Abstract
Development of efficient nanoparticles (NPs) for cancer therapy remains a challenge. NPs are required to have high stability, uniform size, sufficient drug loading, targeting capability, and ability to overcome drug resistance. In this study, we report the development of a nanoparticle formulation that can meet all these challenging requirements for targeted glioblastoma multiform (GBM) therapy. This multifunctional nanoparticle is composed of a polyethylene glycol (PEG) coated magnetic iron oxide NP conjugated with cyclodextrin (CD) and chlorotoxin (CTX) and loaded with fluorescein and paclitaxel (PTX) (IONP-PTX-CTX-FL). The physicochemical properties of the IONP-PTX-CTX-FL were characterized by TEM, dynamic light scattering (DLS), and HPLC. The cellular uptake of NPs was studied using flow cytometry and confocal microscopy. Cell viability and apoptosis were assessed with the Alamar Blue viability assay and flow cytometry, respectively. The IONP-PTX-CTX-FL had a uniform size of ~44 nm and high stability in cell culture medium. Importantly, the presence of CTX on NPs enhanced the uptake of the NPs by GBM cells and improved the efficacy of PTX in killing both GBM and GBM drug-resistant cells. The IONP-PTX-CTX-FL has demonstrated its great potential for brain cancer therapy and may also be used to deliver PTX to treat other cancers.
Keywords: iron oxide nanoparticles, paclitaxel, glioblastoma, chlorotoxin, β-cyclodextrin
1. Introduction
Glioblastoma multiform (GBM) is the most common and aggressive malignant primary brain tumor and accounts for ~70% of new cases of primary brain tumors in the US annually.[1] Although temozommide (TMZ) in combination with radiation is now the contemporary standard of care for GBMs, the prognosis for GBM remains dismal with most patients dying within 12 months.[1] The poor prognosis is primarily attributed to the following factors: (1) the majority of GBMs are not responsive to TMZ due to the resistance mediated by O6-methylguanine-DNA methyltransferase (MGMT), a DNA repair protein that limits the cytotoxic effects of TMZ;[2] (2) TMZ is poorly soluble in physiological conditions and is subject to rapid hydrolysis that further limits its anti-tumor efficacy;[3] (3) TMZ has high IC50 values to GBM cells and a large dose of TMZ is needed for the treatment, causing high systemic toxicity.[4] A number of NP formulations have been developed to deliver TMZ for treatment of brain cancers.[4–5] Nevertheless, it remains a challenge to load a large amount of hydrophobic TMZ on NPs while making NPs simultaneously meet a number of general requirements for effective cell killing, including high stability, small and uniform size, and tumor targeting capability.
Here we report the development of a highly dispersed and colloidally stable multifunctional NP of small size as a drug delivery carrier to specifically deliver paclitaxel (PTX) to brain tumor cells and effectively kill them. Unlike TMZ which is unstable in physiological solutions, PTX does not degrade in various aqueous conditions. Also, PTX is a mitotic inhibitor to GBM cells and is highly potent, requiring only a small amount to kill tumor cells,[6] and thus can be readily loaded onto NPs. PTX can kill GBM cells with or without MGMT expression. Our nanoparticle is designed to overcome the aformentioned drawbacks of TMZ and improve treatment of GBM. Our multifunctional nanoparticle is made of a polyethylene glycol (PEG)-coated iron oxide NP conjugated with cyclodextrin (CD), CTX, PTX and 1-admantenmethylamide-fluorescein (AD-FL). The PEG-coated iron oxide NPs have demonstrated high monodispersity and colloidally stability in cell culture media as shown in our previous study.[7] Iron oxide nanoparticles (IONPs) have emerged as one of best candidates for cancer therapy.[8] IONPs are biodegradable, and the Fe ions from degraded SPIONs enters the body’s natural iron stores such as hemoglobin in red blood cells.[9] IONPs are also effective contrast agents for MRI because of their intrinsic magnetic properties. The localized interactions of IONPs with protons of water molecules generates contrast by reducing T1 or T2 decay relaxation times.[10] CD was used to load hydrophobic molecules including PTX and AD-FL. CTX was conjugated to facilitate specific targeting. CTX is known to bind to matrix metalloproteinase-2 (MMP-2) overexpressed on the surface of many brain tumors but not on normal brain tissue.[11] We have demonstrated the CTX-mediated delivery of NPs for imaging of brain tumors with magnetic resonance imaging and for gene delivery.[12]
In this study, the size, shape and surface charge of NPs were characterized by dynamic light scattering (DLS), transmission electron microscope (TEM)and ζ-potential measurements, respectively. The drug loading was quantified by high-performance liquid chromatography (HPLC). The NP uptake by target cells was assessed by flow cytometry and confocal laser scanning microscopy (CLSM). A cell line SF-763 with high MGMT expression and a cell line U-118 MG with low MGMT expression, that represent the majority of glioblastoma cells,[13] were used to evaluate the efficacy of NPs in killing cells. PTX-resistant SF-763 and U-118 MG cell lines were established by long-term exposure of the cells to a low dose of PTX. Cell viability of two GBM cell lines and their PTX-resistant cell line counterparts after treatment with NPs were determined by the Alamar Blue viability assay. Finally, apoptosis in four cell lines was investigated by flow cytometry after treatments with PTX, IONP-PTX or IONP-PTX-CTX.
2. Results and Discussion
2.1. Preparation and characterization of NPs
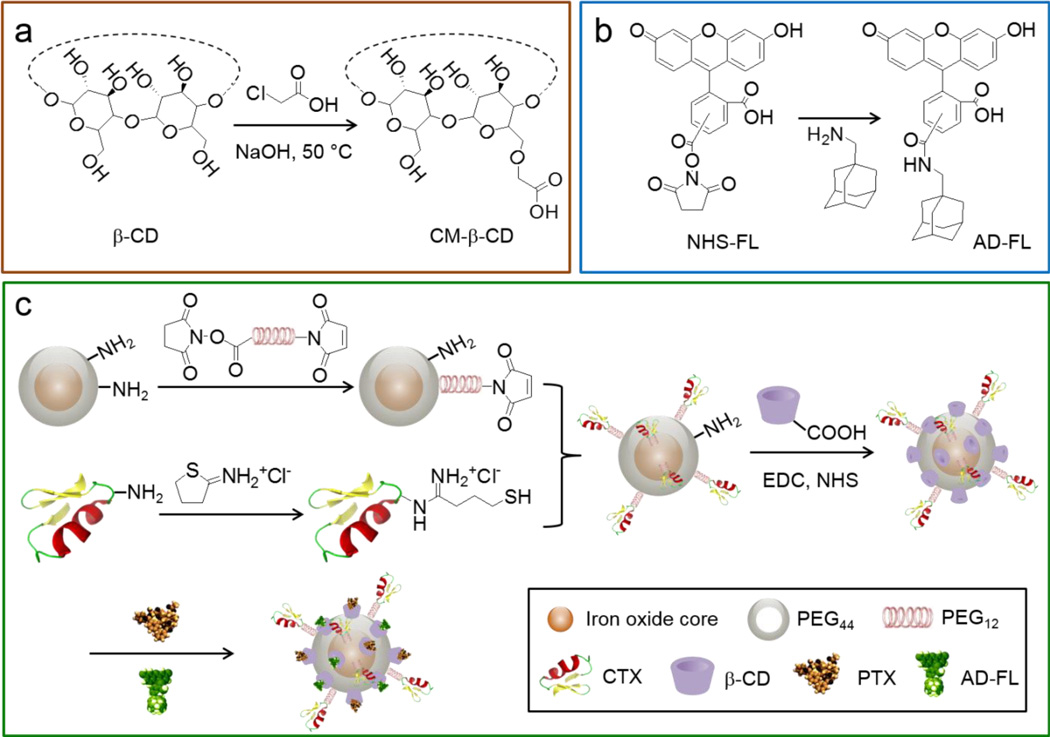
The preparation of IONP-PTX-CTX-FL is illustrated in Scheme 1. CM-β-CD (Scheme 1a) and AD-FL (Scheme 1b) were first prepared following the published methods.[14–15] Iron oxide nanoparticles coated with a monolayer of PEG2000-amine (IONP-PEG-NH2) served as the drug carrier. IONP-PEG-NH2 was conjugated with CTX to obtain IONP-CTX using a heterobifunctinal crosslinker: SM(PEG)12, followed by conjugation of CM-β-CD to IONP-CTX using EDC and NHS. Dimethyl sulfoxide (DMSO) solutions of PTX and fluorophore AD-FL were then mixed with aqueous solution of IONP-CD-CTX. After overnight incubation, the resultant mixture was purified to obtain IONP-PTX-CTX-FL (Scheme 1c). IONP-NH2 conjugated with CM-β-CD, PTX, and AD-FL (without CTX) served as a CTX-free, non-targeted control NP. A PTX-free, CTX-conjugated NP (IONP-CTX) was also prepared as targeted, non-therapeutic control NP.
