Abstract
A vaccine formulation that would be effective against all strains of influenza virus has long been a goal of vaccine developers, but antibodies after infection or vaccination were seen to be strain specific and there was little evidence of cross-reactive antibodies that neutralized across subtypes. Recently a number of broadly neutralizing monoclonal antibodies have been characterized. This review describes the different classes of broadly neutralizing antibodies and discusses the potential of their therapeutic use or for design of immunogens that induce a high proportion of broadly neutralizing antibodies.
Introduction
Influenza vaccines have been used since the 1940s. They are safe but need to be multivalent to protect against the multiple circulating viruses, and the components need to be updated nearly every year in response to mutations of the virus. The holy grail for influenza vaccine would be a single formulation that cross-protects against all current and future strains. Recent discoveries of cross-reactive monoclonal antibodies have given hope that a universal influenza vaccine may be possible.
This review covers recent work (approximately 2009 to 2014) to characterize neutralizing antibodies against influenza with emphasis on those that show some level of cross-reactivity between different subtypes.
Early observations
Human influenza virus was first isolated in 1933. Memories of the devastating death toll of the 1918–1919 epidemic fuelled efforts to develop a vaccine, spurred even more by the advent of the Second World War. By 1936 it had been recognized that influenza viruses are antigenically diverse. Methods to inactivate the virus with formalin overcame the inherent safety concerns of live virus vaccines and the vaccine given to troops in World War II was trivalent, containing A/PR/8/34, A/Weiss/43, and B/Lee/40. This vaccine was shown to provide protection against type A and B viruses until 1947, when it dramatically failed. The 1947 viruses were originally classified as “A prime” but eventually were grouped into the H1N1 subtype, despite the marked change in antigenic properties. By 1954 there were two fundamental questions on antigenic variation [1]. One was whether the virus mutates in response to environment (such as infection of a new host, or presence of antibodies), versus the ideas of G. K. Hirst and J. Y. Sugg that a pre-existing variant is selected out by environmental pressure. The second question was whether there are a limited number of variants of influenza virus that wax and wane in the human population (J. Salk, T. Francis), or whether the virus is continually changing (F. L. Horsfall, F. M. Burnet). A finite number of variants would imply that a vaccine containing all of them would be effective. Unfortunately this is not the case, and we now know that influenza evolves linearly by selection of escape mutants, usually by antibodies, from a small population of variants generated by random mutation from the preceding virus. This means that development of a universal influenza vaccine requires a strategy other than including all known strains.
Antigenic drift and shift, neutralizing antigens, current vaccine strategies
Influenza viruses are classified by serological cross-reactivity, or lack thereof. Types A, B and C do not cross-react by any serological test. Type A viruses all share cross-reactivity of internal proteins, nucleoprotein (NP) and matrix (M1), but the surface glycoproteins hemagglutinin (HA, or H) and neuraminidase (NA or N) are divided into serological subtypes H1 to H16 and N1 to N9 that do not cross-react with serum antibodies. Only H1, H2 and H3 with N1 or N2 circulate in the human population. Recent influenza sequences from bats proposed as H17, H18, N10 and N11 have functionally different glycoproteins and the viruses have not yet been isolated [2]. A new subtype entering the human population is described as antigenic shift, such as when H2N2 viruses replaced H1N1 in 1957 and H3N2 replaced H2N2 in 1968. Antigenic shift is facilitated by the large variety of influenza viruses in bird populations and by the segmented nature of the genome that allows reassortment of genes in a mixed infection. Following antigenic shift, the new virus undergoes progressive changes due to antibody selection, known as antigenic drift.
All the genes of influenza virus undergo some degree of variation, all occurring by the same basic mechanism. Influenza has an RNA genome that codes for its own RNA polymerase. RNA polymerases in general lack the editing feature of DNA polymerases, an exonuclease domain that removes a mismatched 3’ nucleotide before elongation can continue. Without this exonuclease activity, the intrinsic error rate of RNA polymerases is relatively high. Most of the resulting variants are lost in the population, but a few may be fixed by chance (“random drift”). Some mutations are positively selected, for example, to escape from antibody neutralization or for more efficient replication or better interaction with a specific host protein, and these variants rapidly take over the population because they confer an advantage. Changes in proteins selected for improved function may change their antigenic properties. There is a marked distinction between antigenic selection (resistance to the immune system) and antigenic change that is a consequence of some other selective pressure.
