SUMMARY
Building on previous studies, we defined the repertoire of proteins comprising the immuno-proteome of E. coli O157:H7 (O157) cultured in DMEM supplemented with norepinephrine (NE; O157 immuno-proteome), a β-adrenergic hormone that regulates E. coli O157 gene expression in the gastrointestinal tract, using a variation of a novel proteomics-based platform proteome mining tool for antigen discovery, called Proteomics-based Expression Library Screening (PELS; Kudva et al., 2006). The E. coli O157 immuno-proteome (O157-IP) comprised 91 proteins, and included those identified previously using PELS, and also proteins comprising DMEM- and bovine rumen fluid- proteomes. Outer membrane protein A (OmpA), a common component of the above proteomes, and reportedly a contributor to E. coli O157 adherence to cultured Hep-2 epithelial cells, was interestingly found to be a modulator rather than a contributor to E. coli O157 adherence to bovine recto-anal junction squamous epithelial (RSE) cells. Our results point to a role for yet to be identified members of the O157-IP in E. coli O157 adherence to RSE-cells, and additionally implicate a possible role for the OmpA regulator, TdcA, in the expression of such adhesins. Our observations have implications for development of efficacious vaccines for preventing E. coli O157 colonization of the bovine gastrointestinal tract.
Keywords: O157, Cattle, Norepinephrine, Immuno-proteome, GeLC-MS/MS
INTRODUCTION
E. coli O157 was first identified as a human enteric pathogen in 1982 and has since been implicated in several outbreaks and sporadic infections [1, 2]. Currently this pathogen ranks fourth after Campylobacter, Salmonella, and Shigella among the etiologic agents causing diarrhea in North America [3, 4]. Cattle are the primary reservoirs for this human pathogen and hence, food that is of bovine origin (beef, milk), or that is contaminated with manure (water, produce), and undercooked can transmit infection to humans [1, 2].
Cattle demonstrate a characteristic seasonal pattern in E. coli O157 shedding [5, 6, 7], with a shedding rate that peaks in summer and early fall, and ranging from 0% to 61% during this time [7, 8, 9]. The average duration an individual animal is culture-positive for E. coli O157 is 30 days, but can range from a few days to 1 year [10, 11]. Although E. coli O157 colonizes cattle, it does not naturally cause disease in this host. Several factors may restrict the ability of this organism to cause disease in cattle such as, a complex interplay between microbial factors uniquely expressed within the gastrointestinal tract (GIT) of cattle, host responses against such factors, and differences between animal and human host environments. At the same time, these factors may also contribute towards the persistence of E. coli O157 in these animals, especially at the recto-anal junction of their GIT [12, 13].
Several pre-harvest control measures are being evaluated in cattle to control or eliminate E. coli O157 from entering the food chain. Some of these measures include dietary changes, biocontrol through niche engineering, competitive exclusion, use of bacteriophages or colicins and administration of vaccines [14, 15, 16, 17, 18, 19]. Vaccines offer a more targeted approach to the elimination of this human pathogen from the ruminant reservoirs; however the commercially available type III secreted (TTSS) Tir and Esp proteins-based cattle vaccine, as well as the E. coli O157 siderophore receptor and porin targeting vaccine, appear to be limited in efficacy, causing log reductions in the number of colonizing E. coli O157 with no effect on the duration of fecal shedding of this bacteria by the animal following administration of 2 -3 doses of these vaccines (20-23). In addition, our studies have shown that the TTSS proteins considered critical for E. coli O157 adherence to the follicle-associated epithelium (FAE) at the recto-anal junction (RAJ) have no role in E. coli O157 adherence to the squamous epithelial cells also constituting this site, [24-26]. This fact renders it imperative that additional proteins playing a role in E. coli O157 colonization of cattle be identified and included to increase vaccine efficacy.
Based on these observations, we previously evaluated the E. coli O157 proteome as expressed in the minimal medium DMEM (O157 DMEM proteome), and bioinformatically inferred a subset of proteins, different from those encoded on the Locus of Enterocyte Effacement, as potential adhesins (25). In a subsequent study, we demonstrated that pooled bovine hyperimmune sera either completely blocks or significantly reduces adherence of E. coli O157 cultured in DMEM to bovine rectoanal junction squamous epithelial (RSE) cells (26 and unpublished data); however, we did not identify the repertoire of O157 protein targets of polyclonal antibodies in the pooled hyperimmune sera in that study. The identification of such immunogenic proteins and evaluation of the ability of salient proteins to contribute to E. coli O157 adherence to RSE cells was the objective of this study.
To identify the panel of such proteins expressed in sufficient amounts within the bovine GIT to be immunogenic, we employed a variation of a novel platform proteome mining tool for protein antigen/biomarker discovery called Proteomics-based Expression Library Screening (PELS; 27). PELS involves immuno-affinity capture of recombinant proteins encoded by genes on inserts within clones constituting optimized expression libraries constructed using genomic DNA of a pathogen of interest using immobilized “bait” antibodies from diverse sources, including acute/convalescent sera of susceptible hosts, sera from reservoirs hosts or sera generated in experimental hosts. SEQUEST searching of relevant databases with mass spectral data following one dimensional SDS-PAGE and tandem mass spectrometry (Ge-LC-MS/MS) of elutions of specifically immuno-captured proteins results in protein identification [27]. A suite of bioinformatics is employed to assign putative functions to hypothetical/unknown proteins, and then specialized bioinformatics-based algorithms are applied when indicated to infer putative functions (e.g., identification of proteins with adhesin potential) [27]. In this particular variation of PELS, native proteins expressed by a wild type pathogen cultured in vitro under conditions that mimic the in vivo environment (rather than recombinant proteins expressed from genomic DNA expression libraries) are used as a substrate for immuno-affinity capture by “bait” antibodies. Such an approach was anticipated to result in in vivo expressed immunogenic protein targets, including those adhesins contributing towards E. coli O157 adherence to bovine RSE cells, for development of comprehensive E. coli O157-cattle colonization prevention modalities.
EXPERIMENTAL PROCEDURES
Bacterial strains, antibodies, and culture conditions
The E. coli O157 strains and antibodies/antisera used in this study are listed in Table 1. The wild-type E. coli O157 strain EDL933, a sequenced human clinical isolate [28], was used as the prototype in all immuno-proteomics assays. To determine the E. coli O157 proteins induced under select conditions mimicking the host in vivo environmental conditions that are part of the E. coli O157 immuno-proteome in cattle, bacteria were cultured in Dulbecco modified Eagle medium-low glucose, (DMEM; Gibco/lnvitrogen Corporation, Grand island, NY) supplemented with 500 μM NE (DMEM-NE) [25-27]. Specifically, an overnight culture of the wild-type O157 strain in Luria-Bertani (LB) broth was pelleted and washed with sterile phosphate buffered saline (PBS; pH 7.4), and inoculated at an initial OD600 of 0.05 into fresh DMEM-NE. After incubation at 37°C with shaking at 250 rpm to an OD600 of 1.0, cells were harvested by centrifugation at 7,000 rpm, 15 min at 4°C. Cells were washed three times with an equal volume of sterile PBS (pH 7.4), and processed to obtain cell lysate and pellet fractions for immuno-proteomics as described previously [27] and below.
Table 1.
