Abstract
The human APOBEC3G (A3G) DNA cytosine deaminase restricts and hypermutates DNA-based parasites including HIV-1. The viral infectivity factor (Vif) prevents restriction by triggering A3G degradation. While the structure of the A3G catalytic domain is known, the structure of the N-terminal Vif-binding domain has proven more elusive. Here, evolution- and structure-guided mutagenesis was used to solubilize the Vif-binding domain of A3G permitting structural determination by NMR spectroscopy. A smaller zinc-coordinating pocket and altered helical packing distinguish it from catalytic domain structures, and help explain the reported inactivity of this domain. This soluble A3G N-terminal domain is bound by Vif, which enabled mutagenesis and biochemical experiments to identify a unique Vif-interacting surface formed by α1-β1, β2-α2, and β4-α4 loops. This structure sheds new light on the Vif-A3G interaction and provides critical information for future drug development.
INTRODUCTION
APOBEC3 proteins are single-stranded DNA cytosine deaminases that are part of a larger network of innate immune effector proteins that serve to limit the replication of many diverse parasitic elements including the AIDS virus HIV-11–3. Up to seven family members, APOBEC3A, B, C, D, F, G, and H, can be expressed in a single human cell4. A combination of overexpression, knockdown, knockout, and separation-of-function studies have shown that A3D, A3F, A3G, and A3H contribute to HIV-1 restriction and hypermutation in CD4+ T lymphocytes (recent publications5–7 and references therein). A3D, A3F, and A3H catalyze C-to-U deamination in 5′-TC dinucleotide motifs, whereas A3G catalyzes deamination in 5′-CC motifs. These viral cDNA deamination events lead to G-to-A mutations in hallmark GA and GG dinucleotide motifs respectively. A3G has two structural domains, an N-terminal pseudo-catalytic (NTD) and C-terminal catalytic (CTD) domain8,9. The CTD alone dictates the observed local dinucleotide preference10,11,12. The NTD is catalytically inert but still strongly binds RNA and single-stranded DNA and forms a ribonucleoprotein complex with the viral Gag protein, enabling encapsidation [e.g. 13–17].
HIV-1 encodes Vif as a counter-defense mechanism against A3 mediated restriction18,19. Vif interacts with core binding factor β (CBF-β), CUL5, ELOB, ELOC, and RBX2 proteins to form an E3 ubiquitin ligase complex20,21. As a consequence, A3G and other potentially restrictive A3 enzymes are polyubiquitinated and degraded by proteasome before being encapsidated [e.g. 18,19,22–25]. The interaction between Vif and A3G has been studied extensively, and key regions for the interaction have been identified, which include 126-FWDPDYQ-132 of A3G and 22-KSLVK-26, 40-YRHHY-44, 69-YWGL-72 of Vif26–29. However, insolubility has limited structural information of the A3G-Vif complex.
A crystal structure of the Vif-CBF-β-ELOBC-CUL5 complex was reported recently30. Vif has two structural domains called alpha/beta and alpha. A3G-binding residues, including K22, K26, 40-YRHHY-44 and W70, are located in the alpha/beta domain30. Although the structure of catalytically active A3 proteins A3A and A3C, as well as the catalytic domains A3G CTD and A3F CTD, were solved by NMR and X-ray crystallography12,31–38, a structure of the N-terminal pseudo-catalytic, Vif-binding domain of A3G has thus far proven elusive. To address this major gap in knowledge, we have generated a soluble A3G NTD variant (sNTD) that binds the Vif-CBF-β-ElOBC complex. The sNTD has 80% amino acid sequence identity to wildtype A3G NTD. In this paper, we report the three-dimensional structure of sNTD and mutationally interrogate the Vif-binding regions.