Definitions of “neutralizing” and “epitope”
Protection against influenza is mediated by innate systems and by T cells and antibodies. The relative roles of these vary with the patient’s genetic profile and history of influenza, but overall the most important contribution to protection is from neutralizing antibodies, and current vaccines are measured by their power to induce neutralizing antibodies. The classical definition of “neutralizing” is to block the ability of a virus to attach to a cell; i.e. to block the first step in viral infection. Such antibodies sterically interfere with the receptor-binding site on the hemagglutinin (HA) so it cannot bind to its sialic acid receptor on the cell surface. However, there are antibodies that interfere in infection at later stages. While these do not meet the classical definition, they effectively neutralize the infection and in laboratory studies are equally protective.
What are the targets of neutralizing antibodies that do not block attachment to sialic acid? While antibodies can be raised that block various viral activities (RNA-dependent RNA polymerase, assembly of nucleoprotein complexes or of the viral matrix protein), these antibodies are not neutralizing or protective because they cannot access their targets during the normal course of infection. Neutralization targets are those outside the viral membrane and so exposed on the virus; the surface proteins HA, neuraminidase (NA) and M2 ion channel. The HA’s first function is to bind sialic acid receptors, but after internalization, in the low pH environment of the endosome, the HA undergoes a conformational change to enable a fusion activity that allows release of the viral genome. So antibodies that efficiently block either binding to receptor, conformational change or the fusion function will also be neutralizing.
Antibodies can only neutralize if they block a function. It follows that they bind to native proteins. Typically they bind to multiple segments of the polypeptide chain that may be dispersed in the primary sequence but come together in the three-dimensional structure. Therefore most neutralizing antibodies bind to a so-called “conformational” epitope, that is lost if the protein is denatured or even partially unfolded.
Mapping antibody epitopes
For this review, an epitope is defined as the amino acids on the antigen that make contact with antibody. Some of the interactions of antibody with antigen are more important than others. The critical contacts, with highest interaction energy, can be identified by selection of escape mutants, or by exhaustive mutagenesis, or by hydrogen exchange methods. These studies only give a partial view of the epitope, albeit the most energetically important view. Full descriptions of epitopes can be made from X-ray crystal structures of the antigen-antibody complex; this is currently the only available method that shows all the atomic interactions between antigen and antibody. An excellent discussion of broadly neutralizing epitopes characterized by X-ray crystallography is given by Lee and Wilson [3].
How polyclonal is human serum?
Much of our knowledge of neutralizing epitopes comes from studies with monoclonal antibodies (mAbs), that allow selection of escape mutants that can be attributed to a single selecting antibody. A monoclonal antibody also allows complexes of antigen and antibody to be crystallized for structural analysis. The human response is, of course, polyclonal, raising questions about the applicability of monoclonal studies (usually mouse) to human protection. Mouse monoclonal antibodies were used to map broad antigenic regions on the HA of H3N2 and H1N1 viruses by using competition assays and cross-reactivities of the mAbs with escape mutants selected by other mAbs. Five sites were found on the H3 HA (sites A through E, [4,5]) and four on H1 HA (Sa, Sb, Ca, Cb, [6]). An antibody that recognizes a change in Site A, for example, did not recognize changes in sites B-E. The deduction is that changes in all antigenic sites would be needed for a virus to escape the human immune system and begin a new epidemic. For some antigenic drift strains of H3N2 viruses this was the case, but more commonly, especially more recently, only one or two changes are found between epidemic viruses. It has been noted for some time that human H3N2 viruses show a changing pattern of immunodominance and it appears that for any given virus, the human response does not cover all antigenic sites and vaccine failure can be due to one or two mutations in the HA [7].
Broadly neutralizing monoclonal antibodies
The N-terminal ectodomain of the M2 ion channel protein is quite highly conserved, and immunization with recombinant M2 generates mAbs with considerable cross-reactivity between subtypes. These antibodies are rare in infected humans but they might be protective if they could be induced by recombinant M2, or amplified in vitro and given therapeutically. Both approaches have been tested and shown to be effective in animal models [8–10]. There is one report of a Phase 2 clinical trial of antibody TCN-032 which showed some reduction in symptoms but no protection from infection in volunteers challenged with influenza virus [11]. On the whole, the M2 approach has not yet shown efficacy; the neutralizing power of antibodies directed against M2 may be inherently low compared to those against HA.
