filament assembly (c), mapped onto a Vps24-2 structural model that is based on the crystal structure of the human Vps24 ortholog, CHMP3 12. The inset shows the color-coding for different percentages of deuteron incorporation. The HDX experiments were repeated twice. The start and end amino acids of each α-helix are labled in the far left panel of (b). (d) HDX changes for Vps24-2 in filament assembly as compared to soluble monomers, color coded as shown (red and orange for increased HDX in the filament relative to the monomer, blue and green for decreased HDX).
Results
Dynamics of Vps24–2 monomers and filaments
In order to establish the baseline dynamics of Vps24-2 monomers in solution and as assembled in filaments, we performed an HDX characterization of these states in the absence of Vps4. We obtained excellent peptide coverage (Supplementary Fig. 2) and examined peptides representing helices α1–α4. We collected Vps24-2 from the gel filtration peak fraction corresponding to the monomer and immediately subjected to HDX for various times. By 60 s, the majority of the monomeric protein had exchanged at least 50% of its protons (Fig. 1b, Supplementary Fig. 3a). The core α1–α2 helical hairpin was the region least susceptible to exchange (Fig. 1b). Because Vps24-2 spontaneously forms filaments at high concentration, we also performed HDX studies of the monomer at higher concentration utilizing a construct containing maltose binding protein (MBP) fused to the N-terminus in order to inhibit polymerization (Fig. 1a, Supplementary Fig. 1b, Supplementary Fig. 3b). We incorporated a C-terminal ssrA tag into this construct to facilitate control experiments with ClpX that are described below. Similar HDX exchange patterns were observed for MBP-Vps24-2-ssrA as for the monomeric Vps24-2 construct (Supplementary Fig. 3b to Fig. 1b).
We induced Vps24-2 to polymerize by increasing its concentration to above 350 μM followed by incubation at 4 °C overnight 18 (Supplementary Fig. 1a). We carried out HDX with the resulting filamentous material (Fig.1c) and showed sharply reduced exchange for the N-terminus of helix α1 and the entire helices α3 and α4 (Fig. 1c, d) as compared to the soluble monomer. In contrast, HDX of α5 increased in the filament as compared to the soluble monomer (Fig. 1c, d). This is consistent with a role for α5 as an autoinhibitory element in the soluble ESCRT-III monomer 15,16.
We analyzed EX1 and EX2 exchange kinetics to probe the mechanism of exchange of the Vps24-2 monomer and filament (Fig. 2, Supplementary Fig. 4). EX1 kinetics are diagnostic for the persistence of global unfolding for long enough to exchange the majority of labile protons. In constrast, EX2 kinetics are indicative of a folded protein that samples an exchange-competent conformation for a time shorter than the intrinsic exchange rate of the labile protons 40. Six peptides that fall entirely within the folded core of Vps24-2 were selected for analysis (Fig. 2a). In the monomer, four of these peptides, from helix α1, α2 and α4 (Fig. 2a), exchanged gradually by an EX2 mechanism. EX2 behavior is evident from inspection of the spectra, which gradually shift to higher mass over time while remaining unimodal (Fig. 2b-d, and Supplementary Fig. 4c).
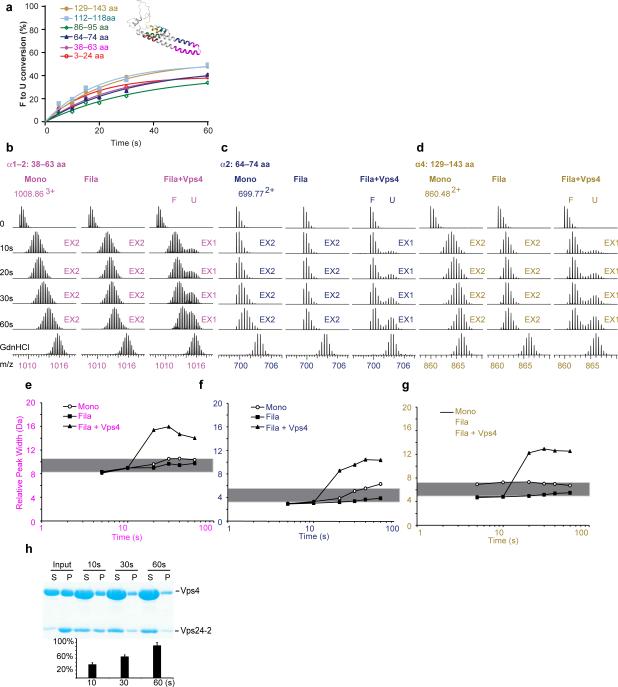
Figure 2. Vps4 completely unfolds ESCRT-III substrate in the course of disassembly.
(a) Folded (F) to unfolded (U) conversion for the peptides in the presence of Vps4, color coded as in the Vps24 structural model shown in the inset. (b-d) Mass spectra of selected peptides from helices α1 (b), α2 (c), and α4 (d) of Vps24-2 monomers (left), filaments (middle), or filament plus Vps4 (right). Controls and time points are indicated. Arrows above the spectra indicate regions of the spectrum representing EX2 and EX1 behavior. (e-g) Peak-width analysis of the selected peptides at 5, 10, 20, 30, 40, 60 s and full deuteration (TD) for selected peptides from helices α1 (e), α2 (f), and α4 (g). Open circles, Vps24-2 monomers; filled squares, Vps24-2 filaments; triangles, Vps24-2 filaments plus Vps4. The grey bar denotes the 2 Da peak-width band for peptides undergoing EX2 kinetics 41. (h) SDS-PAGE gel and quantification bar chart of Vps4WT-mediated disassembly of Vps24-2 filaments as analyzed by sedimentation. Pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE. Error bars are the s. d. of three experiments. WT, wild-type.
