Abstract
Coat color in Holstein dairy cattle is primarily controlled by the melanocortin 1 receptor (MC1R) gene, a central determinant of black (eumelanin) vs. red/brown pheomelanin synthesis across animal species. The major MC1R alleles in Holsteins are Dominant Black (MC1RD) and Recessive Red (MC1Re). A novel form of dominant red coat color was first observed in an animal born in 1980. The mutation underlying this phenotype was named Dominant Red and is epistatic to the constitutively activated MC1RD. Here we show that a missense mutation in the coatomer protein complex, subunit alpha (COPA), a gene with previously no known role in pigmentation synthesis, is completely associated with Dominant Red in Holstein dairy cattle. The mutation results in an arginine to cysteine substitution at an amino acid residue completely conserved across eukaryotes. Despite this high level of conservation we show that both heterozygotes and homozygotes are healthy and viable. Analysis of hair pigment composition shows that the Dominant Red phenotype is similar to the MC1R Recessive Red phenotype, although less effective at reducing eumelanin synthesis. RNA-seq data similarly show that Dominant Red animals achieve predominantly pheomelanin synthesis by downregulating genes normally required for eumelanin synthesis. COPA is a component of the coat protein I seven subunit complex that is involved with retrograde and cis-Golgi intracellular coated vesicle transport of both protein and RNA cargo. This suggests that Dominant Red may be caused by aberrant MC1R protein or mRNA trafficking within the highly compartmentalized melanocyte, mimicking the effect of the Recessive Red loss of function MC1R allele.
Introduction
The vast majority of dairy cattle of the Holstein breed display a black and white spotted coat color while a subset of this breed are red and white spotted. The melanocortin 1 receptor (MC1R) gene determines the basic coat color in cattle and in Holsteins there are four known MC1R alleles: Dominant Black (MC1R D), Black/Red (MC1R BR), the ancestral wild-type allele (MC1R +) and Recessive Red (MC1R e) [1–3]. The order of dominance is MC1R D>MC1R BR>MC1R +>MC1R e. Holsteins homozygous or heterozygous for any combination of the MC1R + or MC1R e allele are red, while animals heterozygous or homozygous for MC1R D are black. Black/Red animals are born red and change to black typically between two to six months of age. The causal mutations for MC1R D and MC1R e have been identified as constitutive activation and loss of function of MC1R, respectively [1,2]. MC1R BR has recently been shown to be a fourth MC1R allele through haplotype and linkage analysis with the causal mutation yet unidentified [3]. In Holsteins, MC1R BR and MC1R + are both rare, such that red vs. black coat color is primarily determined by the segregation of MC1R D and MC1R e. Holstein breeders have become accustomed to the concept of red coat color being recessive to black and extensively utilize genetic testing of these MC1R mutations to mate carriers of MC1R e and/or MC1R + together to produce red offspring.
In 1980, a female Holstein calf (HOCANF3541221, SURINAM SHEIK ROSABEL-RED) was born in Canada that displayed the typical red Holstein coat color phenotype (Fig 1), but from parents that were not thought to carry either of the MC1R alleles associated with red color based on pedigree. Genetic testing verified that this red animal was homozygous for MC1R D but yet approximately 50% of her progeny were phenotypically red, suggesting the presence of a new dominant form of red coat color, which was termed “Variant Red” [4]. At the time, this name reflected the unknown origin and mode of inheritance of this new genetic cause of red coat color. Here, we refer to this form of red coat color as “Dominant Red” (DR) to more clearly describe the mode of inheritance and differentiate it from the Recessive Red form of coat color caused by MC1R. We propose that the Dominant Red locus is designated DR with two alleles, the derivative allele DR DR and the ancestral or wild-type allele DR +. The ability of DR DR to override the production of black pigment caused by a constitutively activated MC1R (MC1R D), the central mechanism of pigment type switching in mammals, indicates that the DR phenotype represents a valuable opportunity to study novel aspects of pigmentation biology.
Fig 1. Dominant Red (DR DR /DR +, MC1RD /-), Dominant Black (DR + /DR +, MC1RD /-) and Recessive Red (DR + /DR +, MC1Re /MC1R e) phenotypes.

(A) The first animal to display the Dominant Red phenotype was SURINAM SHEIK ROSABEL-RED (HOCANF3541221), born in 1980 in Ontario Canada. (B) A Dominant Black Holstein female. (C) A Recessive Red Holstein female. (D) A Dominant Red Holstein female. Reprinted under a CC BY license, with permission from (A) Canadian Livestock Photography Inc., original copyright 1985 and (B, C and D) Cybil Fisher Photography, original copyright 2014.
In a previous study the beta-defensin 103 (DEFB103) gene on Bos taurus autosome (BTA) 27 was suggested as being associated with DR [5]. However, this analysis was limited to five candidate genes with known roles in pigmentation biology and found only marginal evidence for genetic linkage (LOD = 3.26). Here, we use whole genome linkage and association mapping techniques coupled with whole genome sequencing to show that in fact DR is located on BTA3 and is completely associated with a single missense mutation in the coatomer protein complex, subunit alpha (COPA) gene.
Results
Hair Pigment Analysis
Studies of coat color in laboratory mice indicate that the effect of MC1R signaling on hair color represents a switch between the synthesis of two alternative pigment types, red/yellow pheomelanin (caused by loss-of-function MC1R mutations) vs. black/brown eumelanin (caused by mutations that constitutively activate MC1R).
Hair samples collected from Dominant Black (DR +/DR +, MC1R D/-), Dominant Red (DR DR/DR +, MC1R D/MC1R D) and Recessive Red (DR +/DR +; MC1R e/MC1R e) animals were analyzed for pigment content and composition [6]. Absorbance of solubilized hair pigment at 500 nm (Soluene-350, A500/mg) is an indicator of total melanin [7]. By this criterion, samples from Dominant Black animals had the highest melanin levels, Dominant Red samples were intermediate, and Recessive Red samples had the lowest levels (Table 1). We used HPLC analysis of melanin degradation products to measure individual pigment types. Pheomelanin content as assessed by 4-amino-3-hydroxyphenylalanine (4-AHP) or thiazole-2,4,5-tricarboxylic acid (TTCA) was approximately 50- to 60-fold and 12-fold higher respectively in Dominant Red and Recessive Red hair samples as compared to Dominant Black [8,9]. Eumelanin content, as assessed by pyrrole-2,3,5-tricarboxylic acid (PTCA) content [8], was highest in Dominant Black hair samples; Dominant Red samples exhibited PTCA levels that were approximately 8-fold lower, and Recessive Red levels were 23-fold lower (Table 1). Although the difference between Dominant Red and Recessive Red PTCA levels (approximately 3-fold) did not achieve statistical significance (P = 0.09), when the ratios of 4-AHP/PTCA or TTCA/PTCA were compared between Dominant Red and Recessive Red, the differences were significant (P<0.001). The Soluene-350 A650/A500 ratio, with higher values representing black/brown color and lower values representing yellow/red color were also significantly different for all three groups with Dominant Black samples having the highest value and Dominant Red being intermediate.
