Summary
Chromatin-binding proteins play vital roles in the assembly and recruitment of multi-subunit complexes harboring effector proteins to specific genomic loci. MRG15, a chromodomain-containing chromatin-binding protein, recruits diverse chromatin-associated complexes that regulate gene transcription, DNA repair, and RNA splicing. Previous studies with Pf1, another chromatin-binding subunit of the Sin3S/Rpd3S histone deacetylase complex, defined the sequence and structural requirements for interactions with the MRG15 MRG domain, a common target of diverse subunits in the aforementioned complexes. We now show that MRGBP, a member of the Tip60/NuA4 histone acetyltransferase complex, engages the same two surfaces of the MRG domain as Pf1. High-affinity interactions occur via a bipartite structural motif including an FxLP sequence motif. MRGBP shares little sequence and structural similarity with Pf1, yet, targets similar pockets on the surface of the MRG domain, mimicking Pf1 in its interactions. Our studies shed light into how MRG domains have evolved to bind diverse targets.
Introduction
The chromatin not only serves to compact the genome but also functions as a signaling platform, regulating many essential processes, including transcription, replication, recombination, and repair. Chromatin-binding proteins, including ‘readers’ of diverse post-translational histone modifications, play a critical role in interpreting these signals and in recruiting downstream effector proteins (Gayatri and Bedford, 2014; Lalonde et al., 2014; Musselman et al., 2014). The readers often feature one or more of a relatively small number of recurring structural motifs that function as chromatin-binding modules. These modules, either in isolation or in combination with similar or disparate motifs within the same protein or within associated proteins of a multi-protein complex, facilitate the recognition of a broad array of histone signals (Wang and Patel, 2011). A few chromatin-binding proteins, including the chromodomain-containing protein MRG15, are co-opted by disparate multi-protein complexes to regulate diverse chromatin-centric processes (Chen et al., 2010), yet the structural basis for this is poorly understood.
The mortality family of transcription factors (MORF) comprises three proteins, two of which (MRG15 and MRGX) promote cell proliferation while the third, MORF4, is involved in replicative senescence (Chen et al., 2011; Chen et al., 2009; Tominaga et al., 2005). MRG15 is found in a number of transcriptional coregulator complexes including the Rb-associated MAF1 complex, the histone acetyltransferase (HAT)-associated Tip60/NuA4 and MAF2 complexes and the histone deacetylase (HDAC)-associated Sin3S/Rpd3S complex (Carrozza et al., 2005; Doyon et al., 2004; Jelinic et al., 2011; Pardo et al., 2002; Yochum and Ayer, 2002). Going beyond its role in transcription regulation, MRG15 also associates with a BRCA complex that is involved in homologous recombination-mediated DNA repair and a complex that promotes alternative RNA splicing (Hayakawa et al., 2010; Luco et al., 2010; Sy et al., 2009). Additionally, MRG15 has been implicated in interactions with condensin to regulate chromosome condensation during interphase (Smith et al., 2013). In line with its varied roles, MRG15 deletion in mouse causes embryonic lethality with profound defects in cell proliferation, differentiation, and organ development and impaired DNA-damage response (Garcia et al., 2007; Tominaga et al., 2005). In the worm, proper control of the levels of MRG-1 (homologue of MRG15) is required for regulating stem cell proliferation (Gupta et al., 2015).
MRG15 harbors two conserved domains including a chromodomain and an MRG domain at the N- and C-termini, respectively (Figure 1A). The chromodomain is atypical and is absent in the other two MORF family members. This domain recognizes the H3K36me2/3 signal found in the intragenic regions of actively transcribed genes, albeit with millimolar affinity (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005; Kumar et al., 2012; Sun et al., 2008; Xu et al., 2008).
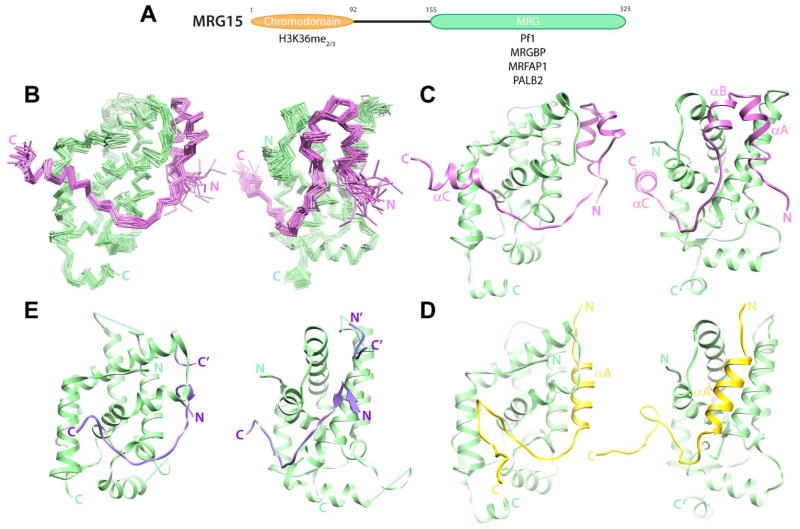
Figure 1.
Structure of the human MRG15 MRG-MRGBP MBD complex and comparison with other protein-protein complexes involving MRG domains. (A) Domain structure of MRG15. Proteins and histone signals recognized by the individual domains are shown. (B) Backbone Cα traces of an ensemble of 20 NMR conformers following a best-fit superposition of secondary structural elements of MRG15 MRG (green) and MRGBP MBD (magenta). The two views are rotated by 70° to highlight the two MRG surfaces (Site I, right; Site II, left) engaged by MRGBP. (C) A representative conformer from the ensemble is shown with the backbone rendered as ribbons. Comparable views of (D) the MRG15 MRG-Pf1 MBD (PDB ID: 2LKM; (Xie et al., 2012)) and (E) the MSL3 MRG-MSL1 MBD (PDB ID: 2Y0N; (Kadlec et al., 2011)) complexes. Note that in the MSL3-MSL1 complex, two discrete MSL1 segments from molecules in adjacent complexes make contacts with Site I in the MRG domain; these segments are distinguished by N/C and N′/C′ labels. See also Figure S1 for biochemical data establishing the minimal MBD of MRGBP and Figure S2 for the quality of the NMR spectra obtained in the complex for both MRG15 MRG and MRGBP MBD.
