Abstract
Aggregatibacter actinomycetemcomitans is an important pathogen in the etiology of human periodontal and systemic diseases. Inactivation of the gene coding for the inner membrane protein, morphogenesis protein C (MorC), results is pleotropic effects pertaining to the membrane structure and function of this bacterium. The role of this protein in membrane biogenesis is unknown. To begin to understand the role of this conserved protein, stable isotope dimethyl labeling in conjunction with mass spectrometry was used to quantitatively analyze differences in the membrane proteomes of the isogenic mutant and wild-type strain. A total of 613 proteins were quantified and 601 of these proteins were found to be equal in abundance between the two strains. The remaining 12 proteins were found in lesser (10) or greater (2) abundance in the membrane preparation of the mutant strain compared with the wild-type strain. The 12 proteins were ascribed functions associated with protein quality control systems, oxidative stress responses, and protein secretion. The potential relationship between these proteins and the phenotypes of the morC mutant strain is discussed.
Keywords: Chaperone proteins, Leukotoxin secretion, Type I secretion
1. INTRODUCTION
The cell envelope of Gram-negative bacteria is an essential structure required for cellular homeostasis, protection from environmental stressors, and interactions with other microorganisms or eukaryotic tissues. Three physiologically distinct but contiguous compartments compose this structure: the inner membrane, a phospholipid bilayer adjacent to the cytosol; the periplasm, a viscous aqueous compartment containing the peptidoglycan cell wall; and the outer membrane, an asymmetric bilayer composed of lipids and lipopolysaccharide exposed to the extracellular environment [1]. Each compartment contains conserved proteins with general cellular functions and proteins exclusively associated with the specific organism. Protein functions include roles in transport, which allows the envelope to serve as a barrier to exclude toxic compounds from the cell while simultaneously facilitating import of nutrients, energy generation, and secretion of proteins [1].
Canonical secretion systems have been well characterized in model organisms. These systems are composed of individual proteins or a complex of proteins spanning the lipid bilayer [2–5]. A conserved type I secretion system is present in the periodontal pathogen A. actinomycetemcomitans and is associated with the secretion of a leukotoxin (LtxA) that destroys white blood cells [6]. This system is composed of an inner membrane ATPase (LtxB), a periplasmic channel protein (LtxD) and a pore protein (TolC, also known as TdeA in A. actinomycetemcomitans) that allows for toxin secretion across the outer membrane. The leukotoxin secretion system is homologous to that used for E. coli hemolysin (HlyA)[7]. In E. coli, the secretion of HlyA does not appear to require accessory proteins. However, our laboratory has identified an A. actinomycetemcomitans mutant that displays a decrease in LtxA production through disruption of a gene not typically associated with the type I secretion system [8]. This mutation maps to a gene coding for a 141 kDa inner membrane protein, MorC (Morphogenesis protein C).
Homologs of the MorC protein are suggested to be present in many other bacterial cells based on genomic sequences but relatively little is known about this protein’s function in other systems [8, 9]. In addition to the defect in LtxA secretion in A. actinomycetemcomitans, loss of MorC results in a change in outer membrane morphology from a rugose to smooth appearance, a decrease in size of the organism and an increase in autoaggregation [8, 10]. All of these membrane-related phenotypes suggest a change in the membrane protein composition of the bacterium.
In this study, isotope dimethyl labeling quantitative proteomics was employed on cell envelope preparations to determine if the pleiotropic effects associated with the inactivation of morC are due to a generalized reduction in membrane proteins or quantitative changes in specific proteins in the mutant compared with the parent strain. 613 proteins were consistently quantified in whole membrane preparations of A. actinomycetemcomitans, >98% of which were found to be in equal abundance between the wild-type and mutant strains. The LtxA secretion apparatus was found to be among these proteins. A specific subset of 12 proteins was shown to be consistently changed. These included leukotoxin, chaperone proteins, a fimbrial secretion system subunit, and oxidative stress response proteins. The potential function of these proteins in the modulation of leukotoxin secretion is discussed.
2. MATERIALS AND METHODS
2.1 Bacterial strains and growth conditions
The wild-type strain (WT), VT1169, is an afimbriated serotype b strain of A. actinomycetemcomitans [11]. The morC mutant (MT) strain is an isogenic mutant of VT1169 [8]. A. actinomycetemcomitans strains were routinely cultured in TSBYE medium (0.3% tryptic soy broth, 0.6% yeast extract; Beckton Dickinson, Franklin Lakes, NJ). Incubation was static at 37°C in a humidified 10% CO2 atmosphere. Spectinomycin was added at a concentration of 50μg ml−1 for maintenance of the morC mutant.