There are recent reports of antibodies that have broad reactivity against NA. One epitope is within a conserved linear sequence in the active site of all type A subtypes and type B NA, but protection is less than 50% [12,13]. Others target a conformational epitope shared by seasonal H1N1, pandemic H1N1 and H5N1 viruses and show prophylactic activity [14]. Antibodies against NA do not block infection but they do block release of virus and so ameliorate disease (reviewed in [15]). The most effective neutralizing antibodies are against the HA.
Initial attempts to raise antibodies against conserved regions of the HA used small peptides as immunogens. The resulting antibodies were cross-reactive, but usually bound only to denatured HA and so were not neutralizing, reviewed in [16]. A mouse mAb designated C179 was shown in 1993 to neutralize viruses of the H1 and H2 subtypes, and partial mapping showed that the epitope was located on the stem of the HA, later fully mapped by X-ray crystallography [17,18]. Sixteen years later two reports appeared describing broadly neutralizing antibodies directed against the stem region of the HA, fully characterized by X-ray crystallography of HA-antibody complexes [19,20]. Many such stem-binding mAbs were reported in the next few years. There have also been descriptions of broadly neutralizing antibodies recognizing the receptor binding region of the HA head [21–23].
It has now become clear that there are varying definitions of “broadly neutralizing” antibodies. In influenza studies the traditional term was “cross-reactive” and it meant binding to more than one subtype of influenza. “Broadly neutralizing” has been applied to mAbs that neutralize multiple subtypes, but is also used by some authors to describe neutralization of more than one strain within a subtype. So one investigator’s “broadly neutralizing” mAb may bind only some clades of H5 HAs while another’s may bind representatives of all 16 HA subtypes. Table 1 shows the well-characterized epitopes of antibodies that bind to several strains of influenza virus published up to December 2014. The entries are sorted from most cross-reactive (14 of 16 subtypes of influenza A and influenza B), through those that bind more than one subtype within the phylogenetically-defined HA Group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16) or Group 2 (H3, H4, H7, H10, H14, H15), to those that bind multiple viruses of a single subtype. It should be noted that testing may not have included all subtypes so the sorting is only approximate. However, it does show that the most cross-reactive antibodies (e.g. CR9114, F16, F10, CR6261) bind to the stem of the HA while receptor site binding antibodies are more selective (e.g. S139/1 and C05). Antibodies that bind within a single subtype all have epitopes on the HA globular head, perhaps reflecting higher neutralizing power on the head domain but limited by higher sequence variation due to antigenic drift and shift.
Table 1.
Broadly neutralizing or cross-reactive epitopes on influenza virus HA
| Antibody | Source | IEDB1 ID | HA subtype | Contacts on HA12 | Contacts on HA2 | Crystal structure (PDB3 ID and viral HA) | Reference |
|---|---|---|---|---|---|---|---|