The MRG domain is found only in a handful of proteins including all three members of the MORF family and the MSL3 protein, which is a subunit of the dosage compensation complex that regulates expression of X-linked genes. The MRG15 MRG domain is the known site of interaction with diverse proteins including MRGBP, MRFAP1, Pf1, and PALB2 in the aforementioned complexes (Cai et al., 2003; Hayakawa et al., 2010; Leung et al., 2001; Sy et al., 2009; Xie et al., 2012; Yochum and Ayer, 2002). Crystallographic and NMR studies have shown that the domain is predominantly helical with a core comprising two orthogonal helical hairpins (Bowman et al., 2006; Xie et al., 2012; Zhang et al., 2006). We previously characterized the interaction between MRG15 and Pf1, and in the process, defined the sequence and structural requirements for efficient interactions with MRG domains (Xie et al., 2012). Our studies revealed a bipartite structural motif within Pf1 comprising a helix and a segment containing an FxLP sequence in an extended conformation that target adjacent surfaces of the MRG domain. Both structural motifs are critical for the nanomolar affinity interaction, since each segment contributes equivalently to the total binding energy. Although all the aforementioned MRG interactors share the FxLP motif, a Pf1-like helical motif is not obvious from sequence analysis, implying potential diversity in MRG-recognition mechanisms and, conversely, the multi-specificity of the MRG domain for engaging diverse targets.
Here we structurally and functionally characterize the interaction between MRG15 and MRGBP, a subunit of the Tip60/NuA4 complex (Cai et al., 2003). Like the Sin3S/Rpd3S complex, the Tip60/NuA4 complex is conserved from yeast to human (Doyon and Cote, 2004). Pf1 and MRGBP orthologs in these species exhibit distinct patterns of sequence conservation but share little sequence similarity with each other at the level of individual domains or sequence motifs (with the exception of the FxLP motif). Our studies reveal that, like Pf1, MRGBP not only interacts with the MRG domain of MRG15 with high affinity, but also engages the same two surfaces of the MRG domain. Despite sharing little similarity with Pf1 at both the sequence and structural levels, MRGBP targets similar pockets on the surface of the MRG domain like Pf1, engaging in a type of molecular mimicry.
Results and Discussion
Biochemical and NMR Studies of the MRG15-MRGBP Interaction
Analysis of the sequences of MRGBP orthologs revealed the presence of two conserved domains in the protein with the more C-terminal domain spanning approximately 50 residues and harboring an FxLP motif. Two helical segments were predicted in this region on either side of the motif (Figure S1A). Because the ~40-residue N-terminal domain was also predicted to be poor in secondary structural elements with only two helices, constructs spanning both domains or only the C-terminal domain were co-expressed with the MRG15 MRG domain, co-purified via affinity chromatography, and subjected to size exclusion chromatography (SEC). Since both constructs eluted in lock-step with the MRG15 MRG domain (Figure S1B), implying the formation of stable complexes, the C-terminal domain was deemed the minimal MRG-binding domain (MBD). The NMR spectrum of MRG15 MRG exhibited significant chemical shift perturbations for many resonances in the presence of an equivalent amount of MRGBP MBD, consistent with the formation of a specific complex (Figure S2). The changes in the NMR spectrum of MRGBP MRG were even more striking with most resonances, barring a few at the N-terminus, showing large-scale chemical shift perturbations. These changes considerably improve the chemical shift dispersion of the amide proton resonances in the bound state (Figure S2), characteristic of a folding transition occurring within the MBD upon binding the MRG domain.
The NMR spectra of the MRG15 MRG-MRGBP MBD complex were of sufficiently high quality to permit almost complete resonance assignments using standard approaches. The structure of the complex was determined via solution NMR making use of distance restraints derived from 1H-1H NOEs and dihedral angle restraints from backbone chemical shifts. Structures of reasonable precision and good covalent geometry with no significant distance or dihedral angle violations (>0.5 Å and >5°, respectively) were obtained (Figure 1B; Table 1). Except for the lower precision at the N- and C-termini of both proteins and an internal loop spanning R204 to K210 for the MRG15 MRG domain, the structures were well-defined.
Table 1.
NMR Structure Determination Statistics for the MRG15 MRG-MRGBP MBD Complex
| Restraint Statistics | |
| NOE-based distance restraints | 5093 |
| Unambiguous NOE-based restraints | 4042 |
| Intra-residue | 1876 |
| Sequential (|i − j| = 1) | 769 |
| Medium-range (1 < |i − j| ≤ 4) | 542 |
| Long-range (|i − j| > 4) | 539 |
| Inter-molecular restraints | 316 |
| Ambiguous NOE-based restraints | 929 |
| Hydrogen bonding distance restraints | 122 |
| Torsion angle restraints | 348 (174 ϕ, 174 ψ) |
| Structure Quality of NMR Ensemble | |
| Restraint satisfaction | |
| RMS differences for distances (Å) | 0.014 ± 0.001 |
| RMS differences for torsion angles (°) | 0.255 ± 0.054 |
| Deviations from ideal covalent geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (°) | 0.489 ± 0.009 |
| Impropers (°) | 1.389 ± 0.066 |
| Ramachandran plot statistics (%) | |
| Residues in most favored regions | 86.2 |
| Residues in additional allowed regions | 12.9 |
| Residues in generously allowed regions | 0.6 |
| Residues in disallowed regions | 0.3 |
| Average Atomic RMSDs from Average Structure (Å) | |
| All atoms | 1.96 |
| All atoms except in disordered regionsa | 1.36 |
| Backbone atoms (N, Cα, C′) | |
| All residues | 1.57 |
| All residues except disordered regionsa | 0.75 |
| All residues in secondary structure elements | 0.71 |
disordered regions include residues 69-74 and 117-118 of MRGBP, the three non-native residues at the N-termini of each protein, residues 204-210 and residue 323 of MRG15
Structure of the MRG Domain in the MRG15-MRGBP Complex
The overall structure of the MRG domain in complex with MRGBP is similar to those reported previously in complex with Pf1 or in the apo-form and comprises nine helices and two short β-strands (Figure 1B; (Bowman et al., 2006; Xie et al., 2012; Zhang et al., 2006)). Further underscoring this point, the backbone atomic root-mean-square deviations (RMSDs) following best-fit superposition of the MRGBP-loaded MRG with the Pf1-loaded and the apo MRGs were 1.45 and 1.30 Å, respectively; for comparison, the RMSD between the Pf1-loaded and the apo MRG was 1.28 Å. These best-fit superpositions excluded loops exhibiting conformational diversity including those between α2 and α3, α4 and α5, and α6 and α7. Among these, the latter loop is situated in close proximity to or is part of the protein-protein interface and its conformation diversity likely plays a role in multi-specificity (see below).