Table 1. Hair pigment content and composition.
| Dominant Black | Dominant Red | Recessive Red | |
|---|---|---|---|
| Assay | n = 5 | n = 8 | n = 9 |
| Soluene-350 (A500/mg) | 0.549 A | 0.247 B | 0.173 C |
| 0.027 | 0.021 | 0.020 | |
| PTCA (ng/mg) | 1918.0 A | 246.0 B | 82.8 B |
| 85.1 | 67.3 | 63.4 | |
| 4-AHP (ng/mg) | 32.8 A | 1943.3 B | 1610.8 B |
| 227.0 | 179.5 | 169.2 | |
| TTCA (ng/mg) | 49.9 A | 623.9 B | 587.6 B |
| 37.6 | 29.7 | 28.0 | |
| 4-AHP/PTCA | 0.0 A | 9.2 B | 19.6 C |
| 1.5 | 1.2 | 1.1 | |
| TTCA/PTCA | 0.0 A | 3.3 B | 7.7 C |
| 1.0 | 0.8 | 0.7 | |
| Soluene-350 A650/A500 | 0.275 A | 0.159 B | 0.132 C |
| 0.008 | 0.007 | 0.006 |
Standard error in italics below respective mean.
A B C Means with significant difference (P<0.05) indicated by different superscript letters.
These results indicate that similarities in hair color phenotype between Dominant Red and Recessive Red reflect similarities in pigment type synthesis, namely, production of pheomelanin rather than eumelanin, although suppression of eumelanin synthesis by Dominant Red is not as effective as in Recessive Red.
Genetic Mapping
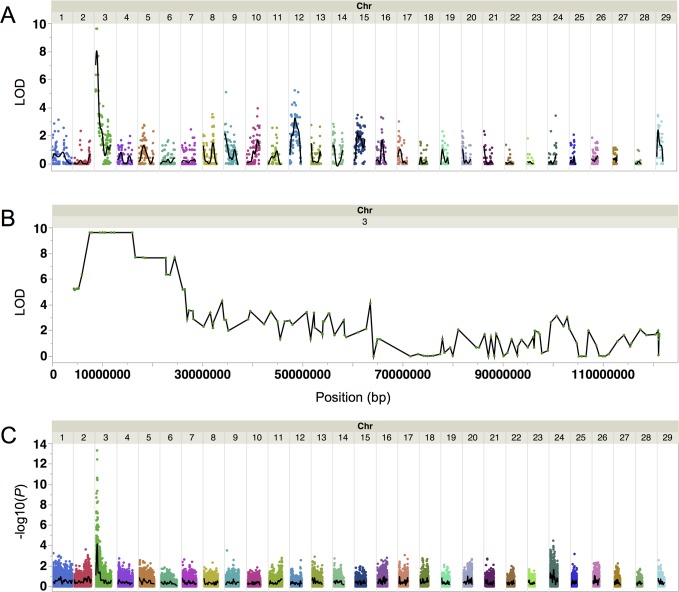
Genetic mapping of DR was performed using a heterozygous DR bull (HOCANM9626808, MORSAN RED GOLD) as well as 15 DR and 17 non-DR progeny, including their dams, comprising a single half-sib family. Two point linkage analysis between coat color and 2,752 autosomal markers, which were heterozygous in the DR sire, was performed using CRI-MAP 2.503 [10]. The highest genome-wide LOD score of 9.6 was found on BTA3 (Fig 2A). Eight contiguous SNPs on BTA3 had this same LOD score and showed complete linkage with the DR phenotype in this half-sib family (Fig 2B). The nearest flanking markers showing recombination to DR define a 10.7 Mb region containing the DR causal variant by linkage analysis, from BTA-21472-no-rs @ 5,886,143 bp to ARS-BFGL-NGS-20167 @ 16,557,950 bp. The UMD3.1/bosTau6 genome assembly was used for all analyses [11].
Fig 2. Linkage and genome-wide association mapping of Dominant Red.
(A) Whole genome linkage mapping using a single half-sib family of 15 DR and 17 WT animals shows Dominant Red is located on BTA3. The black line is a cubic spline smoother with a lambda of 0.05. (B) Linkage mapping shows eight contiguous SNPs on BTA3 in complete association with Dominant Red across a 10.7 Mb region. (C) Genome-wide association analysis in 95 DR and more than 500 WT animals identifies a significant association on BTA3 within the 10.7 Mb region identified by linkage mapping. The most significant marker is ARS-BFGL-NGS-94819 @ 9,722,400 bp. The black line is a cubic spline smoother with a lambda of 0.05.
A genome-wide association analysis (GWAS) was performed using 95 DR heterozygous animals, 55 non-DR animals closely related to the 95 DR individuals and 500 non-DR animals randomly selected from the population. The 500 non-DR randomly selected animals had previously been genotyped for genomic selection purposes. The DFAM procedure of the PLINK [12] software package was used to perform a family-based association test for disease traits, a method which accommodates the use of unrelated individuals and case/control traits. A single peak on BTA3 was identified as being most highly associated with the DR phenotype (Fig 2C). A single SNP (ARS-BFGL-NGS-94819 @ 9,722,400 bp) showed the strongest genome-wide association of-log10(P) = 13.3 with all 95 DR animals having at least one copy of the DR associated allele. However, the DR associated allele of ARS-BFGL-NGS-94819 had a frequency of 6.7% in the 500 non-DR animals, indicating that it would result in a high false positive rate if this SNP were used as a genetic marker for DR.
We searched for a haplotype that was identical by descent (IBD) in DR animals within the 10.7 Mb region defined by linkage mapping and confirmed by GWAS results in a larger group of animals. The major allele at each SNP across this region in 95 DR animals was considered to represent the allele present on the putative IBD haplotype. The minimal IBD haplotype was defined as the region where all of the 95 DR animals had at least one copy of the major allele. A region satisfying this criterion was identified from ARS-BFGL-NGS-112616 @ 7,906,099 bp to ARS-BFGL-NGS-1429 @ 10,462,387 bp, spanning 2.6 Mb of BTA3 (S1 Fig). A single individual did not conform to this rule, possibly due to recombination events that we could not resolve. When possible, more than one animal in discordance with this definition was used to define the boundary of the IBD haplotype.