RESULTS
Engineering a soluble A3G Vif-binding domain
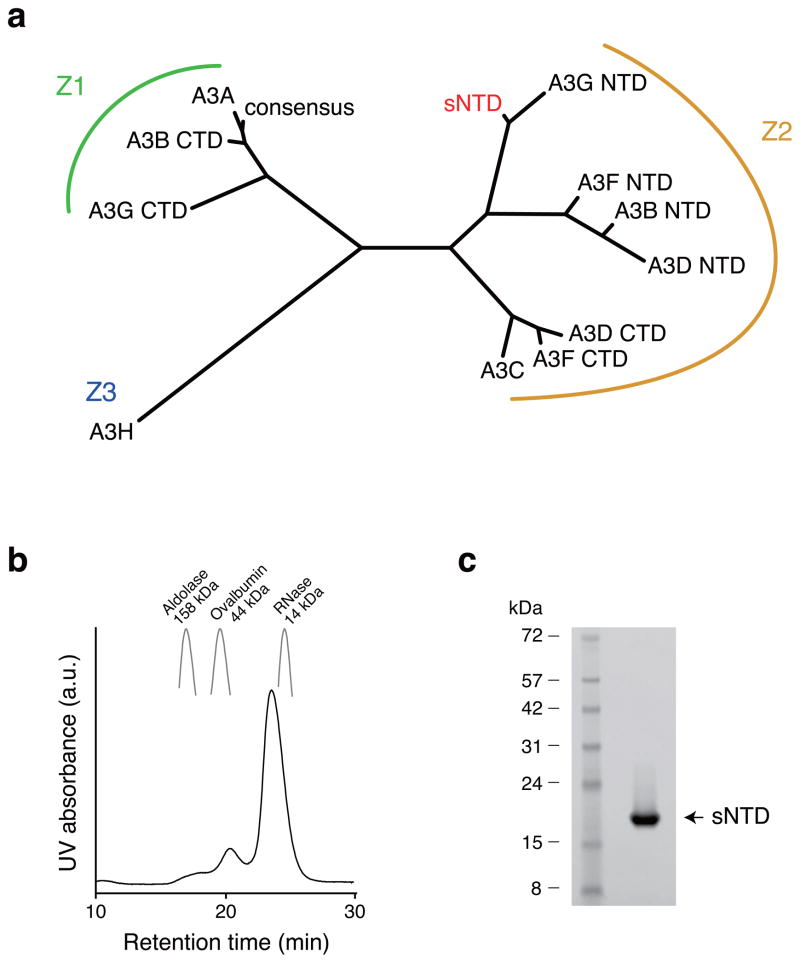
The wildtype A3G NTD was insoluble in E. coli. To overcome this technical hurdle, we compared the amino acid sequence of A3G NTD with A3A, A3B CTD, and A3G CTD, which were soluble and amenable to structural studies12,31–34,37. A3A, A3B CTD, and A3G CTD were catalytically active, but did not interact with Vif, and belong to the same phylogenetic subgroup, i.e., the Z1 group39 (Fig. 1a). An alignment of A3A, A3B CTD, and A3G CTD allowed us to generate a consensus Z1 amino acid sequence (Supplementary Fig. 1a). We expressed and purified the consensus Z1 protein from E. coli, and found that it was soluble and folded. Wildtype A3G NTD amino acid residues were used to replace consensus Z1 residues one at a time, and all of the resulting constructs were tested for protein expression, solubility, and stability. We used NMR to evaluate soluble derivatives for signal dispersion and homogeneity. Ultimately, 80% amino acid sequence identity to wildtype NTD was achieved without substantially compromising solubility or quality of NMR spectra (Fig. 1b, c). Additional wildtype A3G amino acids were not tolerated (Supplementary Fig. 1b, c). We henceforth refer to this soluble A3G NTD construct as sNTD.
Figure 1.
Generation of soluble N-terminal Vif-binding domain of A3G. (a) Amino acid sequence similarities of APOBEC3 proteins. Amino acid sequences of APOBEC3 proteins are aligned by ClustalW60, and the branch length represents similarity of amino acid sequences. A3 proteins are grouped, as described previously39. A3C, A3F CTD and A3G NTD bind HIV-1 Vif, and all of them belong to the Z2 group. (b) Elution pattern of sNTD by size-exclusion chromatography. Elution peaks of Aldolase (158 kDa), Ovalbumine (44kDa) and RNase (14kDa) are shown as markers of retention times. (c) sNTD was purified to a single band in an SDS-PAGE gel. Supplementary Data Set 1 provides uncropped form of the SDS-PAGE gel.
Solution structure of sNTD
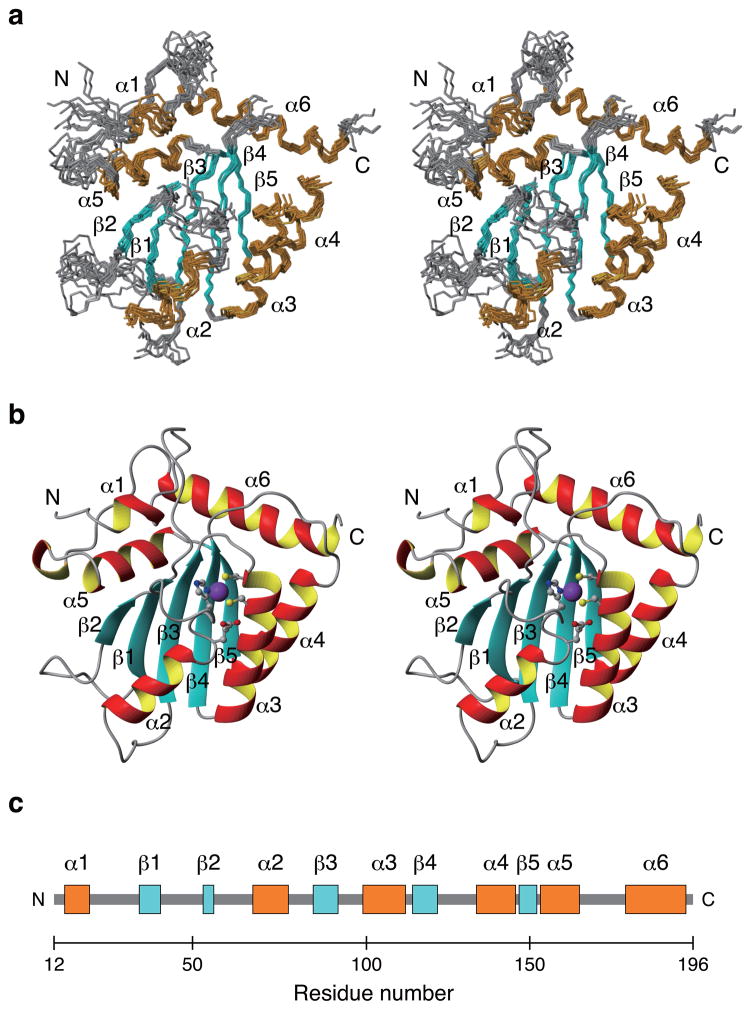
We generated a series of 15N, 13C, and/or 2H isotope-labeled protein samples to determine the solution structure of sNTD using state-of-the-art NMR techniques combined with amino acid-specific labeling techniques (representative NMR spectra are shown in Supplementary Fig. 2)31,40. Each sample was subjected to size-exclusion chromatography for purification, and the monomeric status was evaluated by the elution profile. sNTD eluted with a major peak corresponding to a molecular weight of 17 kDa and a minor peak corresponding to 47 kDa (Fig. 1b); the theoretical molecular weight of sNTD is 21 kDa. sNTD exhibited fair quality of NMR spectra with most amide proton signals resolved in a transverse relaxation-optimized spectroscopy (TROSY)41 spectrum, although signal intensities were inhomogeneous, suggesting diverse dynamics throughout sNTD (Supplementary Fig. 3). Using conventional triple resonance NMR spectra40, we assigned backbone signals (1HN, 15N, 1Hα, 13Cα and 13C′) for 146 residues (81.1%) of sNTD. 24 residues were missing from TROSY spectra due to severe line broadening, suggesting conformational exchange. Nuclear Overhauser effect (NOE) signals and a 1H/2H-exchange experiment depicted sNTD to contain a β-sheet of five β-strands arranged in the order of β2_(54–56)-β1_(36–42)-β3_(85–92)-β4_(113–120)-β5_(148–152) with β2-β1-β3 in an anti-parallel orientation, and β3-β4-β5 in a parallel orientation (Supplementary Fig. 4). 