MRGBP Binds to Similar Surfaces of MRG15 MRG Domain Targeted by Pf1
The MRGBP MBD, on the other hand, forms three helices designated αA, αB, and αC spanning residues W78–T83, Q88–S93, and E109–R116, respectively (Figure 1C). The αA and αB helices pack against each other to form a helical sub-domain while the segment C-terminal to it leading up to and including the FxLP motif adopts an extended conformation followed immediately by the αC helix. The helical sub-domain and the C-terminal segment thus form a bipartite structural motif that wraps around the MRG domain engaging two discrete surfaces, designated Site I and Site II, respectively. These are broadly the same surfaces targeted by the Pf1 MBD in the MRG15-Pf1 complex (Figure 1D; (Xie et al., 2012)). However, unlike the single helix used by Pf1 to engage Site I, the MRG15 MRG deploys two helices to engage Site I and an additional helix to engage Site II.
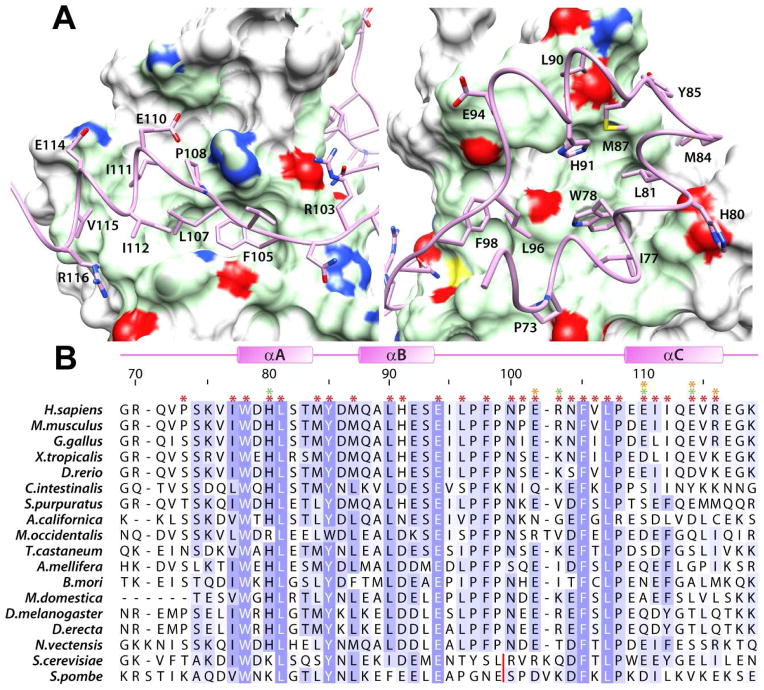
The MRG15-MRGBP complex is stabilized primarily by hydrophobic interactions (Figure S3). The W78 side chain in the MRGBP helical sub-domain functions as a key organizing center (Figure 2A), not only engaging a deep hydrophobic pocket in the MRG domain but also nucleating a hydrophobic core formed within the sub-domain by the side chains of I77, L81, M87, H91, L96, and F98, all of which also engage the predominantly hydrophobic MRG surface. The protein-protein interface is extended by the side chains of MRGBP H80, M84, Y85, and L90, which target a more peripheral surface of the MRG domain; these interactions are hydrophobic in character with the exception of H80, which engages in electrostatic interactions with E216 (Figures 2 & S3). Consistent with their critical roles in stabilizing the complex, each of the aforementioned residues are highly or completely conserved in MRGBP orthologs (Figure 2B).
Figure 2.
Non-covalent interactions in the MRG15 MRG-MRGBP MBD complex. (A) Close-up views of the protein-protein interface highlighting the key residues within MRGBP MBD making contacts with the two MRG surfaces (Site I, right; Site II, left). The molecular surface of MRG domain in contact with MRGBP is rendered in green; the side chain nitrogen, sulfur, and oxygen atoms are colored blue, yellow, and red, respectively. (B) A multiple sequence alignment of MRGBP orthologs from a variety of species. The asterisks on top identify residues in the human protein involved in intermolecular interactions (red: hydrophobic; orange: hydrogen bonding; green: electrostatic). Residues are colored according to the degree of sequence conservation using JalView (Waterhouse et al., 2009). The red line denotes the location of multi-residue insertions in the yeast proteins. See also Figure S3 for the catalog of intermolecular interactions deduced using MONSTER (Salerno et al., 2004).
As expected, residues comprising the MRGBP FxLP motif including F105, L107, and P108 target an identical sub-surface of the MRG domain within Site II as in the MRG15-Pf1 complex. The side chain of F105 is completely buried in a deep pocket along with the side chain of L107 while P108 interacts with the surface immediately adjacent to this pocket (Figures 2 & S3). Residues in the MRGBP αC helix including I111, I112, and V115 engage in hydrophobic interactions while E110, E114, and R116 engage in electrostatic and/or hydrogen bonding interactions with K157, K165, and D173, respectively (Figures 2 & S3). Although some of the residues in this region are conserved in MRGBP orthologs, residues beyond I112 show greater sequence diversity (Figure 2B).