Whole Genome Sequencing
A heterozygous DR individual (HOCANF9845383, BROEDERDALE MALTBY CAMILLA) was selected for whole genome sequencing in order to identify all candidate causal sequence variants within the 2.6 Mb IBD region. This individual was selected based on maximum homozygosity across the IBD region (S1 Fig, top row) in order to minimize the number of non-DR associated variants. This means that we selected an animal whose DR + chromosome was closely related to its DR DR chromosome. At the time of sequencing no DR DR /DR DR homozygotes were yet known. From the whole genome sequencing data 8,485 SNPs were detected within the IBD region, 77 of which cause an amino acid change and 51 of which were heterozygous. Sequence data from the 1000 Bull Genomes Project [13] was used to exclude variants that have been detected in non-DR animals, which further narrowed the list of candidate causal SNPs to 25. Variant coverage, quality scores, and manual inspection of the aligned reads were used to evaluate which SNPs had the most evidence of being true variants. SNP calling criteria was purposefully set at a low threshold to minimize exclusion of true variants.
The six candidate causal SNPs with the highest evidence of being true variants were genotyped in a panel of 20 DR heterozygous animals (S1 Table). Three of the SNPs were excluded as false positives as they were not detected by conventional sequencing methods in the same animal used for whole genome sequencing. This suggests that all other remaining candidate causal SNPs with lower quality scores were also likely to be false positives. Two of the SNPs were excluded due to homozygotes of alternative alleles being found in the panel of 20 DR heterozygotes. The one remaining SNP (C>T @ BTA3:9,479,761 bp) had the highest quality score out of the 25 candidate causal SNPs detected within the entire IBD region and was found to be heterozygous in all 20 DR heterozygotes in the panel (S1 Table). No insertions or deletions that were also heterozygous were detected in the IBD region.
Analysis of Structural Variants in the IBD Region
To determine if a structural variant is associated with the Dominant Red phenotype read depth and deviant mate-pairs within the IBD region were analyzed for insert size, read order and strand orientation. The sequence from the Dominant Red individual was compared to a Dominant Black Holstein reference individual (HOCANM10705608, BRAEDALE GOLDWYN) that had been sequenced to the same average depth. Normalized read depth in windows was analyzed first within both individuals separately using CNVnator and then between individuals, using CNV-seq. The result of the SV analysis is summarized in S2 Fig. Using CNVnator, two regions (BTA3:8,508,750–8,539,500 and BTA3:8,611,500–8,679,000) were called as deletions in both individuals and likely represent assembly/mapping artifacts. In addition, one region (BTA3:8,731,500–8,752,500) was called as a deletion in the reference individual whereas the Dominant Red individual displayed a normal read depth. However, this region does not have support from the CNV-seq analysis or any deviant reads and is likely a false positive. No regions were found to significantly deviate in read depth among individuals using CNV-seq. Analysis of deviant mate-pairs with SVDetect found one putative duplication in Dominant Red (BTA3:9,007,145–9,008,020). This duplication is also supported in the reference individual and may be an assembly artifact. Taken together, no likely candidate causative structural variants within the IBD region specific to the DR individual were identified.
Causal Variant Validation and Conservation
Due to the lack of structural variants and insertions or deletions associated with DR in the IBD region the C>T SNP at BTA3:9,479,761 bp was considered to be the most likely DR candidate causal variant. A diagnostic test for this SNP was developed and used to genotype 78 known DR heterozygotes. All 78 animals were found to be heterozygous at this SNP. Of 189 randomly selected non-DR animals and 94 non-DR relatives of DR animals, all were homozygous for the wild-type allele. These results demonstrate the complete association of the DR candidate causal variant with DR phenotype.
The candidate causal variant at BTA3:9,479,761 bp is located in the coding region of COPA, a gene not previously known to have a direct role in pigmentation biology. In the COPA mRNA transcript NM_001105645 this SNP is located at coding position 478 and results in an arginine to cysteine substitution at amino acid position 160 in NP_001099115 (c.478C>T p.Arg160Cys). Protein sequence alignment to all COPA orthologs present in NCBI databases reveal that an arginine amino acid is completely conserved at this position from mammals to yeast, residing in a highly conserved WD40 repeat motif (Fig 3). Sequence conservation across such a broad range of species suggests evolutionary constraint due to an important function; indeed, the Arg160Cys substitution in human COPA is predicted to be probably damaging or damaging by PolyPhen-2 [14] and SIFT [15], respectively. The DR candidate causal variant is therefore likely to have a significant effect on the function of this conserved region.
Fig 3. Evolutionary conservation of Dominant Red associated COPA mutation.
Multiple sequence alignment using MUSCLE [16] of COPA and orthologues indicates high conservation across 17 species from mammals to yeast. The arginine (R) to cysteine (C) amino acid substitution caused by the Dominant Red SNP is indicated with the arrow and is completely conserved in all species with available sequences. The WD40 repeat motif is highlighted in yellow.
Due to the high evolutionary conservation of the amino acid residue affected by the DR candidate causal variant and the fact that no homozygotes for DR had previously been documented, we considered the possibility that the DR variant may be detrimental in the homozygous state. The DR diagnostic test was used to investigate this scenario by genotyping red individuals that were progeny of two DR parents. Out of 14 animals tested to date four individuals were determined to be homozygous for the DR candidate causal variant. All animals appeared healthy indicating that DR homozygotes are viable.
RNA-Seq Analysis of Dominant Red Skin
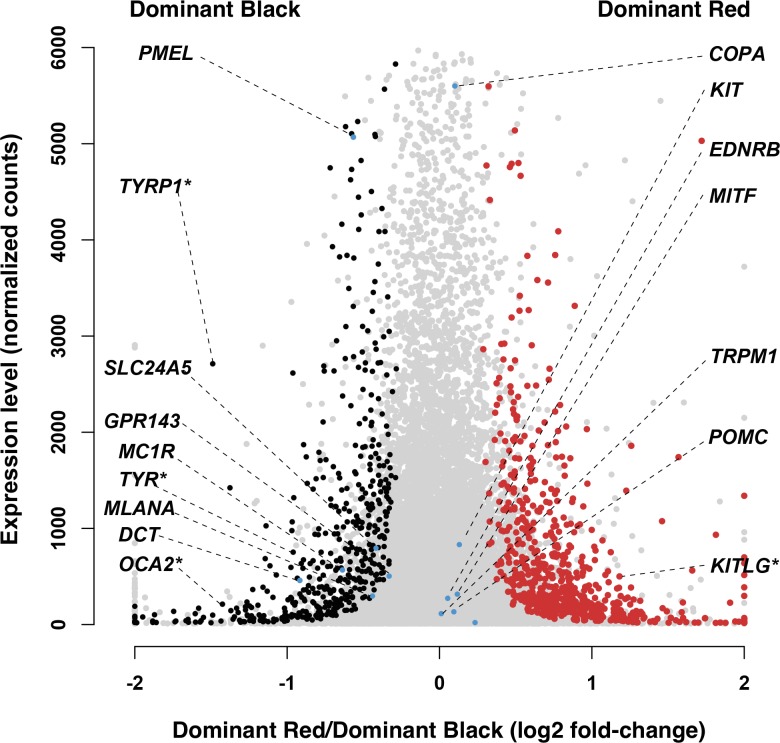
To gain additional insight into the mechanisms underlying Dominant Red gene action, we compared gene expression profiles between skin from Dominant Red (DR DR/DR +, MC1R D/MC1R D) and Dominant Black (DR +/DR +, MC1R D/-) animals. Skin biopsies from three individuals of each genotype were used as the source of cDNA libraries that were sequenced on an Illumina HiSeq instrument and then aligned against the bovine reference genome (UMD3.1/bosTau6). Differentially expressed genes were identified with DESeq2 [17]; among 15,143 genes that were assayed, 111 and 1104 exhibited significantly different levels of expression at a FDR<0.05 and a FDR<0.1, respectively (Fig 4, S2 Table).