2.2 Whole membrane isolation
Bacteria for each of the three biological replicates were streaked for isolation on fresh TSBYE plates from a stock frozen at −80°C. Several colonies were inoculated into liquid media and grown overnight. The overnight cultures were diluted in 250 ml broth and incubated until they reached mid-logarithmic phase (OD495 = 0.3). An aliquot of cells (~5 ml) was removed and subjected to testing for contamination by Gram-staining and inspection of growth characteristics by streaking to an agar plate. This aliquot was also used to verify the phenotypes of the wild-type and morC mutant cells. The remainder was utilized for membrane isolation based on the method of Smith et al. [12]. Briefly, cells were lysed using a French pressure cell (Thermo Scientific, Waltham, MA) and cell debris removed by centrifugation at 10,000 g for 30 minutes. Membranes were recovered by centrifugation at 100,000 g and the pellet suspended in PBS. The procedure was repeated three times and membrane pellets were stored dry at −80°C.
2.3 Dimethyl labeling, fractionation, nanoscale liquid chromatography-mass spectrometry (LC/MS)
Whole envelope fractions were subjected to LC/MS analyses based on the methods of Smith et al. [12] with modifications. The WT and MT membrane pellets (~ 0.5 mg) were suspended in 0.5 mL of 6 M urea/2M thiourea/20 mM HEPES (pH 8) and sonicated on ice with 3 bursts of 20 seconds each, at 1-min intervals. Reduction and alkylation of disulfides was achieved by addition of dithiothreitol (final concentration of 4.5 mM) at 55°C for 30 min., followed by the addition of iodoacetamide (10 mM final concentration) and incubated in the dark at room temperature for 15 min. The solution was diluted 4-fold (20 mM HEPES, pH 8.0) followed by addition of sequencing grade trypsin (Promega, WI) with an enzyme to substrate ratio of 1:50. Following overnight incubation at 37°C, the reaction was terminated by the addition of trifluoracetic acid (TFA) to 1%. The acidified tryptic peptides were purified using Sep-Pak tC18 cartridge (WAT023590, 1cc, 100 mg; Waters, MA) and lyophilized. The lyophilized peptides were reconstituted in 100 uL 1M HEPES (pH 7.5) and subjected to dimethyl labeling as previously described [13, 14]. Briefly, 2 μL of 10% formaldehyde (Sigma, St. Louis, MO) and 4 μL of 500 mM sodium cyanoborohydride (NaCNBH3) (Sigma St. Louis, MO) were added to the WT peptides. 2 μL of 10% d2-formaldehyde (diluted from a 20% solution, 98% D2; Cambridge Isotope Laboratories, MA) and 4 μL of 500 mM sodium cyanoborodeuteride NaCNBD3 in 1 M NaOH (98.7% D2; CDN Isotopes, Canada) were added to the MT peptides. The peptide mixtures were incubated at room temperature for 1 h. The labeling and incubation steps were repeated and it was confirmed that > 99% of the peptides were labeled. The resulting peptides were acidified by addition of TFA to 7%, combined, purified by Sep-Pak tC18, and lyophilized. The lyophilized peptides were suspended in 150 μL of buffer A (10 mM potassium phosphate (pH 2.8) and 25% acetonitrile (CH3CN)). Fifty μL of the peptide solution was fractionated on a PolySULFOETHYL A (silica SCX) cation exchange TopTip (0–200 μL sized, PolyLC, MD) by step elution (in 100 μL aliquots) with solutions of 0, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100, 200, and 350 mM of KCl in buffer A. The eluates were lyophilized, ziptipped (Pierce, Rockford, IL) and dried again. The peptides were stored at −80°C until liquid chromatography/mass spectrometry (LC/MS) analysis.
2.4 Protein identification and data analysis
Database searches for protein identification were conducted following the methods of Smith et al. [12]. Product ion spectra were searched against the OralGen annotation of the A. actinomycetemcomitans HK1651 genome (www.oralgen.org) containing sequences in forward and reverse orientations using the SEQUEST search engines in Proteome Discoverer 1.4 (Thermo Fisher Scientific, MA). The 20 raw files from each experiment were processed as one contiguous input file and a single result file (.msf) was generated. Search parameters were as follows: fully enzymatic activity and two missed cleavage sites allowed for trypsin; peptide MW of 350–5000.; mass tolerance of 20 ppm and 0.8 Da for precursor and fragment ions, respectively; dynamic modifications on methionine (+15.9949 Da: oxidation) (4 maximum dynamic modifications allowed per peptide), static modification on cysteine (+57.0215 Da: carbamidomethylation) as well as static isomeric dimethyl modifications on lysine and N-terminus (+28.0312984 Da: for WT and +34.068961 Da for MT). The raw files were searched separately with “light” or “heavy” labels in the same workflow. Cross-correlation (XCorr) values were applied to limit the false positive (FP) rates to less than 1% in the data sets (with the Target/Decoy PSM Validator node). The relative abundances of peptides between WT and MT (isotopologues) were quantitated by integrating the intensities of peptide ion elution profiles with the Precursor Ions Quantifier node in the Proteome Discoverer (Parameters: Mass precision: 2 ppm, Protein quantification using all peptides, and Single-peak Quan channels were used). The XCorr cut-off values for doubly (2+) and triply (3+) charged peptides are as follows: replicate 1: 2.115 (2+); 2.25 (3+); replicate 2: 2.035 (2+); 2.245 (3+); replicate 3: 2.1 (2+); 2.205 (3+). All the protein identification and quantification information (<1% FP; with protein grouping enabled) was exported from the .msf result files to Excel spreadsheets, which are included as Supplementary Information 1–3.