Scheme 1.
Schematic of IONP-PTX-CTX-FL preparation. (a) Preparation of CM-β-CD; (b) preparation of AD-FL; (c) conjugation of CTX and CM-β-CD onto NP and loading of PTX and AD-FL to form IONP-PTX-CTX-FL.
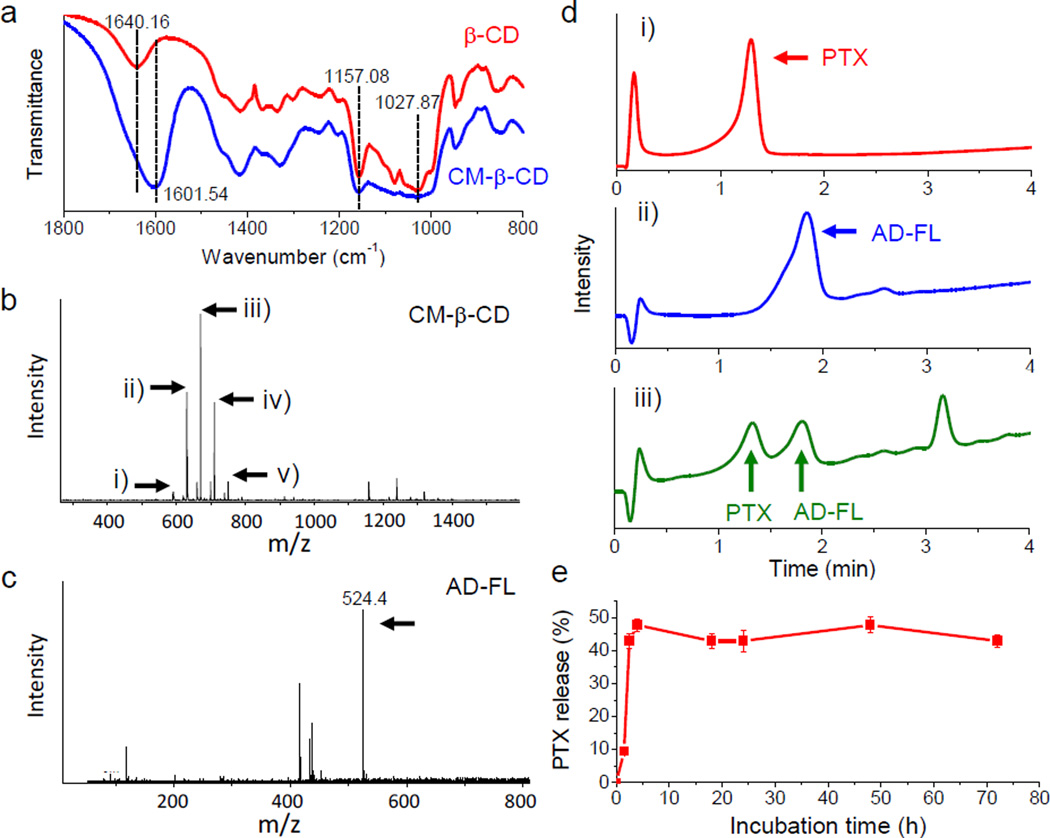
Modification of β-CD with carboxymethyl groups was characterized by FT-IR and mass spectroscopy (MS). FT-IR showed the presence of carboxylate (1601.54 cm−1) after the modification (Figure 1a). The peak at 1640.16 cm−1 corresponds to the bending vibration of adsorbed H-O-H on β-CD. However, such peak may be overlapped with the peak of carboxylate on CM-β-CD because of strong absorption of the carboxylate. The peaks at 1157.08 cm−1 and 1027.87 cm−1 correspond to stretching of C-O-C on both β-CD and CM-β-CD. The MS spectra showed that β-CD had 5 different levels of substitution as shown by the m/z values (591.5, 631.1, 670.7, 710.5 and 750.4). These m/z values correlate to double-charged and sodium-associated β-CD possessing 0, 1, 2, 3 and 4 substitutions, respectively. According to the peak intensities, the majority of CM-β-CD has 2 carboxymethyl groups per CD (Figure 1b). Modification of NHS-fluorescein by AD was confirmed by MS (Mw of 524.4, positive ion mode) (Figure 1c).
Figure 1.
Chemical analysis of NP surface components. (a) FT-IR spectra of β-CD (red) and CM-β-CD (blue). (b) MS spectrum of CM-β-CD. Arrows indicate m/z of double-pronated β-CD with different numbers of carboxymethyl substitutions: i) 0, ii) 1, iii) 2, iv) 3, v) 4. (c) MS spectrum of AD-FL. Arrow indicates m/zof intact protonated AD-FL; (d) HPLC analysis of (i) free PTX, (ii) free AD-FL, and (iii) PTX and AD-FL extracted from NPs; (e) Release of PTX from IONP-PTX-CTX-FL in solution determined by HPLC.
HPLC was used to quantitatively examine the presence and amounts of drug and fluorophore on NPs. As shown in Figure 1d (i and ii), free PTX or AD-FL had defined and separated elution profiles. To assess PTX and AD-FL on NPs, they were extracted with chloroform. The chloroform was then evaporated and the remainder re-dissolved in acetonitrile for HPLC analysis. The elution profiles of PTX and AD-FL extracted from NPs were consistent with free PTX and AD-FL, respectively, indicating both PTX and AD-FL were successfully loaded onto NPs (Figure 1d, iii). An additional peak is likely caused by the oleic acid residue from NPs. The number of PTX molecules per NP varied from 48 to 109 and the average number of CTX molecules per NP was ~6, as determined by micro-BCA protein quantification assay (Table 1). HPLC was also used to quantify release of PTX from NPs in solution (Figure 1e). The results showed that although there was an initial quick release in the first 4 hrs, ~50% of PTX still remained on NPs after 3 days of incubation. This indicates that the majority of the NP-PTX would not be released into the blood of a neutral pH. After NP-PTX are taken into cells, the acidic pH in endosomes and lysosomes helps cleave the amide bond between CM-β-CD and NPs, and thus release the PTX into cytoplasm.[16]
Table 1.