Fig 4. Differential expression results comparing Dominant Red (DR DR /DR +, MC1R D /MC1R D) and Dominant Black (DR + /DR +, MC1R D /-) skin samples.
Gene expression levels (normalized gene counts averaged across the six analyzed samples) are represented as a function of log2-fold changes of Dominant Red versus Dominant Black samples. Genes that are significantly overexpressed in Dominant Black skin samples are depicted in black, those overexpressed in Dominant Red samples are illustrated in red, while non-significant changes are illustrated in gray. Genes previously implicated in melanocyte biology and/or pigment type-switching are annotated (selected genes that do not exhibit a significant difference at FDR<0.1 are highlighted in blue while the ones showing significant expression changes are marked additionally with an asterisk associated to the gene symbol).
Fig 4 depicts those results with the level of gene expression plotted as a function of the log2 ratio of Dominant Red/Dominant Black; thus, genes overexpressed in Dominant Black compared to Dominant Red skin are on the left side of the plot, and vice versa, with genes exhibiting a significant difference (FDR<0.1) highlighted in black or red, respectively. Genes previously implicated in melanocyte biology and/or pigment type-switching are annotated (selected genes that do not exhibit a significant difference at FDR<0.1 are highlighted in blue). Notably, TYRP1, TYR, and OCA2, which encode proteins previously known to be required for and upregulated during eumelanogenesis, are overexpressed in Dominant Black compared to Dominant Red skin. Two additional genes (PMEL and DCT), also required for and upregulated during eumelanogenesis, are also overexpressed in Dominant Black compared to Dominant Red skin but at an FDR>0.1. We also considered the three major paracrine ligand-receptor systems involved in melanocyte biology and pigment type-switching: ASIP and POMC—MC1R, KITLG—KIT, and EDN3—ENDRB; these genes were either not detected or not differentially expressed, with the exception of KITLG, which is overexpressed in Dominant Red compared to Dominant Black skin. Taken together, these results suggest that the biochemical mechanism of pheomelanin production in Dominant Red skin is identical to that caused by MC1R deficiency, and that the phenotype arises due to melanocyte-autonomous alterations in gene expression.
We also carried out a functional annotation of differentially expressed genes using the FunNet tool [18]. The most striking result, apparent from a KEGG analysis is that the four pathways most overrepresented in differentially expressed genes are Endocytosis, MAPK signaling, ER protein processing, and Antigen processing and presentation, all of which are increased in Dominant Red compared to Dominant Black skin (Table 2). These observations are consistent with impairment of protein trafficking due to the COPA missense alteration, and upregulation of compensatory pathways.
Table 2. KEGG pathways significantly enriched in genes that are overexpressed in Dominant Red skin samples.
| KEGG Pathway | Enrichment P-value |
|---|---|
| Endocytosis | 1.65x10-3 |
| Protein processing in endoplasmic reticulum | 2.19x10-2 |
| Antigen processing and presentation | 2.01x10-2 |
| Phosphatidylinositol signaling system | 2.71x10-2 |
| Natural killer cell mediated cytotoxicity | 5.39x10-3 |
| Jak-STAT signaling pathway | 4.45x10-2 |
| Oocyte meiosis | 1.75x10-2 |
| Lysine degradation | 3.15x10-2 |
Origin of the Dominant Red Mutation
The DR DR mutation must have occurred on the IBD haplotype described earlier in the germline of either the sire or dam of the DR founder animal ROSABEL. Whole genome genotyping data was available on some of the ancestors of ROSABEL: A PUGET-SOUND SHEIK (sire), PROVIN MTN IVANHOE JEWEL (paternal grand sire), OSBORNDALE IVANHOE (paternal great grand sire) and SEILING ROCKMAN (maternal great grand sire). Using the IBD haplotype data we were able to exclude the sire of ROSABEL as the germline source of the DR variant as this individual did not carry the DR IBD haplotype. This indicates that the DR mutation occurred in either the germline of ROSABEL’s dam or very early in embryonic development of ROSABEL herself. A DNA sample from ROSABEL’s dam was not available, however the maternal great grand sire had previously been genotyped and was found to carry the DR IBD haplotype. This individual (HOCANM275932, SEILING ROCKMAN) had at least one copy of the major allele among the 95 Dominant Red heterozygotes at each locus over an expanded 18 Mb region (S1 Fig). This individual would be expected to be wild-type for the DR associated mutation in COPA since he was phenotypically black, although a sample was not available for confirmation. Further genotyping of ROSABEL’s maternal ancestors would be needed to conclusively determine which individual contributed the haplotype on which the DR variant originated. However, due to the size of the region concordant between SEILING ROCKMAN and the major allele in 95 DR heterozygotes it is likely that this individual contributed the chromosome on which the DR DR mutation later occurred.
A bull thought to be unrelated to ROSABEL (HOUSAM2141664, MAPLE-VANE SURPRISE-RED) has been reported to transmit a similar dominant red phenotype. A semen sample of this bull was obtained from the USDA National Animal Germplasm Program and genotyped for the COPA mutation using the diagnostic test. This individual was in fact heterozygous for the DR-associated missense mutation. Haplotype analysis found that SURPRISE was concordant for the DR associated allele at 19 out of 21 SNPs in the IBD region for which data was available. The same mutation may have occurred independently; alternatively, it is possible that there is an error in the pedigree of SURPRISE, and that the DR-associated allele in SURPRISE originated in ROSABEL.