Functional Analysis of MRG15-MRGBP Interactions
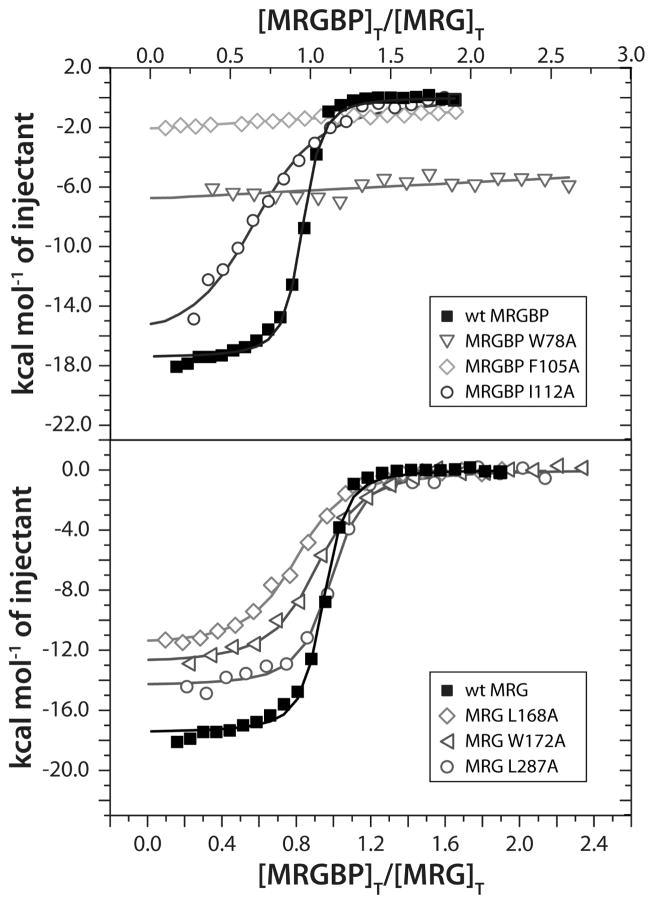
To clarify the role of individual residues towards the stability of the MRG15-MRGBP complex, residues in both proteins at the interface were mutated and evaluated for binding via isothermal titration calorimetry (ITC) assays (Figure 3). The wild-type MRG15 MRG and MRGBP MBD proteins bound with an equilibrium dissociation constant (Kd) ~60 nM in this assay which is comparable to that measured for the wild-type proteins in the MRG15 MRG-Pf1 MBD complex (Kd ~20 nM; (Xie et al., 2012)). The high-affinity of the MRG15-MRGBP interaction is consistent with the size of protein-protein interface (~1260 Å2), which is comparable to that in the MRG15-Pf1 complex. Mutations of the invariant MRGBP residues W78 and F105 to alanine had the most dramatic consequences, with little or virtually no detectable binding, implying almost complete abrogation of the interaction (Figure 3 & Table S1). These results are consistent with their critical roles suggested by the NMR structure. On the other hand, alanine mutations of M84 and I112, which engage in more peripheral interactions, were less deleterious with decreases in binding affinities of about three- and thirteen-fold, respectively, relative to wild-type.
Figure 3.
ITC analysis of binding of wild-type and various mutant MRG15 MRG and MRGBP MBD proteins. Representative binding curves from titrations of MRGBP MBD (in the syringe) with MRG15 MRG (in the cell) for the indicated proteins are shown. Titration data for mutant MRGBP MBD and MRG15 MRG are shown in the top and bottom panels, respectively. All experiments were conducted at 25 °C. See Supplementary Table S1 for the dissociation constants inferred from these measurements.
Alanine mutations of MRG15 L168 and W172, which form the pocket targeted by MRGBP L107 in addition to serving as interaction surfaces for MRGBP I111 and I112 (Figure 2 & S3), were most deleterious with reductions in binding affinity of about seven- to eight-fold (Figure 3 & Table S1). Mutation of M273, which is located at the periphery of Site I and interacts marginally with F98, exhibited binding comparable to the wild-type protein. Rather unexpectedly, the alanine mutation of L287, which forms the wall of the pocket targeted by MRGBP W78 side chain had only a little over two-fold effect on binding (Figure 3 and Table S1). We surmise that the bulky tryptophan side chain adapts its conformation to take advantage of the additional space created by the mutation in the pocket, resulting in a closely (re)packed interface. Collectively, the results of our functional analysis of mutant MRG15 MRG and MRGBP MBD proteins provide support for our structural model for the MRG15-MRGBP interaction.