| CR9114 | human VH 1-69 | 178112 | 14/16 A subtypes and B | H5: H44, Q46, D47, I48, S304, M305, P306 | V364, D365, G366, W367, A382, K384, T387, Q388, I391, D392, V394, T395, V398, I402 | 4FQI A/Vietnam/1203/2004 | [32] |
| CR9114 | human VH1-69 | 178113 | 14/16 A subtypes and B | H7: N53, T55, E56, T57, N307, L308, P309 | I366, D367, G368, W369, A384, Y386, T389, Q390, I393, D394, I396, T397, L400, I404 | 4FQV A/Netherlands/219/2003(H7 N7) | [32] |
| CR9114 | human VH1-69 | 178114 | 14/16 A subtypes and B | H3: N54, T56, E57, L58, D307, K308, P309 | I363, D364, G365, W366, A381, L383, T386, Q387, I390, D391, I393, N394, L397, I401 | 4FQY (Hong Kong/68 H3) | [32] |
| F16 | human VH3-30*18 | 164106 | H1, H2, H5, H6, H8, H9, H3, H4, H7, H10 | H1: H45, S46, S306, T333 | V362, D363, G364, W365, L382, K385, T386, Q387, N388, I389, D390, T393, N397, I400, E401 | 3ZTN (Calif/04/2009 H1), 3ZTJ (Aichi/68 H3) | [33] |
| F10 | human phage display | 229014 | H1, H2, H5, H6, H8, H9, H11 | H24, H44, Q46, S304, M305 | V364, D365, G366, W367, K384, T387, Q388, I391, T395, V398, N399, I402 | 3FKU Vietnam/1203/2004 | [20] |
| CR6261 | human VH1-69 | 226309 | Group 1 (H1, H2, H5, H6, H8, H9) | H25, H45, V46, N47, L496, S306, L307, P308, T333 | D363, G364, W365, Q382, T385, Q386, I389, D390, T393, V396, N397, I400 | 3GBN (SouthCarolina/1918, H1), 3GBM (H5). Only H chain is bound | [19,34] |
| S139/1 | mouse | 176782 | H1, H2, H3, H13, and H16 receptor site | H3: Y115, T148, G151, G152, S153, S154, W170, Y172, K173, S174, G175, S176, K206, E207, N210, L211, V213, L243 | 4GMS Victoria/3/75 | [22] | |
| C179 | mouse | 186645 | H1, H2, H5, H6, H9 | H2: H43, K45, I47, T302, L303, P304, T329 | V358, D359, G360, W361, K378, T381, Q382, K383, F385, D386, V392, I396 I362, D363, G364, W365, Q382, | 4HLZ Japan/305/1957 | [17,18] |
| 3.1 | human VH3-30 | 227882 | H1, H2, H5, H6 | H45, V47, N48, L49, N304, S305, S306, L307, P308, T333; | T385, Q386, I389, T393, V396, N397, I400; | 4PY8 South Carolina/1/1918 | [35] |
| C05 | human | 180352 | H1, H2, H3, H9 | Y114, T147, N149, G150, G151, S152, N153, S161, W169, T171, K172, Q205, E206, T208, S209, L210, L242 | 4FQR Aichi/1968 H3) | [36] | |
| 1F2, 1F4, and 1E1 | human | 177664 | H1 H3 H9 | FIEGGWTGMVDGWYGYHH, part of fusion peptide | [37] | ||
| CR8020 | human | 167844 | H3, H7, H10 | E341; | E360, G361, I363, D364, R370, E375, T377, G378, Q379, A380, A381, L383, N491, E495 | 3SDY (Hong Kong/1/68 H3) | [38] |
| CR8043 (similar to CR8020) | human | 196975 | H3, H10, less H7 | P37, E341, K342 | E360, G361, I363, D364, R370, T377, Q379, L383 | 4NM8 HK/1/1968 | [39] |
| F045-092 VH1-69 | human | 227712 | H1, H2 H3 | Y115, T148, N150, G151, G152, S153, S154, S162, W170, Y172, K173, S174, G175, N210, L211, L243 | 4O58 Victoria/3/75 | [40] [23] | |
| PN-SIA49 | human VH3-23 | 0 | H1, H2, H5 Group 1 | Mutations that reduce binding: H25A, N336A, P338A | M360A,D362A, G363A, W364A, T384A, V395A,N396A, E400A (sequence numbering refers to A/PR/8/34 | [41] | |
| 39.29 | human | 189321 | H1, H3 | H34, V36, N54, S70, K292, N294, P305, D307, | T334, V363, D364, G365, W366, Q379, A380, D382, L383, K384, T386, Q387, I390, D391, I393, N394, G395, L397, N398, I401, K403, T404, N405, R498 | 4KVN Perth/16/2009 | [42] |
| 1F1 | human | 179938 | Various H1 | K147, V149, S159, W167, K170, S173, H197, P200, T201, T203, D204, Q206, S207, L208, Q210, K236, D239, A241, G242 | 4GXU South Carolina/1/1918 | [43] | |