2.5 Statistical analyses
Quantitative and statistical metrics were used to identify proteins with differential abundance between WT and MT. All MT:WT ratios were log2 transformed to normalize data. Proteins present in all three biological replicates with log2 ratios consistently less than −1 or greater than 1 (corresponding to a two-fold change) were considered for analysis. For each replicate, z-scores were calculated for individual protein ratios based on the mean and standard deviation from that experiment. Average z-scores across all replicates were calculated and proteins with an average z-score of ±2 (corresponding to a p-value of less than 0.05) were classified as increased or decreased. Exact p-values were calculated based on the normal distribution. All analyses were conducted in Excel 2013 (Microsoft, Redmond, WA) and JMP 11.0 (SAS Institute, Cary, NC).
2.6 Bioinformatic analyses
Prediction of subcellular localization of changed proteins was based on multiple bioinformatic tools according to the method of Smith et al. [12]. Functional annotations were assigned to each protein using the WebMGA server to identify cluster of orthologous groups (COG) assignments, gene ontology (GO) terms, and enzyme commission (EC) numbers [15–17]. The BLAST tool was used to identify similar Escherichia coli K-12 or A. actinomycetemcomitans ANH9381 sequences using the EcoGene and Uniprot databases respectively (e-value cutoff = 1e−20, www.ecogene.org, www.uniprot.org). Transporters were identified by querying the transporter classification database with an e-value cutoff of 1e−10 [18]. All available information for each protein was compiled and used to manually synthesize a descriptive annotation and biological function.
2.7 Generation and purification of MorC antibodies
Polyclonal antibodies were generated in rabbits (Cocalico Biologicals, Reamstown, PA) using a 63 kDa expressed fragment of MorC, corresponding to amino acids 46-620. The protein was expressed in E. coli BL21(DE3) cells as a maltose-binding protein (MBP) fusion protein from the pMAL-c2x plasmid (New England Biolabs, Ipswich, MA). The 1725 bp fragment was amplified using primers containing engineered restriction sites for directional cloning into the BamHI and HindIII sites of the plasmid with high fidelity polymerase (Roche, Basel, Switzerland). Maximal protein expression was achieved by induction of exponential-phase E. coli cells with 0.3 mM IPTG for 2 h at 37°C. The MBP-fusion protein, obtained following lysis and centrifugation [19], was purified by amylose affinity chromatography (New England Biolabs, Ipswich, MA) using a batch/column method. The immobilized MBP fusion protein was eluted by the addition of 10 mM maltose to the column buffer. The recombinant protein was collected and concentrated by ultrafiltration (Millipore, Billerica, MA) with a 10,000 kDa cut off and used as the immunogen.
Serum antibodies were concentrated by the addition of 50% ammonium sulfate, dialyzed overnight with phosphate buffered saline (PBS) and incubated with a glutathione s-transferase (GST) fusion protein of MorC immobilized to cyanogen bromide-activated-Sepharose 4B (Sigma Aldrich, St. Louis, MO). The GST-63 kDa protein was generated by expression of the morC fragment, described above, using the pGEX-6P-1 expression plasmid (GE Healthcare BioSciences, Pittsburgh, PA). Purified antibodies were eluted by 0.2M glycine, pH 2.8 and neutralized with 3M Tris, pH 8.8. Antibodies were stored as a 50% glycerol stock at −20°C.
2.8 Generation of 6x Histidine Tagged Leukotoxin B (LtxB)
The ltxB gene was amplified by PCR from genomic DNA using primers corresponding to the 5′ (AAAGTCGACATGCACCACCACCACCACCACGACTCACAGAAAAATACTAATC) and 3′ (AATCTAGATTAATTTACTTGTAATTGGTG) termini of the gene with engineered SalI and XbaI (underlined) endonuclease restriction sites, respectively, for directional cloning into an A. actinomycetemcomitans shuttle plasmid [8]. A 6x His sequence was engineered into the 5′ primer following the endonuclease restriction site (italics). The PCR product was cloned and the 6xHisltxB construct containing plasmid was introduced into the wild-type and morC mutant cells by electroporation.