Physicochemical properties of various NP formulations used in this study.
| NP conjugates | Core size [nm] |
Z-average size [nm] |
PDI | Zeta-potential (pH 7.0) [mV] |
Approximate number per NP | ||
|---|---|---|---|---|---|---|---|
| PTX | CTX | AD-FL | |||||
| IONP-CTX | 15 ± 0.9 | 36.75 | 0.095 | −5.05 | 0 | 6 | 0 |
| IONP-PTX-FL | 15 ± 0.9 | 35.94 | 0.107 | −6.04 | 65 | 0 | 81 |
| IONP-PTX-CTX-FL | 15 ± 0.9 | 44.45 | 0.107 | −0.673 | 48 | 6 | 69 |
| IONP-PTX | 15 ± 0.9 | 30.76 | 0.055 | −9.88 | 109 | 0 | 0 |
| IONP-PTX-CTX | 15 ± 0.9 | 34.91 | 0.048 | −0.415 | 84 | 6 | 0 |
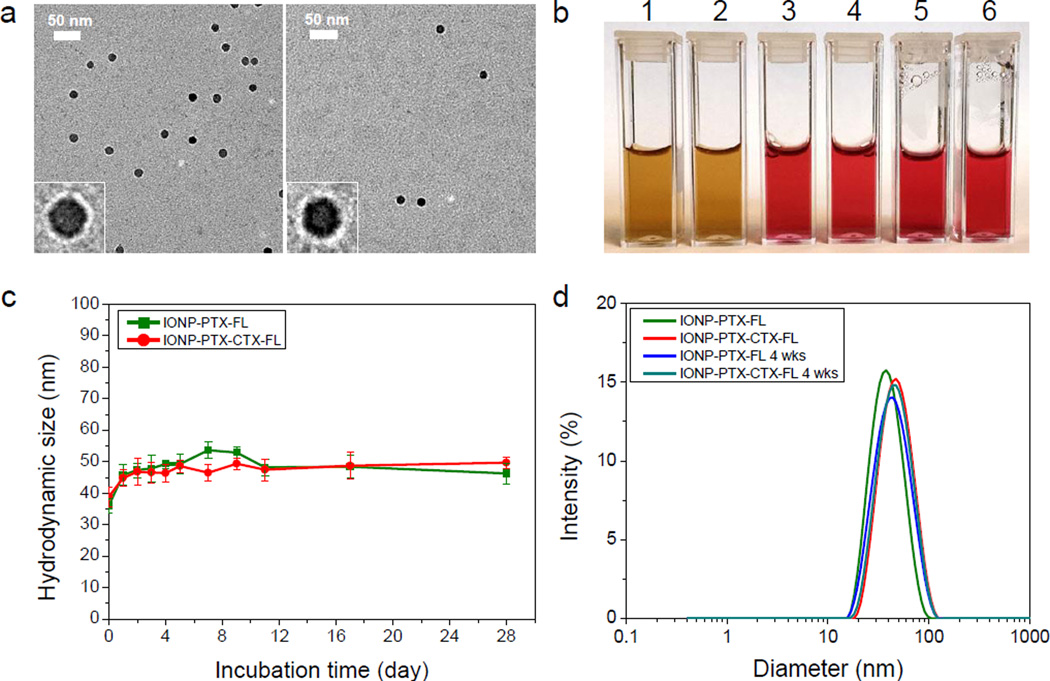
The physicochemical properties of NPs were characterized by TEM, HPLC, DLS and ζ-potential measurements (Figure 2 and Table 1). TEM images of NPs with negative staining showed that the NP had a core diameter of ~15 nm (Figure 2a). This size dimension is suitable for T2 relaxation in magnetic resonance imaging applications.[17] The surface coating (the white shell surrounding the NP core) on the NP is clearly observed in this TEM image, which is possible only with negative staining (Figure 2a, insets). The stability of drug-bearing NPs were examined by diluting NPs in cell culture medium placed in a 37°C water bath and monitoring the change of their hydrodynamic size over a 4-week period. The stability of these NPs in different media can be visualized on the photographs taken before and after incubation (Figure 2b). No precipitation in particle solutions in vials was observed after incubation for 4 weeks. An example of a unstable NP with precipitation as a negative control is shown in Figure S2 as a comparison. DLS analysis also showed that both IONP-PTX-FL and IONP-PTX-CTX-FL had a similar size and a narrow size distribution, and there was no apparent difference in mean size or size distribution before and after 4 wks of incubation (Figure 2c and d). These results indicated that these NPs were very stable in biological fluid where protein, amino acids and various types of ions are present. Sizes of NPs increased after 1 day incubation, likely due to serum protein binding.[18] NPs in all the media studied were stable for over 3 months when stored in a 4°C refrigerator. These NPs were remarkably stable, probably owing to the presence of the highly dense and stable PEG coating through strong affinity of triethoxysilylpropylsuccinic anhydride to iron oxide surfaces and the formation of siloxane bonds between PEG chains.[7] It should be noted that NPs reduce the optical intensity of FLs conjugated on NPs by a mechanism known as quenching. However, all the NPs used in this study would produce nearly equal influence on FLs and thus the results of relative cellular uptakes would not be affected by the quenching.
Figure 2.
Size, morphology and stability of IONP-PTX-FL and IONP-PTX-CTX-FL. (a) TEM micrographs of IONP-PTX-FL (left) and IONP-PTX-CTX-FL (right) with negative staining. Scale bars represent 50 nm. Insets are enlarged images of single particles. (b) Photographs of IONP-PTX-FL and IONP-PTX-CTX-FL in PBS (1 and 2) and complete DMEM cell culture medium at 0 day (3 and 4) and at 4 wks after incubation at 37°C (5 and 6). (c) Hydrodynamic sizes of NPs incubated in cell culture medium at 37°C as a function of incubation time over 4 wks. (d) DLS analysis of NPs in PBS before and 4 wks after incubation with cell culture medium at 37°C.