It has been proposed that a peak width change that remains within 2 Da of its initial value throughout the course of exchange is diagnostic for the EX2 mechanism 41. To provide a quantitative assessment of the exchange mechanism, peak widths were evaluated as a function of time. The width of the peak as defined at a threshhold of 20% of the maximum valued remained within or near 2 Da of its initial value for the four peptides from helices α1, α2 and α4 at all time points measured (Fig. 2e-g and Supplementary Fig. 4d.). Even though the peak width change for peptides from the distal part of α1 (Supplementary Fig. 4b) and α3 (Supplementary Fig. 4f) remained within 2 Da at all time points measured, the deuteron incorporation for these two peptides (Supplementary Fig. 4a, e) was so rapid (around 80% deuteron incoporation at 5 s, and >90% deuteron incorporation by 30 s), that neither EX1 nor EX2 kinetics could be unambiguously assigned to these two peptides for the Vps24-2 monomer.
For the Vps24-2 filament, exchange was more gradual for all six peptides examined (Fig. 2c, e, h and Supplementary Fig. 4a, c, e). All six spectra were clearly unimodal over time. Monitoring of the peak width confirmed that the spectra for all six peptides remained narrow, characteristic of an EX2 mechanism (Fig. 2e-g, Supplementary Fig. 4b, d, f). These data show that the Vps24-2 monomer is a dynamic protein that has folded but marginally stable helical core, while filamentous Vps24-2 has a rigid, folded α1–α4 core. None of the Vps24-2 filament peptides from helical regions showed any EX1 behavior, as evaluated by peak width analysis and the lack of a bimodal spectrum over the time course of exchange.
Vps4 disassembles ESCRT-III by unfolding entire subunits
We quantified disassembly of Vps24-2 filaments by Vps4 by monitoring the fraction of solubilized Vps24-2 in the supernatant after ultracentrifugation (Fig. 2h). Vps4 solubilized about 30% of the Vps24-2 within 10 s, and 80% by 60 s (Fig. 2h). We therefore monitored HDX of Vps24-2 over the interval 5-60 s (Fig. 2b-g and Supplementary Fig. 4). All six peptides from helices α1, α2, and α4 manifested clearly visible bimodal distributions during exchange (Fig. 2b-d and Supplementary Fig. 4a,c,e). We carried out peak width analysis for all six peptides in the presence of Vps4 (Fig. 2e-g and Supplementary Fig. 4b, d, f). Maximum peak widths increase dramatically, to as much as 10 Da higher than the initial value for peptide 3-24 aa (Supplementary Fig. 4b). We intrepret this as compelling evidence for an EX1 exchange mechanism that is dependent on the presence of Vps4. This provides strong evidence that Vps4 unfolds Vps24-2 in the process of solubilizing it. We obtained the rate of Vps24-2 conversion from the folded to the unfolded state by fitting two Gaussian peaks to the bimodal isotope cluster and dividing the areas of the bimodal isotope cluster with the isotope cluster area of the unfolded state (U/(F+U)). Similar conversion rates were observed for all six peptides (Fig. 2a), which is consistent with their dependence on a single global unfolding process.
Vps4 and ClpX unfold MBP-Vps24-2-ssrA by a common mechanism
The ClpX unfoldase is among the most extensively characterized members of the AAA+ ATPase superfamily 37,38,42-46. ClpX engages soluble, folded protein substrates and denatures them by processively translocating them through its central pore 37. In the context of the ClpXP protease, ClpX threads these denatured substrates directly into the internal degradation chamber of the associated peptidase ClpP for proteolytic cleavage, whereas in isolation ClpX releases translocated polypeptides on the other side of the ring. We included an 11-residue ssrA tag at the C-terminus of MBP-Vps24-2-ssrA to allow its specific recognition by ClpX 47. We carried out parallel HDX studies of the processing of soluble MBP-Vps24-2-ssrA monomers by Vps4 and ClpX in order to make a side-by-side comparison of the unfolding mechanisms.
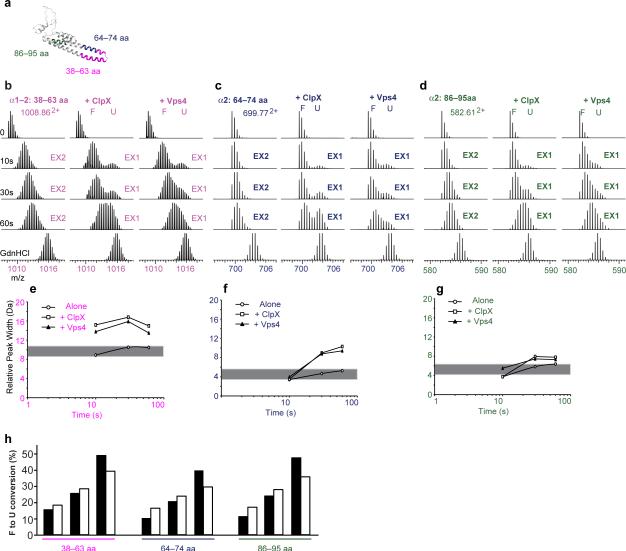
Either Vps4 or ClpX at similar molar concentrations were incubated with soluble MBP-Vps24-2-ssrA, and substrate processing and proton exchange were triggered simultaneously by dilution to 5 % of its previous concentration into ATP and 100% D2O. Under these conditions, we observed robust EX1-dominated exchange for peptides from the Vps24-2 portion of MBP-Vps24-2-ssrA (Fig. 3) at 10–60 s. Vps4 and ClpX unfolded the Vps24-2 portion of the construct to a similar extent as judged by the bimodal spectra (Fig. 3b-d) and peak width analysis (Fig. 3e-g). The total extent of unfolding was similar for ClpX and Vps4 when compared for all three peptides (Fig. 3h).