2.9 Immunoblotting of A. actinomycetemcomitans proteins
Immunoblots were performed to validate the proteomics results. Protein concentrations in whole cell lysates were calculated by the BCA method (Thermo Scientific, Waltham, MA). Secreted leukotoxin was prepared based on the method of Tang et al. [20]. Spent liquid media from A. actinomycetemcomitans strains was concentrated by ultrafiltration using a 50,000 MW cutoff (Millipore, Billerica, MA). Equal amounts of protein were separated by SDS-PAGE on 4–15% gradient gels (Bio-Rad, Hercules, CA) and transferred to an Immobilon-FL PVDF membrane (Millipore, Billerica, MA). Membranes were incubated with Odyssey blocking buffer (LiCor Biosciences, Lincoln, NE), washed three times with PBS-T (Dulbecco’s phosphate buffered saline pH, 7.4 + 0.1% tween-20) and probed with either anti-Aae [18], anti-MorC, anti-LtxA [21, 22] or antibodies specific to the 6xHis epitope tag (Abcam, Cambridge, UK). Binding of primary antibodies was detected by species-specific secondary antibodies conjugated with fluorescent tags according to the manufacturer’s instructions (Li-Cor Biosciences, Lincoln, NE). Fluorescence was detected on an Odyssey Clx infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
2.10 Lipid analysis
Lipid analysis was performed based on the method of Whittaker et al. using cells grown in liquid culture [23]. Mid-logrithmic phase A. actinomycetemcomitans was grown in 1L broth, collected by centrifugation and washed once with PBS. Fatty acids were saponified, methylated and extracted with methyl tert-butyl ether (MTBE) followed by a second extraction of the organic phase with 0.3N NaOH. Analysis was performed using a gas chromatograph (GC2010, Shimadzu, Kyoto, Japan) equipped with a split injector (temperature: 260°C, injection volume: 1 μL, split ratio: 1:20) and a mass spectrometer (GCMS-QP2010 Plus, Shimadzu, Kyoto, Japan) using a Rtx-2330 column (30 m length × 0.25 mm diameter × 0.20 μm film thickness; Restek Bellefonte, PA). FAME were detected in full scan mode (m/z 45 to 500) and identification was achieved using authentic standard mixtures (BAME mix, Supelco Inc., Bellefonte, PA; GLC-463 and GLC-603 from Nu-Chek Prep. Elysian, MN). Mass spectra were based on the web-accessible mass spectra database at American Oil Chemists’ Society (AOCS) lipid library (http://lipidlibrary.aocs.org/).
RESULTS AND DISCUSSION
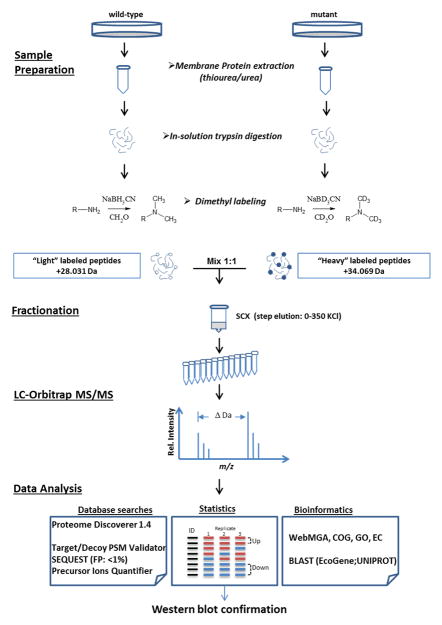
Heterogeneity in individual protein abundance exists in bacterial cells [24]. Some structural proteins, enzymes, and ribosomal subunits are found in high abundance while other proteins are found at much lower levels [25]. In this study, we used a dimethyl labeling strategy (outlined in Figure 1) to quantitatively assess the differences in the membrane proteomes of the isogenic mutant and wild-type strain. To detect peptides derived from the 141 kDa MorC, hypothesized to be a low abundance protein the fractionation procedure was carefully optimized to detect MorC as well as to maximize the proteome coverage. Following trypsin digestion and dimethyl labeling, peptides were separated into 20 fractions on a strong cationic exchanger [12]. Only after this extensive fractionation of the membrane extract were peptides from MorC detected, suggesting that MorC is either present in low abundance in the membrane or is not easily extracted under the conditions used here.
Figure 1. Quantitative proteomics workflow.
Bacterial cells were grown to mid-logarithmic phase and whole envelopes were isolated by differential ultracentrifugation. Envelope-associated proteins were solubilized in a urea/thiourea buffer, incubated with trypsin, isotope labeled and fractionated by cation exchange chromatography. Fractions were subject to LC/MS analysis and data analyzed using various bioinformatics techniques.
We identified 751, 923, and 817 proteins (< 1% FP) in biological replicates 1, 2, and 3 respectively, with MT:WT abundance ratios determined for around 95% of the identified proteins (replicate 1: 715 ratios; replicate 2: 888 ratios; replicate 3: 771 ratios). Proteins that were identified and quantified in all three replicates are listed with their associated MT:WT ratios in Supplementary Information 4. In this study, 613 proteins were identified with quantifiable ratios in all replicates (Figure 2A). Subsequent statistical analyses were performed on the dataset including these 613 proteins. MT:WT ratios were subject to log2 transformation to normalize the ratios. Values of ±1 indicate a two-fold decrease or increase in abundance in the morC mutant. Average values were plotted for each protein to visualize the entire dataset and the distribution was found to be clustered around a log2 ratio of zero (Figure 2B). The average log2 ratio across all experiments was −0.175 (SD = 0.79) with 88% of protein ratios falling within one and >95% within two SD of the mean. These data demonstrate that the majority of proteins identified are equal in abundance between wild-type and morC mutant cells, implying that the phenotypes of the morC mutant are due to specific protein changes rather than a general decrease in membrane proteins.