The surface charges of these NPs were determined through ζ-potential measurements. Except for IONP-PTX-CTX-FL, which had a near-neutral charge (−0.673 mV) because of CTX coating, the CTX-free NPs showed slightly negative charges (Table 1).
2.2. Enhanced cell uptake of CTX-conjugated NPs
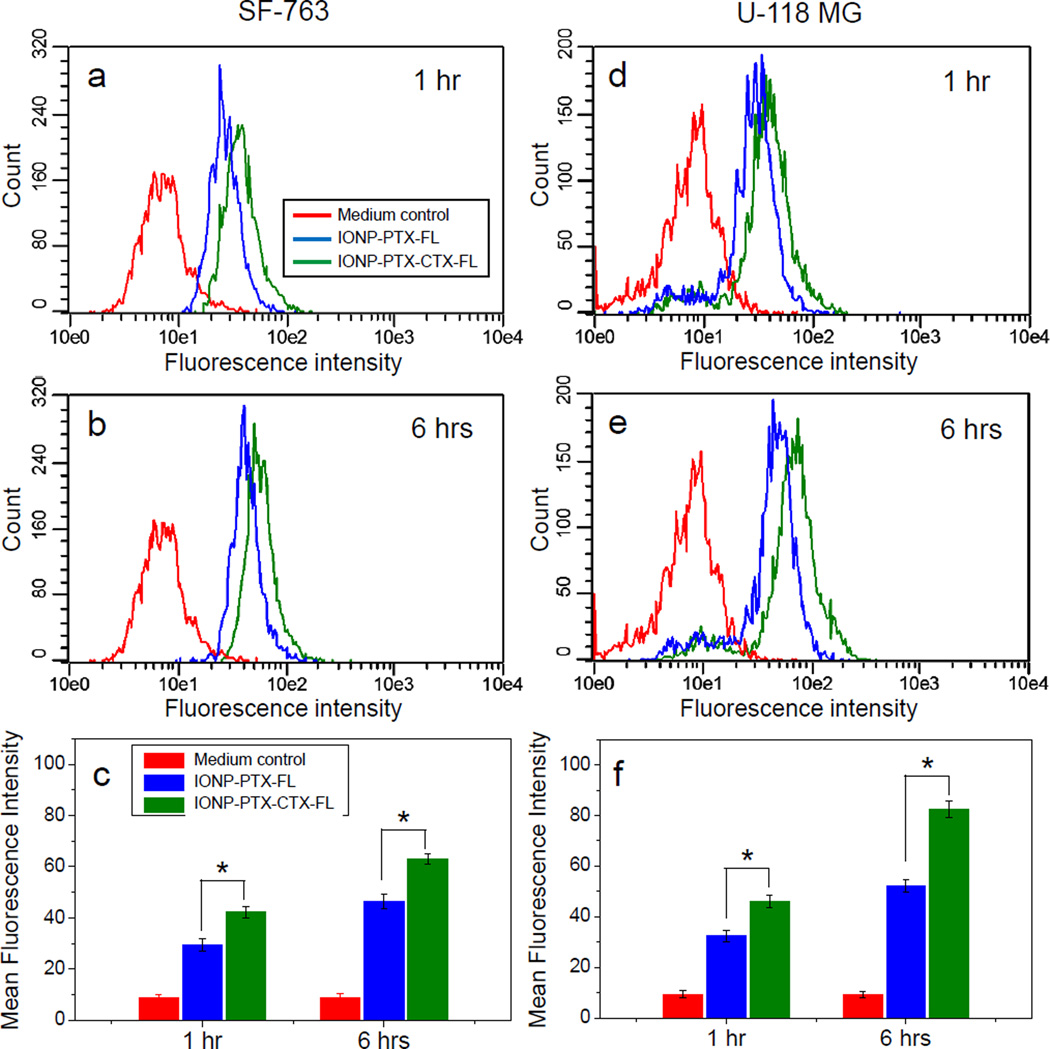
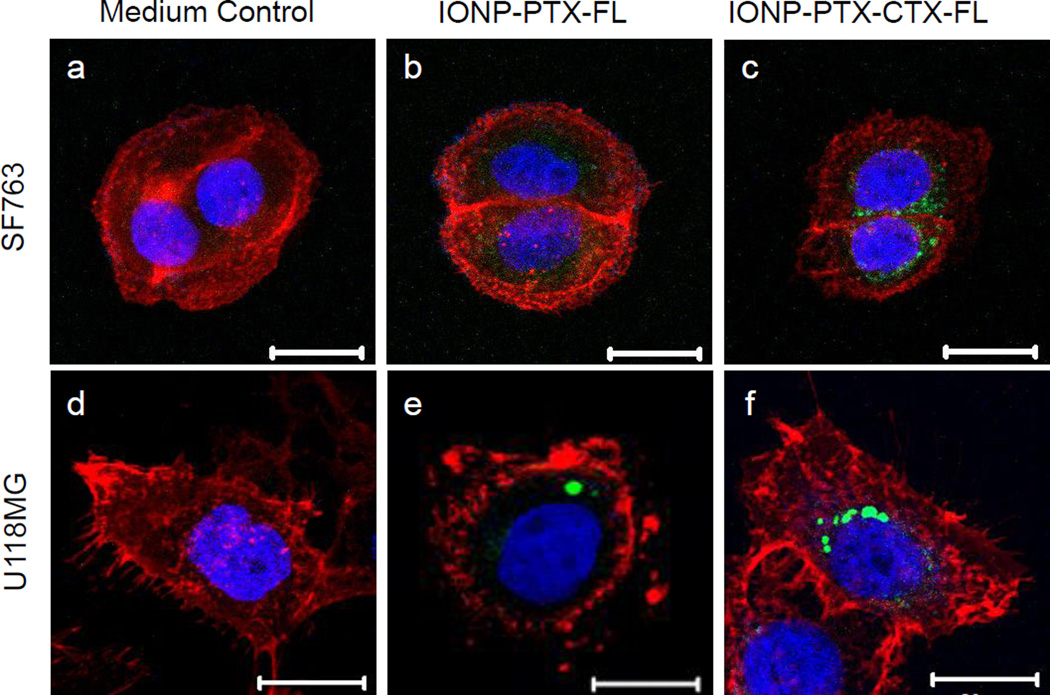
To study the cellular uptake of NPs, SF-763 and U-118 MG were incubated with 40 µg/mL of NPs for 1 hr and 6 hrs and analyzed by flow cytometry. Flow cytometry results showed that although non-specific uptake of these NPs was observed, CTX-conjugation enhanced the uptake of NPs by target cells by ~30% (Figure 3). Compared to SF-763 cells, U-118 MG cells showed higher uptake of NPs after 6 hr incubation. Such a difference in uptake of NPs might affect cellular activity of PTX. We also quantified cellular iron and protein contents by colorimetric ferrozine and Bradford assays, respectively. The iron content in SF-763 cells after 1 hr incubation with IONP-PTX-CTX was ~0.36 µg Fe/µg protein, which was higher than iron content in cells incubated with IONP-PTX (~0.23 µg Fe/µg protein, close to medium background of ~0.24 µg Fe/µg protein). Similar pattern was also observed in U-118 MG cells (Figure S1, Supporting Information). Confocal microscopy was used to observe intracellular localization of NPs (Figure 4b, c, e and f). The images showed that IONP-PTX-CTX-FL located in the area of the cytoplasm near the nucleus. As the MMP-2 on cell membrane is a receptor of CTX, the CTX-conjugated NPs were very likely internalized into cells through a receptor-mediated manner. It was also observed that more IONP-PTX-CTX-FL was present in cells than IONP-PTX-FL.
Figure 3.
Flow cytometric analysis of cellular uptake of 40 µg/mL IONP-PTX-FL and IONP-PTX-CTX-FL by (a–c) SF-763 cells and (d–f) U118-MG cells, (a, d) after 1 hr and (b, e) after 6 hrs incubation. (c, f) Quantitative mean fluorescence intensities derived from the flow cytometric analysis (* p < 0.05 by Student’s t-test).
Figure 4.
Confocal microscopic analysis of cellular uptake of 40 µg/mL IONP-PTX-FL and IONP-PTX-CTX-FL after 6-hour incubation in (a–c) SF-763 and (d–f) U-118 MG cells. Cells were incubated in (a, d) medium control (no NPs), (b, e) with IONP-PTX-FL, and (c, f) with IONP-PTX-CTX-FL. Scale bars: 20 µm.
2.3. CTX enhanced tumor cell killing by NPs
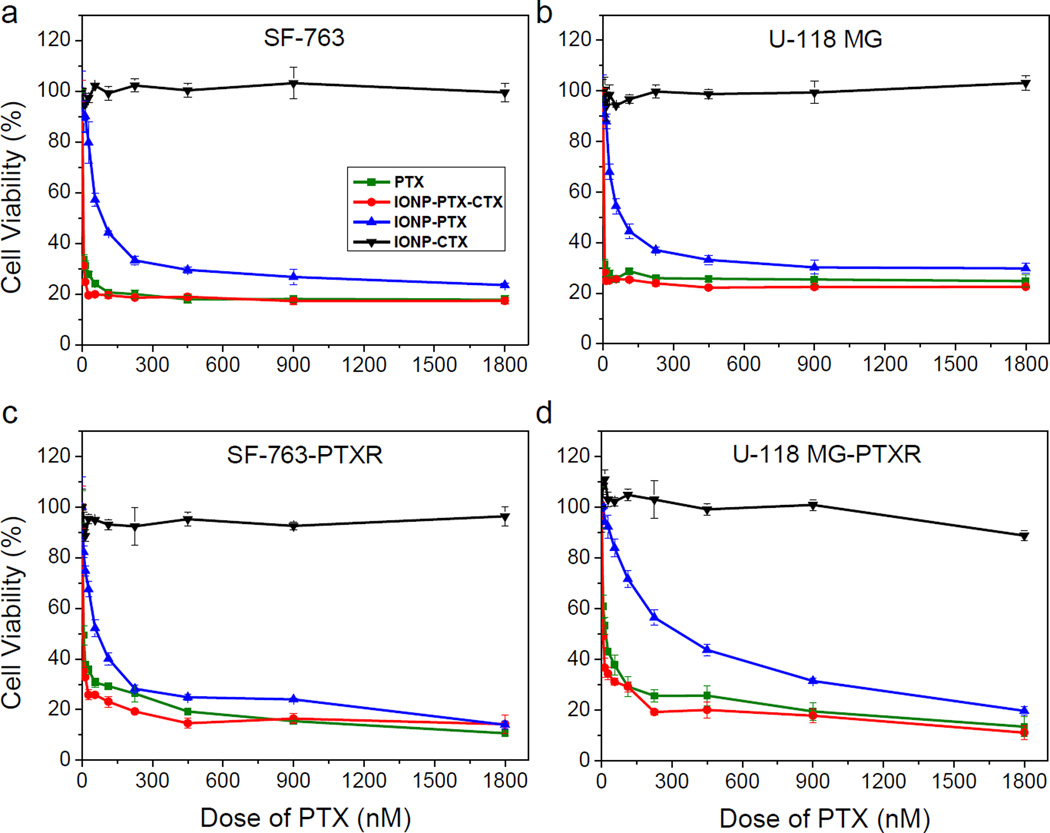
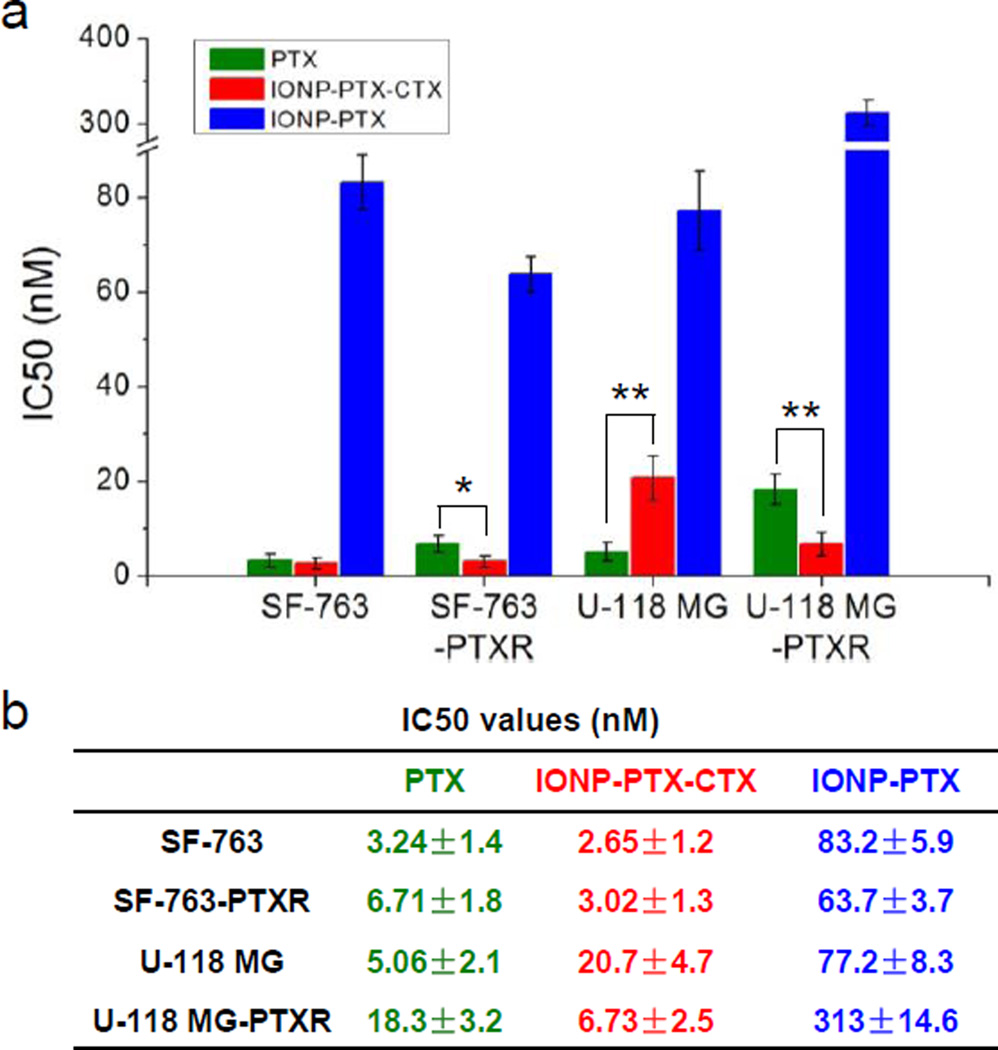
To test the effect of CTX targeted NPs on cell killing, SF-763 and U-118 MG were treated with free PTX, IONP-CTX, IONP-PTX or IONP-PTX-CTX for 72 hrs and cell viability was evaluated by the Alamar Blue assay. FL was not conjugated on these NPs to minimize its interference on the fluorescence-based cell viability assay. As shown in Figure 5, free PTX showed strong therapeutic potency toward both cell lines and no significant difference could be seen. NPs conjugated with CTX alone showed no cell killing, indicating that the NPs themselves were non-toxic to cells and CTX did not affect cell viability. The non-targeted IONP-PTX showed reduced potency compared to free PTX, probably due to delayed drug release from NPs compared to free PTX. In contrast, the conjugation of CTX to NPs significantly enhanced cell killing by NPs, as shown in the IC50 chart (Figure 6). The enhancement effect might be due to the increase of cellular uptake of CTX-conjugated NPs (Figure 3 and Figure 4). CTX has been reported to bind MMP-2, Annexin A2 and a chloride channel which are all located on cell membranes.