Figure 3. Vps4 unfolds the Vps24-2 part of MBP-Vps24-2-ssrA as efficiently as ClpX.
(a) Ribbon presentation of the Vps24 structural model with the location of three peptides from the helical core. (b-d) Respective peak-width analysis of the three peptides at 10, 30, 60 s and full deuteration (TD). Open circles, MBP-Vps24-2-ssrA alone; open squares, with ClpX; filled triangles, with Vps4. The grey bar denotes the 2 Da peak-width change allowance. (e-g) Mass spectra of the three selected peptides alone (left), and with ClpX (middle) or Vps4 (right). Controls and time points are indicated. (h) Bar graphs indicate the unfolded proportion of each peptide at different time points in the presence of ClpX (black) or Vps4 (white).
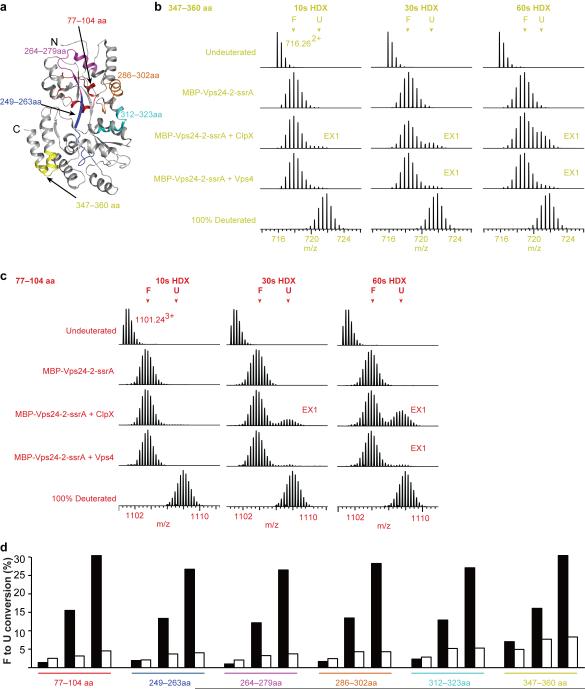
To address the relative potency of Vps4 and ClpX as generic unfoldases, we examined their ability to unfold regions of the MBP portion of the fusion construct. When we treated substrate with ClpX, a peptide from the C-terminal portion of MBP (residue 347–360), encompassing helix XIII and helix XIV (Fig. 4a)48, showed around 7%, 16 %, and 30% unfolding at 10, 30, and 60 s (Fig. 4b, d). On the other hand, the same peptide showed around 5%, 8% and 8% unfolding at 10, 30 and 60 s when Vps4 was present. (Fig. 4b, d). An N-terminal peptide of MBP (residue 77–104), which maps to the center of the MBP fold (Fig. 4a,), showed no visually detectable unfolding at 10 s by either Vps4 or ClpX (Fig. 4c,). ClpX robustly unfolded the same peptide by 60 s (30%,), with trace unfolding by Vps4 over the same time period (Fig. 4c,). Another four peptides in between these two peptides were also examined and all showed detectable bimodal spectra by 30-60 s in the presence of either ClpX or Vps4 (Supplementary Fig. 5,). At the earliest time examined, 10 s, ClpX unfolded a 2–3 times greater fraction of the C-terminal peptide of MBP as compared to the N-terminus (Fig. 4d). By 60 s, the difference disappeared for ClpX, consistent with the known processive C-to-N unfolding mechanism. In the case of Vps4, the difference persists, suggesting that Vps4 might be stalling after failing to disassemble the central region of MBP. Compared to Vps4, ClpX unfolds MBP to a 200% greater extent in the C-terminal region and a 500% greater extent for the central and N-terminal regions by 60 s. This seems in line with expectation, given that ClpX is a relatively non-specific unfoldase (provided only that the substrate contains an ssrA motif), while Vps4 is specialized for ESCRT-III disassembly. While ClpX is, unsurprisingly, the more robust unfoldase, the main inference drawn from the MBP experiments is that Vps4 and ClpX are qualitatively similar in their C-to-N-terminal unfolding of the model substrate.
Figure 4. Unfolding of MBP-Vps24-2-ssrA by ClpX and Vps4.
(a) Cartoon presentation of the MBP stucture (EPDB ID: 1DMB (48)) with the location of six color-coded peptides from MBP-Vps24-2-ssrA. (b, c) Mass spectrate for selected peptides from the C terminus of MBP (resi. 347–360 (b)) and from the N terminus of MBP (resi. 77–104 (c)). (d) Bar graphs indicated the unfolded proportion of each peptide at different time points in the presence of ClpX (black) or Vps4 (white).