Figure 2. Characterization of the dataset.
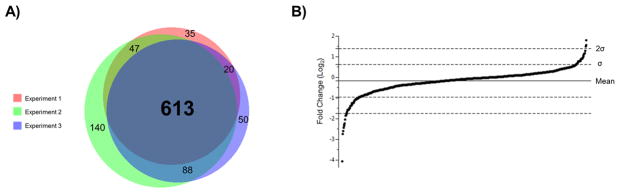
A) A Venn diagram was constructed by inputting identified proteins into the BioVenn website (http://www.cmbi.ru.nl/cdd/biovenn/). Each circle represents a single biological replicate and numbers indicate how many proteins are present in each section. B) Ratios for each consistently identified protein were log2 transformed and plotted in ascending order using JMP 11.0. Each point in the graph represents a single protein.
Biological and technical variation between replicates was controlled for by stringent statistical criteria for identification of differentially abundant proteins. Proteins were initially filtered based on MT:WT ratios and z-scores were calculated if their ratios were consistently changed by at least two-fold between all three experiments. Those which were differentially expressed in MT vs. WT were defined as having average ratios >2 standard deviations from the mean (corresponding to a p-value of < 0.05). Based on these criteria, we deduced that 601 out of 613 proteins (>98%) were unchanged in abundance between WT and MT and 12 proteins were significantly increased or decreased in the morC mutant cells (Table 1, Figure 2B).
Table 1.
Statistical analysis of differentially abundant proteins.
| Oralgen ID | Average Ratio (log2) | SD (log2) | Average z-score | p-value |
|---|---|---|---|---|
| AA01213 | −4.07 | 1.21 | −6.07 | <0.0001 |
| AA00086 | −2.74 | 1.27 | −4.06 | <0.0001 |
| AA00561 | −2.62 | 0.69 | −3.88 | <0.001 |
| AA00470 | −2.44 | 0.92 | −3.6 | <0.001 |
| AA02012 | −2.35 | 0.12 | −3.47 | <0.001 |
| AA00068 | −2.31 | 1.08 | −3.4 | <0.001 |
| AA02806 | −2.23 | 0.69 | −3.29 | <0.01 |
| AA00875 | −1.84 | 0.41 | −2.69 | <0.01 |
| AA00401 | −1.76 | 0.62 | −2.57 | <0.05 |
| AA01280 | −1.53 | 0.26 | −2.22 | <0.05 |
| AA02212 | 1.48 | 0.47 | 2.33 | <0.05 |
| AA01869 | 1.79 | 0.33 | 2.8 | <0.01 |
Changes in the membrane protein profiles were confirmed using available antibodies to known A. actinomycetemcomitans proteins. In all immunoblots, protein concentrations from both strains were normalized before separation by gel electrophoresis to account for the increased aggregation phenotype of the mutant strain. The abundance of most of the proteins identified were shown to be unchanged between the two strains by quantitative proteomics and subsequent statistical analyses. The epithelial cell adhesin Aae (AA02347), an outer membrane protein secreted via the type V autotransporter pathway [22], was included in this cohort of proteins. No difference in abundance between the strains was seen using antibodies specific for Aae (Figure 3A). MorC (AA00961) was not detected in the envelope preparation of the mutant strain compared with the wild-type strain. The absence of MorC in the mutant strain was confirmed by immunoblotting (Figure 3B). A reduction in leukotoxin secretion has been described as a phenotype for the morC mutant strain [8]. Quantification of immunoblots detecting leukotoxin from spent medium demonstrated an 82% reduction in the secretion of the toxin from the mutant compared with the wild-type strain (Figure 3C). A similar decrease (77%) in leukotoxin secretion was observed across all replicates as detected by our statistical analysis of the MS data.
Figure 3. Western blotting and Bioinformatic analyses of significantly changed proteins.