Discussion
Here we provide genetic evidence that a missense mutation in COPA causes Dominant Red, a novel dominant red coat color phenotype that recently evolved in the Holstein dairy cattle breed. The identification of the same region of BTA3 as being associated with DR using two different genetic mapping methods indicates that the previous results suggesting DR is located on BTA27 and possibly caused by a mutation in DEFB103 are incorrect [5]. Haplotype analysis narrowed the region containing the DR causal variant to a 2.6 Mb IBD region of BTA3. The sire of the DR founder animal did not carry this IBD haplotype indicating that the causal mutation originated in the maternal germline of the dam or early during embryonic development of the DR founder, ROSABEL. Whole genome sequencing identified six candidate causal variants in the IBD region, five of which were excluded after screening a panel of 20 known DR heterozygotes. The one remaining SNP at BTA3:9,479,761 bp was concordant with expected DR genotype in a panel of 78 DR and 283 non-DR animals. This SNP results in an arginine to cysteine substitution at amino acid 160 of COPA, predicted to impair COPA function, and is consistent with differential gene expression detected using RNA-Seq analysis of Dominant Red compared to Dominant Black skin. Taken together with additional results from hair biochemical and RNA-Seq analyses, our results provide strong evidence that a reduction of COPA activity causes a red coat color by interfering with the effects of melanocortin receptor signaling.
COPA encodes the α-COP subunit of the coat protein I (COPI) seven subunit complex that is involved with intracellular coated vesicle transport. Originally characterized as a critical component for retrograde transport of luminal and membrane proteins from the Golgi to the endoplasmic reticulum and between Golgi compartments [19–21], COPI participates in a range of macromolecular complexes with diverse cargoes, including ribonucleoproteins that modulate trafficking of RNA in neuronal cells [22,23]. Because the pattern of mRNA expression in Dominant Red skin mimics that observed in Recessive Red skin, we speculate that reduced COPA activity caused by the Arg160Cys substitution impairs MC1R signaling via altered localization of its mRNA and/or protein. Like neurons, melanocytes are highly compartmentalized cells, and it is possible that MC1R mRNA is normally transported to specific locations in dendrites, a process disrupted by the DR mutation. Alternatively, or in addition, the DR mutation may interfere with MC1R receptor recycling between plasma membrane and endosomal compartments (reviewed in [24]).
Previous studies of COPA function have focused mostly on cell biology and biochemical approaches; indeed, heritable variation of COPA has not been described previously in mammals, and Dominant Red cattle represent a unique opportunity to study COPI function in an organismal context. Loss-of-function for COPA, COPB, or COPB2 in zebrafish causes embryonic lethality [24], and it therefore seems likely that the Arg160Cys mutation is hypomorphic. An analogous argument has been made for a missense mutation of Archain 1 (Arcn1) in laboratory mice, which causes neurodegeneration and pigmentary dilution; Arcn1 encodes the delta-subunit of COPI [23]. Pigmentary dilution in the mouse mutant was suggested to involve defects in trafficking of proteins required for melanogenesis; our results suggest the additional possibility that Arcn1-related pigmentary changes represent a reduction of MC1R signaling, an idea that could be tested by examination of Arcn1; Mc1r double mutants.
In cattle, both Recessive Red and Dominant Red animals display variation in the intensity of red coloration over the entire body as well as a tendency to darken in the extremities, possibly due to modifier loci or non-genetic effects. However, some have suggested that Dominant Red animals are more often a darker shade of red than Recessive Red animals, consistent with our observation that eumelanin is reduced more in Recessive Red than in Dominant Red.
Visually we observed that Dominant Red animals had a tendency to have darker hairs intermixed with lighter hairs while Recessive Red animals displayed a more uniform intra-hair pigmentation color. However, we observed variation in shade of red within both groups of red animals. We also observed that, at birth, Dominant Red animals were indistinguishable from Recessive Red and that any darkening that might occur in some Dominant Red animals typically takes place after four to six months of age. These observations on color are all based on Dominant Red heterozygotes since very few homozygotes are in existence due to the recent occurrence of this mutation. The work reported here provides the basis for careful evaluation of potential additive and epistatic relationships between Dominant Red and Recessive Red in livestock, an opportunity that is usually limited to model organisms.
During the preparation of this manuscript it has come to our attention that another group has independently identified the very same COPA mutation as associated with Dominant Red color in Holsteins, providing further support that this missense mutation is causal (Bourneuf E, Otz E, Michot P, Grohs P, Piton C, Deloche M.-C., Cornier M, Delahaye M, Fritz S, Leclerc H, Longin C, Boukadiri A, Saintilan R, Créchet F, Mosca M, Guillaume F, Bouet S, Baur A, Vasilescu A, Genestout L, Allais-Bonnet A, Rocha D, Colle M.-A., Klopp C, Esquerré D, Barbey S, Fayolle G, Danchin-Burge C, Bed'Hom B, Daetwyler H D, Boichard D, Pin D and Capitan A, in preparation).
Conclusions
Here we show that a mutation causing an arginine to cysteine substitution at a highly conserved position in the fourth WD40 repeat motif of COPA causes the striking Dominant Red phenotype in Holstein cattle. The mutation affects a key component of the fundamental cellular process of coated vesicle transport with no known adverse effects on other traits. Hair chemical analyses indicate a shift from eumelanin to pheomelanin production in Dominant Red individuals, similar to what is seen in the Recessive Red phenotype caused by a loss-of-function mutation in MC1R. RNA-Seq analyses indicate that Dominant Red skin samples have decreased expression of genes necessary for eumelanin synthesis (TYRP1, TYR, and OCA2). Expression of these genes is upregulated by MC1R signaling, which suggests that altered COPA activity in Dominant Red skin samples impairs eumelanin synthesis by selectively inhibiting MC1R signaling. The COPA mutation associated with Dominant Red provides an opportunity to study the potential relationship between coated vesicle transport and MC1R signaling.
Materials and Methods
Genetic nomenclature
In cattle as well as in many other domestic animals, variation at MC1R was originally recognized and described as the “Extension” (E) locus. In accord with the HUGO Gene Nomenclature Committee Recommendations [25], we use MC1R as the gene name instead of E, with superscripts D, BR, +, and e to refer to alleles. As described here, we provide very strong evidence that Dominant Red represents allelic variation in COPA; however, since there are no known COPA alleles in other animals yet, we refer to the ancestral and derivative Dominant Red alleles as DR DR and DR +, respectively.
Animals
Holstein cattle hair samples were obtained from dairy farmers in Canada and the USA unless stated otherwise. Where relevant and available, individuals are identified by their breed association registration number and name to facilitate lookup of additional information on breed association websites. Animals used for chemical hair pigment and RNA-Seq analyses were selected based on pedigree information so as to maximize within-group genotype similarity. Specifically, none of the animals used came from backgrounds that include MC1R BR or MC1R + (which are both rare in the Holstein breed), all Dominant Red and Dominant Black animals had no record of Recessive Red in their background, and all Recessive Red animals had no record of Dominant Red in their background. Additionally, all Dominant Red animals had one black and one red parent; thus, the phenotypic designations (and inferred genotypes) are: Dominant Black (DR +/DR +; MC1R D/MC1R D), Dominant Red (DR DR/DR +; MC1R D/MC1R D), and Recessive Red (DR +/DR +; MC1R e/MC1R e). This study utilized tissues and/or DNA obtained from domestic animals maintained by their owners. All samples were collected following best practice husbandry and veterinary procedures. No laboratory animals or laboratory animal research subject to IACUC regulations were used.