| 2D1 | human | 164527 | H1 1918 and 2009 (not seasonal H1) | S138, S139, P141, N142, K171, G172, S173, S174, Y175, P176, K177, S179, K180, S181, V183, N211, E260, T262 | 3LZF (S.Carolina/1918 H1) 3LZG (Cal/04/2009 H1) H+L binding | [27] [44] | |
| 5J8 | human | 191110 | Seasonal and 2009 H1 except NewCal & Brisbane | K147, V149, T150, A151, G157, A158, K159, W167, A203, D204, Q206, S207, L208, K236, D239, Q240 | 4M5Z California/07/2009 | [26] | |
| CH65 | human | 159269 | most H1 receptor site | G147, V148, S149, A150, W166, T168, G169, N171, G172, L173, N200, G202, D203, R205, A206, L207, K235, D238, R239, E240 | 3SM5 (Solomon Is/3/06 H1) | [21] | |
| CH67 | human | 180309 | H1N1 1986–2007 | G147, V148, S149, A150, W166, T168, N171, G172, L173, N200, G202, D203, R205, A206, L207, H209, K232, D238 | 4HKX Solomon Islands/3/2006 | [45] | |
| GC0587 | mouse | 226441 | H1 | T89, A90, S127, E129, R130, E132, P135, S138, K180, K186, K188, Y270, A273 | 4LVH Korea/1/2009 H1N1 | [46] | |
| GC0757 | mouse | 190190 | E129, R130, F131, E132, P135, T137, S138, K180, S181, I183, Y270 | 4F15 Korea/1/2009 H1N1 | [47] | ||
| 2G1 | human VH1-69 | 181066 | H2 human and swine | T138, Q139, T141, G144, G145, S146, R147, P155, T165, K166, K167, G168, S169, T203, L204 | 4HG4 Japan/305/1957 | [24] | |
| 8F8 | human VH3-33 | 181071 | H2 human and swine | T141, T143, G144, G145, S146, R147, A150, G153, N154, P155, W163, T165, E166, G168, T199, E200, T203, L204, Q236 | 4HF5 Japan/305/1957 | [24] | |
| 8M2 | human VH1-69 | 181067 | H2 human and swine | T141, T143, G144, G145, S146, R147, P155, W163, T165, K166, G168, S169, N196, D197, E198, T199, E200, R202, T203, L204, K232, G235, L236, G237, S238 | 4HFU Japan/305/1957 | [24] | |
| B-1, D-1 | human | 115630, 115652 | H3 many strains | Within 189–197 NFDKLYIWG and 227–239 SSRISIYWTIVKP | [48] | ||
| F005-126 | human | 233323 | H3 1968–2004 | S107, K108, N187, D188, N189, P255, G256, S286, D287, A288, P289, P300, N301 | 3WHE Aichi/68 | [49] | |
| 100F4 | human | 0 | H5 most clades | 68, 112, 137, 143, 251, 254, and 255 | [50] | ||
| 65C6 | human | 164481 | H5 all clades except 7.2 | 118, 121, 161, 164, and 167 (mature H5 numbering) P134, S137, K177, Y180, T183 (IEDB) | [51] | ||
| 9F4 | mouse | 136074 | H5 clades | 271–281, centered on 272–275 | [52,53] | ||
| AVFluIgG 01 | human | 170144 | H5 | I132, I133, P134, W138, S139, Y180, T183 | [54] | ||
| H5M9 | mouse | 194677 | most H5N1 clades | D59, D61, V63, K64, R69, E85, N88, P90, E91, H126, E128, E286, Y287, N289, N291 | 4MHH Vietnam/1203/2004 | [55] | |
| HA-7 | mouse | 186786 | H5 several clades | 88–90 NVP and 126–131 HFEKIQ | cryoEM | [56] | |
| CR8059 | human VH1-18 | 178109 | B strains | T52, K53, S54, H55, F56, K67, N74, T76, D77, H100, E101, G300, S301, L302, P303 | 4FQK B/Brisbane/60/2008 | [32] | |
| CR8071 | human VH1-18 | 178108 | Most B strains | T52, K53, Y55, F56, K67, L73, N74, C75, T76, D77, H100, E101, V105, G299, S300, L301, P302, I304 | 4FQJ B/Florida/4/2006 | [32] |
Immune Epitope Database www.iedb.org
Numbering is from the initiating Met and is continuous through the signal peptide and across the HA1-HA2 junction. In most crystal structures the numbering is separate for HA1 and HA2, and it often follows homology to H3 HA.