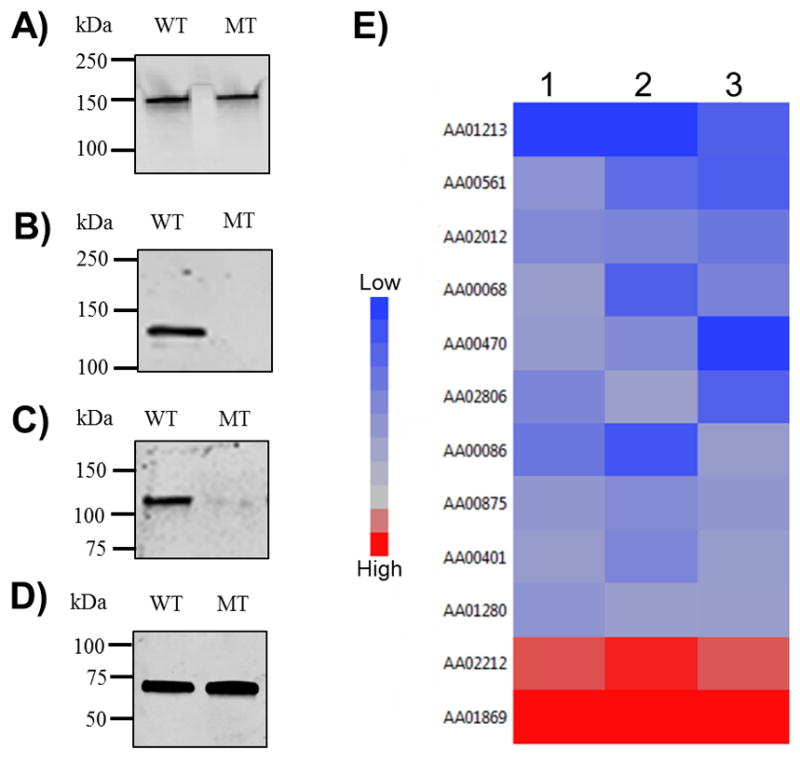
A–C) Immunoblots of membrane protein using antibodies specific to: A) Aae; B) MorC; C) leukotoxin (LtxA); and D) 6xHis tagged leukotoxin B (LtxB). E) Heat map of the log2 transformed ratios for each differentially abundant protein using JMP11.0. Each column is a single replicate and each row represents a single protein. Cool colors (blue) indicate proteins reduced in the mutant whereas warm colors (red) indicates an increase.
The defect in leukotoxin secretion is hypothesized to result from reduction in abundance of one or more proteins directly associated with the type I secretion apparatus in the membrane. The secretion system required to transport leukotoxin from the cytoplasm to the environment consists of three components. An ATPase (LtxB, AA2805) that transports the toxin across the inner membrane to the periplasmic channel (LtxD, AA2803) and an outer membrane pore (TolC, AA02077) to release the toxin to the environment [6, 26]. The average abundance of each of these three proteins was found to be equal in both strains (Supplementary material). The abundance of LtxB was verified to be equal between wild-type and morC mutant cells as determined by immunoblotting (Figure 3D).
As previously mentioned, TolC forms the outer membrane pore for LtxA secretion. This protein is also associated with drug efflux systems and is required for export of specific antimicrobials [27]. We have tested the morC mutant strain for differences in susceptibility to these antimicrobial agents and have not found any differences between the wild-type and mutant strains (data not shown). This finding suggests that TolC is still functional in the morC mutant and implies that the defect in LtxA secretion is attributed to the loss of functionality associated with either LtxB, D or both proteins.
The decrease of leukotoxin secretion from the morC mutant is not attributed to a decrease in the proteins composing the secretion apparatus. This finding raises the possibility of a change in the 3-dimensional structure of one or more of the protein components. Lipids are known to be important for the correct folding of membrane proteins [28]. Therefore, analysis of the fatty acid composition of the two strains was conducted by GC-MS to identify potential changes. The types and percentages of fatty acids identified in this study were in close agreement with previously published data from a different strain of A. actinomycetemcomitans [29]. However, no obvious differences in fatty acid composition were detected between the wild-type and mutant strains (Supplementary Table 1). The absence of a difference in fatty acids implies that the lipid composition of the membrane is not affected by the absence of MorC in the membrane.
Proteins differentially expressed between WT and MT across replicates are visualized by a heat map in Figure 3E. Bioinformatic analysis (Table 2) of these sequences provided information about the cellular location, conserved domains, enzyme activity and predicted functions [12]. Four of the proteins are predicted to be chaperones, which are critical to the maintenance of cellular homeostasis. Chaperones interact with other proteins to maintain the proper tertiary structure for secretion and also serve to degrade misfolded or aggregated proteins [30]. Three chaperones were found to be reduced (HtpX, SlyD, GroES) in the morC mutant strain. An increase in misfolded or aggregated proteins in E. coli [31] results in a membrane stress response and the up-regulation of specific chaperones including DegP. Although the A. actinomycetemcomitans genome does not appear to encode DegP, a protein with overlapping function, DegQ, was found to be increased in the mutant strain (Table 2). The overall decrease in abundance of known chaperones may lead to an increase in the incorporation of misfolded proteins into the membrane of the mutant strain resulting in the phenotypes associated with this mutant.
Table 2.
Bioinformatic analysis of proteins significantly changed in abundance.