Pigment Analysis
Hair was collected from the paralumbar fossa region of females approximately one year of age. Hair was cut immediately adjacent to the skin and wrapped in aluminum foil until analysis. Hair samples (15–20 mg) were homogenized with a Ten-Broeck homogenizer at a concentration of 10 mg/ml and 100 μl aliquots were subjected to Soluene-350 solubilization [7], alkaline hydrogen peroxide oxidation [8], and hydroiodic acid hydrolysis [9].
Genotyping
The sire, progeny and respective dams of the half-sib family used for linkage analysis were genotyped on the BovineLD Genotyping BeadChip (Illumina) [26]. Animals used for the GWAS were genotyped on the BovineSNP50 Genotyping BeadChip (Illumina) [27] or genotypes were imputed to this level from a lower density SNP panel. Additional animals were targeted for genotyping through the established genomic selection system with assistance from Canadian Dairy Network, Holstein Association of Canada, and Holstein Association USA. Owners provided hair samples directly to commercial genotyping labs. A subset of samples was also sent by the owners to the researcher’s lab for follow up genotyping. These samples were composed of either hair or blood and were isolated using the Gentra Puregene kit (Qiagen). Animals that could be both Recessive Red and/or Dominant Red based on pedigree analysis were excluded from all analyses unless a negative genetic test for Recessive Red was obtained.
Sequencing
SOLiD mate pair libraries were prepared from a Dominant Red individual (HOCANF9845383, BROEDERDALE MALTBY CAMILLA) according to the SOLiD System Mate-paired Library Preparation protocol of the Applied Biosystems SOLiD System (ABI). The libraries were sequenced on two flowchips using the SOLiD 5500xl platform. The initial libraries had a large number of duplicate reads so a paired-end library was then prepared and sequenced on the same machine. The resulting data was of good quality. The sequence data have been uploaded to the NCBI sequence read archive (SRA: SRS834811).
Short reads were aligned in color space against the UMD3.1/bosTau6 genome assembly using BWA v0.5.9 [28]. After mapping, the duplicates within each BAM file were marked using Picard v1.54 (http://broadinstitute.github.io/picard). Because the first two runs were using the same mate-pair library, the BAM files were merged and duplicates were marked again for removal.
Variants (SNPs and indels) were called using SAMtools v0.1.18 [29] mpileup with no quality filtering in order to not exclude any true positives with low quality scores. Genotypes from the 50K SNP chip were used to validate sequencing results. Genotypes were concordant between variant calling results and existing genotype data for all SNPs present in the IBD region that are also on the 50K SNP chip, 12 homozygous non-reference SNPs and 10 heterozygous SNPs.
SNPs and indels were annotated with predicted functional consequences using NGS-SNP [30]. Overlapping genes, transcripts, proteins, protein domains, and variants were included among the annotations, along with any known pathways or phenotypes linked to the genes in cattle or to their orthologues in humans. Sequence alignments were constructed between missense SNP-altered proteins their orthologues and scored using the SIFT [15] algorithm to predict the functional significance of protein substitutions. Variants were classified as “known” if the non-reference allele was present in the dbSNP database and “novel” otherwise. The source databases used by NGS-SNP during annotation include Ensembl release 68, dbSNP build 133, Entrez Gene, and UniProt release 2012_09.
Detection of Structural Variations
In order to detect putative causative structural variations within the IBD region the two SOLiD mate-pair libraries for the Dominant Red individual was mapped against the UMD3.1/bosTau6 reference assembly. For comparison, two SOLiD mate-pair libraries from a Holstein bull (HOCANM10705608, BRAEDALE GOLDWYN) were downloaded from the NCBI sequence read archive (SRA: SRR592656, SRR592657) [31] and used as a reference since this individual does not have the Dominant Red phenotype. The reads from both individuals were mapped with MOSAIK v2.2 [32] using a hash size of 14 (-hs 14), allowing four mismatches per read (-mm 4), an alignment candidate threshold of 20 (-act 20) and otherwise default settings. Non-unique alignments were excluded and duplicated read pairs were removed with the Picard v1.123 (http://broadinstitute.github.io/picard) package’s MarkDuplicates utility before subsequent analyses.
Putative copy number variants (CNVs) were analyzed across BTA3 but only called within the IBD region. First the program CNV-seq [33] was used to compare read depth between the Dominant Red and reference individual in overlapping windows across the chromosome. Based on the default significance threshold of P<0.001 and log2-fold change threshold of ±0.6, CNV-seq automatically calculated a suitable window size of 2,979 bp. In order to call a significant CNV at least two consecutive windows needed to be significant. Second, the program CNVnator [34] was used to call putative CNVs based on read depth within both individuals separately in sliding windows of 750 bp across chromosome three. CNVnator combines nearby windows that display a similar CNV signal and calculates statistics for the resulting regions. A significance threshold of P<0.001 was used for the CNVnator analysis.
In addition to the read depth based analysis, the program SVDetect [35] was used to detect putative SVs based on deviant read mate-pairs. A read mate-pair is called deviant if it has an abnormal insert size, read order orientation or read strand orientation. This method makes it possible to detect copy number balanced variants such as inversions and translocations in addition to duplication and deletion events that affect read depth. SVDetect clusters reads into links that corresponds to a potential SV and classifies it based on how the mate-pairs deviates. Only high quality alignments were used by excluding reads with a mapping quality less than 20. For SVDetect, default settings were used to detect the abnormal mate-pairs. Deviant reads were detected and clustered into links separately for the Dominant Red and reference individual. The putative SVs were selected in the Dominant Red individual from links that were supported by at least five reads and had a final score of one (highest confidence). Any links crossing an assembly gap were removed. These candidates were then compared with the reference individual and overlapping links of the same type were deemed as assembly/alignment artifacts or not associated with the Dominant Red phenotype.
Diagnostic Test
A diagnostic test was developed for the causal Dominant Red SNP (COPA c.478C>T p.Arg160Cys) at BTA3:9,479,761 bp using a Custom TaqMan SNP Genotyping Assays (ABI). Primers and probes were Forward: 5’-TCAGAAGACCTGGTCGTGTCA-3’, WT allele: 5’-VIC-CAAACGCGCACAGTC-NFQ-3’, DR allele: 5’-FAM-CCAAACGCACACAGTC-NFQ-3’ and Reverse: 5’-TGGGCAGCTCACCAGAAATATC-3’. Genotyping was performed according to the standard protocol using TaqMan Genotyping Master Mix (ABI).