Bacteria and antibodies/antisera used in this study.
| Bacteria | Source | Antibodies/Antisera | Source |
|---|---|---|---|
| E. coli O157:H7 strain EDL 933 (Stx1+, Stx2+, Intimin+) | American Type Culture Collection (ATCC), Manassas, VA | Mouse monoclonal antibody (McAb) 1. anti-EspA 2. anti-EspB 3. anti-Tir |
National Animal Disease Center (NADC), USDA, ARS, Ames, IA collection; Obtained from Dr. Frank Ebel, Institut für Medizinische Mikrobiologie, Giessen, Germany |
| E. coli O157:H7 strain 86-24 SmR Streptomycin resistant derivative of the human E. coli O157:H7 isolate 86-24 from the Washington State Outbreak [32] (SmR, Stx1−, Stx2+, Intimin+) | National Animal Disease Center (NADC), USDA, ARS, Ames, IA collection; Obtained from Dr. Alison O'Brien, USUHS, Bethesda, MD. | ||
| E. coli O157:H7 isolate RM 6049; Human isolate associated with the 2006 spinach outbreak [33, 34] (Stx1−, Stx2+, Intimin+) | Dr. Robert Mandrell, Produce Safety and Microbiology, USDA, ARS, WRRC, Albany, CA. | ||
| E. coli O157:H7 strain 86-24 SmR, NalR; Naladixic acid resistant of E. coli O157:H7 SmR [35, 36] (SmR, NalR, Stx1−, Stx2+, Intimin+) | Dr. Alfredo Torres, UTMB, Galveston, TX. | Pooled Polyclonal Antisera comprising of: 1. Rabbit anti-EspA 2. Rabbit anti-EspB 3. Rabbit anti-Tir 4. Bovine anti-Intimin 5. Rabbit anti-H7 |
National Animal Disease Center (NADC), USDA, ARS, Ames, IA collection; Obtained from, 1+2. Dr. Jorge Giron, UF-Gainesville, FL 3.Dr. A. O'Brien, USUHS, Bethesda, MD 4.NADC, Ames, IA 5.Becton,Dickinson, and Company, Franklin Lakes, NJ |
| OmpA, TdcA mutants and complemented derivatives of E. coli O157:H7 strain 86-24 SmR,NalR: AGT601: 86-24 ompA::cat, SmR CmR AGT601S: 86-24ΔompA, SmR AGT601S (pOmpA) P9C8F2: 86-24 tdcA::Tnp; SmrKmr AGT602: P9C8F2 ompA::cat, tdcA::Tnp; Smr Cmr Kmr. |
E. coli O157 adherence inhibition assay
The adherence of E. coli O157 to the bovine rectoanal junction squamous epithelial (RSE) cells has been demonstrated and exploited for development of a standardized assay to study adherence [24-26]. In this study, the ability of monoclonal antibodies against each of the TTSS proteins, including EspA, EspB and Tir [2], pooled polyclonal antisera [26] elicited against purified TTSS proteins, namely, EspA, EspB, Tir and Intimin, and the flagellar antigen H7 [2], and bovine hyperimmune sera [26], to block adherence of E. coli O157 to RSE cells (National Animal Disease Center [NADC] stocks) was evaluated (Tables 1-3). All antibodies/antisera were tested at 1:5, 1:10, 1:50 and 1:100 dilutions, except for final assays which were performed at 1:50/1:100 dilutions to conserve sera. Results were consistent across dilutions. Specificity of the antisera was verified in western blots using E. coli O157 cell lysates and individual recombinant proteins (data not shown). Normal rabbit sera (Sigma) or cattle sera (NADC) from healthy, non-infected animals, at a 1:5, 1:50 or 1:100 dilutions, were used as controls.
Table 3.
Quantitation of RSE and HEp-2 cells with adherent E. coli O157 strains in the absence and presence of antisera.
| E.coli O157 strains tested | Percent Mean +/− standard error of mean, of eukaryotic cells with adherent bacteria in the ranges shown(1) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No Antisera | Pooled Polyclonal Antisera(2) | Pooled Hyperimmune Bovine Sera(2) | |||||||
| Bacterial Adherence Pattern | >10 | 1-10(3) | Bacterial Adherence Pattern | >10 | 1-10 | Bacterial Adherence Pattern | >10 | 1-10 | |
| RSE cells: | |||||||||
| EDL 933 | Aggregative, Moderate | 33.5 ± 14.5 | 67 ± 14 | Aggregative, Moderate | 36 ± 9 | 64 ± 9 | Non-adherent | 0 | 48 ± 1 |
| 86-24 SmR (4) | Diffuse, Strong | 66 ±2.5 | 35 ±2.5 | Diffuse, Strong | 60 ±0 | 40 ±0 | Non-adherent | 0 | 44 ±1.5 |
| RM 6049 | Aggregative, Strong | 100 ±0 | 0 | Aggregative, Strong | 83 ±0 | 17 ±0 | Aggregative, Moderate | 42 ±0.5 | 59 ±0.5 |
| HEp-2 cells: | |||||||||
| EDL 933 | Diffuse, Moderate | 3 ± 3 | 63 ±0 | Non-adherent | 0 | 38.5±3 | Non-adherent | 7 ± 4 | 19.5 ± 4 |
| 86-24 SmR | Diffuse, Moderate | 0 | 78 ±8 | Non-adherent | 0 | 41 ±1 | Non-adherent | 0 | 41 ±7.5 |
| RM 6049 | Diffuse, Moderate | 5 ±5 | 56 ±4 | Non-adherent | 0 | 32 ±6 | Non-adherent | 2 ±2 | 45 ±5 |
Average of two trials conducted for each strain is shown. Each trial had one slide per bacterial group. Each slide in turn had 4 technical replicates spotted on it or in chambers; 15-20 well-dispersed cells were evaluated per spot or chamber. Percent means for ranges used to determine “moderate or strong” adherence are in bold.
All polyclonal antisera were used at 1:50 dilution.
Number of bacteria adhering to each cell is shown as a range of >10, and 1-10. Number of cells without bacteria is not shown.
SmR: Streptomycin-resistant
The RSE adherence assay was conducted as described previously [24-26] with some modifications. Specifically, RSE cells were washed and resuspended in 1 ml DMEM-No Glucose, in 16 x 100mm glass tubes, to a final concentration of 105cells/ml. Bacterial pellets from overnight cultures in DMEM-Low Glucose, incubated at 37°C without aeration, were resuspended in sterile saline with or without sera (control), and incubated at 37°C for 30 mins. The bacteria-antibody mix was then added to the RSE cell suspension to final bacteria:cell ratio of 10:1, and the mixture incubated with aeration (37°C, 110 rpm, for 4 h). At the end of 4 h, the mixture was pelleted and washed thoroughly, once with 14 ml DMEM-NG and twice with 14 ml of sterile, distilled water (dH2O) before reconstituting in 100μl dH2O. Eight 2μl drops of this suspension were placed on Polysine (Thermo Scientific Pierce) slides and dried overnight under direct light to quench non-specific fluorescence, before fixing in cold 95% ethanol for 10 min. The slides were then stained with 1 % toluidine blue, and fluorescence-tagged antibodies that target the O157-antigen and the RSE cell cytokeratins as described previously [24-26].
E. coli O157 adherence patterns on RSE cells were qualitatively recorded as diffuse, or aggregative (clumps) for all positive interactions that involved direct association of bacteria with the cells , and well-dispersed RSE cells (15-20 per drop) were quantitatively analyzed for number of adhering bacteria as previously described [24-26]. If more than 50% of RSE cells had >10 bacteria attached, the adherence was recorded as strongly positive. If more than 50% of RSE cells had 1-10 adherent bacteria, the adherence was recorded as moderately positive. For less than 50% RSE cells with 1-5 adherent bacteria, the adherence was recorded as non-adherent.
For comparision, the adherence-inhibition assays were also performed with HEp-2 cells (Human epidermoid carcinoma of the larynx cells with HeLa contamination; ATCC CCL-23; American Type Culture Collection, Manassas, VA), along the lines of the protocol used for RSE cells as described before [24-26]. Slides were stained with fluorescence-tagged antibodies that target the O157-antigen and the HEp-2 cell actin filaments as described previously [24-26] and qualitatively-quantitatively recorded as indicated above. In addition, RSE/Hep2 adherence assays were performed with diverse wild type or mutant E. coli O157 strains using the same protocol described above but without the addition of antisera/antibodies.
Hyper-immune anti-E. coli O157 cattle sera
Hyper-immune bovine serum samples were generated as described previously [29]. Cattle were experimentally inoculated with a mixture of E. coli O157 strains of cattle and human origin, including strain EDL933, and serum samples collected from nine cattle that had remained culture positive for up to two months (hyper-immune sera) [29]. The hyper-immune sera possessed high titer antibodies against previously identified O157 proteins [25, 27]. Hyper-immune sera were pooled together to compensate for variations in individual immune responses [25, 27], for subsequent use either in the RSE adherence assay or in immuno-proteomics.