1H, 1H-NOE patterns indicated that β2-β1 form an antiparallel β-sheet, however, only Y37 NH from β1 showed slow 2H exchange with solvent, whereas F54 NH and G56 NH from β2 exchanged completely with solvent within one hour (Supplementary Fig. 4 and data not shown). Thus, hydrogen bonds between F54 NH and V39 CO and G56 NH and Y37 CO were weaker than that between Y37 NH and G56 CO, indicating that the structure and motions of β 2 is likely to be less restrained compared to the other β strands.
The sNTD solution structure was determined with 1609 distance and 106 dihedral angle restraints (structural statistics summarized in Table 1). The structure of sNTD has six alpha-helices, including α1_(16–19), α2_(68–75), α3_(98–108), α4_(131–140), α5_(154–164), and α6_(178–194), organized around the β-strands described above (Fig. 2). Secondary structures were well defined, although α1, β2 and α2 were less converged compared to other secondary structures (Fig. 2a). The poorer convergence for β2 was indicative of weaker hydrogen bonding with β1. The loop between β1 and β2 (β1-β2 loop) was disordered because of insufficient NOE restraints. Residues located in the β1-β2 loop (E38, V39, K40, K42, G43, S45, R46) and the α2-β3 loop (Q84, E85, Y86) each showed two weak signals in TROSY spectra (Supplementary Fig. 3). Since the β1-β2 and α2-β3 loops face each other in the sNTD structure, these two loops may exchange between two conformations (Fig. 2b).
Table 1.
NMR and refinement statistics for protein structures
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 1543 |
| Intra-residue | 676 |
| Inter-residue | |
| Sequential (|i – j| = 1) | 338 |
| Medium-range (|i – j| < 4) | 183 |
| Long-range (|i – j| > 5) | 346 |
| Intermolecular | 0 |
| Hydrogen bonds | 33 (66) |
| Total dihedral angle restraints | |
| ϕ | 106 |
| ψ | 0 |
| Structure statistics | |
| Violations (mean ± s.d.) | |
| Distance constraints (Å) | 0.07 ± 0.06 |
| Dihedral angle constraints (°) | 0.90 ± 0.77 |
| Max. dihedral angle violation (°) | 3.70 |
| Max. distance constraint violation (Å) | 0.30 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.004546 ± 0.000087 |
| Bond angles (°) | 0.6737 ± 0.0076 |
| Impropers (°) | 0.4877 ± 0.0153 |
| Average pairwise r.m.s. deviation** (Å) | |
| Heavy | 1.57 ± 0.17 |
| Backbone | 0.82 ± 0.17 |
Pairwise r.m.s. deviation was calculated among ## refined structures.
Total RDCs
Qfree (%)
Q and Qfree is recommended in the case where there are many RDCs, often though there are not enough to make this statistically meaningful
Figure 2.
Solution structure of the Vif-binding domain of A3G. (a) Stereoview of the ten lower-energy calculated sNTD structures superimposed using the backbone atoms (N, Cα, and C) of all residues except those absent in TROSY spectra, including 19–24, 48–49, 55–56, 67–69, 81–86, 112–118. These residues are not shown in this figure. (b) Stereoview of a cartoon representation of the energy-minimized average structure from ten lowest energy calculated sNTD structures. A zinc ion (purple), side chain atoms of zinc coordinating residues, including H65, C97, and C100, and of a conserved glutamic acid, E67, are shown with ball-and-stick representation. (c) Schematic of sNTD secondary structure with helices and strands colored orange and blue, respectively.