[11, 19] Although IONP-CTX did not affect cell viability, the interactions of CTX with these cell membrane components might affect efficacy of PTX. Therefore, we expected that the enhanced cell killing might result from increased cell uptake and a synergistic effect of CTX and PTX. It is worth noting that the loading of PTX was only ~84 molecules /per NP (~3.3% w/w drug/NP) for IONP-PTX-CTX. Compared to nanoformulations of TMZ, which had the loading of as high as 28.65% w/w,[4] the nanoformulation of PTX presented here was more effective and requires much less drug loading.
Figure 5.
Cell viability as function of PTX dose. Viability of (a) SF-763, (b) U-118 MG, (c) SF-763-PTXR and (d) U-118 MG-PTXR cells 72 hrs after treatment with various doses of free PTX, IONP-PTX, IONP-PTX-CTX or PTX-free IONP-CTX.
Figure 6.
IC50s of SF-763, SF-763-PTXR, U-118 MG and U-118 MG-PTXR cells 72 hrs after treatment with PTX, IONP-PTX, or IONP-PTX-CTX (* p < 0.05; ** p < 0.01, Student’s t-test).
It is known that many tumors, including brain tumors, can develop resistance to PTX treatment. In this study, we established two PTX-resistant glioblastoma cell lines and tested anti-resistance effects of our NPs. SF-763 or U-118 MG cells were exposed continuously to 2 nM of PTX for over 1 month. The two PTX-resistant cell lines were named SF-763-PTXR and U-118 MG-PTXR, respectively. These cell lines were treated with PTX and cell viability was determined by the Alamar Blue assay. The result showed that the two cell lines responded differently to the long-term exposure to PTX. Unlike U-118 MG-PTXR, which showed significant resistance to PTX (IC50 value changed from 5.06 nM to 18.3 nM), SF-763-PTXR showed only slight resistance (3.24 nM to 6.71 nM) (Figure 6). The higher IC50 values in U-118 MG and U-118 MG-PTXR cells than in SF-763 and SF-763-PTXR cells indicated the stronger resistance of U-118 MG to PTX than SF-763. However, as expected, both PTX-resistant cell lines did not show resistance to IONP-PTX-CTX treatment. The U-118 MG-PTXR cells were even more sensitive than U-118 MG to IONP-PTX-CTX treatment (IC50 was 6.73 nM). It is known that multiple mechanisms contribute to the resistance of PTX, including overexpression of efflux proteins (e.g., P-glycoprotein (P-gp)) and multidrug resistance genes (e.g. MDR-1), molecular changes of beta-tubulin, and changes in apoptotic regulatory. [20, 21] The drug resistance of cancer cells makes chemotherapy ineffective and reduces survival. Applications of NPs in drug delivery may overcome drug resistance by bypassing efflux proteins (e.g. P-gp for PTX resistance[20]) as many NPs enter cells through receptor-mediated endocytosis. In this study, the SF-763-PTXR and U-118 MG-PTXR cells that survived the long-term low-dose exposure to PTX might overexpress P-gp that exported PTX from cells and therefore reduced its efficacy. As PTX is the substrate of an efflux protein P-gp, the IONP-PTX-CTX might enter cells through MMP-2 and bypass P-gp, and therefore overcome the resistance of GBM cells. The result shows promise for improved treatment of drug-resistant brain cancer.