Analysis of the E. coli O157 immuno-proteome (O157-IP): (i) Affinity-purification of polyclonal antibodies (pAbs) from pooled hyper-immune cattle sera and coupling of pAbs (“bait” pAbs) to HiTrap NHS-activated HP columns
Polyclonal antibodies (pAbs) from pooled, hyper-immune cattle sera were affinity-purified using HiTrap Protein G HP (1 ml) columns (Amersham Biosciences, Piscataway, NJ), as recommended by the manufacturer and described previously [27]. Briefly, 1 ml of pooled hyper-immune cattle sera was diluted 1:4 in binding buffer (0.02 M Sodium phosphate buffer, pH 7.0), and then loaded using a syringe onto the protein G column equilibrated with ten volumes of binding buffer. Following a wash with ten volumes of binding buffer, bound IgG pAbs (Protein G binds all IgG subclasses but not other Ig isotypes) [30] were eluted with 4 ml of elution buffer (0.1 M Glycine, pH 2.7), directly into five tubes each containing 200 μl of 1M Tris-HCl, pH 9.0. Following quantitation using a nomograph [31], affinity-purified IgG pAbs (“bait” pAbs) were cross-linked to HiTrap NHS-activated HP columns (“charged”), as described previously [27].
(ii) Capture of NE-induced native E. coli O157 proteins by “bait” pAbs on “charged” columns
The capture was performed in a manner similar to that described previously [27]. Charged columns with cross-linked bait pAbs were equilibrated with ten column volumes of binding buffer consisting of PBS (pH 7.4), 0.2% n-octyl-β-D-glucopyranoside (NOG). Cell lysate and pellet fractions of wild type O157 cultured in DMEM supplemented with NE were generated by three cycles of freeze-thaw, diluted with PBS-0.2% NOG containing 2x concentration of “Complete” protease inhibitor cocktail (Roche Diagnostics) in 20 ml, and loaded on charged columns with a syringe at a flow rate of 1 ml/min. Loosely bound proteins were removed by rinsing with 20 volumes of loading buffer. Captured NE-induced native O157 proteins were eluted with 10 ml of 1 M acetic acid directly into a 15 ml Falcon tube containing 500 μl of ammonium hydroxide. Separate charged columns were used for lysate and pellet fractions. Each charged column was reused four times, and eluted fractions after each use pooled to maximize the amount of captured proteins. Elutions of cell lysate and pellet fractions of wild type O157 cultured in DMEM supplemented with NE loaded on columns with quenched active groups and without coupled bait PAbs (“uncharged”) served as controls for assessing nonspecific adsorption to the column matrix. Following confirmation of specificity of immuno-affinity capture as described previously [27], elutions of cell lysates and the cognate pellet fractions from charged and uncharged columns were subjected to 1D SDS-PAGE liquid chromatography tandem mass spectrometry (GeLCMS/MS) as described below.
(iii) 1D SDS-PAGE liquid chromatography tandem mass spectrometry (GeLC-MS/MS)
Protein samples prepared using immuno-proteomics were analyzed at the Harvard Partners Center for Genetics and Genomics, Cambridge, Massachusetts [25, 27]. Eluted native E. coli O157 proteins from the previous steps were concentrated using spin filters (MW cutoff 5000 Daltons; Vivascience Inc., Englewood, NY), fractionated on 1D SDS-PAGE, and digested in-gel with trypsin prior to tandem MS/MS as described earlier [27]. The rationale for incorporating a 1D SDS-PAGE fractionation step is that this modification reduces complexity of protein mixtures, permits a larger dynamic range of protein identification, and allows for significantly better reproducibility [32].
For MS, samples were subjected to three different runs on an LCQ DECA XP plus Proteome X workstation from Thermo Finnigan as described earlier [32]. For each run, 10 μL of each reconstituted sample was injected with a Famos Autosampler, and the separation was done on a 75 μm (inner diameter) x 20 cm column packed C18 media running at a flow rate of 0.25 μl/min provided from a Surveyor MS pump with a flow splitter with a gradient of water, 0.1% formic acid and then 5% acetonitrile, 0.1% formic acid (5%-72%) over the course of 480 min (8.0 hour run). Between each set of samples, a standard of a 5 Angiotensin mix of peptides (Michrom BioResources) was run to ascertain column performance, and observe any potential carryover that might have occurred. The LCQ was run in a top five configurations, with one MS scan and five MS/MS scans. Dynamic exclusion was set to 1 with a limit of 30 seconds.
Peptide identifications were made using SEQUEST (Thermo Finnigan) through the Bioworks Browser 3.2, as described previously [27]. Sequential database searches were performed using the O157 strains EDL933 and Sakai FASTA database from European Bioinformatics institute http://www.ebi.ac.uk/newt/display using static carbamidomethyl-modified cysteines and differential oxidized methionines. A reverse O157 strain EDL933 FASTA database was spiked in to provide noise and determine validity of the peptide hits, so that known and theoretical protein hits can be determined without compromising the statistical relevance of all the data [32]. LCQ data were searched with a 2-Dalton window on the MS precursor with a 0.8 Dalton on the fragment ions. Peptide score cutoff values were chosen at cross-correlation values (Xcorr) of 1.8 for singly charged ions, 2.5 for doubly charged ions, and 3.0 for triply charged ions, along with delta rank scoring preliminary cutoff (deltaCN) values of 0.1, and cross-correlation normalized values (RSp) of 1. The cross-correlation values chosen for each peptide assured a high confidence match for the different charge states, while the (deltaCN) ensured the uniqueness of the peptide hit. The RSp value of 1 ensured that the peptide matched the top hit in the preliminary scoring. At these peptide filter values, very few reverse database hits were observed, which permitted a higher confidence on the few single peptide protein identifications. Furthermore, single hit proteins were also manually validated to ensure relevance.
Bioinformatics
NE-induced immunogenic native O157 proteins that were designated as unknown/hypothetical upon querying the O157 strain EDL933 FASTA database at European Bioinformatics Institute were analyzed via bioinformatics. To ascribe a cellular location, amino acid sequences of cognate proteins were obtained from the O157 strain EDL 933 database at http://www.tigr.org, and used to first identify extracytoplasmic proteins containing signal sequences using the program SignalP 3.0 at http://www.cbs.dtu.dk/services/SignalP. Next, extracytoplasmic proteins secreted by pathways not involving a signal peptide (non-classical protein secretion) were screened for using programs SecretomeP 2.0 at http://www.cbs.dtu.dk/services/SecretomeP/ and TatP 1.0 at http://www.cbs.dtu.dk/services/TatP/. Then, subcellular protein localization of extracytoplasmic proteins was predicted using the PSORT/PSORT-B program (http://psort.nibb.ac.jp/). Putative functions were determined by querying the Conserved Domain Database (CDD) at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
RESULTS AND DISCUSSION
Polyclonal and monoclonal antibodies targeting the TTSS proteins do not block or decrease E. coli O157 adherence to RSE cells
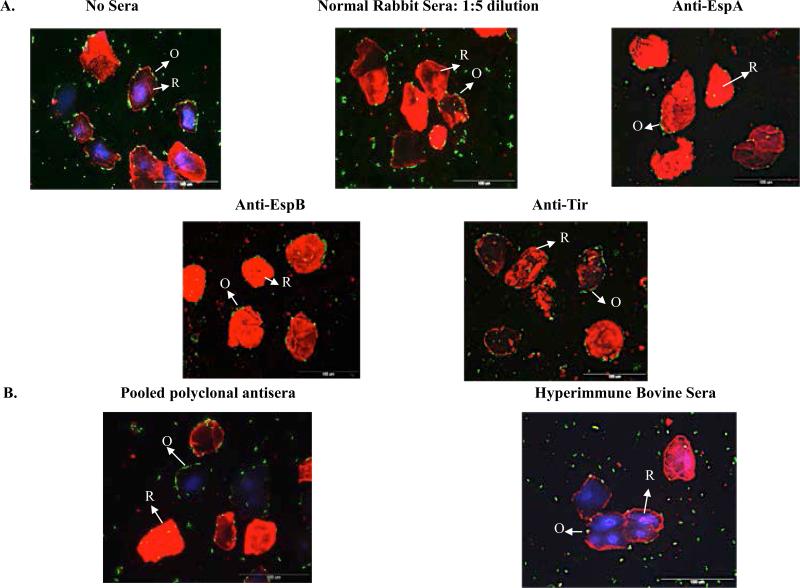
E. coli O157 strain EDL 933 (Table 1) pre-incubated with normal rabbit/bovine sera, at 1:5 dilution in sterile saline, showed the same RSE cell-adherence pattern (aggregative, moderately positive) as observed for this bacterial strain in the absence of any sera (Figure 1, panel A) [24-27]. Likewise, bacteria pre-incubated with pooled, polyclonal antisera generated against the TTSS proteins (Intimin, Tir, EspA and EspB), and flagellar H7 antigen (Table 2), as well as mouse monoclonal antibodies specifically generated against individual TTSS proteins (EspA, EspB, Tir; Figure 1, panel A; Table 2), did not show any changes in the RSE cell-adherence patterns/numbers, confirming our previous observations that none of these proteins influenced O157 adherence to RSE cells [26, 27]. Similar results were obtained with E. coli O157 strains 86-24SmR and RM 6049, associated with different outbreaks (Tables 1 & 3) [2, 33, 34, 35]. The pooled polyclonal antisera and monoclonal antibodies were only effective in blocking adherence of all the E. coli O157 strains tested to HEp-2 cells (Tables 2-3). These results clearly support alternate mechanisms of adherence being used by E. coli O157 to bind RSE cells at the bovine RAJ.