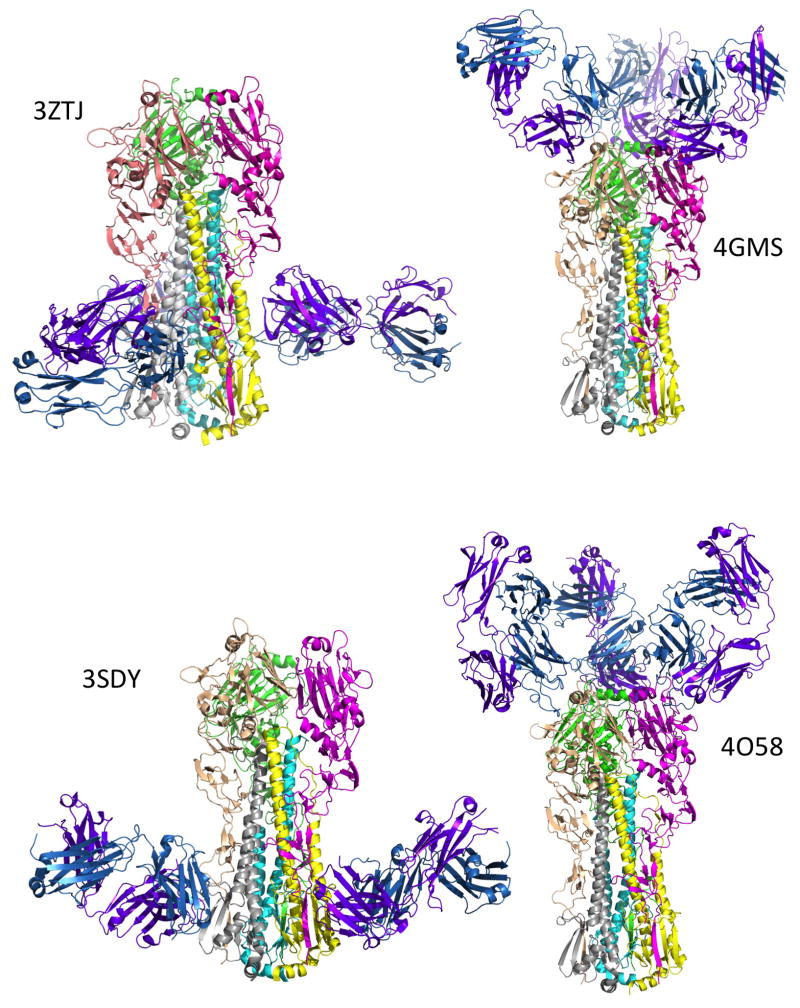
The binding sites for representative mAbs that bind to the stem and receptor binding domains are shown in Figure 1. Note that although the stem antibodies bind in the same general region, the binding sites vary up and down and around the stem and in the angle of the antibody Fab relative to the HA. The antibodies that bind to conserved elements of the receptor binding site similarly vary in placement of the Fab [24]. Antibodies that are not cross-reactive bind to variable loops that surround the receptor binding pocket [25] while those with intra-subunit cross-reactivity show binding to somewhat conserved epitopes on the globular HA head [26,27].
Figure 1.
Binding sites of representative broadly neutralizing antibodies on HA seen in X-ray crystal structures. The HA is shown as the trimer in approximately the same orientation in all panels. Monomer 1 colors are magenta (HA1) and yellow (HA2); monomer 2 are wheat (HA1) and gray (HA2); monomer 3 are green (HA1) and cyan (HA2). In all cases there are three antibody Fabs bound per HA trimer although the third is hidden from view in some cases. The antibody Heavy chain is colored purple and the Light chain is sky blue. The two stem-binding antibodies (F16 and CR8020, see Table 1) bind at different sites on the stem (PDB IDs 3ZTJ and 3SDY respectively). The antibodies that bind to the receptor binding region (S139/1 and F045-092) bind at different angles (PDB IDs 4GMS and 4O58 respectively). The figures were made using PyMOL Molecular Graphics System, Schrödinger, LLC.
The structural studies have also shown that antibodies can bind across the canonical antigenic sites A – E of H3 and Sa – Sb of H1. This is the case with broadly neutralizing antibodies that bind around the receptor binding site (Table 1) but also with some of the original mouse mAbs made against Aichi/68 H3 that are more specific (see structures referenced in [25]).
Conclusions and Future Prospects
For many years investigators searched for broadly-neutralizing antibodies in sera and did not find significant evidence, suggesting they are rare. There are several reports that breadth of the response can be widened by changing the immunization route or by using adjuvant, but the protective effect in humans is not clear. The recent advances in mAb technology, particularly in making human mAbs have resulted in many examples of antibodies that are truly broadly neutralizing, and these can be enriched in serum of mice by immunization with “headless” HA constructs [28,29] or by using HAs with different head domains for prime and boost [30,31].
The broadly-neutralizing mAbs are also of potential use therapeutically. Antibodies CR8020 and CR6261 completed Phase 1 clinical trials but a small challenge study did not show efficacy and a phase 2 trial was halted before enrollment. Another mAb called VIS410 is currently in Phase 1 trial but its epitope has not been published, at least not under that name (www.clinicaltrials.gov).
At this time there is a lot of activity to develop broadly neutralizing antibody technology against the variable influenza viruses, but challenges remain. Vaccines could be further refined to induce anti-stem antibodies but the neutralizing power when fusion is blocked may be inherently lower than that of antibodies that block receptor binding, as seen in the lack of efficacy in the challenge study. However, mAb technology allows engineering to improve avidity of antibodies and that may be a path to successful therapeutic cross-reactive mAbs.
Highlights.