2.4. CTX enhanced IONP-PTX-induced cellular apoptosis
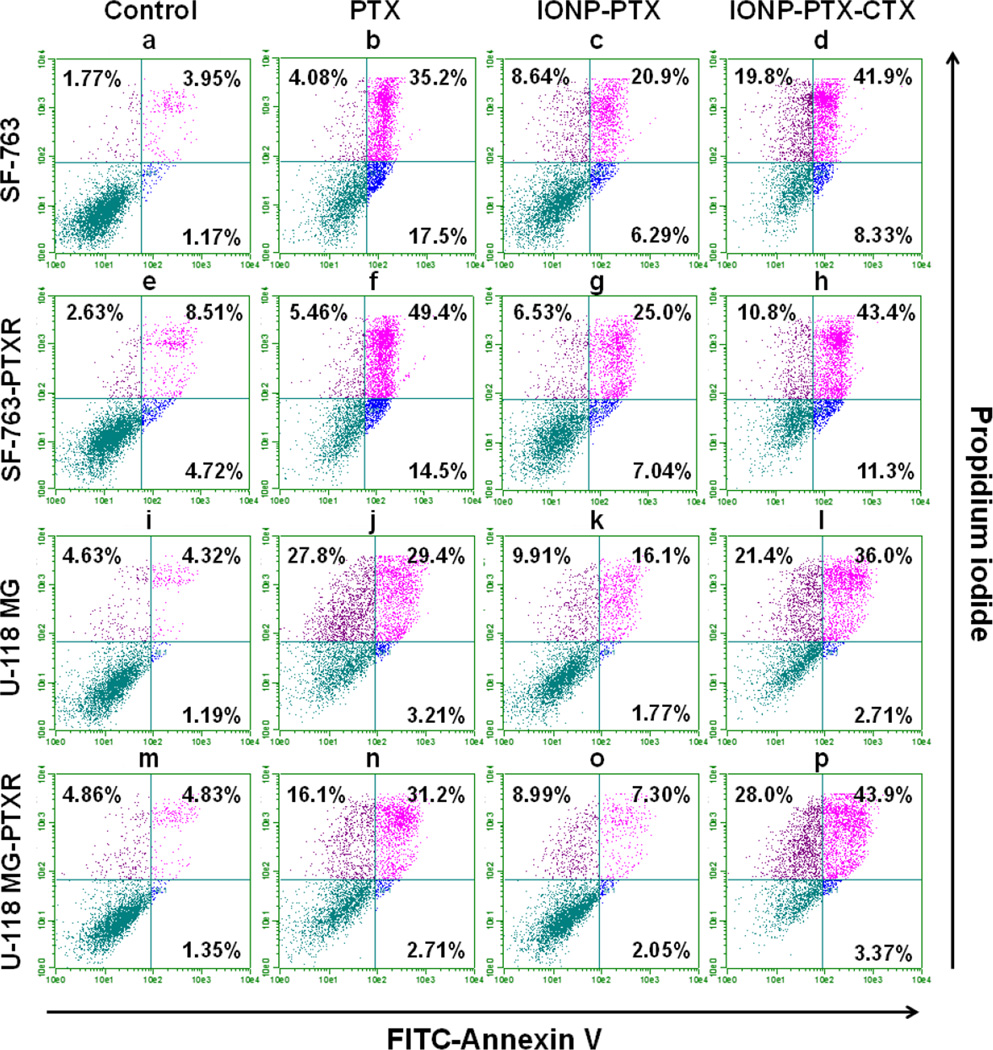
As a microtubule inhibitor, PTX perturbs cell division and eventually causes cell apoptosis.[22] To understand whether the enhanced cell killing was caused by increased apoptosis, we examined cell apoptosis after NP treatment using flow cytometry. During early apoptosis, phosphatidylserine flips to the outside of the cell membrane and can be labeled with Annexin V.[23] Propidium iodide was used to label the nucleus of dead cells as viable cells extrude it. Four cell lines were treated with 100 nM PTX or PTX-equivalent IONP-PTX or IONP-PTX-CTX for three days. Cells labeled with FITC-Annexin V and propidium iodide were analyzed by flow cytometry. As shown in Figure 7 and Figure S3 (Supporting Information), PTX-resistant cell lines SF-763-PTXR and U-118 MG-PTXR had slightly more late apoptotic cells (8.51% and 4.83%) compared to their original cell line counterparts (3.95% and 4.32%), respectively (Figure 7, e vs. a, and m vs. i, upper-right quadrant). That was probably because of continuous exposure to low-dose PTX. Consistent with the cell behavior shown in the cell viability assay, IONP-PTX induced less apoptotic cell death than free PTX (population in upper-right quadrant decreased from 31.2% to 7.3% for U-118 MG-PTXR and from 49.4% to 25% for SF-763-PTXR). However, CTX-conjugated IONP-PTX showed enhanced apoptotic cell death induction compared to both IONP-PTX and free PTX, especially in U-118 MG-PTXR cells (population increased in upper-right quadrant from 31.2% to 43.9%) (Figure 7, d, h, i and p). In addition to the cell viability assay which showed the enhancement in tumor cell killing by IONP-PTX-CTX, the flow cytometry analysis further confirmed that such enhancement was apoptosis-dependent. As many brain tumors develop resistance to drugs, the targeted nanoparticle might have an advantage in treating such drug-resistant brain tumors.
Figure 7.
Annexin V and propidium iodide-based flow cytometric analysis of apoptosis induced by PTX, IONP-PTX, and IONP-PTX-CTX. Cells were treated with 100 nM of PTX or PTX-equivalent NPs for 72 hrs. (a–d) SF-763 cells; (e–h) SF-763-PTXR cells; (i–l) U-118 MG cells; (m–p) U-118 MG-PTXR cells. (a, e, i, m) medium control; (b, f, j, n) free PTX; (c, g, k, o) IONP-PTX; (d, h, l, p) IONP-PTX-CTX.
3. Conclusions
We constructed CTX-conjugated and PTX loaded IONPs and tested their efficacy against drug-resistant and non-resistant GBM cell lines with and without MGMT expression. These NPs have a uniform size and shape, and exhibit high stability in PBS and cell culture medium. Unlike CTX-free NPs (i.e., IONP-PTX), which showed decreased potency compared to free PTX, the CTX conjugated NPs (i.e., IONP-PTX-CTX) greatly improved potency of PTX against GBM cells. Flow cytometry analysis also indicated that the enhanced tumor cell killing by IONP-PTX-CTX in both PTX-resistant and non-resistant GBM cell lines, and the enhanced cell killing was apoptosis-dependent. As many GBMs generate resistance to chemotherapy, the IONP-PTX-CTX might be a potential solution for treating drug-resistant GBMs. Furthermore, the efficacy of IONP-PTX-CTX was also independent of MGMT expression. Although more investigations, such as in vivo studies, are needed to further validate the applicability, the current results have shown the great potential of CTX-conjugated, PTX-loaded IONPs for treatment of drug-resistant GBMs.