Figure 1.
Adherence patterns of E. coli O157 strains EDL 933 on RSE cells, in the absence and presence of antisera. Panel A, in the absence of sera and presence of normal rabbit serum (NRS; 1:5 dilution), and different monoclonal antibodies. Panel B, in the presence of pooled polyclonal antisera against TTSS proteins and the H7 flagellar antigen, and the pooled bovine hyperimmune sera. The monoclonal and polyclonal antisera were used at 1:50 dilution in these adherence-inhibition assays. The immunofluorescence stained slides are shown at 40x magnification. Bacteria (O, O157) have green fluorescence, and RSE cells’ (R) cytokeratins have orange-red fluorescence and their nuclei have blue fluorescence.
Table 2.
Quantitation of RSE and HEp-2 cells with adherent E. coli O157 strain EDL933 in the absence and presence of monoclonal antibodies.
| Bacteria Tested | Bacterial Adherence Pattern | Eukaryotic cells with adherent bacteria, in the ranges shown, for two different trials(1) (MOI(2) = 106 bacteria:105 cells) |
Percent Mean +/− standard error of mean, of eukaryotic cells with adherent bacteria in the ranges shown(6) | Percent Mean +/− standard error of mean, of eukaryotic cells with adherent bacteria in the ranges shown(6) | |||
|---|---|---|---|---|---|---|---|
| Trial I | Trial II | ||||||
| >10 | 1-10(3) | >10 | 1-10 | >10 | 1-10 | ||
| RSE cells(4): | |||||||
| EDL 933 | Aggregative, Moderate | 38 (80)(5) | 42 (80) | 15 (80) | 65 (80) | 33.5 ± 14.5 | 67 ± 14 |
| EDL 933 + McAb anti-EspA | Aggregative, Moderate | 23 (80) | 57 (80) | 33 (80) | 47 (80) | 35 ± 6 | 65 ± 6 |
| EDL 933 + McAb anti-EspB | Aggregative, Moderate | 36 (75) | 39 (75) | 12 (76) | 64 (76) | 32 ± 16 | 68 ± 16 |
| EDL 933 + McAb anti-Tir | Aggregative, Strong | 54 (65) | 11 (65) | 37 (70) | 32 (70) | 68 ± 15 | 31.5 ± 14.5 |
| HEp-2 cells(4): | |||||||
| EDL 933 | Diffuse, Moderate | 0 (80) | 50 (80) | 5 (80) | 50 (80) | 3 ± 3 | 63 ±0 |
| EDL 933 + McAb anti-EspA | Non-adherent | 0 (80) | 43 (80) | 0 (80) | 33 (80) | 24 ±3.0 | 47.5 ±6.5 |
| EDL 933 + McAb anti-EspB | Diffuse, Moderate | 5 (80) | 75 (80) | 7 (80) | 60 (80) | 7.5 ± 1.5 | 84.5 ± 9.5 |
| EDL 933 + McAb anti-Tir | Non-adherent | 0 (80) | 35 (80) | 0 (80) | 41 (80) | 0 | 47.5 ± 3.5 |
Each trial had one slide per bacterial group. Each slide in turn had 4 technical replicates spotted on it or in chambers; 15-20 well-dispersed cells were evaluated per spot or chamber. Monoclonal antibodies and polyclonal hyperimmune antisera were used at 1:50 dilution.
MOI, multiplicity of infection.
Number of bacteria adhering to each cell is shown as a range of >10, and 1-10. Number of cells without bacteria is not shown.
Total number of cells evaluated in each trial is shown in parenthesis.
Percent means for ranges used to determine “moderate or strong” adherence are in bold.
Bovine hyperimmune sera abrogates/decreases E. coli O157 adherence to RSE cells
Compared to the pooled polyclonal and monoclonal antibodies tested, only the pooled bovine hyperimmune sera effectively blocked/decreased E. coli O157 adherence to both the RSE and HEp-2 cells. The ability of the hyperimmune sera to negatively impact adherence was also observed with diverse E. coli O157 strains (Tables 1 & 3; Figure 1, panel B) further confirming our previous observations that this pooled serum contained antibodies to bacterial proteins other than the TTSS proteins [25, 27].
The NE-induced E. coli O157 protein repertoire includes targets of polyclonal antibodies in pooled cattle hyper immune sera (the E. coli O157 bovine immuno-proteome or O157-IP)
To rapidly determine the E. coli O157 bovine immuno-proteome (O157-IP), we utilized a modification of the platform proteome mining tool for protein antigen/biomarker discovery called Proteomics-based Expression Library Screening (27). In this particular variation of PELS (Figure 2), native O157 proteins (rather than recombinant proteins) served as the substrate for immuno-affinity capture by “bait” polyclonal antibodies. These substrate proteins were generated by culturing O157 in vitro under conditions that approximated the in vivo host environment. In particular, the growth medium was supplemented with the β-adrenergic hormone, NE, which provided the in vivo environmental cues. NE, an enteric neurotransmitter, is present in the lumen of the gastrointestinal tract due to neuronal release following stress and trauma, and is a mimic of autoinducer 3 (AI-3), a cell-to-cell signaling molecule produced by resident enteric microflora [36-38]. AI-3 is used as a signal by O157 to determine arrival at its intestinal niche, and also as a signal to commence expression of a variety of genes, including those related to virulence, via quorum sensing [36-38]. Furthermore, NE reportedly augments adherence of O157 to ligated bovine ileal loops [39], and to diverse mucosal epithelial cells [40, 41].
Figure 2.
Principle of technology used to identify the E. coli O157-IP.
To define the E. coli O157 -PA in cattle, we harvested and lysed cells cultured in DMEM-NE, and captured immunogenic native proteins by passing pellet and lysate fractions through Hi Trap NHS-activated columns coupled to polyclonal antibodies affinity purified from previously generated hyperimmune sera of cattle colonized with a diverse collection of O157 strains [27; Figure 2]. GeLC-MS/MS followed by SEQUEST database searching was then used to identify specifically captured native O157 proteins contained in pooled elution fractions (Figure 2). Sequential database searches performed using the O157 strains EDL933 and Sakai FASTA database from European Bioinformatics institute yielded minimal false-positive rates for the proteins identified in the three separate GeLC-MS/MS runs used for this study and were: 2.8% for Run I, 7.1% for Run II, and 12.6% for Run III (See Supplemental Data Sheet 1 and Single peptide spectra).