A vaccine that induces broadly-neutralizing antibodies would not need annual updates.
Neutralizing epitopes are almost always conformational and discontinuous, not linear.
Recent crystal structures show the location of cross-neutralizing epitopes on HA.
Many broadly-neutralizing antibodies bind the HA stem, others bind the receptor pocket.
Acknowledgments
Work in the author’s laboratory was supported by the National Institute of Allergy and Infectious Diseases (R01 AI050933).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hilleman MR. Antigenic variation of influenza viruses. Annu Rev Microbiol. 1954;8:311–332. doi: 10.1146/annurev.mi.08.100154.001523. [DOI] [PubMed] [Google Scholar]
- 2.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *3.Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol. 2015;386:323–341. doi: 10.1007/82_2014_413. An excellent review of the structural features of broadly neutralizing antibodies that bind HA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster RG, Laver WG. Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology. 1980;104:139–148. doi: 10.1016/0042-6822(80)90372-4. [DOI] [PubMed] [Google Scholar]
- **5.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. A classic paper that put together the results of escape mutant analysis of mAb epitopes and natural variants seen during antigenic drift with the newly determined X-ray crystal structure of the HA. [DOI] [PubMed] [Google Scholar]
- 6.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 7.Popova L, Smith K, West AH, Wilson PC, James JA, Thompson LF, Air GM. Immunodominance of Antigenic Site B over Site A of Hemagglutinin of Recent H3N2 Influenza Viruses. PLoS One. 2012;7:e41895. doi: 10.1371/journal.pone.0041895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebrahimi SM, Dabaghian M, Tebianian M, Jazi MHZ. In contrast to conventional inactivated influenza vaccines, 4xM2e.HSP70c fusion protein fully protected mice against lethal dose of H1, H3 and H9 influenza A isolates circulating in Iran. Virology. 2012;430:63–72. doi: 10.1016/j.virol.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Ma J-H, Yang F-R, Yu H, Zhou Y-J, Li G-X, Huang M, Wen F, Tong G. An M2e-based synthetic peptide vaccine for influenza A virus confers heterosubtypic protection from lethal virus challenge. Virology journal. 2013;10:227. doi: 10.1186/1743-422X-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pissawong T, Maneewatch S, Thueng-In K, Srimanote P, Dong-din-on F, Thanongsaksrikul J, Songserm T, Tongtawe P, Bangphoomi K, Chaicumpa W. Human monoclonal ScFv that bind to different functional domains of M2 and inhibit H5N1 influenza virus replication. Virol J. 2013;10:148. doi: 10.1186/1743-422X-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos EL, Mitcham JL, Koller TD, Bonavia A, Usner DW, Balaratnam G, Fredlund P, Swiderek KM. Efficacy and Safety of Treatment with an Anti-M2e Monoclonal Antibody in Experimental Human Influenza. J Infect Dis. 2014:jiu539. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- 12.Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res. 2013;100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Doyle TM, Li C, Bucher DJ, Hashem AM, Van Domselaar G, Wang J, Farnsworth A, She YM, Cyr T, He R, et al. A monoclonal antibody targeting a highly conserved epitope in influenza B neuraminidase provides protection against drug resistant strains. Biochem Biophys Res Commun. 2013;441:226–229. doi: 10.1016/j.bbrc.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 14.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, et al. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol. 2013;87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Air GM. Influenza neuraminidase. Influenza Other Respi Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laver WG, Air GM, Webster RG, Smith-Gill SJ. Epitopes on protein antigens: misconceptions and realities. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 17.Dreyfus C, Ekiert DC, Wilson IA. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol. 2013;87:7149–7154. doi: 10.1128/JVI.02975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekiert DC, Bhabha G, Elsliger M-A, Friesen RHE, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A. 2012;109:17040–17045. doi: 10.1073/pnas.1212371109. Description of a highly cross-reactive antibody that targets the receptor binding site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **28.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1 doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **29.Mallajosyula VVA, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, Sarma SP, Flynn JA, Temperton NJ, Liang X, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A. 2014;111:E2514–2523. doi: 10.1073/pnas.1402766111. These two papers describe approaches to increase the proportion of anti-stem antibodies after immunization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellebedy AH, Krammer F, Li G-M, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A. 2014;111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nachbagauer R, Wohlbold TJ, Hirsh A, Hai R, Sjursen H, Palese P, Cox RJ, Krammer F. Induction of Broadly Reactive Anti-Hemagglutinin Stalk Antibodies by an H5N1 Vaccine in Humans. J Virol. 