Substrates enter the central pore of Vps4
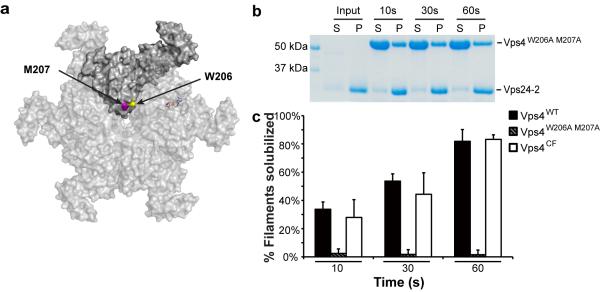
The similarities in the kinetics of unfolding behavior seemed consistent with the previous suggestion that Vps4 might, by analogy to ClpX and other AAA+ unfoldases, translocate unfolded ESCRT-III subunits through the central pore of the hexamer 28 (Fig. 5a). In order to directly test the role of pore loop residues in substrate disassembly, we generated the double pore loop mutant Vps4W206A M207A (Fig. 5a). The basal ATPase activity of Vps4W206A M207A was slightly greater than half that of Vps4WT (Supplementary Fig. 6b). However, Vps4W206A M207A was completely unable to solubilize Vps24-2 filaments (Fig. 5b, c). These data confirm that mutation of the pore loop directly blocks ESCRT-III disassembly, explaining the previously reported loss of function of human VPS4 pore loop mutants in HIV-1 budding 28.
Figure 5. Pore loop residues are required for disassembly.
(a) Surface presentation of the Vps4 hexamer model based on the p97 hexameric structure (PDBID: 1E32 (34)). Residues 206 and 207 of one of the protomers were colored yellow and magenta, respectively, to highlight their locations in the pore. (b) SDS-PAGE gel of Vps4W206A M207A-mediated disassembly of Vps24-2 filament. (c) Quantification of Vps4WT, Vps4W206A M207A and Vps4CF mediated disassembly of Vps24-2 filament by sedimentation. Error bars are the s. d. of three measurements.
In order to determine whether the ESCRT core becomes exposed and directly interacts with the Vps pore during processive translocation, we applied site-directed cross-linking between an engineered cysteine in the Vps4 pore loop and cysteines introduced at positions 92 or 136 of Vps24-2. These two cysteine in Vps24-2 are located in α2 and α4 of the core domain, respectively (Supplementary Fig. 6c). We generated a cysteine-free Vps4 variant (Vps4CF will be used here to refer to Vps4C317A C376A) and used it to introduce a single cysteine at position 206 in the central pore (Vps4CF 206C). Vps4CF had greater than half the ATPase activity and essentially identical Vps24-2 solubilization activity compared to wild type (Supplementary Fig. 6a,b). The ATPase activity of wild-type Vps4 and variants increased upon exposure to the Vps24-2 substrate, as found for other Vps4-substrate combinations 49.
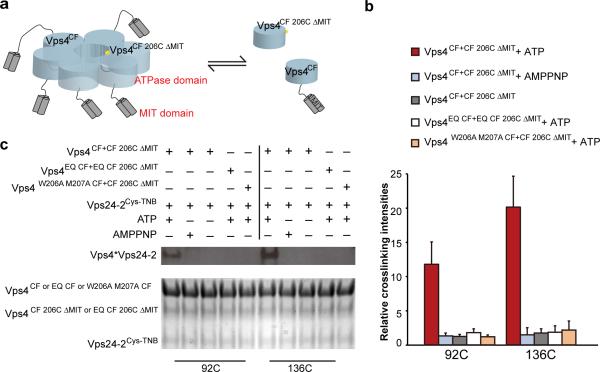
We carried out cross-linking out in the presence and absence of ATP or the non-hydrolyzable ATP analogue AMPPNP. In initial experiments, some cross-linking occurred between Vps4CF 206C and Vps24-2136C when no nucleotide or when AMPPNP was present (data not shown). We hypothesized that this behavior was due to the equilibration of Vps4 between the hexamer and the monomeric state, in which the pore-loop Cys206 is solvent-exposed and accessible for low-efficiency crosslinking even with folded Vsp24-2. In order to protect monomeric Vps4CF 206C from reaction, we truncated its N-terminal Vps2-binding MIT domain. The presence of the MIT domain is required for recruitment of ESCRT-III, therefore the monomeric MIT-deleted construct (henceforward Vps4CF 206C ΔMIT) cannot, by design, bind to ESCRT-III. We combined Vps4CF 206C ΔMIT at a 1:8 molar ratio with full-length Vps4CF to generate mixed hexamers that contain on average less than one crosslinkable Vps4CF 206C ΔMIT subunit (Fig. 6a). The hexamers used in the cross-linking studies described below contain, on average, five MIT domains contributed by the five full-length subunits. These mixed hexamers had basal and substrate-stimulated ATPase activity identical to homohexameric Vps4CF (Supplementary Fig. 6b).
Figure 6. ATP-dependent cross-linking of Vps24-2 to the central pore.