The O157-IP in cattle included 91 proteins (1.7% of the proteome of the sequenced O157 strain EDL 933) [28; Supplemental data-Table 1]. The O157-IP included proteins of unknown function, as well as those diverse functions that localized to all compartments of the bacterial cell, and encoded by genes situated on the backbone (genomic sequences common to both the non-pathogenic E. coli K12 and O157), O-islands (genomic sequences unique to O157), and pO157 (the virulence plasmid present in all O157 strains) [28]. Given that O157 only colonizes the gastrointestinal tract of cattle but causes neither the symptoms nor the extraintestinal sequelae seen in humans, we anticipated the identification of several previously defined adhesins for colonization of O157 and related pathogens. In accordance, we identified Iha, an iron-regulated, outer membrane ferric siderophore receptor that reportedly contributes to the adherence of E. coli O157 and other pathogens to cultured intestinal epithelial cells [42, 43]. Interestingly, Iha has been recently reported to be expressed at higher levels in bovine reservoirs than in humans [44]. Another immunogenic protein identified in this study was OmpA, an outer membrane porin implicated in the adherence of E. coli O157 to cultured intestinal epithelial cells [45-47]. Both Iha and OmpA were also identified by PELS [27], which presents a strong rationale for further evaluation of the role of these proteins in O157 colonization of the bovine gastrointestinal tract.
This methodology further identified Stx1, a bacteriophage-encoded toxin implicated in the pathogenesis of complicated E. coli O157 disease in humans. This toxin was recently reported to contribute to E. coli O157 adherence to intestinal epithelial cells by increasing surface expression of nucleolin on host epithelial cells, thereby augmenting intimin-γ (see below)-mediated adherence [48]. Interestingly, the O157-IP did not include intimin-γ, the primary E. coli O157 adhesin that is encoded by a gene on a pathogenicity island called the locus of enterocyte effacement (LEE) [49], despite the fact that this protein was part of the NE-induced E. coli O157 proteome (data not shown). Intimin-γ was not part of the PELS-defined E. coli O157 IP in cattle as well [27]. The reasons for this are unclear; perhaps the humoral adaptive immune response to this adhesin in cattle is suboptimal following colonization after experimental oral inoculation and/or the protein is expressed at low levels, in a site-specific manner.
EspP, a component of the PELS-identified IP was also identified in this study [27]. An immune response to this secreted plasmid (pO157)-encoded protein decreases fecal shedding of O157 following oral challenge when it is optimally administered as a constituent of an experimental, parenteral multi-component vaccine formulated with an adjuvant to cattle [50]. However, none of the LEE-encoded secreted proteins such as EspA, EspB, and Tir, shown to be targeted by immune responses of cattle and administered as part of a bovine vaccine [50] were identified in this study. This was unexpected since one of these vaccine components, namely EspB, was part of the NE-defined E. coli O157 proteome (data not shown), and also identified by PELS in an earlier study [27]. The reason for this observation is unclear; a plausible explanation is that these proteins, under conditions of in vitro culture of wild type E. coli O157 adopted in this study, are expressed below the threshold for efficient immuno-affinity capture. Yet another immunogenic protein identified in this study that merits evaluation for development of novel strategies for preventing E. coli O157 colonization of the bovine gastrointestinal tract is the outer membrane enterobactin receptor, FepA. Studies evaluating the use of FepA vaccines against coliforms causing bovine mastitis have shown that bovine anti-FepA IgG inhibited growth of coliforms by interfering with the binding of the ferric enterobactin complex to this receptor [51].
Also part of the O157-IP were proteins previously reported to be part of the quorum sensing regulatory network (Supplemental data-Table 1) in E. coli or other pathogens. One of these was ArcB, an inner membrane sensor-kinase with global regulatory function under microaerobic and anaerobic conditions, which has demonstrated homology to LuxQ of the AI-2 pathway and acts in concert with LuxS to enable the growth of Actinobacillus actinomycetemcomitans under iron-limiting conditions [52, 53]. ArcB is also responsible for transducing stress signals to the phage shock protein PspF regulon in E. coli [54]. Another protein, PspA, is a part of the PspF regulon, and regulates energy usage in the cell by promoting anaerobic growth and reducing motility. Also identified was MopA (GroEL), a heat shock chaperone protein that has a role in the synthesis of N-acyl homoserine lactone used in quorum sensing by Sinorhizobium meliloti strain AK631 [55]. This protein also regulates the Vibrio fischeri lux operon in E. coli cells via the folding of the transcriptional regulator of the operon, LuxR [56]. This proteome mining tool further identified Dps, a DNA binding protein involved in DNA protection during starvation and oxidative stress, shown to be regulated by quorum sensing in Burkholderia pseudomallei [57], besides the outer membrane protein Lpp, a murein lipoprotein, which along with OmpA, is induced by AI-2 in E. coli [58]. Also identified as part of the O157-IP were OmpC, an outer membrane protein that plays a role in bacterial adaptation to hyperosmotic environments, and reportedly is induced by autoclaved stationary phase culture supernatant of E. coli via a quorum sensing pathway different from that of AI-2 [59], and an inner membrane catalase KatG. The regulation of catalase expression via quorum sensing has been demonstrated previously in Pseudomonas aeruginosa [60]. These results were predicted since NE is a mimic of AI-3, the cell-to-cell signaling molecule used by E. coli O157 [40-42], and other pathogens [61] to signal arrival at a specific niche within the host and regulate expression of genes encoding diverse functions, including virulence.
Interestingly, the O157-IP included twenty-five proteins that had been previously identified as being part of the bovine rumen fluid-proteome [62; Supplemental data-Table 1]. These 25 proteins could be associated either (i) with a role in transport, metabolism and anaerobic respiration or, (ii) with a role in bacterial adaptation to stress and adherence or, (iii) were encoded within genomes of bacteriophages. These results were anticipated since they are consistent with the requirements for adaptation to, propagation and survival within the hostile host environment; such proteins were also part of the PELS-defined E. coli O157 immuno-proteome in cattle (27) (Supplemental data-Table 1)
A point that needs emphasis is that since this technology operates on the principle of immuno-proteomics, not all proteins encoded by genes within a multigene family will be identified despite induction of such proteins following growth of the pathogen under conditions that reflect the in vivo environment. Hence, the O157-IP reported in this study only included those E. coli O157 proteins that were both induced during growth of this pathogen in the presence of NE and immunogenic in cattle. In accordance, our results reveal that the technology identified only immunogenic but not the non-immunogenic E. coli O157 proteins encoded by multigene families that were either functionally or genetically linked. This is exemplified by the inclusion of RhlB, the DEAD-box RNA helicase and Eno, an enolase, and exclusion of PNPase, a polynucleotide phosphorylase (all of which belong to a multi-protein family involved in the degradation of RNA [63], from the O157-IP.
TdcA, a plausible regulator of effectors of RSE cell adherence
Considering that the outer membrane protein, OmpA, was a target both in this and other studies (27), and also identified in vitro in the DMEM/DMEM-NE- and the rumen fluid- associated E. coli O157 proteomes [25, 62], we evaluated the role of this protein in RSE cell adherence. OmpA has also been shown to enable E. coli O157 adherence to human epithelial cells [45-47]. We analyzed previously characterized isogenic mutants of E. coli O157 strain 86-24 SmR, NalR (Table 1) with either an insertional (AGT601) or deletion (AGT601S) event interrupting ompA expression and compared their adherence to the parent and ompA complemented (AGT601S (pOmpA)) strains in both RSE and HEp-2 cell adherence assays (Tables 1 & 5) [45-47]. Likewise, we also evaluated a transposon insertional mutant causing the inactivation of tdcA associated primarily with the degradation of threonine and causing OmpA over expression (P9C8F2) by itself or, with an additional isogenic mutation in ompA (AGT602) (Table 1), in the same parent strain [45-47]. Although all the strains adhered to HeLa cells in the other studies, quantitative differences were observed among strains in the percent of adhering bacteria: the TdcA mutant was hyperadherent, whereas all OmpA mutants demonstrated slightly decreased adherence compared to the parent strain; pattern of adherence of the complemented strain was similar to that of the parent strain [45-47].