4. Experimental section
Materials
The 2-iminothiolane (Traut's reagent) was purchased from Molecular Biosciences (Boulder, CO). NHS-PEG12-maleimide and NHS-Fluorescein were purchased from Thermo Fisher Scientific (Rockford, IL). PTX was purchased from LC Laboratories (Woburn, MA). Wheat germ agglutinin-Alexa Fluor 555 conjugate was purchased from Life technologies (Grand Island, NY). All other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Synthesis of IONP-PEG-NH2 and surface functionalization
PEG2000-NH2 monolayer-coated IONPs (IONP-PEG-NH2) were synthesized using the method reported previously.[7] To prepare IONP-PTX-CTX-FL, CTX was conjugated to IONP-PEG-NH2 using a heterobifunctional PEG linker. To functionalize IONP-PEG-NH2 (2 mg), CTX (286 µg, 1 mg/mL in PBS) was thiolated by 2-iminothiolane water solution (5.92 µL, 1 mg/mL) for 1 h to form CTX–Traut’s. Concurrently, a solution of NHS-PEG12–maleimide (1.42 µL, 250 mM) was added to IONP-PEG-NH2 (1 mg of Fe/mL in PBS) and allowed to react for 30 min. Unreacted PEG was removed using a PD-10 desalting column (GE Healthcare, Piscataway, NJ) equilibrated with PBS. PEG–maleimide modified IONP-PEG-NH2 were then mixed with the CTX–Traut’s and allowed to react for 30 min before removing unreacted CTX using S-200 sephacryl resin equilibrated with 10 mM of MES buffer (pH 4.6) to obtain IONP-CTX. Then the conjugation of carboxymethylated beta-cyclodextrin (CM-β-CD) onto IONP-CTX proceeded as follows. First, CM-β-CD was obtained from modifying β-CD using a previously reported method.[14] IONP-CTX (~2 mg, 1 mg/mL in MES buffer) was then allowed to react with CM-β-CD (1.7 mg) for 3 hrs in the presence of EDC (0.7 mg) and NHS (0.2 mg) before purification with S-200 column equilibrated with distilled water to obtain IONP-CD-CTX. Finally, PTX and AD-FL were loaded onto IONP-CD-CTX to obtain IONP-CTX-PTX-FL. AD-modified fluorescein (AD-FL) was made by reacting 1-AD with NHS-Fluorescein in DMSO for 24 hrs at 1:1 molar ratio. PTX and AD-FL dissolved in DMSO were added into IONP-CD-CTX (1 mg of Fe/mL in PBS) (final concentrations of PTX and AD-FL were 500 µM and 200 µM, respectively), and incubated for 3 hrs at room temperature. The mixture was then centrifuged to remove aggregates and purified by S-200 column equilibrated with distilled water. IONP-CD-CTX without PTX and AD-FL was used as drug-free control. NPs with no CTX (1 mg of Fe/mL in PBS) were loaded with PTX and AD-FL, and served as non-targeted therapeutic control. All NPs were filtered through 0.2 µm polytetrafluoroethylene syringe filters and stored in a refrigerator before use.
Characterization of chemical molecules and NP conjugates
The FT-IR spectra of CM-β-CD were recorded on a Nicolet 6700 spectrometer (Thermo Scientific Inc., Waltham, MA). Absorbance spectra were acquired at 4 cm−1 resolution and the signal was averaged over 32 scans. CM-β-CD were mixed with KBr and pressed into a pellet for analysis. Mass spectroscopy (MS) spectra of CM-β-CD and AD-FL were collected on a Bruker Esquire ion trap mass spectrometer (Bruker Daltonics, Billerica, MA) using the positive ion mode. PTX and AD-FL bound on NPs were quantified by HPLC. The column was Magic Bullet C-18 Column (Michrom Bioresources, Auburn, CA) (25 mm, tapered ID 2.1 to 0.5 mm, 5 µm). The A solvent was 5% ACN in 0.1% v/v acetic acid and the B solvent was 95% ACN in 0.1% v/v acetic acid. The gradient was 25–95% B in 5 min. The equilibration time was 3 min at 25% B. The column temperature was set at 30°C and the flow rate was 0.4 mL/min. The injection volume was set at 10 µL and 254 nm was used as a detection wavelength. These conditions were used for both PTX and AD-FL. The loading of CTX onto NPs was quantified by a Micro BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Briefly, the concentrations of free CTX before and after conjugation reaction was quantified using a CTX standard curve. The amount of conjugated CTX were obtained by subtracting unreacted CTX from total CTX.
For negative-staining TEM images of NPs, NP solution (4 µL) was transferred onto a TEM grid (copper grid, 300-mesh, coated with carbon and Formvar film) and stained with 2% uranyl acetate. After drying the solution in air using a filter paper, negative-staining TEM images were acquired on a Tecnai G2 F20 electron microscope (FEI, Hillsboro, OR) operating at a voltage of 200 kV. The hydrodynamic size and ζ-potential of IONPs were determined using Zetasizer Nano-ZS (Malvern Instruments, Worcestershire, UK). The analyses were performed at the room temperature. The pH value of all NP solutions was 7.0. The medium stability test of IONPs was performed by diluting IONPs 10 times in complete DMEM cell culture medium and incubated in a 37°C water bath. The hydrodynamic sizes were measured at various incubation time points.
Cell culture
SF-763 and U-118 MG human GBM cells were purchased from the American Type Culture Collection (ATCC) and grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic (Life technologies, Grand Island, NY). Cells were cultured in an incubator maintained at 37°C and 5% CO2 with 95% humidity. SF-763-PTXR and U-118 MG-PTXR were established by continuously exposing cells to 2 nM PTX for 1 month.
Cellular uptake of NPs by flow cytometry analysis
SF-763 or U-118 MG cells were incubated with NPs for 1 hr or 6 hrs, followed by washing with cold PBS for 3 times. Cells were then digested by Trypsin and washed with cold PBS. Cells were then fixed in 2% paraformaldehyde at 4°C overnight. Cells were centrifuged to remove fixative, washed once with PBS and resuspended in PBS for flow cytometry analysis. Data acquisition and analysis were performed on a Guava EasyCyte Mini flow cytometry system (Millipore, Billerica, MA) using Guava Express Pro software module.
Characterization of cellular uptake of NPs by confocal laser scanning microscopy (CLSM)
SF-763 or U-118 MG cells were seeded onto glass cover slips in a 6-well plate. After overnight incubation, cells were incubated with NPs (40 µg/mL) for 6 hrs. Cells were then fixed with 4% paraformaldehyde for 15 min at 37°C and stained with WGA-Alexa Fluor 555 (5 µg/mL) (Invitrogen, Carlsbad, CA) for 5 min at 37°C, followed by 3 times of PBS washing (5 min each). Cells were then incubated with DAPI for 5 min at 37°C, followed by PBS washing. After PBS washing, cells were mounted with VECTASHIELD mounting medium (Vector Laboratories, Inc. Burlingame, CA). The images of cells were acquired using a Laser Scanning Microscope LSM 510, Version 3.2 SP2 (Carl Zeiss GmbH, Germany).
Cell viability by Alamar Blue assay
Cells were seeded in a 96-well plate and incubated overnight in the aforementioned growth conditions. In the following day, the medium was replaced with a medium containing PTX (PTX was dissolved in DMSO to a concentration of 180 µM as stock solution), IONP-CTX, IONP-PTX-FL or IONP-PTX-CTX-FL or with medium control. Nine different drug concentrations (1800, 900, 450, 225, 112.5, 56.25, 28.13, 14.06 and 7.03 nM) were used, and samples at each concentration were ran in triplicate. The cells were incubated with PTX, IONP-CTX, IONP-PTX-FL or IONP-PTX-FL for 72 h. Cell viability was assessed using the Alamar Blue assay. Briefly, the medium was replaced with cell culture medium containing reagent and allowed to incubate for 2 hrs. Following the incubation, a microplate reader (SpectraMax i3, Molecular Devices, Sunnyvale, CA) was used to determine the fluorescence intensity of the dye (550ex/590em). The fluorescence intensity from NP or free drug treated cells was compared to those from untreated control cells to determine percent viability.
Apoptosis by flow cytometry
Cells were seeded into 6-well plates and incubated overnight. NP stock solutions were added into cell cultures at a PTX concentration of 100 nM. Cells were incubated with NPs for 72 h. Cells were then trypsinized, aspirated, and washed once with PBS. Cells were then counted and suspended in 0.5 mL Annexin V binding buffer containing 50 µg/mL propidium iodide and 0.5 µL FITC-Annexin V reagent (BioLegend, San Diego, CA). Cells were further incubated for 15 min at room temperature in dark and analyzed by flow cytometry. Data acquisition was performed on a Guava EasyCyte Mini flow cytometry system (Millipore, Billerica, MA) and analysis was performed using Guava Express Pro software module.