| Oralgen ID | Annotation | Biological Function | Protein Name | COG | GO | EC | TCDB | Predicted Localization |
|---|---|---|---|---|---|---|---|---|
| AA01213 | Competence protein | Transport of DNA | ComEA | COG2931 | 0005509 | 3.A.11.1.4 | OM | |
| AA00086 | SEPHCHC synthase | Vitamin K biosynthesis | MenD | COG0225 | 0030976 | IM | ||
| AA00561 | Transcription factor | Competence gene expression | TfoX | COG0208 | C | |||
| AA00470 | Chaperone protein (metalloprotease) | Protein folding and transport | HtpX | COG0234 | 0004222 | 3.4.24 | P | |
| AA02012 | Peptide methionine sulfoxide reductase | Resistance to oxidative stress | MsrB | COG0265 | 0016671 | 1.8.4 | P | |
| AA00068 | Superoxide dismutase | Resistance to oxidative stress | SodA | COG4965 | 0004784 | 1.15.1.1 | P | |
| AA02806 | Leukotoxin A | Toxin | LtxA | COG1555 | 1.C.11.1.3 | E | ||
| AA00875 | Tight adherence protein B | Fimbrial secretion system subunit | TadB | COG3070 | 3.A.7.15.1 | IM | ||
| AA00401 | Peptidyl-prolyl cis-trans isomerase | Protein folding and transport | SlyD | COG0501 | 0006457 | P | ||
| AA01280 | Chaperone protein | Protein folding and transport | GroES | COG1047 | 0006457 | C | ||
| AA02212 | Ribonucleoside diphosphate reductase | Ribonucleotide biosynthesis | NrdF | COG1165 | 0004748 | 1.17.4.1 | C | |
| AA01869 | Chaperone protein | Protein folding and transport | DegQ | COG0605 | 9.B.160.1.10 | P |
COG: Cluster of orthologous groups, GO: Gene ontology, EC: Enzyme commission, TCDB: Transporter classification database, C: cytoplasmic, E: extracellular, IM: inner membrane, OM: outer membrane, P: periplasmic
Two other proteins that were demonstrated to be reduced in abundance in the mutant strain are involved in an oxidative stress response: SodA, an enzyme that detoxifies superoxide radicals [32] and MsrB, responsible for reducing oxidized methionines [33]. Interestingly, a mutation in quinol peroxidase (qpo), another oxidative stress response protein reduces leukotoxin secretion in A. actinomycetemcomitans [34]. Leukotoxin is also inactivated by reactive oxygen species and interacts directly with a superoxide dismutase in A. actinomycetemcomitans [35]. Our findings support a link between oxidative stress and secretion of LtxA, however the precise mechanism is not known.
Three protein secretion systems have been identified in A. actinomycetemcomitans: Type I, II, and V [12, 36]. Our analysis suggests the absence of MorC in the membrane affects the secretion of proteins involved with two types of secretion systems: leukotoxin (LtxA) secreted via the type I secretion pathway and TadB, is a structural component of a subtype of the type II secretion pathway. Proteins secreted via the type V systems are not affected by the absence of MorC in the membrane. This class of proteins includes the monomeric (Aae) and trimeric (EmaA, ApiA) autotransporter proteins [19, 22, 37]. This finding is in contrast to the observation that secretion of the monomeric autotransporter Flu (Ag43) is impacted in a strain of E. coli in which the morC homolog (tamB or ytfN) was inactivated [9].
A. actinomycetemcomitans expresses pili (fimbriae) that are essential for biofilm formation, adhesion and pathogenesis [38]. An 11 protein complex (RcpCAB–TadZABCDEFG) is associated with secretion of the fimbriae subunit (Flp1). We have previously identified this secretion system in the strain used in this study, which does not possess fimbriae [12]. In this study, we observed a consistent decrease in the abundance of ten of these proteins in the morC mutant, of which only TadB (AA00875) achieved statistical significance (see Table 1 for TadB and supplemental material for RcpCAB-TadZACDEFG, AA00870-874 and AA00876-880). TadB is predicted to localize to the inner membrane with six transmembrane domains [39]. The rationale for the decrease of these proteins in an afimbriated strain is not apparent. This observation does raise the possibility that MorC is required for fimbrial subunit secretion in strains that express fimbriae.
CONCLUSION
In this study, we have presented quantitative changes in the abundance of membrane proteins in an A. actinomycetemcomitans morC mutant strain. The majority of membrane proteins remained unchanged in mutant cells as compared with the parent strain. This suggests that the phenotypes associated with the mutant are not related to a generalized decrease in the abundance of membrane proteins. Proteins composing the type I secretion apparatus for leukotoxin were included in this group, which indicates that the defect in toxin secretion is not due to the change in abundance of these proteins nor the fatty acid composition of the membrane. The findings of this study support the hypothesis that the pleotropic phenotypes of the morC mutant are associated with the decrease in the abundance of specific proteins. These proteins are associated with functions involving cellular homeostasis and oxidative stress response. The role of these proteins in relationship to the phenotypes displayed by the morC mutant strain are under investigation.
Supplementary Material
Acknowledgments
We would like to thank Jana Kraft (University of Vermont) for the lipid analyses, Edward T. Lally (University of Pennsylvania) for providing the anti-leukotoxin antibody and Thomas Freeman (University of Vermont) for generating the His-tagged protein and immunoblot. We also thank James Vincent (Vermont Genetics Network Bioinformatics Core) for helpful discussions about this work. This study was supported by NIH grant RO1-DE018889 (KPM). The Vermont Genetics Network Proteomics Facility is supported through NIH grant P20GM103449 from the INBRE Program of the National Institute of General Medical Sciences.