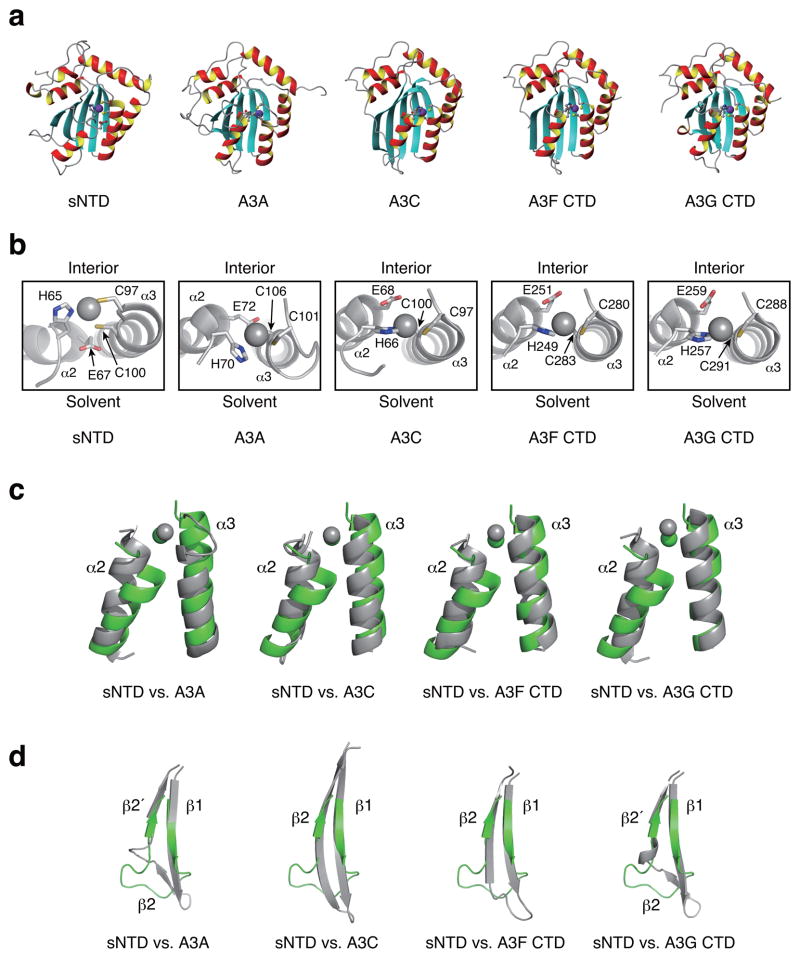
The overall structure of sNTD was similar to other A3 proteins, including A3A, A3C, A3F CTD, and A3G CTD12,31–38 (Fig. 3a). However, the sNTD structure also had several unique features. First, the start of α2 is residue 68 of sNTD, one residue after the conserved glutamic acid residue (E67), whereas in α2’s of catalytically active A3 domains the helix starts one residue prior to the conserved glutamic acid. This structural difference substantially affected the spatial positioning of E67, locating the residue too far from the Zn2+ ion to participate in catalysis (Fig. 3b). Second, the α2 N-terminus was tilted toward the C-terminus of α3, an orientation not seen in any reported A3 catalytic domain structures (Fig. 3c). These two features provided a likely explanation for prior studies that assigned catalytic activity exclusively to the C-terminal domain8,9,42. Third, sNTD lacked a bulge structure present in the β2 region of A3A and A3G CTD (Fig. 3d). The β2 strand of sNTD was shorter than in other A3s, and located in the position of β2′ of the β2-bulge-β2′ structure of A3A and A3G CTD (Fig. 3d)12,31,32,34,37.
Figure 3.
Structure comparison of sNTD with other APOBEC3 proteins. (a) Cartoon representations of structures of the energy-minimized average sNTD (this study), A3A (PDB code, 2M65)37, A3C (3VOW)38, A3F CTD (4IOU)35 and A3G CTD (3IR2)34 with zinc displayed in purple. (b) Enlarged view of the APOBEC3 zinc-binding sites from (a). The side-chains of zinc coordinating residues are rendered with ball-and-stick representation. For each structure, the top faces to the protein interior, and bottom is accessible to solvent. Zinc is shown in gray sphere. (c) Comparison of the α2 and α3 helices. α2 and α3 of sNTD (green) are superimposed to that of another APOBEC3 protein, including A3A (2M65)37, A3C (3VOW)38, A3F CTD (4IOU)35 and A3G CTD (3IR2)34. Zinc is displayed in green (sNTD) or gray (other A3s). (d) The β1-bulge-β2 region of sNTD is compared with other APOBEC3 proteins. Each image shows sNTD (green) superimposed on another A3 domain protein (gray).
We substituted 20% of wildtype residues to solubilize NTD. Two thirds of the substituted residues are located in loops, including Δ(M1-R11), Y13D, R14P, Y22N, L62D, Δ(R78), A109Q, D110P, P111T, K112H, F126A, K141A, R142G, Δ(D143-R146), R169G, E170A, L171P, E173Q, N176D and N177G, whereas the remaining substitution are in helices, including F71L, H72S, W73L and F74V in α2, T101A in α3, C139A in α4, P179D, K180E, Y181H, Y182S, I183Q, L184A, H186S, I187G and M189R in α6, with the exception of M149I in β2.
sNTD is catalytically inactive in vitro
Because our solublization strategy started with the consensus sequence of several active deaminases, we next asked if sNTD possessed catalytic activity using one-dimensional 1H NMR spectra11,12. As a positive control, catalytically active A3G CTD variant (CTD-2K3A)31 was incubated with substrate DNA (5′-ATTCCCAATT-3′)12, and a signal corresponding to the uridine H5 proton emerged within 30 minutes at 5.72 ppm (Supplementary Fig. 5d). This result indicated that CTD-2K3A had catalyzed the first deamination reaction (5′-CCC to 5′-CCU). Within 24 hours, CTD-2K3A completed the second deamination reaction, 5′-CCU to 5′-CUU, resulting two H5 proton signals, one at 5.74ppm and the other at 5.81 ppm (Supplementary Fig. 5e). By contrast, sNTD did not cause changes to the NMR spectrum of the substrate DNA even after 24 hours (Supplementary Fig. 5f). Thus, sNTD lacks detectable deaminase activity, consistent with prior studies demonstrating the catalytic inactivity of this domain8,9,42.