Supplementary Material
Acknowledgments
The work is supported by NIH grants R01CA161953 and R01CA134213. Q. M. acknowledges support from an NIH Ruth L. Kirschstein T32 Fellowship (T32CA138312). We thank Dr. Martin Sadilek from Department of Chemistry for the assistance of HPLC and Dr. Greg Martin from Keck Microscopy Facility in the University of Washington for the assistance of confocal microscopy.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Qingxin Mu, Department of Materials Science and Engineering, University of Washington, Seattle, Washington, 98195, USA; Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, 98109, USA.
Mike Jeon, Department of Materials Science and Engineering, University of Washington, Seattle, Washington, 98195, USA.
Meng-Hsuan Hsiao, Department of Materials Science and Engineering, National Chiao Tung University, Hsinchu City, 300 Taiwan.
Victoria K. Patton, Department of Chemical Engineering, University of Washington, Seattle, Washington, 98195, USA
Kui Wang, Department of Materials Science and Engineering, University of Washington, Seattle, Washington, 98195, USA.
Oliver W. Press, Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, 98109, USA
Miqin Zhang, Email: mzhang@uw.edu, Department of Materials Science and Engineering, University of Washington, Seattle, Washington, 98195, USA.
References
- 1.Wen PY, Kesari S. N. Engl. J. Med. 2008;359:492. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.a) Donson AM, Addo-Yobo SO, Handler MH, Gore L, Foreman NK. Pediatr. Blood Cancer. 2007;48:403. doi: 10.1002/pbc.20803. [DOI] [PubMed] [Google Scholar]; b) Sharma S, Salehi F, Scheithauer BW, Rotondo F, Syro LV, Kovacs K. Anticancer Res. 2009;29:3759. [PubMed] [Google Scholar]
- 3.Kim H, Likhari P, Parker D, Statkevich P, Marco A, Lin CC, Nomeir AA. J. Pharm. Biomed. Anal. 2001;24:461. doi: 10.1016/s0731-7085(00)00466-0. [DOI] [PubMed] [Google Scholar]
- 4.Ling Y, Wei K, Zou F, Zhong SZ. Int. J. Pharm. 2012;430:266. doi: 10.1016/j.ijpharm.2012.03.047. [DOI] [PubMed] [Google Scholar]
- 5.a) Tian X-H, Lin X-N, Wei F, Feng W, Huang Z-C, Wang P, Ren L, Diao Y. International Journal of Nanomedicine. 2011;6:445. doi: 10.2147/IJN.S16570. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Patil R, Portilla-Arias J, Ding H, Inoue S, Konda B, Hu J, Wawrowsky K, Shin P, Black K, Holler E, Ljubimova J. Pharm Res. 2010;27:2317. doi: 10.1007/s11095-010-0091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jain A, Singhai P, Gurnany E, Updhayay S, Mody N. J Nanopart Res. 2013;15:1. [Google Scholar]
- 6.Panchagnula R. Int. J. Pharm. 1998;172:1. [Google Scholar]
- 7.Fang C, Bhattarai N, Sun C, Zhang MQ. Small. 2009;5:1637. doi: 10.1002/smll.200801647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kievit FM, Zhang MQ. Accounts Chem. Res. 2011;44:853. doi: 10.1021/ar2000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Sun C, Du K, Fang C, Bhattarai N, Veiseh O, Kievit F, Stephen Z, Lee D, Ellenbogen RG, Ratner B, Zhang M. ACS Nano. 2010;4:2402. doi: 10.1021/nn100190v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Weissleder R, Stark DD, Engelstad BL, Bacon BR, Compton CC, White DL, Jacobs P, Lewis J. AJR Am J Roentgenol. 1989;152:167. doi: 10.2214/ajr.152.1.167. [DOI] [PubMed] [Google Scholar]
- 10.a) Qiao R, Yang C, Gao M. J. Mater. Chem. 2009;19:6274. [Google Scholar]; b) Yigit MV, Moore A, Medarova Z. Pharm. Res. 2012;29:1180. doi: 10.1007/s11095-012-0679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Veiseh O, Gunn J, Zhang M. Adv. Drug Deliver. Rev. 2010;62:284. doi: 10.1016/j.addr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshane J, Garner CC, Sontheimer H. J. Biol. Chem. 2003;278:4135. doi: 10.1074/jbc.M205662200. [DOI] [PubMed] [Google Scholar]
- 12.a) Veiseh O, Sun C, Gunn J, Kohler N, Gabikian P, Lee D, Bhattarai N, Ellenbogen R, Sze R, Hallahan A, Olson J, Zhang M. Nano Lett. 2005;5:1003. doi: 10.1021/nl0502569. [DOI] [PubMed] [Google Scholar]; b) Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, Du K, Pullar B, Lee D, Ellenbogen RG, Olson J, Zhang M. Cancer Res. 2009;69:6200. doi: 10.1158/0008-5472.CAN-09-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kievit FM, Veiseh O, Fang C, Bhattarai N, Lee D, Ellenbogen RG, Zhang MQ. ACS Nano. 2010;4:4587. doi: 10.1021/nn1008512. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Veiseh O, Kievit FM, Fang C, Mu N, Jana S, Leung MC, Mok H, Ellenbogen RG, Park JO, Zhang MQ. Biomaterials. 2010;31:8032. doi: 10.1016/j.biomaterials.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Bobola MS, Varadarajan S, Smith NW, Goff RD, Kolstoe DD, Blank A, Gold B, Silber JR. Clin. Cancer Res. 2007;13:612. doi: 10.1158/1078-0432.CCR-06-1127. [DOI] [PubMed] [Google Scholar]; b) Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, Sharp SY, Vassal G, Pearson ADJ, Reis RM, Hargrave D, Workman P, Jones C. Cancer Res. 2010;70:9243. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furusaki E, Ueno Y, Sakairi N, Nishi N, Tokura S. Carbohydrate Polymers. 1996;29:29. [Google Scholar]
- 15.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Meng F, Zhong Y, Cheng R, Deng C, Zhong Z. Nanomedicine. 2014;9:487. doi: 10.2217/nnm.13.212. [DOI] [PubMed] [Google Scholar]; b) Kolhe P, Khandare J, Pillai O, Kannan S, Lieh-Lai M, Kannan RM. Biomaterials. 2006;27:660. doi: 10.1016/j.biomaterials.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Lee N, Hyeon T. Chem. Soc. Rev. 2012;41:2575. doi: 10.1039/c1cs15248c. [DOI] [PubMed] [Google Scholar]
- 18.Mu QX, Li ZW, Li X, Mishra SR, Zhang B, Si ZK, Yang L, Jiang W, Yan B. J. Phys. Chem. C. 2009;113:5390. [Google Scholar]
- 19.a) Kesavan K, Ratliff J, Johnson EW, Dahlberg W, Asara JM, Misra P, Frangioni JV, Jacoby DB. J. Biol. Chem. 2010;285:4366. doi: 10.1074/jbc.M109.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Debin JA, Maggio JE, Strichartz GR. Am. J. Physiol. 1993;264:C361. doi: 10.1152/ajpcell.1993.264.2.C361. [DOI] [PubMed] [Google Scholar]
- 20.Yusuf RZ, Duan Z, Lamendola DE, Penson RT, Seiden MV. Curr. Cancer Drug Targets. 2003;3:1. doi: 10.2174/1568009033333754. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Zhai S, Li L, Li X, Jiang C, Zhang C, Yan B. J. Pharmacol. Sci. 2014;126:66. doi: 10.1254/jphs.14050fp. [DOI] [PubMed] [Google Scholar]
- 22.Foa R, Norton L, Seidman AD. Int. J. Clin. Lab. Res. 1994;24:6. doi: 10.1007/BF02592403. [DOI] [PubMed] [Google Scholar]
- 23.Koopman G, Reutelingsperger CPM, Kuijten GAM, Keehnen RMJ, Pals ST, Vanoers MHJ. Blood. 1994;84:1415. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.