Conserved and Disparate Themes in MRG Recognition and the Basis for Multi-Specificity
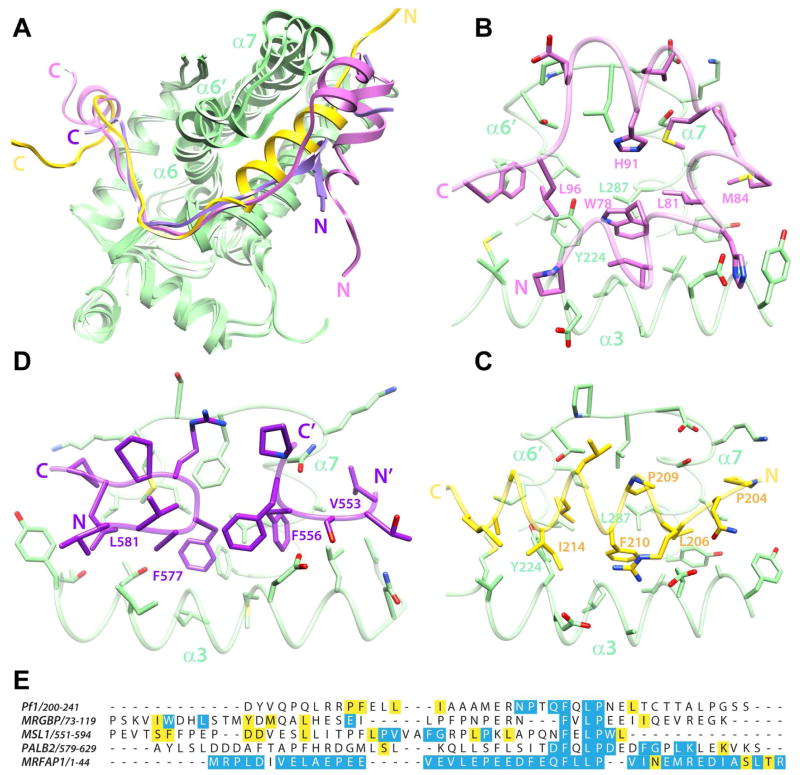
Thus far, three high-resolution structures of MRG domains in complex with three disparate targets have been described. These include the structures of human MRG15 MRG domain in complex with Pf1 and MRGBP MBDs and that of human MSL3 MRG domain in complex with MSL1 MBD (Figure 1C–E; (Kadlec et al., 2011; Xie et al., 2012)). MRG15 and MSL3 MRG domains share 35% sequence identity and 56% sequence similarity. However, they share close structural similarity with backbone RMSD of 1.37 Å following a best-fit superposition. Like Pf1 and MRGBP, MSL1 binds the MSL3 MRG domain via a bi-partite structural motif that includes an FxLP sequence motif that engages Site II in an extended conformation. However, unlike Pf1 and MRGBP, which deploy helices for engaging Site I, MSL1 deploys a turn-like conformation and a beta-hairpin motif to engage Site I. Another striking feature of these complexes is the diversity of conformations adopted by the α6′ helix, the loop following this helix, and the N-terminus of α7 that contribute to Site I (helix designations follow Xie et al. 2012, Figure S3). This segment is relatively closed in the MSL3-MSL1 complex and also in the ‘apo’ MRG15 structure where it engages the same segment of another protomer, but is progressively more open in the MRG15-Pf1 and MRG15-MRGBP complexes, correlating with the bulkiness of the MBD making close contacts with the surface (Figure 4A). In contrast, Site I is largely conformationally homogeneous with the exception of only a few residues in the α6′ helix adopting a slightly altered conformation in the different structures. The conformational homogeneity perhaps explains why this region has evolved to bind a considerably restricted set of sequences, all bearing the FxLP motif.
Figure 4.
A comparative analysis of the structures of different MRG complexes. (A) Overlays of the MRG15 MRG-MRGBP MBD, MRG15 MRG-Pf1 MBD, and MSL3 MRG-MSL1 MBD complexes following best-fit superposition of backbone atoms in the MRG domains. The coloring scheme is the same as that in figure 1. Notice how the segment between α6′ and the N-terminus of α7 adopts different conformations in the three structures. This segment is relatively closed in the MSL1 complex but is progressively more open in the Pf1 and MRGBP complexes. Side chain interactions with various Site I residues in the respective MRG domains of the (B) MRGBP, (C) Pf1 and (D) MSL1 complexes are shown. The side chains of residues making comparable contacts in the various complexes are annotated. Note that in the MSL3-MSL1 complex, two discrete MSL1 segments from molecules in adjacent complexes make contacts with Site I in the MRG domain. (E) A CLUSTAL Ω-guided multiple sequence alignment of established MRG-interactors to emphasize the lack of sequence similarity between these domains. Conserved and invariant residues in the respective orthologous proteins are highlighted in yellow and blue, respectively. The high degree of sequence conservation for MRFAP1 is likely due to its occurrence in a narrow range of species. Notice the poor sequence conservation N-terminal to the FxLP motif in PALB2 orthologs. See also Figure S4 demonstrating a high-affinity interaction between PALB2 MBD and MRG15 MRG.
Notwithstanding the diversity in backbone conformations of the structural motifs engaging Site I in the different MRG complexes, conserved side chains maintain similar dispositions targeting the same pockets on the surface of the MRG domain (Figure 4B–C). The following pairs of residues in MRGBP and Pf1 thus appear to be structurally equivalent: W78/F210; L81/L206; M84/P204; H91/P209; L96/I214. Although most of the side chains that comprise Site I maintain similar conformations, the side chain of MRG15 Y224 adopts strikingly different conformations in the two complexes, presumably to eliminate clashes with L213 in the Pf1 complex and to allow for optimal side chain packing in the MRGBP complex.
The aforementioned similarities extend even to the MSL3-MSL1 complex (Kadlec et al., 2011), despite the fact that only a small subset of residues in Site I are invariant in the MRG15 and MSL3 MRG domains. Equivalencies can be established for the following pairs of MRGBP and MSL1 residues: W78/F577; L81/F556; M84/V553; L96/L581 (Figure 4D). This suggests that some of the key features of MRG domains for engaging targets are shared despite the early evolutionary divergence of the paralogs.
Implications for Other MRG15 Complexes
To gain insight into the possible mode of MRG15 MRG recognition by these proteins, we analyzed the sequence conservation profiles of the four known interactors of the MRG15 MRG domain (Figure 4E). As expected, the sequences of Pf1 and MRGBP orthologs from yeast to human showed very disparate patterns of conservation, consistent with their different modes of engaging the MRG domain. By way of contrast, we analyzed the sequences of MSL1 orthologs (which interact with the MSL3 MRG domain) from fly to human and these sequences also showed a distinct pattern of sequence conservation. However, all five interactors shared an FxLP motif with residues in three out of the four positions essentially invariant in all the orthologs examined.