RNA-Seq
Punch biopsies were performed to obtain skin samples from six individuals (three Dominant Red, three Dominant Black), and then stored in RNALater at -80°C. Total RNA was isolated using RNeasy Fibrous Tissue Mini Kit (Qiagen), assessed for integrity with an Agilent Bioanalyzer instrument, and cDNA libraries then prepared with the TruSeq RNA Sample Preparation Kit v2 (Illumina). Individual libraries were multiplexed as six per lane and sequenced as paired-end 50 base pair reads on an Illumina HiSeq 2000 instrument at the Genome Sequencing Laboratory of the HudsonAlpha Institute.
RNA-Seq reads obtained for each sample were aligned against the bovine reference genome (UMD3.1/bosTau6) with TopHat2 software, using genomic sequence and transcript annotations obtained from Ensembl (release 72). On average ≈34 million paired-end reads were obtained for each sample, out of which ≈66% were mapped individually and ≈62% were mapped as concordant pairs against the reference transcriptome. Gene counts were computed from read alignments with GenomicRanges and GenomicFeatures packages from Bioconductor (release 2.14), and then used to test for differential expression between Dominant Red and Dominant Black skin samples with DESeq2 package from Bioconductor. DESeq2 relies on negative binomial generalized linear models to determine whether the number of counts for a transcript or gene is significantly different across a range of experimental conditions.
Supporting Information
A 2.6 Mb IBD haplotype in Dominant Red animals is located on BTA3 from ARS-BFGL-NGS-112616 @ 7,906,099 bp to ARS-BFGL-NGS-1429 @ 10,462,387 bp. Animals are in rows and SNPs are in columns. Phenotype is indicated in the first column with Dominant Red animals shaded in red and wild-type animals shaded in gray. SNPs are shown that span the entire 10.7 Mb region identified in the linkage analysis. Orange shading indicates homozygotes for the major allele, yellow shading indicates heterozygotes and blue shading indicates homozygotes for the minor allele. The IBD haplotype is shaded in gray and the SNP with the highest GWAS significance shaded in green (ARS-BFGL-NGS-94819 @ 9,722,400 bp). The four ancestors of the founder animal ROSABEL are indicated at the left of the figure. SEILING ROCKMAN shows a haplotype concordant with the major allele from Dominant Red animals extending beyond the 2.6 Mb IBD haplotype.
(TIFF)
The IBD region was subject to analysis of candidate structural variations using whole genome sequencing reads from a Dominant Red (HOCANF9845383, BROEDERDALE MALTBY CAMILLA) and reference individual (HOCANM10705608, BRAEDALE GOLDWYN). The top track represents RefSeq Gene annotation. Normalized and GC corrected read depth (RD) for the two individuals are found in the bottom two tracks with the mean RD marked with a red line. Significant deletions from CNVnator (orange) and duplications from SVDetect (green) within each individual are overlaid on the RD graph. No other significant structural variations were detected, including analysis with CNV-seq to compare RD between individuals.
(TIFF)
Six SNPs identified by whole genome sequencing within the IBD region were screened on a panel of 20 DR animals. A “.” indicates the same genotype as the reference genome. Variant calling quality scores from whole genome sequencing analysis are listed in the top row of the table. Three SNPs (8,028,785 bp, 9,385,285 bp, and 9,943,108 bp) were found to be false positives from whole genome sequencing after Sanger sequencing using the same individual. Two SNPs (10,295,754 bp and 10,295,797 bp) were excluded as candidate causal mutations since not all DR individuals carried the variant allele. The remaining SNP (9,479,761 bp) was found to be heterozygous in all 20 DR individuals.
(XLSX)
Genes are labeled using Ensembl identifiers, as well as associated gene symbols in all cases in which they could be retrieved from Ensembl databases. Differential expression analyses were performed using DESeq2 package (see text for details). A threshold of FDR<0.1 was used to evaluate significant expression changes distinguishing skin samples associated with the two compared phenotypes.
(XLSX)
Acknowledgments
Major contributors of samples were made by Morsan Farms (Canada), Clint Broeders (Canada), Cody Orton (USA), Jason Kearns (USA) and Matt Dorshorst (USA). Sample collection was coordinated by Canadian Dairy Network, Holstein Association of Canada, and Holstein Association USA. GP thanks Jackson Mah for sample analysis at the University of Alberta, and Michelle Miller and Yee Ying Lock at Delta Genomics (Edmonton, AB) for the sequencing of HOCANF9845383 BROEDERDALE MALTBY CAMILLA. The CriMap 2.503 software update was courtesy of Jill Maddox and Ian Evans.
Data Availability
Whole genome sequencing data has been uploaded to the NCBI sequence read archive (SRA: SRS834811).
Funding Statement
This project was financially supported by a grant from the Dairy Cattle Genetics Research and Development (DairyGen) Council of Canadian Dairy Network. SI and KW were supported, in part, by a Japan Society for the Promotion of Science (JSPC) grant (No. 23591659 and 26461705). Additional funding was provided by Genome Canada and the Alberta Livestock and Meat Agency. PS and GP thank Alberta Innovates Bio Solutions for support.
References
- 1. Klungland H, Vage DI, Gomez-Raya L, Adalsteinsson S, Lien S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm Genome. 1995;6: 636–639. 10.1007/BF00352371 [DOI] [PubMed] [Google Scholar]
- 2. Joerg H, Fries HR, Meijerink E, Stranzinger GF. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm Genome. 1996;7: 317–318. 10.1007/s003359900090 [DOI] [PubMed] [Google Scholar]
- 3.Lawlor TJ, VanRaden PM, Null D, Levisee J, Dorshorst B. Using haplotypes to unravel the inheritance of Holstein coat color. Proceedings, 10th World Congress of Genetics Applied to Livestock Production. 2014.
- 4.Leduc M. The Various Mechanisms of Red Colour Transmission in the Holstein Breed. Holstein Journal. 2006;: 45–47.