sNTD binds Vif
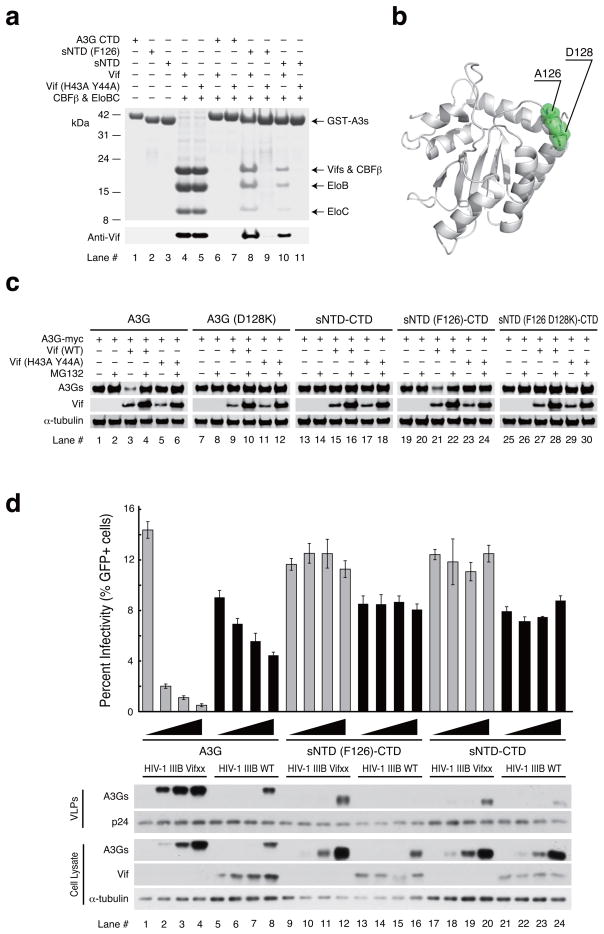
We used GST-pull down experiments to monitor interaction between sNTD and Vif-CBF-β-ELOBC (VCBC) co-expressed in E. coli. sNTD bound to the VCBC complex (Fig. 4a, lane 10). H43A and Y44A substitutions in Vif reduced binding to undetectable levels (Fig. 4a, lane 11). Since F126 of wildtype NTD had been substituted to an alanine in sNTD in order to increase solubility, we reversed this mutation and tested binding to the VCBC complex. sNTD(F126) bound more strongly to the VCBC complex than the original sNTD construct (Fig. 4a, lane 8), and this interaction became undetectable when Vif(H43A Y44A) was used instead of wildtype Vif (Fig. 4a, lane 9). The location of A126 appeared to be in the middle of the loop between β4 and α4, where it was exposed to solvent (Fig. 4b). This surface location was important because the A126F substitution was unlikely to alter either secondary or tertiary structural elements.
Figure 4.
Functional assays of sNTD. (a) E. coli co-expression and GST-pull down assay of sNTD and the Vif, CBFβ, elongin B, elongin C complex (VCBC complex). Samples are GST-fused A3G CTD (lanes 6 and 7), GST-fused sNTD(F126) (lanes 8 and 9) or GST-fused sNTD (lanes 10 and 11) expressed together with untagged CBF-β, elonginBC and wild type HIV-1 Vif or Vif(H43A Y44A). Lanes 1–5 show reference bands for the expressed protein. (b) Locations of D128 and A126 (displayed in green) in the sNTD structure. (c) Vif-mediated degradation assay using wildtype A3G (A3G) (lanes 1–6), A3G with D128K substitution [A3G(D128K)] (lanes 7–12), sNTD fused with wild type CTD (sNTD-CTD) (lanes 13–18), sNTD with A126F substitution fused with wild type CTD [sNTD(F126)-CTD] (lanes 19–24) and sNTD with A126F and D128K double-substitutions fused with wild type CTD [sNTD(F126 D128K)-CTD] (lanes 25–30). Alpha-isoform of tubulin (α-tubulin) was used as a loading control. Vif (wt) is wildtype Vif, and Vif (H43A Y44A) contains alanine substitutions of H43 and Y44. (d) Single cycle infectivity experiment comparing wildtype A3G and full-length variants with sNTD(F126)-CTD and sNTD-CTD. The histogram reports the infectivity of Vif-deficient (Vifxx) or Vif-proficient (WT) HIV-1 IIIB produced in 293T cells expressing the indicated A3G-myc construct (0, 25, 50, 100 ng) (graph shows mean values ± s. d. n = 3 independent cell cultures). The corresponding immunoblots show expression levels of relevant proteins in cell lysates (anti-myc for A3G-myc, anti-Vif, and anti-Tubulin) and virus-like particles (VLPs; anti-myc for A3G-myc and anti-p24). alpha-isoform of tubulin (α-tubulin) was used as a loading control.