Analysis of PALB2 orthologs from zebrafish to human revealed a high level of sequence conservation for residues in the vicinity of the FxLP motif (Figure 4E). Unlike in Pf1 and MRGBP, conservation N-terminal to this segment is remarkably poor, suggesting either that this segment is not required for binding or that PALB2 binds MRG15 with much lower affinity. To evaluate the affinity of PALB2 MBD (spanning residues 579 to 629) for MRG15 MRG domain, we conducted ITC analyses, which revealed a high-affinity interaction between these polypeptides (Kd ~20 nM; Figure S4; Table S1). This implies that high-affinity binding is possible exclusively via Site II alone or that interactions involving Site I are diverse even among PALB2 orthologs. This contrasts with the findings for the Pf1 MBD interaction with MRG15 MRG domain in which only about half of the total binding energy was provided by the interaction of Site II with the C-terminal segment bearing the FxLP motif (Xie et al., 2012).
Analysis of MRFAP1 orthologs indicated extremely high levels of sequence conservation throughout the ~120 residue protein (Figure 4E). However, this is probably due to the fact that the protein is found within a narrow range of species, being restricted only to placentals. Although the segment N-terminal to the FxLP motif contains several non-polar residues, the presence of prolines and a general lack of predicted helical structure in this region suggest a novel mode of MRG interaction. Thus, none of the MRG15 MRG interactors exhibit any significant degree of similarity with any other interactor at the level of primary structure except in the region encompassing the FxLP motif. The motif thus plays a critical role in MRG interactions both as affinity and specificity determinants.
Biological Implications
The involvement of analogous surfaces of the MRG domain in the interactions that have thus far been structurally characterized suggests that interactors compete with each other for MRG15-binding. Since all of the interactions measured thus far are in the nanomolar range, this not only implies that these interactions occur mutually exclusively but also that multiple MRG15-containing complexes likely exist simultaneously. Indeed, these predictions are borne out by proteomic studies conducted using MRG15 or its interactors as the ‘bait’ (Hayakawa et al., 2007; Kirkwood et al., 2013; Nishibuchi et al., 2014).
Recent studies in yeast and in mammals suggest that MRG15 and MRGBP exist as a separate sub-complex and serve to recruit the remaining subunits of the Tip60/NuA4 complex to intragenic regions of genes (Cheng and Cote, 2014; Rossetto et al., 2014), facilitating the acetylation of histones in these regions to create a permissive chromatin environment for transcription. This is counteracted by the actions of the HDAC-containing Sin3S/Rpd3S complex which harbors MRG15 and Pf1 as the chromatin binding subunits (Kumar et al., 2012; Li et al., 2007). Interestingly, PALB2 is another chromatin-binding subunit in the BRCA complex that is targeted by MRG15 (Bleuyard et al., 2012). Since MRG15 appears to bind its chromatin target H3K36me2/3 with only millimolar affinity (when measured in a peptide context), it raises the intriguing possibility that MRG15 in concert with diverse MRG interactors have evolved to recognize a variety of signals on chromatin due, in part, to the stable association mediated by the MRG domain. In this regard, the MRGBP protein harbors a highly-conserved domain of unknown function near the N-terminus of the protein. Further studies are required to test whether this domain could function in chromatin binding. We note that MBD-loaded MRG complexes can also function in higher-order assembly of multi-subunit complexes including the Sin3S/Rpd3S and the dosage compensation complexes (Kadlec et al., 2011; Kumar et al., 2012), further demonstrating the versatility of MRG domains to engage diverse protein targets.
Conclusions
Our studies provide insights into how MRG domains have evolved to engage their targets in high-affinity interactions while concurrently preserving their ability to recognize structurally unrelated targets over billions of years of evolution. The multi-specificity of MRG domains relies on the one hand on the plasticity of one of the two surfaces that is typically involved in these interactions. Structural diversity of the interactors notwithstanding, conserved residues within these proteins often target the same pockets on the surface of the MRG domains, thereby engaging in a form of molecular mimicry. This type of mimicry is reminiscent of several protein-protein complexes featuring apparently promiscuous (multi-specific) interactors (Sahu et al., 2008; Schreiber and Keating, 2011; Swanson et al., 2004). Rather intriguingly, the second MRG surface, which is comparatively rigid, serves as the docking site for a conserved tetrapeptide motif in all of the MRG-interactors described thus far, contributing critically to the affinity and specificity of these interactions. Yet, there is considerable sequence diversity even in the vicinity of the tetrapeptide motif and the MRG complexes feature intermolecular interactions that appear to be opportunistic and not always well-conserved in orthologs, perhaps tuning the stability of the interaction to the needs of the biological process.
Experimental Procedures
Production of MRG15 MRG and MRGBP MBD
The human MRG15 MRG (residues 155-323) was expressed and purified as described before (Kumar et al., 2011). The sequences encoding human MRGBP (residues 32-119 and 69-119) were amplified by PCR and inserted into the pMCSG7 expression vector, for expression as His6-tagged fusion protein. The cloned gene segments were confirmed by DNA sequencing. E.coli BL21(DE3) cells (Novagen, WI) transformed with the plasmid were grown in Terrific Broth (Invitrogen, CA). Protein expression was induced using 1 mM isopropyl-β-D-thio-galactopyranoside (IPTG) at 20 °C when the OD600 nm reached about ~1.3 and the cells were harvested 15–17 h later. Cell pellets were suspended in 50 mM Tris-HCl buffer (pH 7.9) containing 0.2 M NaCl, 8 M Urea, 1 mM Tris (2-carboxy-ethyl) phosphine hydrochloride (TCEP), 1 mM phenylmethylsulfonylfluoride (PMSF), 1 μM leupeptin, 1 mM pepstatin, and 0.1% Triton X-100, and lysed via sonication followed by high-speed centrifugation for 30 min. The supernatant was incubated with His-Select Ni2+-resin (Sigma-Aldrich, MO) for 40 min, followed by extensive washing with 50 mM Tris buffer (pH 7.9) containing 0.2 M NaCl, and 1 mM TCEP and 50 mM Tris buffer (pH 7.9) containing 1 M NaCl, and 1 mM TCEP. The protein was eluted with 50 mM Tris buffer (pH 7.9) containing 0.2 M NaCl, 1 mM TCEP, and 0.25 M Imidazole, followed by dialysis against 50 mM Tris buffer (pH 7.9) containing 0.2 M NaCl, and 1 mM TCEP and subsequent incubation with tobacco etch virus (TEV) protease at room temperature for 4 h and then at 4 °C overnight. The cleaved protein was further purified by His-Select Ni2+-resin (Sigma-Aldrich, MO) and reverse-phase HPLC using a C18 column (Grace Vydac, CA) and a linear gradient of 0.1% trifluoroacetic acid and 0.1% trifluoroacetic acid in 80% acetonitrile, and protein-containing fractions were collected and lyophilized. Uniformly 15N- and/or 13C-labeled proteins were produced using the same procedure except that the cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose (Cambridge Isotopes, MA), respectively. Protein identity and integrity was assessed by ESI-MS and SDS-PAGE gel electrophoresis.