- 5.Dreger DL, Schmutz S. The variant red coat colour phenotype of Holstein cattle maps to BTA27. Anim Genet. 2009. 10.1111/j.1365-2052.2009.01969.x [DOI] [PubMed]
- 6. d'Ischia M, Wakamatsu K, Napolitano A, Briganti S, Garcia Borron JC, Kovacs D, et al. Melanins and melanogenesis: methods, standards, protocols. Pigment Cell Melanoma Res. 2013;26: 616–633. 10.1111/pcmr.12121 [DOI] [PubMed] [Google Scholar]
- 7. Ozeki H, Ito S, Wakamatsu K, Thody AJ. Spectrophotometric characterization of eumelanin and pheomelanin in hair. Pigment Cell Res. 1996;9: 265–270. 10.1111/j.1600-0749.1996.tb00116.x [DOI] [PubMed] [Google Scholar]
- 8. Ito S, Nakanishi Y, Valenzuela RK, Brilliant MH, Kolbe L, Wakamatsu K. Usefulness of alkaline hydrogen peroxide oxidation to analyze eumelanin and pheomelanin in various tissue samples: application to chemical analysis of human hair melanins. Pigment Cell Melanoma Res. 2011;24: 605–613. 10.1111/j.1755-148X.2011.00864.x [DOI] [PubMed] [Google Scholar]
- 9. Wakamatsu K, Ito S, Rees JL. The usefulness of 4-amino-3-hydroxyphenylalanine as a specific marker of pheomelanin. Pigment Cell Res. 2002;15: 225–232. 10.1034/j.1600-0749.2002.02009.x [DOI] [PubMed] [Google Scholar]
- 10. Green P, Falls K, Crooks S. Documentation for CRIMAP, version 2.4. Washington University School of Medicine, St Louis: 1990. [Google Scholar]
- 11. Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biology. 2009;10: R42 10.1186/gb-2009-10-4-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brøndum RF, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46: 858–865. 10.1038/ng.3034 [DOI] [PubMed] [Google Scholar]
- 14. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4: 1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 16. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. Oxford University Press; 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. BioMed Central Ltd; 2014;15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prifti E, Zucker J-D, Clement K, Henegar C. FunNet: an integrative tool for exploring transcriptional interactions. Bioinformatics (Oxford, England). 2008;24: 2636–2638. 10.1093/bioinformatics/btn492 [DOI] [PubMed] [Google Scholar]
- 19. Cosson P, Letourneur F. Coatomer (COPI)-coated vesicles: role in intracellular transport and protein sorting. Curr Opin Cell Biol. 1997;9: 484–487. 10.1016/S0955-0674(97)80023-3 [DOI] [PubMed] [Google Scholar]
- 20. Lowe M, Kreis TE. Regulation of membrane traffic in animal cells by COPI. Biochim Biophys Acta. 1998;1404: 53–66. 10.1016/S0167-4889(98)00046-9 [DOI] [PubMed] [Google Scholar]
- 21. Beck R, Rawet M, Ravet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583: 2701–2709. 10.1016/j.febslet.2009.07.032 [DOI] [PubMed] [Google Scholar]
- 22. Bi J, Tsai N-P, Lu H-Y, Loh HH, Wei L-N. Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. Proc Natl Acad Sci USA. 2007;104: 13810–13815. 10.1073/pnas.0703805104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peter CJ, Evans M, Thayanithy V, Taniguchi-Ishigaki N, Bach I, Kolpak A, et al. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum Mol Genet. 2011;20: 1701–1711. 10.1093/hmg/ddr046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. García-Borrón JC, Abdel-Malek Z, Jiménez-Cervantes C. MC1R, the cAMP pathway, and the response to solar UV: extending the horizon beyond pigmentation. Pigment Cell Melanoma Res. 2014;27: 699–720. 10.1111/pcmr.12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bruford EA. Highlights of the “Gene Nomenclature Across Species” Meeting. Hum Genomics. 2010;4: 213 10.1186/1479-7364-4-3-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boichard D, Chung H, Dassonneville R, David X, Eggen A, Fritz S, et al. Design of a bovine low-density SNP array optimized for imputation. PLoS ONE. 2012;7: e34130 10.1371/journal.pone.0034130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE. 2009;4: e5350 10.1371/journal.pone.0005350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools.—PubMed—NCBI. Bioinformatics. Oxford University Press; 2009;25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grant JR, Arantes AS, Liao X, Stothard P. In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics (Oxford, England). Oxford University Press; 2011;27: 2300–2301. 10.1093/bioinformatics/btr372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stothard P, Choi J-W, Basu U, Sumner-Thomson JM, Meng Y, Liao X, et al. Whole genome resequencing of black Angus and Holstein cattle for SNP and CNV discovery. BMC Genomics. BioMed Central Ltd; 2011;12: 559 10.1186/1471-2164-12-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee W-P, Stromberg MP, Ward A, Stewart C, Garrison EP, Marth GT. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS ONE. 2014;9: e90581 10.1371/journal.pone.0090581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10: 80 10.1186/1471-2105-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21: 974–984. 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeitouni B, Boeva V, Janoueix-Lerosey I, Loeillet S, Legoix-né P, Nicolas A, et al. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics (Oxford, England). 2010;26: 1895–1896. 10.1093/bioinformatics/btq293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A 2.6 Mb IBD haplotype in Dominant Red animals is located on BTA3 from ARS-BFGL-NGS-112616 @ 7,906,099 bp to ARS-BFGL-NGS-1429 @ 10,462,387 bp. Animals are in rows and SNPs are in columns. Phenotype is indicated in the first column with Dominant Red animals shaded in red and wild-type animals shaded in gray. SNPs are shown that span the entire 10.7 Mb region identified in the linkage analysis. Orange shading indicates homozygotes for the major allele, yellow shading indicates heterozygotes and blue shading indicates homozygotes for the minor allele. The IBD haplotype is shaded in gray and the SNP with the highest GWAS significance shaded in green (ARS-BFGL-NGS-94819 @ 9,722,400 bp). The four ancestors of the founder animal ROSABEL are indicated at the left of the figure. SEILING ROCKMAN shows a haplotype concordant with the major allele from Dominant Red animals extending beyond the 2.6 Mb IBD haplotype.
(TIFF)
The IBD region was subject to analysis of candidate structural variations using whole genome sequencing reads from a Dominant Red (HOCANF9845383, BROEDERDALE MALTBY CAMILLA) and reference individual (HOCANM10705608, BRAEDALE GOLDWYN). The top track represents RefSeq Gene annotation. Normalized and GC corrected read depth (RD) for the two individuals are found in the bottom two tracks with the mean RD marked with a red line. Significant deletions from CNVnator (orange) and duplications from SVDetect (green) within each individual are overlaid on the RD graph. No other significant structural variations were detected, including analysis with CNV-seq to compare RD between individuals.
(TIFF)
Six SNPs identified by whole genome sequencing within the IBD region were screened on a panel of 20 DR animals. A “.” indicates the same genotype as the reference genome. Variant calling quality scores from whole genome sequencing analysis are listed in the top row of the table. Three SNPs (8,028,785 bp, 9,385,285 bp, and 9,943,108 bp) were found to be false positives from whole genome sequencing after Sanger sequencing using the same individual. Two SNPs (10,295,754 bp and 10,295,797 bp) were excluded as candidate causal mutations since not all DR individuals carried the variant allele. The remaining SNP (9,479,761 bp) was found to be heterozygous in all 20 DR individuals.
(XLSX)
Genes are labeled using Ensembl identifiers, as well as associated gene symbols in all cases in which they could be retrieved from Ensembl databases. Differential expression analyses were performed using DESeq2 package (see text for details). A threshold of FDR<0.1 was used to evaluate significant expression changes distinguishing skin samples associated with the two compared phenotypes.
(XLSX)
Data Availability Statement
Whole genome sequencing data has been uploaded to the NCBI sequence read archive (SRA: SRS834811).