Results of HEp-2 adherence assays were concordant with those observed with HeLa cells (Table 4); under our assay conditions the complemented strain AGT601Sp(OmpA) and the parent strain exhibited similar patterns of adherence. A higher percentage of HEp-2 cells with adhering TdcA mutant (P9C8F2) strain, and a slightly lower percentage of HEp-2 cells with adhering OmpA mutants, AGT601 and AGT602, were observed when compared to the parent and complemented strains. Adherence of the OmpA-deletion mutant AGT601S was low enough to be categorized as non-adherent to HEp-2 cells (Table 4).
Table 4.
Quantitation of E. coli O157 wild-type, TdcA mutant, isogenic OmpA mutants and complemented strains adherence to RSE and HEp-2 cells.
| Bacteria Tested | Bacterial Adherence Pattern | Eukaryotic cells with adherent bacteria, in the ranges shown, for two different trials(1) (MOI(2) = 106 bacteria:105 cells) |
Percent Mean +/− standard error of mean, of eukaryotic cells with adherent bacteria in the ranges shown(6) | ||||
|---|---|---|---|---|---|---|---|
| Trial I | Trial II | ||||||
| >10 | 1-10(3) | >10 | 1-10 | >10 | 1-10 | ||
| RSE cells (4): | |||||||
| 86-24 SmR, NalR | Diffuse, Strong | 46 (80)(5) | 34 (80) | 67 (80) | 13 (80) | 70.5± 13 | 29.5 ± 13 |
| AGT601 | Aggregative, Strong | 61 (80) | 19 (80) | 62 (80) | 18 (80) | 76.5± 0.5 | 23.5 ± 0.5 |
| AGT601S | Aggregative, Strong | 70 (80) | 39 (80) | 74 (80) | 6 (80) | 90± 2 | 10 ± 2 |
| AGT601S (pOmpA) | Diffuse, Strong | 55 (80) | 25 (80) | 46 (80) | 34 (80) | 62.5 ± 5.5 | 37.5 ± 5.5 |
| P9C8F2 | Diffuse, Moderate | 13 (80) | 67 (80) | 0 (80) | 80 (80) | 8 ± 8 | 92 ±8 |
| AGT602 | Diffuse, Moderate | 8 (80) | 72 (80) | 0 (0) | 80 (80) | 5 ± 5 | 95 ± 5 |
| HEp-2 cells (4): | |||||||
| 86-24 SmR, NalR | Diffuse, Moderate | 0 (80) | 55 (80) | 0 (80) | 46 (80) | 0 | 63.5 ± 5.5 |
| AGT601 | Diffuse, Moderate | 0 (80) | 43 (80) | 0 (80) | 55 (80) | 0 | 61.5 ±6.5 |
| AGT601S | Non-adherent | 0 (80) | 36 (80) | 0 (80) | 37 (80) | 0 | 45.5 ± 0.5 |
| AGT601S (pOmpA) | Diffuse, Moderate | 0 (70) | 50 (70) | 0 (70) | 59 (70) | 0 | 77.5 ± 6.5 |
| P9C8F2 | Diffuse, Moderate | 0 (75) | 66 (75) | 15 (80) | 65 (80) | 9.5± 9.5 | 84.5 ± 3.5 |
| AGT602 | Diffuse, Moderate | 8 (75) | 38 (75) | 1 (80) | 48 (80) | 6 ± 5 | 55.5 ± 4.5 |
Each trial had one slide per bacterial group. Each slide in turn had 4 technical replicates spotted on it or in chambers; 15-20 well-dispersed cells were evaluated per spot or chamber.
MOI, multiplicity of infection.
Number of bacteria adhering to each cell is shown as a range of >10, and 1-10. Number of cells without bacteria is not shown.
Total number of cells evaluated in each trial is shown in parenthesis.
Percent means for ranges used to determine “moderate or strong” adherence are in bold.
In contrast, the results were considerably different in the RSE cell adherence assays (Table 4, Figure 3). The AGT601 and AGT601S strains demonstrated an aggregative, strong adherence phenotype compared to the parent and OmpA complemented strains that were diffuse, strong in their adherence (Table 4). On the other hand, the P9C8F2 and AGT602 strains adhered in a diffuse, moderate manner and in much lower numbers (1-5 bacteria per RSE cell) than the other strains (Tables 1 & 4). This suggests that (i) OmpA, although not central to the RSE-O157 interaction, modulates E. coli O157 adherence to RSE cells, and its absence allows other unidentified adhesins to exert stronger (aggregative) bacterial interactions with the RSE cells, while its over expression interferes with this interaction; and (ii) TdcA may have a regulatory effect on an hitherto unidentified adhesin, the decreased expression of which in a tdcA mutant (P9C8F2) is the likely cause for its decreased adherence to RSE cells. Neither the over expression of OmpA in this mutant (P9C8F2), nor the isogenic insertional mutation of OmpA in the same (AGT602) changed its RSE cell-adherence phenotype (Table 4; Figure 3). These observations strongly support a modulating role for OmpA and the involvement of other adhesins in the adherence of O157 to bovine RSE cells. While the exact mechanism of modulation is unclear, a likely role for alterations in membrane architecture is an interesting possibility. This is a hypothesis we plan to test in future experiments.
Figure 3.
Adherence patterns of E. coli O157 strains 86-24 SmR, NalR, its isogenic ompA (AGT601, AGT601S) and/or tdcA (P9C8F2, AGT602) mutants, and OmpA complemented (AGT 601S (pOmpA)) strains on RSE cells. The immunofluorescence stained slides are shown at 40x magnification. Bacteria (O, O157) have green fluorescence, and RSE cells’ (R) cytokeratins have orange-red fluorescence and their nuclei have blue fluorescence.
Studies with the antisera, antibodies and these mutants clearly show that with RSE cell adherence, factors of no relevance (eg EspA, EspB) cause no change in the adherence patterns while factors that possibly modulate (eg. OmpA) cause quantitative/qualitative increases in adherence in their absence; the absence of factors (eg. in the tdcA mutant) that may have a key role in adherence results in decreased or abrogated adherence. Inferences gleaned from these interesting observations mandate further analysis of tdcA mutants for identification of adhesins other than OmpA that facilitate O157 interactions with RSE cells.
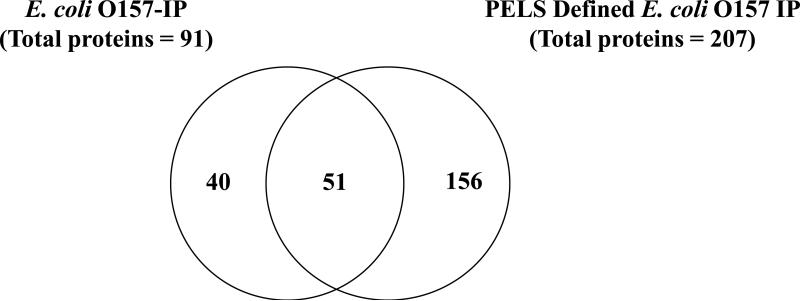
The proteomics-based tool described in this study presents a convenient strategy for rapidly defining native immuno-proteomes of diverse microorganisms cultured in vitro by provision of cues the microorganism is likely to encounter within the in vivo host environment. To corroborate the methodology, we compared the O157-IP determined in this study with the recently described PELS-defined E. coli O157 IP. Although the O157 IP expectedly was narrower than the PELS-defined E. coli O157 IP, given the source of the substrate for immuno-affinity capture, 51 (56%) immunogenic proteins were identified by both methodologies, whereas 40/91 (44%) were uniquely identified in this study (Figure 4). The 51 core proteins likely represent those that play an important role in E. coli O157 colonization of cattle, and hence are currently under further evaluation.
Figure 4.
The E. coli O157-IP complements PELS defined-E. coli O157-IP.