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were performed on a Microcal iTC200 calorimeter (Malvern Instruments, MA) at 25 °C in 25 mM sodium phosphate buffer (pH 7.8) containing 0.15 M NaCl, and 1 mM TCEP. Proteins were extensively dialyzed against the buffer used for the measurements. Protein concentrations were determined spectrophotometrically (Gill and von Hippel, 1989). The wild-type and mutant MRG15 MRG proteins were kept in the cell while the wild-type and mutant MRGBP MBD polypeptides were in the syringe at initial concentrations of 15–20 μM and 0.15–0.2 mM respectively; similar starting protein concentrations were used for the experiments with human PALB2 MBD (residues 579-629). Binding curves were fitted assuming a single-site binding model using the Origin 7.0 software provided by the manufacturer.
MRG15 MRG-MRGBP MBD Complex Generation and NMR Sample Preparation
The complex was generated by mixing 15N,13C,-labeled MRG15 MRG or MRGBP MBD with an equivalent amount of unlabeled MRGBP MBD or MRG15 MRG, respectively, at a low concentration (~20 μM) in 50 mM Tris buffer (pH 7.9) containing 0.2 M NaCl, and 3 mM dithiothreitol, and was concentrated to ~0.2–0.3 mM, followed by size exclusion chromatography purification using a Superdex 75 column (GE Healthcare, New Jersey) pre-equilibrated with 50 mM Tris buffer (pH 7.9) containing 0.2 M NaCl, and 3 mM dithiothreitol. The complex-containing fractions were concentrated and then exchanged into 50 mM sodium phosphate buffer (pH 6.9) containing 3 mM dithiothreitol-d10, 10% D2O and 0.2% NaN3. Protein concentrations were determined spectrophotometrically prior to the addition of NaN3. Samples of the protein-protein complex were lyophilized and re-dissolved in 99.996% D2O for NMR experiments performed in D2O (Sigma-Aldrich, MO).
NMR Spectroscopy and Structure Determination
All NMR data were acquired on an Agilent Inova 600 MHz spectrometer equipped with a pulsed-field-gradient triple-resonance cold probe at 35 °C. NMR data processing and analysis were performed using Felix 98.0 (Accelrys, CA) and Sparky (Goddard and Kneller, 2004). Sample concentrations used for the structure determination were 0.8–0.9 mM. Backbone and side chain 1H, 15N, and 13C resonance assignments for each of the interacting proteins in the MRG15 MRG-MRGBP69–119 complex were obtained by analyzing 3D HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, HCACO, HCCH-COSY and HCCH-TOCSY spectra (Bax and Grzesiek, 1993; Ferentz and Wagner, 2000). Aromatic resonances were assigned based on a careful analysis of 2D 1H-13C aromatic HSQC, 3D 13C-edited NOESY and 15N,13C-double half-filtered NOESY spectra (Otting and Wüthrich, 1990).
For structure determination, backbone φ and ψ torsion angle restraints were derived from analysis of 1Hα, 13Cα, 13Cβ, 13C′, and backbone 15N chemical shifts using TALOS+ (Shen et al., 2009). Restraints were imposed only for those residues with TALOS+ reliability scores of 10. NOE-based distance restraints for each protein in the complex were derived from 3D 15N-edited NOESY (mixing time, τm=80 ms) spectra recorded in H2O and 3D 15N, 13C-filtered, 15N, 13C-edited NOESY (τm=140–150 ms), 3D 13C-edited aliphatic NOESY (τm = 65 ms) and 2D 15N, 13C-double half-filtered NOESY (τm = 70 ms) spectra recorded in D2O.
Structures were determined using ARIA 1.2 in conjunction with CNS 1.1 starting from an initial structure with extended backbone conformations (Brünger et al., 1998; Linge et al., 2003). All NOEs were calibrated automatically and assigned iteratively by ARIA; the assignments were checked manually for errors after every run. Eighty conformers were calculated in the final iteration out of which the 40 conformers with the lowest restraint energies were refined in a shell of explicit water and the 20 conformers with the lowest restraint energies, restraint violations, and RMS deviations from the ideal covalent geometry were selected for further analysis. The final conformers were analyzed using CNS (Brünger et al., 1998), PROCHECK (Laskowski et al., 1996), MONSTER (Salerno et al., 2004). Molecular images were generated using CHIMERA (Pettersen et al., 2004). Structural comparisons were performed with Swiss-PDBViewer 3.7 (Guex and Peitsch, 1997).
Supplementary Material
Highlights.
Disparate nanomolar affinity complexes conserved from yeast to human
Two discrete interaction surfaces on the MRG domain, one plastic, the other, rigid
MRG-binding motifs: a helical domain and an extended conformation ending in a helix
Molecular mimicry in engaging the plastic surface; FxLP motif for the rigid surface
Acknowledgments
Funding for this work was provided by NIH grant R01GM64715 and American Heart Association grant 14GRNT20170003 to I.R. We are grateful to Drs. Kaoru Tominaga and Donald Ayer for generously sharing cDNAs for MRGBP and MRG15, respectively. A.M.Z. was supported by a Weinberg Undergraduate Research Grant from Northwestern. We gratefully acknowledge support for structural biology research from the Lurie Comprehensive Cancer Center at Northwestern.