(a) Mixed Vps4 hexamers are composed on average of five VpsCF subunit and one VpsCF 206C ΔMIT , such that VpsCF 206C ΔMIT , the only subunit competent for cross-linking, is incapable of binding Vps24-2 when it is in the monomeric form. (b) Non-reducing SDS-PAGE gel of Vps24-292C (lane 1-5) and Vps24-2136C (lane 6-10) crosslinked to Vps4 single cysteine mutants. The disulfide cross-linked Vps24-2*Vps4 complexes were detected by anti-His western blot (top panel). Input Vps4 and Vps24-2 was visualized by comassie blue staining (bottom panel). (c) Bar graphs show the relative amounts of crosslinked product formed between Vps24-2 92C and 136C mutants and Vps4 single cysteine mutant under different conditions. Error bars are the s. d. of three measurements. For normalization, the lowest mean intensity was assigned a value of 1.
Vps24-2136C cross-linked efficiently to the Vps4CF 206C ΔMIT subunits in mixed hexamers, but only in the presence of ATP (Fig. 6b, c). No reaction was evident in the absence of nucleotide or in the presence of AMPPNP. We mutated the catalytic amino acid Glu233 in the Walker B motif to Gln to produce a ATP hydrolysis-deficient mutant in the cysteine-free background (Vps4EQ CF). As a further control, we presented Vps4EQ CF 206C ΔMIT in mixed hexamers with full-length Vps4EQ CF, which lacks ATPase activity (Supplementary Fig. 6b) 50. We observed no cross-linking with this ATPase-inactivated Vps4 even in the presence of ATP. Finally, in order to explore whether the cross-linking behavior was general to other regions of the Vps24-2 structure, we replaced Ile92 of α2 with a unique cysteine. In the CHMP3 crystal structure the corresponding residue points inward into the hydrophobic core (Supplementary Fig. 6c) and is thus shielded from solvent. Cys92 reacted with about 60% of the efficiency of Cys136 (Fig. 6b, c), which is consistent with its more N-terminal position. The control reactions with no nucleotide, in the presence of AMPPNP, or the mixture with ATP and the Walker B mutant Vps4EQ CF yielded no reaction with Vps24-292C (Fig. 6b, c). Finally, when we presented Vps4CF 206C ΔMIT in mixed hexamers with Vps4W206A M207A CF, no crosslinking was observed to either Vps24-292C or Vps24-2136C in the presence of ATP (Fig. 6b, c). These data showed that two different parts of the ESCRT-III structural core come into direct contact with the hexamer pore in a manner that depends on ATP hydrolysis and intact pore loops.
Discussion
The HDX and cross-linking data provide strong evidence for unfolding and processive translocation of a model ESCRT-III subunit by Vps4. These findings provide direct confirmation for what has been a favored model in the field for a decade 28, yet has eluded direct verification. The global denaturation mechanism might seem at first glance like overkill. In principle, less ATP might be consumed if only localized regions of assembly contacts in ESCRT-III were destabilized. However, Vps4 functions in the disassembly of many different ESCRT-III subunits. It seems likely that the early acting subunit Vps20 might form one type of contact with Snf7, which comprises the bulk of the complex. Late acting and less abundant subunits such as Vps2, Did2, and Ist1 likely have yet other types of contacts. ESCRT-III forms a range of filaments and tubes of varying diameters, with the ones observed in vivo ranging from 1.5 μm diameter midbodies in cytokinesis to 25 nm intralumenal bud necks in yeast multivesicular bodies 3,9,13,18-22. Although it has not yet been established that all ESCRT-III assemblies are functional or competent for disassembly, it is unlikely that their structural nature is identical. Global denaturation has a clear advantage in providing a single mechanism to deal with all of these situations.
In the context of ESCRT-III disassembly at sites of vesicle budding on membranes, the translocation mechanism offers some apparent benefits. The ESCRT complexes are likely to be densely packed at vesicle budding and scission sites. Available EM images are consistent with the concept that the vicinity of these sites is sterically crowded 21. The translocation mechanism allows the newly liberated ESCRT-III subunit to emerge on the side of Vps4 that is distal to the membrane. This relocalization has the virtue of preventing the extracted ESCRT-III subunit from simply re-assembling into the newly vacant site created by its own departure from the assembly.
Vps4 belongs to the meiotic clade of AAA+ ATPases 51. The other members of this clade include the microtubule severing enzymes spastin and katanin, which are thought to function by removing individual tubulin subunits from microtubules. A small angle x-ray scattering reconstruction of spastin showed that it is a hexamer with a central pore, similar to models proposed for Vps4 52. Based on the observation that spastin possesses a central pore, it has been proposed that at least a portion of the tubulin monomer is unfolded and threaded through this pore in the course of disassembly 52. Given the close parallels between Vps4 and spastin, together with the results presented here, it seems reasonable to speculate that spastin could completely denature tubulin subunits and extrude them all the way through the central pore.
Some ESCRT-III subunits, such as Vps24, do not bind directly to Vps4, and the question remains as to how these subunits are removed. Once the Vps4-binding subunits are denatured, any non-covalent interactions with the remaining Vps24 subunits would be lost. One possibility is that Vps4 recruitment to specifically binding subunits also brings non-binding ESCRT-III subunits like Vps24 into close enough proximity of the central processing pore to be engaged and translocated. A similar mechanism has been described for the degradation of heterodimeric substrates by the ClpAP protease, where only one of the substrate monomers contains a degradation signal but delivers the second monomer by tethering it to the protease 53. An alternative, simple explanation is that not all of the ESCRT-III subunits actually need to be processed through the central pore. Once a critical number of subunits were released from the lattice, remaining subunits might spontaneously or cooperatively dissociate. Such a mechanism would be consistent with our observation that over the same 1-minute time frame in which ~75 % of Vps24-2 is disassembled, only ~35–40% of the Vps24-2 subunits undergo unfolding. This suggests that Vps4 may have to process only as few as one-half of the subunits in the assembly in order to completely break it apart.