Abbreviations
- WT
Wild-type
- MT
Mutant
REFERENCES CITED
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delepelaire P. Type I secretion in gram-negative bacteria. Biochim Biophys Acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane--distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kachlany SC, Fine DH, Figurski DH. Secretion of RTX leukotoxin by Actinobacillus actinomycetemcomitans. Infect Immun. 2000;68:6094–6100. doi: 10.1128/iai.68.11.6094-6100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lally ET, Golub EE, Kieba IR, Taichman NS, et al. Analysis of the Actinobacillus actinomycetemcomitans leukotoxin gene. Delineation of unique features and comparison to homologous toxins. J Biol Chem. 1989;264:15451–15456. [PubMed] [Google Scholar]
- 8.Gallant CV, Sedic M, Chicoine EA, Ruiz T, Mintz KP. Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. J Bacteriol. 2008;190:5972–5980. doi: 10.1128/JB.00548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selkrig J, Mosbahi K, Webb CT, Belousoff MJ, et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol. 2012;19:506–510. S501. doi: 10.1038/nsmb.2261. [DOI] [PubMed] [Google Scholar]
- 10.Azari F, Nyland L, Yu C, Radermacher M, et al. Ultrastructural analysis of the rugose cell envelope of a member of the Pasteurellaceae family. J Bacteriol. 2013;195:1680–1688. doi: 10.1128/JB.02149-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mintz KP, Fives-Taylor PM. Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line. Infect Immun. 1994;62:3672–3678. doi: 10.1128/iai.62.9.3672-3678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KP, Fields JG, Voogt RD, Deng B, et al. The cell envelope proteome of Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol. 2014 doi: 10.1111/omi.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu JL, Huang SY, Chow NH, Chen SH. Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 14.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bairoch A. The ENZYME data bank in 1999. Nucleic Acids Res. 1999;27:310–311. doi: 10.1093/nar/27.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mintz KP. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology. 2004;150:2677–2688. doi: 10.1099/mic.0.27110-0. [DOI] [PubMed] [Google Scholar]
- 20.Tang G, Kawai T, Komatsuzawa H, Mintz KP. Lipopolysaccharides mediate leukotoxin secretion in Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol. 2012;27:70–82. doi: 10.1111/j.2041-1014.2011.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lally ET, Kieba IR, Demuth DR, Rosenbloom J, et al. Identification and expression of the Actinobacillus actinomycetemcomitans leukotoxin gene. Biochem Biophys Res Commun. 1989;159:256–262. doi: 10.1016/0006-291x(89)92431-5. [DOI] [PubMed] [Google Scholar]
- 22.Rose JE, Meyer DH, Fives-Taylor PM. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect Immun. 2003;71:2384–2393. doi: 10.1128/IAI.71.5.2384-2393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittaker P, Keys CE, Brown EW, Fry FS. Differentiation of Enterobacter sakazakii from closely related Enterobacter and Citrobacter species using fatty acid profiles. J Agric Food Chem. 2007;55:4617–4623. doi: 10.1021/jf070193a. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi Y, Choi PJ, Li GW, Chen H, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihama Y, Schmidt T, Rappsilber J, Mann M, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lally ET, Golub EE, Kieba IR, Taichman NS, et al. Structure and function of the B and D genes of the Actinobacillus actinomycetemcomitans leukotoxin complex. Microb Pathog. 1991;11:111–121. doi: 10.1016/0882-4010(91)90004-t. [DOI] [PubMed] [Google Scholar]
- 27.Crosby JA, Kachlany SC. TdeA, a TolC-like protein required for toxin and drug export in Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene. 2007;388:83–92. doi: 10.1016/j.gene.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell DC. Progress in understanding the role of lipids in membrane protein folding. Biochim Biophys Acta. 2012;1818:951–956. doi: 10.1016/j.bbamem.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 29.Braunthal SD, Holt SC, Tanner AC, Socransky SS. Cellular fatty acid composition of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J Clin Microbiol. 1980;11:625–630. doi: 10.1128/jcm.11.6.625-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 31.Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 32.Touati D. Molecular genetics of superoxide dismutases. Free Radic Biol Med. 1988;5:393–402. doi: 10.1016/0891-5849(88)90113-x. [DOI] [PubMed] [Google Scholar]
- 33.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Takashima E, Konishi K. Characterization of a quinol peroxidase mutant in Aggregatibacter actinomycetemcomitans. FEMS Microbiol Lett. 2008;286:66–70. doi: 10.1111/j.1574-6968.2008.01253.x. [DOI] [PubMed] [Google Scholar]
- 35.Balashova NV, Park DH, Patel JK, Figurski DH, Kachlany SC. Interaction between leukotoxin and Cu, Zn superoxide dismutase in Aggregatibacter actinomycetemcomitans. Infect Immun. 2007;75:4490–4497. doi: 10.1128/IAI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zijnge V, Kieselbach T, Oscarsson J. Proteomics of protein secretion by Aggregatibacter actinomycetemcomitans. PLoS One. 2012;7:e41662. doi: 10.1371/journal.pone.0041662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asakawa R, Komatsuzawa H, Kawai T, Yamada S, et al. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2003;50:1125–1139. doi: 10.1046/j.1365-2958.2003.03748.x. [DOI] [PubMed] [Google Scholar]
- 38.Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 39.Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, et al. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.