Footnotes
Accession code - The RCSB PDB accession code for the atomic coordinates of the MRG15 MRG-MRGBP MBD complex is 2n1d. The BMRB code for NMR chemical shifts is 25556.
Author Contributions
T.X. and I.R. designed all the experiments; T.X. conducted all the biochemical experiments and together with Y.Z. collected all the NMR data; T.X. and I.R. analyzed the NMR data and determined the structure; T.X. and A.M.Z. conducted the ITC experiments and together with I.R. analyzed all the data; T.X., A.M.Z. and I.R. wrote the manuscript.
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem Res. 1993;26:131–138. [Google Scholar]
- Bleuyard JY, Buisson R, Masson JY, Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13:135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BR, Moure CM, Kirtane BM, Welschhans RL, Tominaga K, Pereira-Smith OM, Quiocho FA. Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure. 2006;14:151–158. doi: 10.1016/j.str.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chen M, Pereira-Smith OM, Tominaga K. Loss of the chromatin regulator MRG15 limits neural stem/progenitor cell proliferation via increased expression of the p21 Cdk inhibitor. Stem Cell Res. 2011;7:75–88. doi: 10.1016/j.scr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Takano-Maruyama M, Pereira-Smith OM, Gaufo GO, Tominaga K. MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. J Neurosci Res. 2009;87:1522–1531. doi: 10.1002/jnr.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tominaga K, Pereira-Smith OM. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann N Y Acad Sci. 2010;1197:134–141. doi: 10.1111/j.1749-6632.2010.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Cote J. A new companion of elongating RNA Polymerase II: TINTIN, an independent sub-module of NuA4/TIP60 for nucleosome transactions. Transcription. 2014;5:e995571. doi: 10.1080/21541264.2014.995571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Garcia SN, Kirtane BM, Podlutsky AJ, Pereira-Smith OM, Tominaga K. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 2007;581:5275–5281. doi: 10.1016/j.febslet.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayatri S, Bedford MT. Readers of histone methylarginine marks. Biochim Biophys Acta. 2014;1839:702–710. doi: 10.1016/j.bbagrm.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. 2004 http://www.cgl.ucsf.edu/home/sparky/
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gupta P, Leahul L, Wang X, Wang C, Bakos B, Jasper K, Hansen D. Proteasome regulation of the chromodomain protein MRG-1 controls the balance between proliferative fate and differentiation in the C. elegans germ line. Development. 2015;142:291–302. doi: 10.1242/dev.115147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Zhang F, Hayakawa N, Ohtani Y, Shinmyozu K, Nakayama J, Andreassen PR. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123:1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol Cell Biol. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kadlec J, Hallacli E, Lipp M, Holz H, Sanchez-Weatherby J, Cusack S, Akhtar A. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol. 2011;18:142–149. doi: 10.1038/nsmb.1960. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kirkwood KJ, Ahmad Y, Larance M, Lamond AI. Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics. Mol Cell Proteomics. 2013;12:3851–3873. doi: 10.1074/mcp.M113.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GS, Chang W, Xie T, Patel A, Zhang Y, Wang GG, David G, Radhakrishnan I. Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. J Mol Biol. 2012;422:519–531. doi: 10.1016/j.jmb.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GS, Xie T, Zhang Y, Radhakrishnan I. Solution structure of the mSin3A PAH2-Pf1 SID1 complex: a Mad1/Mxd1-like interaction disrupted by MRG15 in the Rpd3S/Sin3S complex. J Mol Biol. 2011;408:987–1000. doi: 10.1016/j.jmb.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde ME, Cheng X, Cote J. Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev. 2014;28:1029–1041. doi: 10.1101/gad.236331.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Leung JK, Berube N, Venable S, Ahmed S, Timchenko N, Pereira-Smith OM. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J Biol Chem. 2001;276:39171–39178. doi: 10.1074/jbc.M103435200. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Khorasanizadeh S, Kutateladze TG. Towards understanding methyllysine readout. Biochim Biophys Acta. 2014;1839:686–693. doi: 10.1016/j.bbagrm.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi G, Shibata Y, Hayakawa T, Hayakawa N, Ohtani Y, Sinmyozu K, Tagami H, Nakayama J. Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2014;289:28956–28970. doi: 10.1074/jbc.M114.573725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G, Wüthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q Rev Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rossetto D, Cramet M, Wang AY, Steunou AL, Lacoste N, Schulze JM, Cote V, Monnet-Saksouk J, Piquet S, Nourani A, et al. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J. 2014;33:1397–1415. doi: 10.15252/embj.201386433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu SC, Swanson KA, Kang RS, Huang K, Brubaker K, Ratcliff K, Radhakrishnan I. Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. J Mol Biol. 2008;375:1444–1456. doi: 10.1016/j.jmb.2007.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno WJ, Seaver SM, Armstrong BR, Radhakrishnan I. MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–568. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr Opin Struct Biol. 2011;21:50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HF, Roberts MA, Nguyen HQ, Peterson M, Hartl TA, Wang XJ, Klebba JE, Rogers GC, Bosco G. Maintenance of interphase chromosome compaction and homolog pairing in Drosophila is regulated by the condensin cap-h2 and its partner Mrg15. Genetics. 2013;195:127–146. doi: 10.1534/genetics.113.153544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284:21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Patel DJ. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. J Biol Chem. 2011;286:18363–18368. doi: 10.1074/jbc.R111.219139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Graveline R, Kumar GS, Zhang Y, Krishnan A, David G, Radhakrishnan I. Structural Basis for Molecular Interactions Involving MRG Domains: Implications in Chromatin Biology. Structure. 2012;20:151–160. doi: 10.1016/j.str.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhao J, Wang B, Du J, Lu Y, Chen J, Ding J. The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. Protein Sci. 2006;15:2423–2434. doi: 10.1110/ps.062397806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.