Methods
Protein expression and purification
We generated a sequence encoding Saccharomyces cerevisiae Vps24 (1–179aa) and Vps2 (181–232aa) through overlapping PCR and cloned into the pMBP-HIS2 vector. We expressed the resulting Vps24-2 construct as an N-terminal TEV-cleavable His6-MBP fusion in E. coli Rosetta2 (DE3) at 20 °C overnight in LB medium. We initially purified the protein using His-Talon resin (Clontech). We mixed the eluted fractions from the His-Talon column with TEV (tobacco etch virus) protease (1:200, w/w) and dialyzed the pooled fractions against buffer QA(20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM β-mercaptoethanol) overnight. We then loaded the cleaved samples onto a 5 ml HiTrap Q-sepharose FF column (GE Healthcare) and eluted the material with a gradient of 0%-100% buffer QB (20 mM Tris-HCl, pH 8.0, 500 mM NaCl) in 12 column volumes. We pooled and gel filtered the peak fractions containing untagged Vps24-2 in buffer A (20 mM Tris, pH 8.0,100 mM NaCl) on a HiLoad 16/60 Superdex 75 (GE Healthcare) column. We expressed full-length Saccharomyces cerevisiae Vps4 (1–437) as a fusion protein with an N-terminal TEV-cleavable His6 tag. We carried out the expression and purification of Vps4 in the same way as for Vps24-2. We generated the cysteine-free Vps4 (Vps4 CF) Vps4W206A M207A mutant and single cysteine mutants of Vps4 and Vps24-2 through site directed mutageneis and purified them as for Vps24-2 except that their N-terminal His-tag was not cleaved off. K. Nyquist (Martin lab, UC Berkeley, CA) gave us the ClpX and ClpP proteins.
We generated the MBP-Vps24-2-ssrA construct by adding an ssrA tag (AANDENYALAA) to the C-terminus of Vps24-2 through overlapping PCR. We induced expression of the resulting fusion protein with 1 mM IPTG (isopropyl β-D-1 thiogalactopyranoside) at 30 °C for 90 minutes. We purified MBP-Vps24-2-ssrA protein by His-Talon resin affinity chromatography (Clontech), gel filtration on a HiLoad 16/60 Superdex 200 (GE Healthcare) column and ion-exchange on a HiTrap Q-sepharose FF column (GE Healthcare).
Sedimentation Analysis of Filament Disassembly
We assembled Vps24-2 filaments by concentrating the protein to at least 350 μM and incubating it at 4 °C overnight. We confirmed filament formation by ultracentrifugation of protein samples in a TLA100 rotor at 50,000 rpm for 30 min at 10 °C. After ultracentifugation, the pellet (P) and supernatant (S) were diluted with equal volumes of SDS-PAGE loading buffer, and equal volumes of S and P fractions were analyzed on a 4%–12% Bis-Tris gel run with MES (2-(N-morpholino) ethanesulfonic acid) buffer. Gels were visualized by Coomassie Blue staining and the intensity of bands was quantified with ImageJ.
For sedimentation analysis of Vps24-2 filament disassembly by Vps4, Vps4CF and Vps4W206A M207A mutant, we mixed 3.6 μM Vps24-2 in the form of filament assembly with 10.2 μM Vps4 or mutant in buffer B (50 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)-Na, pH 7.5, 100 mM KCl, 5 mM MgCl2). We initiated reactions by adding ATP to a final concentration of 1 mM. After incubation at 30 °C for 10, 30, and 60 s, we quenched the reactions at 0 °C by adding EDTA to a final concentration of 50 mM. We subjected the samples to ultracentrifugation and analyzed them as described above. We obtained data from three independent repeats.
Hydrogen–Deuterium Exchange Mass Spectrometry Measurements
For MBP-Vps24-2-ssrA, Vps24-2 monomer, and Vps24-2 filament assembly HDX-MS experiments, we initiated amide hydrogen exchange by diluting 5 μl protein samples (67 μM for MBP-Vps24-2-ssrA, 25 μM for Vps24-2 monomer and 72 μM for filaments) into 95 μl D2O buffer (50 mM HEPES, pH 7.5, 100 mM KCl and 5 mM MgCl2) containing 1 mM ATP at 30°C. At various time points, we quenched the exchange reaction by cooling to 0 °C and adjusting pH to 2.2 through adding an equal volume of ice-cold quench buffer 1 (400 mM KH2PO4/K2HPO4, pH 2.2, 6 M guanidine HCl). After incubation on ice for 20 s, we further diluted the quenched samples to 20% of their previous concentration through addition of ice-cold quench buffer 2 (400 mM KH2PO4/K2HPO4, pH 2.2) and injected into an HPLC-MS (Agilent 1100) system with in-line peptic digestion and desalting immersed in ice bath. We eluted the desalted peptides and directly analyzed them with an Orbitrap Discovery mass spectrometer (Thermo). We extensively cleaned the HPLC system between samples. We prepared fully deuterated control samples by 3 cycles of drying and resolubilization in D2O buffer and 6M guanidine HCl. We analyzed both non-deuterated and fully deuterated Vps24-2 samples (or MBP-Vps24-2-ssrA) to serve as 0% and 100% controls.