A limitation of this technique is the requirement for handling wild type pathogens, especially those categorized under Category “A” by the Centers for Disease Control and Prevention, which necessitates appropriately trained personnel and biocontainment facilities. Another limitation is that a prior knowledge of relevant in vivo environmental conditions/cues that induce virulence gene expression and the respective surrogates for supplementing in vitro cultures of the cognate pathogen is required; however, such conditions/surrogates of in vivo signals may be defined empirically based on those for related pathogens occupying similar anatomical niches. For example, in addition to a role in regulation of virulence gene expression in E. coli O157, NE has been recently reported to augment virulence properties of Campylobacter jejuni, following supplementation of an in vitro culture of this gastrointestinal pathogen in iron-limited medium [61].
In conclusion, this study corroborates the observations made by us and other researchers that proteins besides the TTSS proteins play a significant role in E. coli O157 colonization of cattle. We are presently evaluating the protective role of the E. coli O157 immuno-proteome in both the mouse and cattle models of colonization, while evaluating E. coli O157 mutants, especially the tdcA mutant, for adhesins of relevance to bovine rectal cell adherence.
Supplementary Material
ACKNOWLEDGEMENTS
US National Institutes of Health (NIH) Grants R21 AI055963 to I.T.K supported part of this work. Excellent technical assistance provided by Bryan Wheeler at the National Animal Disease Center (NADC), Ames, IA, with the eukaryotic cell adherence/adherence-inhibition assays is acknowledged. The Animal Resource Unit personnel at NADC, especially Dr. Rebecca Madison and Denise Chapman, are gratefully acknowledged for assisting with the collection of the bovine rectoanal junction tissues and swabs. C.J.H was supported in part by the National Research Initiative of the US Department of Agriculture Cooperative State Research, Education and Extension Service grants 99-35201-8539 and 04-04562, and US NIH Grants NO1-HD-0-3309, U54-AI-57141, P20-RR16454, P20-RR15587 and P20-GM103408.
Abbreviations
- RAJ
recto-anal junction
- RSE
recto-anal junction squamous epithelial cells
- PELS
Proteomics-based Expression Library Screening
- IP
Immuno-proteome
- NE
norepinephrine
- NADC
National Animal Disease Center
Footnotes
Disclaimer: Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
USDA is an equal opportunity provider and employer.
SUPPLEMENTAL DATA
Table 1: Proteins comprising the O157-IP in cattle.
Data Sheet 1: O157-IP MS/MS data
Single peptide spectra: Single peptide spectra of O157-IP
CONFLICT OF INTEREST STATEMENT
The authors declare no competing financial/commercial conflicts of interest.
REFERENCES
- 1.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli 0157:H7 infections. A broad clinical spectrum. Ann.Intern.Med. 1998;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, O'Brien AD. Escherichia coli O157:H7 and other Shiga Toxin-Producing E. coli Strains. ASM Press; Washington, D.C.: 1998. [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States –Major pathogens. Emerg, Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morbidity and Mortality Weekly Report. 2011 Jun 10;60:749–755. [PubMed] [Google Scholar]
- 5.Blaser MF, Smith PD, Ravdin JI, Greenberg HB, Guerrant RL, editors. Infections of the Gastrointestinal Tract. Raven Press, Ltd.; New York: 1995. pp. 739–761. [Google Scholar]
- 6.Hancock DD, Besser TE, Rice DH. Macro- and micro- epidemiology of E. coli O157:H7-prospects for pre-harvest control.. Presented at the 97th Annual Meeting of the U.S. Animal Health Association; Las Vegas, NV.. 1993. [Google Scholar]
- 7.Hancock DD, Besser TE, Kinsel ML, Tarr PI, Rice DH, Paros MG. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington State. J. Epidemiol. Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland RE. Some infectious causes of diarrhea in young farm animals. Clin Microbiol Rev. 1990;3:345–375. doi: 10.1128/cmr.3.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besser TE, Hancock DD, Pritchett LC, McRae EM, Rice DH, Tarr PI. Duration of Detection of Fecal Excretion of Escherichia coli O157:H7 in Cattle. J. Infect. Dis. 1997;175:726–729. doi: 10.1093/infdis/175.3.726. [DOI] [PubMed] [Google Scholar]
- 11.Zhao T, Doyle MP, Shere J, Garber L. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 1995;61:1290–1293. doi: 10.1128/aem.61.4.1290-1293.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DG, Gally DL. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naylor SW, Roe AJ, Nart P, Spears K, Smith DG, Low JC, Gally DL. Escherichia coli O157: H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiol. 2005;151:2773–2781. doi: 10.1099/mic.0.28060-0. [DOI] [PubMed] [Google Scholar]
- 14.Buchko SJ, Holley RA, Olson WO, Gannon VP, Veira DM. The effect of different grain diets on fecal shedding of Escherichia coli O157:H7 by steers. J Food Prot. 2000;63:1467–74. doi: 10.4315/0362-028x-63.11.1467. [DOI] [PubMed] [Google Scholar]
- 15.Kudva IT, Hatfield PG, Hovde CJ. Effect of Diet on the Shedding of Escherichia coli O157:H7 in a Sheep Model. Appl. Environ. Microbiol. 1995;61:1363–1370. doi: 10.1128/aem.61.4.1363-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudva IT, Jelacic S, Tarr PI, Youderian PA, Hovde CJ. Biocontrol of Escherichia coli O157 with O157-specific Bacteriophages. Appl. Environ. Microbiol. 1999;65:3767–3773. doi: 10.1128/aem.65.9.3767-3773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murinda SE, Roberts RF, Wilson RA. Evaluation of colicins for inhibitory activity against diarrheagenic Escherichia coli strains including serotype O157:H7. Appl Environ Microbiol. 1996;62:3196–202. doi: 10.1128/aem.62.9.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurmi E, Nuotio L, Schneitz C. The competitive exclusion concept: development and future. Int J Food Microbiol. 1992;15:237–40. doi: 10.1016/0168-1605(92)90054-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhao T, Doyle MP, Harmon BG, Brown CA, Mueller PO, Parks AH. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J Clin Microbiol. 1998;36:641–7. doi: 10.1128/jcm.36.3.641-647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter AA, Klashinsky S, Li Y, Frey E, Townsend H, Rogan D, Erickson G, Hinkley S, Klopfenstein T, Moxley RA, Smith DR, Finlay BB. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine. 2004;22:362–369. doi: 10.1016/j.vaccine.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Allen KJ, Rogan D, Finlay BB, Potter AA, Asper DJ. Vaccination with type III secreted proteins leads to decreased shedding in calves after experimental infection with Escherichia coli O157. Can. J. Vet. Res. 2011;75:98–105. [PMC free article] [PubMed] [Google Scholar]
- 22.Fox JT, Thomson DU, Drouillard JS, Thornton AB, Burkhardt DT, Emery DA, Nagaraja TG. Efficacy of Escherichia coli O157:H7 siderophore receptor/porin proteins–based vaccine in feedlot cattle naturally shedding E. coli O157. Foodborne Path. Dis. 2009;6:893–899. doi: 10.1089/fpd.2009.0336. [DOI] [PubMed] [Google Scholar]
- 23.Thomson DU, Loneragan GH, Thornton AB, Lechtenberg KF, Emery DA, Burkhardt DT, Nagaraja TG. Use of a Siderophore Receptor and Porin Proteins-Based Vaccine to Control the Burden of Escherichia coli O157:H7 in Feedlot Cattle. Food. Path. Dis. 2009;6:871–877. doi: 10.1089/fpd.2009.0290. [DOI] [PubMed] [Google Scholar]
- 24.Kudva IT, Dean-Nystrom E. Bovine Recto-anal Junction Squamous Epithelial (RSE) cell Adhesion Assay for Studying Escherichia coli O157 Adherence. J. App. Microbiol. 2011;111:1283–1294. doi: 10.1111/j.1365-2672.2011.05139.x. [DOI] [PubMed] [Google Scholar]