2014;88:13260–13268. doi: 10.1128/JVI.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. Crystal structures of several antibody-HA complexes, including antibody C9114 complexed with influenza B HA and influenza A H5 and H7 and cryo-electron microscopy image reconstrction of the same antibody complexed with H1, H3 and H7 HAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 34.Throsby M, van den Brink E, Jongeneelen M, Poon LLM, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyrzucki A, Dreyfus C, Kohler I, Steck M, Wilson IA, Hangartner L. Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody. J Virol. 2014;88:7083–7092. doi: 10.1128/JVI.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu W, Chen A, Miao Y, Xia S, Ling Z, Xu K, Wang T, Xu Y, Cui J, Wu H, et al. Fully human broadly neutralizing monoclonal antibodies against influenza A viruses generated from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient. Virology. 2013;435:320–328. doi: 10.1016/j.virol.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJWM, Brandenburg B, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A. 2014;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol. 2011;85:11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Marco D, Clementi N, Mancini N, Solforosi L, Moreno GJ, Sun X, Tumpey TM, Gubareva LV, Mishin V, Clementi M, et al. A non-VH1-69 heterosubtypic neutralizing human monoclonal antibody protects mice against H1N1 and H5N1 viruses. PloS one. 2012;7:e34415. doi: 10.1371/journal.pone.0034415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, Fong R, Yan D, Kim J, Zhang J, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE, Wilson IA, Basler CF. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8:e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krause JC, Ekiert DC, Tumpey TM, Smith PB, Wilson IA, Crowe JE., Jr An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody. MBio. 2011;2:e00345–00310. doi: 10.1128/mBio.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt AG, Xu H, Khan AR, O'Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho KJ, Hong KW, Kim SH, Seok JH, Kim S, Lee JH, Saelens X, Kim KH. Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin. PLoS One. 2014;9:e89803. doi: 10.1371/journal.pone.0089803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho KJ, Lee JH, Hong KW, Kim SH, Park Y, Lee JY, Kang S, Kim S, Yang JH, Kim EK, et al. Insight into structural diversity of influenza virus haemagglutinin. J Gen Virol. 2013;94:1712–1722. doi: 10.1099/vir.0.051136-0. [DOI] [PubMed] [Google Scholar]
- 48.Kubota-Koketsu R, Mizuta H, Oshita M, Ideno S, Yunoki M, Kuhara M, Yamamoto N, Okuno Y, Ikuta K. Broad neutralizing human monoclonal antibodies against influenza virus from vaccinated healthy donors. Biochem Biophys Res Commun. 2009;387:180–185. doi: 10.1016/j.bbrc.2009.06.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, Wakiyama M, Shirouzu M, Okada J, Okuno Y, et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol. 2014;88:7130–7144. doi: 10.1128/JVI.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian M, Hu H, Zuo T, Wang G, Zhang L, Zhou P. Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol. 2013;87:3571–3577. doi: 10.1128/JVI.01292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol. 2012;86:2978–2989. doi: 10.1128/JVI.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh H-LJ, Akerström S, Shen S, Bereczky S, Karlberg H, Klingström J, Lal SK, Mirazimi A, Tan Y-J. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J Virol. 2010;84:8275–8286. doi: 10.1128/JVI.02593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mak TM, Hanson BJ, Tan YJ. Chimerization and characterization of a monoclonal antibody with potent neutralizing activity across multiple influenza A H5N1 clades. Antiviral Res. 2014;107:76–83. doi: 10.1016/j.antiviral.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 54.Cao Z, Meng J, Li X, Wu R, Huang Y, He Y. The epitope and neutralization mechanism of AVFluIgG01, a broad-reactive human monoclonal antibody against H5N1 influenza virus. PloS one. 2012;7:e38126. doi: 10.1371/journal.pone.0038126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu X, Guo Y-H, Jiang T, Wang Y-D, Chan K-H, Li X-F, Yu W, McBride R, Paulson JC, Yuen K-Y, et al. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol. 2013;87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du L, Jin L, Zhao G, Sun S, Li J, Yu H, Li Y, Zheng B-J, Liddington RC, Zhou Y, et al. Identification and Structural Characterization of a Broadly Neutralizing Antibody Targeting a Novel Conserved Epitope on the Influenza Virus H5N1 Hemagglutinin. J Virol. 2013;87:2215–2225. doi: 10.1128/JVI.02344-12. [DOI] [PMC free article] [PubMed] [Google Scholar]