For MBP-Vps24-2-ssrA and Vps24-2 filament assembly with Vps4, 67 μM MBP-Vps24-2-ssrA or 72 μM Vps24-2 filament assembly we pre-incubated samples with 200 μM Vps4 on ice for 5 min. We initiated amide hydrogen exchange by dilution of the pre-incubated sample to 5 % of its previous concentration with D2O buffer containing 1 mM ATP at 30°C. The subsequent experimental procedure was as described above. After dilution to 5 % of its previous concentration, the we kept the protein concentration the same as in the sedimentation assays. For MBP-Vps24-2-ssrA with ClpX experiments, we pre-incubated 52.5 μM MBP-Vps24-2-ssrA with 150 μM ClpX on ice for 5 min, and the subsequent experimental procedure was as described above. For deuteron incorporation measurements of undigested protein at 11 different time points ranging from 10 s to 15 min, we performed experiments as above except that the in-line peptic digestion was omited and that the elution was performed without gradient.
HDX data analysis
We performed initial peptide identification by running tandem MS/MS experiments. We identified peptides through a Mascot search 54 in PEAKS Studio 7 (Thermo). We carried out the initial analysis of the peptide centroids using HD-Examiner v1.3 (Sierra Analytics) followed by manual verification of every peptide to check retention time, charge state, m/z range, and the presence of overlapping peptides. We calculated relative deuteration levels (%D) of the peptic peptides covering 95% of Vps24-2 by comparing the centroids of the molecular ion isotope envelope to that of the non-deuterated and fully deuterated samples using the software HD-Examiner v1.3. We adjusted the deuteron content for deuteron gain/loss during digestion and HPLC. For analysis of the EX2 to EX1 transition, we calculated the abundance of the two deuterated mass species by fitting two Gaussian peaks to the bimodal isotope cluster in Prism (GraphPad). The isotope cluster area of the unfolded state (U) was divided by the sum of the areas of the bimodal isotope cluster (U/(F+U)) and plotted versus the exchange time.
For deuteron incopration measurements of undigested protein samples, we calculated the molecular weight (MW) of undigested sample using GPMAW 8.2 (Lighthouse Data), and we calculated the relative Deuteron incorporation at each time points by dividing the maximum Deuteron incorporation (MW fully deuterated – MW undeuterated) with the Deuteron incorporation at each time points (MWi – MW undeuterated).
Crosslinking
We mixed Vps4CF (or Vps4W206A M207A CF) with Vps4CF 206C ΔMIT , and we mixed Vps4EQ CF with Vps4EQ CF 206C ΔMIT, both at a molar ratio of 8:1. We then reduced the resultant mixture at 22 °C for 1 hr with 5 mM DTT (dithiothreitol), concentrated it to 400 μM, and assembled it into hexamers by adding 2 mM AMPPNP and 5 mM EDTA, followed by buffer exchange using Micro Bio-Spin columns (Bio-Rad) to remove excessive DTT. We activated the introduced cysteine in the I92C or M136C mutants of Vps24-2 by formation of a mixed disulfide with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, Sigma). Following buffer exchange using Micro Bio-Spin columns (Bio-Rad) to remove excessive DTNB, we mixed 1 μM DTNB activated Vps24-2 single cysteine mutant with 5 μM pre-assembled Vps4 hexamer mixture in the presence of either 10 mM ATP/MgCl2 or 10 mM AMPPNP/EDTA or no nucleotide in crosslinking buffer (50 mM HEPES, pH 7.5, 200 mM KCl). We allowed the crosslinking reaction to proceed in the dark for 1 min at 30°C before we stopped it by adding 200 mM iodoacetic acid (IAA) in 400 mM Tris-HCl, pH 8.5, 6.2 M urea, 2 mM EDTA. We allowed alkylation with IAA to proceed in the dark at 22°C for 30 min. We then loaded the samples onto nonreducing SDS-PAGE, and disulfide cross-linked Vps24-2*Vps4 complexes were detected bywestern blot using an anti-polyHis antibody (R&D systems, monoclonal mouse IgG1 mlone# AD1.1.10 http://www.rndsystems.com/Products/MAB050, 1:5000).
Electron microscopy
We concentrated Vps24 to 350 μM for overnight self-assembly at 4 °C18 and we then diluted the resulting filaments to 3.5 μM for negative stain EM. We placed 4 μl droplets of the sample on glow-discharged carbon-coated copper grids and we negatively stained them using 2% (w/v) uranyl acetate solution. We examined the negatively stained samples under a transmission electron microscope (Tecnai-12; FEI) operated at 120 keV and recorded on a charge-coupled device camera (4k TemCam-F416; TVIPS) at a magnification of 49,000×.