F126 is important for Vif-mediated degradation
We created a construct in which sNTD was fused to wildtype CTD (A3G sNTD-CTD) and tested whether this construct behaves similarly to wildtype full-length A3G in a Vif-mediated degradation assay (Fig. 4c). Wildtype A3G was used as a positive control and, as expected, demonstrated reduced protein levels in 293T cells when co-expressed with Vif (Fig. 4c, lane 3) compared to expression alone (Fig. 4c, lane 1). The A3G protein level was restored by addition of protease inhibitor MG132 (Fig. 4c lane 4). We used a double mutation of H43A and Y44A in Vif (Fig. 4c lane 5) and D128K in A3G (Fig. 4c lane 9) as negative controls because these mutations have been shown to impair Vif-mediated degradation of A3G18,26,43–47. Interestingly, A3G sNTD-CTD protein levels were not reduced when co-expressed with Vif (Fig. 4c lane 15). Because sNTD(F126) bound to the VCBC complex more strongly than sNTD in vitro (Fig. 4a lane 8), we prepared a construct with sNTD(F126) fused to wildtype CTD [sNTD(F126)-CTD] and tested its protein stability in a Vif-mediated degradation assay. sNTD(F126)-CTD exhibited reduced protein levels in the presence of Vif (Fig. 4c, lane 21), which were restored by co-expression with Vif(H43A Y44A) (Fig. 4c, lane 23) or when cells were treated with MG132 (Fig. 4c, lane 22). Furthermore, adding the Vif-antagonizing single amino acid substitution, D128K, protected the sNTD(F126 D128K)-CTD construct from being degraded by Vif (Fig. 4c, lane 27). Prior studies have implicated the importance of F126 in Vif-binding and Vif-mediated degradation of A3G26,28,48. Thus, these data indicate that a single amino acid reversion (F126) can restore Vif-susceptibility to the A3G sNTD-CTD construct, and that the overall mechanism of Vif-mediated degradation is preserved in this construct.
sNTD lacks antiviral activity
We compared wildtype A3G, sNTD-CTD, and sNTD(F126)-CTD constructs in HIV-1 restriction experiments (Fig. 4d). As reported in previously, wildtype A3G packaged into vif-deficient HIV-1 particles and restricts viral infectivity in a dose dependent manner (Fig. 4d, lanes 1–4). Antiviral activity was strongly counteracted by Vif as evidenced by less A3G in cells and viral particles, and mostly restored viral infectivity (Fig. 4d, lanes 5–8). In contrast, sNTD-CTD resisted Vif-mediated degradation and failed to package substantially into viral particles, despite near wildtype A3G levels of cellular expression (Fig. 4d, compare lanes 17–20 and 21–24). However, as described above in co-expression studies, a single amino acid substitution A126F [(sNTD(F126)-CTD], restored the susceptibility of this construct to Vif-mediated degradation but did not restore packaging and restriction activities (Fig. 4d, compare lanes 9–12 and 13–16). These data confirmed that the sNTD(F126)-CTD construct is functional for Vif degradation but compromised for packaging and restriction. Because packaging requires the formation of a ribonucleoprotein complex with Gag [e.g. 13–17] and similar regions of A3G have been implicated in both the Vif interaction and RNA binding, the sNTD(F126) variant appeared to separate these two activities by preserving the capacity to be bound and degraded by Vif and losing the capacity to bind RNA (which also manifests as packaging and restriction deficiencies).
Vif binding to sNTD differs from A3C and A3F
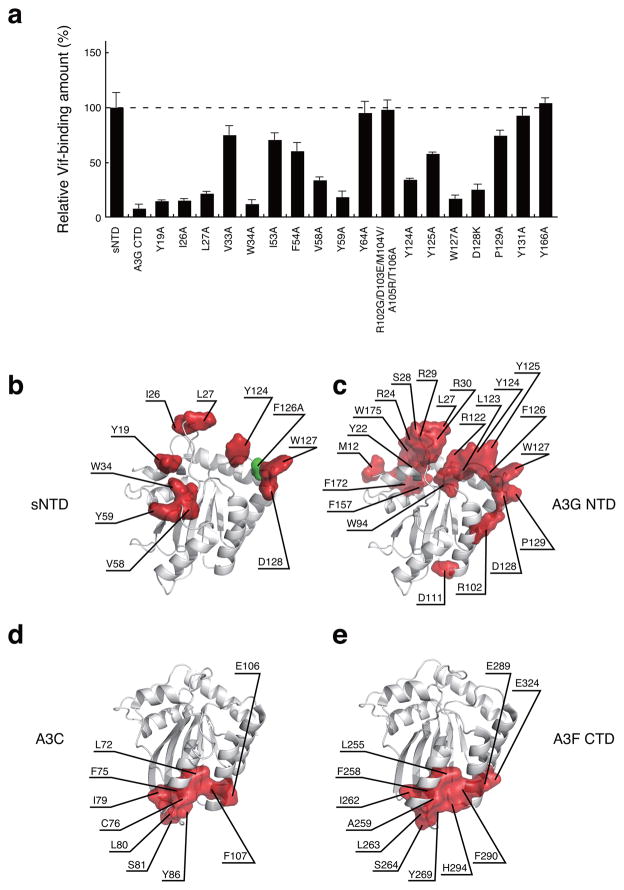
We used sNTD variants, E. coli co-expression and GST co-purification as described above to identify amino acid residues required for interaction of sNTD with the VCBC complex. Substitution Y19 at the C-terminus of α1, I26, L27 and W34 in the α1-β1 loop, V58 and Y59 of the β2-α2 loop, and Y124, W127 and D128 from the β4-α4 loop decreased binding to the VCBC complex by greater than 50% (Fig. 5a, representative SDS PAGE gel images are provided in Supplemental Fig. 5g). These residues substantially expanded the size of the A3G NTD surface area required for Vif interaction (Fig. 5b). Interestingly, using a different experimental approach, Lavens et al. implicated a similar surface area (Fig. 5c)49. Thus, the A3G NTD Vif-binding surfaces appeared to be in distinct areas from that of A3C and A3F CTD, which are formed by residues located in the α2 and α3 helices (Fig. 5d and 5e)38.