- 25.Kudva IT, Griffin RW, Krastins B, Sarracino DA, Calderwood SB, John M. Proteins other than the Locus of Enterocyte Effacement-encoded Proteins Contribute to Escherichia coli O157:H7 Adherence to Bovine Rectoanal Junction Stratified Squamous Epithelial cells. BMC Microbiol. 2012;12:103. doi: 10.1186/1471-2180-12-103. doi:10.1186/1471-2180-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudva IT, Hovde CJ, John M. Adherence of Non-O157 Shiga-Toxin Escherichia coli to Bovine Recto-anal Junction Squamous Epithelial Cells Appears to be Mediated by Mechanisms Distinct from those Used by O157. Food. Path. Dis. 2013;10:375–381. doi: 10.1089/fpd.2012.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudva IT, Krastins B, Sheng H, Griffin RW, Sarracino DA, Tarr PI, Hovde CJ, Calderwood SB, John M. Proteomics-based expression library screening (PELS): a novel method for rapidly defining microbial immunoproteomes. Mol Cell Proteomics. 2006;5:514–9. doi: 10.1074/mcp.T600013-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 29.Rice DH, Sheng HQ, Wynia SA, Hovde CJ. Rectoanal Mucosal Swab Culture Is More Sensitive Than Fecal Culture and Distinguishes Escherichia coli O157:H7-Colonized Cattle and Those Transiently Shedding the Same Organism. J. Clin. Microbiol. 2003;41:4924–4929. doi: 10.1128/JCM.41.11.4924-4929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harlow E, Lane D, editors. Antibodies-A Laboratory Manual. Cold Spring Harbor Laboratory; New York: 1998. [Google Scholar]
- 31.Bollag DM, Rozycki MD, Edelstein JS. Protein Methods. Wiley-Liss, Inc.; New York: 1996. [Google Scholar]
- 32.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman MA, Menge C, Casey TA, Laegrid W, Bosworth BT, Dean-Nystrom E. Bovine immune responses to Shiga-toxigenic Escherichia coli O157:H7. Clin. Vac. Immunol. 2006:1322–1327. doi: 10.1128/CVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooley M, Carychao D, Crawford-Miksza L, Jay MT, Myers C, et al. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS ONE. 2007;2(11):e1159. doi: 10.1371/journal.pone.0001159. doi:10.1371/journal.pone.0001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravva SV, Cooley MB, Sarreal CZ, Mandrell RE. Fitness of outbreak and environmental strains of Escherichia coli O157:H7 in aerosolizable soil and association of clonal variation in stress gene regulation. 2014;3:528–548. doi: 10.3390/pathogens3030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Frey E, Mackenzie AMR, Finlay BB. Human response to Escherichia coli O157:H7 infection: Antibodies to secreted virulence factors. Infect. Immun. 2000;68:5090–5095. doi: 10.1128/iai.68.9.5090-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proceed. Natl. Acad. Sci. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke MB, Sperandio V. Events at the host-microbial interface of the gastrointestinal tract III. Cell to cell signaling among microbial flora, host, and pathogens: there is a whole lot of talking going on. Am. J. Phsiol. Gastrointest. Liver Physiol. 2005;288:G1105–09. doi: 10.1152/ajpgi.00572.2004. [DOI] [PubMed] [Google Scholar]
- 39.Vlisidou I, Lyte M, vanDiemen PM, Hawes P, Monaghan P, Wallis TS, Stevens MP. The neuroendocrine stress hormone norepinephrine augments Escherichia coli O157:H7-induced enteritis and adherence in a bovine ligated ileal loop model of infection. Infect. Immun. 2004;72:5446–5451. doi: 10.1128/IAI.72.9.5446-5451.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, Brown DR, Xie Y, Green BT, Lyte M. Catecholamines modulate Escherichia coli O157:H7 adherence to murine cecal mucosa. Shock. 2003;20:183–188. doi: 10.1097/01.shk.0000073867.66587.e0. [DOI] [PubMed] [Google Scholar]
- 41.Green BT, Lyte M, Chen C, Xie Y, Casey MA, Kulkarni-Narla A, Vulchanova L, Brown DR. Adrenergic modulation of Escherichia coli O157:H7 adherence to the colonic mucosa. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1238–G1246. doi: 10.1152/ajpgi.00471.2003. [DOI] [PubMed] [Google Scholar]
- 42.Tarr PI, Bilge SS, Vary JC, Jr., Jelacic S, Habeeb RL, Ward TR, Baylor MR, Besser TE. Iha: a Novel Escherichia coli O157:H7 Adherence-Conferring Molecule Encoded on a Recently Acquired Chromosomal Island of Conserved Structure. Infect. Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HLT, Tarr PI. The IrgA Homologue Adhesin Iha Is an Escherichia coli Virulence Factor in Murine Urinary Tract Infection. Infect. Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid RA, Tabata TA, Oatley MJ, Besser TE, Tarr PI, Moseley SL. Expression of putative virulence factors of Escherichia coli O157:H7 differs in bovine and human infections. Infect Immun. 2006;74:4142–4148. doi: 10.1128/IAI.00299-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres AG, Kaper JB. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 2003;71:4985–4995. doi: 10.1128/IAI.71.9.4985-4995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres AG, Jeter C, Langley W, Matthysse AG. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl Environ Microbiol. 2005;71:8008–15. doi: 10.1128/AEM.71.12.8008-8015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres AG, Li Y, Tutt CB, Xin L, Eaves-Pyles T, Soong L. Outer membrane protein A of Escherichia coli O157:H7 stimulates dendritic cell activation. Infect. Immun. 2006;74:2676–2685. doi: 10.1128/IAI.74.5.2676-2685.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson CM, Sinclair JF, Smith MJ, O'Brien AD. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc Natl Acad Sci U S A. 2006;103:9667–72. doi: 10.1073/pnas.0602359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 50.Potter AA, Klashinsky S, Li Y, Frey E, Townsend H, Rogan D, Erickson G, Hinkley S, Klopfenstein T, Moxley RA, Smith DR, Finlay BB. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine. 2004;22:362–369. doi: 10.1016/j.vaccine.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Lin J, Hogan JS, Smith KL. Growth responses of coliform bacteria to purified immunoglobulin G from cows immunized with ferric enterobactin receptor FepA. J Dairy Sci. 1999;82:86–92. doi: 10.3168/jds.S0022-0302(99)75212-4. [DOI] [PubMed] [Google Scholar]
- 52.Fong KP, Gao L, Demuth DR. LuxS and arcB control aerobic growth ofActinobacillus actinomycetemcomitans under iron limitation. Infect Immun. 2003;71:298–308. doi: 10.1128/IAI.71.1.298-308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signalautoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 54.Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281:21147–61. doi: 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- 55.Marketon MM, Gonzalez JE. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J Bacteriol. 2002;184:3466–75. doi: 10.1128/JB.184.13.3466-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manukhov IV, Kotova V, Zavil'gel'skii GB. Host factors in the regulation of the Vibrio fischeri lux operon in Escherichia coli cells. Mikrobiologiia. 2006;75:525–531. [PubMed] [Google Scholar]
- 57.Lumjiaktase P, Diggle SP, Loprasert S, Tungpradabkul S, Daykin M, Camara M, Williams P, Kunakorn M. Quorum sensing regulates dpsA and the oxidative stress response in Burkholderia pseudomallei. Microbiol. 2006;52:3651–9. doi: 10.1099/mic.0.29226-0. [DOI] [PubMed] [Google Scholar]
- 58.DeLisa MP, Wu CF, Wang L, Valdes JJ, Bentley WE. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J Bacteriol. 2001;183:5239–47. doi: 10.1128/JB.183.18.5239-5247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl Environ Microbiol. 2004;70:2038–43. doi: 10.1128/AEM.70.4.2038-2043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, Huang CT, Fredericks J, Burnett S, Stewart PS, McFeters G, Passador L, Iglewski BH. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–93. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 61.Cogan TA, Thomas AO, Rees LE, Taylor AH, Jepson MA, Williams PH, Ketley J, Humphrey TJ. Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut. 2006;2007;56:1060–1065. doi: 10.1136/gut.2006.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kudva IT, Stanton TB, Lippolis JD. Escherichia coli O157:H7 bovine rumen fluid proteome reflects adaptive bacterial responses. BMC Microbiol. 2014;14:48. doi: 10.1186/1471-2180-14-48. doi: 10.1186/1471-2180-14-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.