ATPase activity assay
We determined the basal ATPase activities of Vps4 wild-type and mutants using the ADP-Glo Kinase assay kit (Promega) at room temperature. Briefly, we incubated 2 μM Vps4 in 25 μl reaction buffer (50 mM HEPES, pH 7.5, 200 mM KCl, 1 mM ATP and 10 mM MgCl2) with or without 4 uM Vps24-2. At different time points (ranging from 10 s to 1 min), we stopped the reactions by addition of 25 μL ADP-Glo™ reagent. After a 40 min incubation to allow for the complete depletion of unconsumed ATP, we added 50 μL kinase detection reagent to simultaneously convert ADP to ATP and allow the newly synthesized ATP to be measured using a luciferase/luciferin reaction. After another incubation of 60 min, we measured luminescence using a plate-reading luminometer (GloMax, Promega) and correlated to ADP concentrations using a standard ATP-to-ADP conversion curve generated in parallel. We interpreted the data in Prism (GraphPad).
Supplementary Material
Acknowledgements
We thank K. Nyquist (U. C. Berkeley) for samples of E. coli ClpX and ClpP. This work was supported by grants R01AI112442 (J.H.H.) and R01GM094497 (A.M.) from the National Institutes of Health.
Footnotes
Author Contributions B. Y. conceived the project, created reagents, acquired data, analyzed data, and wrote the manuscript; G. S. analyzed data; Q. S. acquired data; A. M. conceived the project and wrote the manuscript; J. H. H. conceived the project, analyzed data, and wrote the manuscript.
References
- 1.Votteler J, Sundquist WI. Virus Budding and the ESCRT Pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson PI, Cashikar A. Multivesicular Body Morphogenesis. Annual Review of Cell and Developmental Biology. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. 28. [DOI] [PubMed] [Google Scholar]
- 3.Guizetti J, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 4.Agromayor M, Martin-Serrano J. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 2013;23:433–441. doi: 10.1016/j.tcb.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Choudhuri K, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez AJ, et al. ESCRT Machinery Is Required for Plasma Membrane Repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 7.Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1307–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lata S, et al. Helical Structures of ESCRT-III are Disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill CP, Babst M. Structure and function of the membrane deformation AAA ATPase Vps4. Biochimica Et Biophysica Acta-Molecular Cell Research. 2012;1823:172–181. doi: 10.1016/j.bbamcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muziol T, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Bajorek M, et al. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao JY, et al. Structural Basis of Ist1 Function and Ist1-Did2 Interaction in the Multivesicular Body Pathway and Cytokinesis. Molecular Biology of the Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamborlini A, et al. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-terminal Domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 17.Lata S, et al. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghazi-Tabatabai S, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The Endosomal Sorting Complex ESCRT-II Mediates the Assembly and Architecture of ESCRT-III Helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 21.Cashikar AG, et al. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife. 2014:e02184. doi: 10.7554/eLife.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen QT, et al. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 24.Stuchell-Brereton M, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 25.Hurley JH, Yang D. MIT domainia. Dev. Cell. 2008;14:6–8. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Kieffer C, et al. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 27.Monroe N, et al. The oligomeric state of the active Vps4 AAA ATPase. J. Mol. Biol. 2013;426:510–525. doi: 10.1016/j.jmb.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott A, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao JY, Xia HC, Yoshino-Koh K, Zhou JH, Xu ZH. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 2007;374:655–670. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartmann C, et al. Vacuolar protein sorting: Two different functional states of the AAA-ATPase Vps4p. J. Mol. Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Gonciarz MD, et al. Biochemical and Structural Studies of Yeast Vps4 Oligomerization. J. Mol. Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu ZH, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2 : 1 complex with Vta1p. J. Mol. Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landsberg MJ, Vajjhala PR, Rothnagel R, Munn AL, Hankamer B. Three-Dimensional Structure of AAA ATPase Vps4: Advancing Structural Insights into the Mechanisms of Endosomal Sorting and Enveloped Virus Budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XD, et al. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 35.Davies BA, et al. Coordination of Substrate Binding and ATP Hydrolysis in Vps4-Mediated ESCRT-III Disassembly. Molecular Biology of the Cell. 2010;21:3396–3408. doi: 10.1091/mbc.E10-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 37.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein- degradation machine. Biochimica Et Biophysica Acta-Molecular Cell Research. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdelhakim AH, Sauer RT, Baker TA. The AAA plus ClpX machine unfolds a keystone subunit to remodel the Mu transpososome. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2437–2442. doi: 10.1073/pnas.0910905106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M, et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 41.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Martin A, Baker TA, Sauer RT. Pore loops of the AAA plus ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA plus ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide Translocation by the AAA plus ClpXP Protease Machine. Chem. Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of Asymmetric ClpX Hexamers Reveal Nucleotide-Dependent Motions in a AAA plus Protein-Unfolding Machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maillard RA, et al. ClpX(P) Generates Mechanical Force to Unfold and Translocate Its Protein Substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA- tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spurlino JC, Lu GY, Quiocho FA. The 2.3-A Resolution Structure of the Maltose-Binding of Maltodextrin-Binding Protein, a Primary Receptor of Bacterial Active Transport and Chemotaxis. J. Biol. Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 49.Azmi IF, et al. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 50.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. Journal of Structural Biology. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 52.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma S, Hoskins JR, Wickner S. Binding and degradation of heterodimeric substrates by ClpAP and ClpXP. J. Biol. Chem. 2005;280:5449–5455. doi: 10.1074/jbc.M412411200. [DOI] [PubMed] [Google Scholar]
- 54.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.