Figure 5.
Identification of Vif-binding regions of sNTD. (a) Relative Vif-binding capabilities of sNTD variants tested using E. coli co-expression and GST-pull down assay. The ratio between protein amounts of [GST-sNTD variant] and [Vif, CBF-β, elonginBC] is plotted (graph shows average values ± s. d. n = 3 independent cell cultures). Representative images of SDS-PAGE gels are provided in supplemental figure 5g (experiments were repeated three times using independent cell cultures). (b) Residues whose substitution reduced sNTD binding to the VCBC complex by greater than 50% are shown in red on the sNTD structure. The position of F126 is shown in green. (c) Residues whose substitution resulted in greater than 50% reduction of A3G Vif-binding as reported49 are shown in red. (d) and (e) highlight A3C and A3F CTD residues that upon substitution cause greater than 50% reduction in Vif-binding, respectively38.
DISCUSSION
We used sequence conservation and restorative mutagenesis to engineer a soluble variant of A3G NTD, which is 80% identical to the wildtype enzyme (Fig 1a). This variant, like wildtype A3G NTD, is not catalytically active but still binds Vif and succumbs to Vif-mediated degradation (Fig. 4 and Supplementary Fig. 5f). This engineering approach, while labor intensive, is applicable to other systems that have evaded structural characterization due to protein solubility issues.
We used NMR spectroscopy to solve the structure of sNTD, representing the first structure of an APOBEC3 non-catalytic domain and the first structure of the Vif-binding domain of A3G. Our results suggest a mechanistic basis for the lack of catalytic activity. Zn2+ is coordinated by residues H65, C97, and C100 at the protein interior (Fig. 3b) and contacts the hydrophobic side chains of L35, W90, I92, and M104 (Supplementary Fig. 6a). These interactions reduce accessibility for a putative target cytosine, as the pocket volume of the Zn2+ coordinating region is reduced compared to A3G CTD (Supplementary Fig. 6b). The active site pocket volume of A3G-CTD is about 400 Å3, while that of sNTD is approximately 100 Å3, which is smaller than the volume of a cytosine nucleobase. This result supports the general trend observed for catalytically active and inactive domains50. In addition, conserved glutamic acid E67 of sNTD is not in a position where Zn2+ coordination is possible, preventing E67 from performing a catalytic role (e. g.51, Fig. 3b and Supplementary Fig. 6a). This altered position can be attributed to changes at the N-terminus of α2 helix, where A3G NTD has a proline at residue 66 just prior to E67 that shortens this helix to begin at M68. In all catalytically active A3 domains the helix begins at A66.
To make soluble NTD we had to make four amino acid substitutions in α2, including F71L, H72S, W73L and F74V, although they are conserved mutations among APOBEC3 proteins (Supplementary Fig. 6c). Residues 71 and 74 interact with F70 (α2), W90 (β3) and F107 (α3), and F70, W90, and F107 interact with Y37 (β1), V39 (β1), and V88 (β3). Together, these residues form a hydrophobic core that stabilizes the β1-α2-β3-α3 structure (Supplementary Fig. 6d). All known A3 protein structures have the similar hydrophobic core involving corresponding residues12,31–38. Residues 72 and 73 are not involved in the hydrophobic core of known A3 structures, and we have shown that these residues can be substituted with alanine without compromising catalytic activity of A3G CTD52. Thus, substitutions at residues 71–74 are unlikely to be responsible for the unique structural features of α2 and α3 of sNTD.
The known variant of Vif(H43A Y44A)26 that disrupts wildtype A3G binding also does not permit binding to sNTD, suggesting that contacts made through H43 and Y44 are maintained between sNTD and Vif. However, sNTD fused to wildtype CTD was largely resistant to the Vif-mediated degradation. We found that A126 was responsible for this reduction, as replacement of this alanine with endogenous phenylalanine increased interaction with the VCBC complex, and restored susceptibility to Vif-mediated degradation. These results confirmed the importance of A3G F126 in Vif-binding, and, together with additional mutant data demonstrated that the sNTD construct is a good structural mimic of wildtype A3G NTD.
A caveat of sNTD function is that neither sNTD-CTD nor sNTD(F126)-CTD were able to incorporate into viral particles or restricted HIV-1 infection. Encapsidation requires interaction with RNA and Gag protein[e.g. 13,14,53–55]. The failure of sNTD variants to encapsidate is most likely due to an abrogated interaction with RNA, because no interaction between sNTD and RNA could be detected in a phosphate buffer containing 100 mM NaCl (data not shown). This loss may be due to deletion and/or substitution of basic residues, including K2, R6, R11, R14, R78, K113, K141, R142, R146, R169 and K180 in sNTD (Figure S1). Future studies may be able to use sNTD as a starting point to determine residues essential for encapsidation and RNA-binding.
The Vif-binding regions of sNTD identified by this study, including α1-β1 β2-α2 and β4-α4 loops provide a structural epitope for Vif binding and validate previous studies with wildtype A3G26,49,56,57 (Fig. 5b, 5c). The β4-α4 loop contains residues 124-YYFWD-128 that are known to be important for Vif-mediated A3G degradation26,58. Structures of the A3C and A3F Vif-binding domains have been solved, and their Vif-binding surfaces were mapped by mutagenesis, co-immunoprecipitation, and degradation assays to a different surface35,36,38,59. In these cases, Vif is proposed to bind a continuous surface located on α2 and α3 helices of A3F CTD and A3C away from the catalytic zinc (Fig. 5d and 5e). Residues F74, L80, Y86, and F107 were found to be critical for the A3C binding to Vif but, consistent with our results, not important for Vif-mediated degradation of A3G38. Previous studies showed that a negatively charged region around D128 appears important for binding to Vif (Supplementary Fig. 6b), as substitution of this amino acid abrogates binding to Vif26–29. A3G-CTD has a more positively charged (blue) profile in the corresponding area that likely interferes with binding to Vif30 (Supplementary Fig. 6b). In summary, we report the first structure of the Vif-binding domain of A3G (sNTD), which further elaborates a novel mode of Vif-binding and provides a substrate for future structural studies of the entire A3G-Vif ligase complex, as well as drug discovery.
ONLINE METHODS
Plasmids, antibodies, and reagents
DNA fragments coding the A3 consensus protein (Supplementary Fig. 1a) or wildtype A3G NTD (residues 1–196) were synthesized with codons optimized for expression in E. coli (Genscript). All DNA fragments coding A3G NTD variants were prepared with a PCR-based method using the synthesized DNAs coding the consensus protein or wildtype A3G NTD as a template. The prepared DNA fragments were inserted into NdeI/XhoI site of pCold-GST vector61. DNA fragment coding soluble A3G CTD variant (A3G CTD-2K3A that contains residues 191–384 with mutations L234K C243A F310K C321A C356A) was obtained with a PCR-based method using the previously prepared expression vector31, and was inserted into Nde I/Xho I site of pCold-GST vector61. For expression of Vif (Vif-176, residue 1–176) and CBF-β (residue 1–141) in E. Coli, codon-optimized DNA fragments coding those proteins (Genscript) were inserted into BamHI/HindIII and NdeI/XhoI sites of pRSFDuet vector (Novagen), respectively. Vif-176(H43A Y44A) expression vector was prepared in the same manner with a PCR-based method using Vif-176. For expression of elongin B (residue 1–118) and elongin C (residue 17–112) in E. coli, DNA fragments coding those proteins were inserted into BamHI/HindIII and NdeI/XhoI sites of pACYCDuet vector (Novagen), respectively. DNA fragments coding Wildtype A3G NTD (residue 1–196), CTD (residue 197–384), or sNTD (residue 1–180) were synthesized with codons optimized for expression in human cells (Genscript). D128K substitution was introduced to the A3G NTD construct with a PCR-based method. Similarly, sNTD variants, including sNTD(D128K), sNTD(F126), and sNTD(F126 D128K), were prepared with a PCR-based method using the DNA fragment encoding sNTD as a template. Each DNA fragment encoding NTD and its variants was combined with the wildtype A3G CTD using a PCR-based method to yield a full-length construct including sNTD-CTD, sNTD(F126)-CTD and sNTD(F126 D128K)-CTD. All full-length constructs were inserted into NheI/HindIII site of pcDNA3.1 myc-His(-) A vector (Invitrogen). pcDNA-HVif expression plasmid was provided by Dr. Klaus Strebel at NIH. A double mutation H43A and Y44A were introduced to pcDNA-HVif with a PCR-based method to generate pcDNA-HVif(H43A Y44A). DNA sequences of all constructs were verified by DNA sequencing. DNA oligomers for PCR and deamination assay were purchased from Integrated DNA Technologies. Mouse monoclonal anti-c-myc antibody (9E10) was purchased from Sigma-Aldrich. Mouse monoclonal anti-Vif antibody (319) and mouse monoclonal anti-α-tubulin antibody (DM1A) were purchased from Abcam. Anti-mouse IgG antibody conjugated with a fluorescent dye was purchased from Rockland Immunochemicals (Code# 610-132-121). All stable-isotope labeled reagents, including [15N]-ammonium chloride, [13C]-D-glucose, [2H, 13C]-D-glucose, deuterium oxide, [15N]-L-phenylalanine (Phe), [15N]-L-isoleucine (Ile), [15N]-L-lysine (Lys), [15N]-L-leucine (Leu), [15N]-L-arginine (Arg), [15N]-L-tyrosine (Tyr), [13C]-L-alanine (Ala), [13C]-Phe, [13C]-Ile, [13C]-Lys, [13C]-Leu, [13C]-L-methyonine (Met), [13C]-Arg, [13C]-L-threonine (Thr), [13C]-Tyr, [13C]-L-valine (Val), [2H, 13C, 15N]-algal amino acid mixture, [2H]-choline o-sulfate, were purchased from Cambridge Isotope Laboratories.
Protein expression and purification
The consensus protein (Supplementary Fig. 1a), sNTD, and their variants were prepared in the similar manner described previously31. BL21 (DE3) cells (Invirogen) were transformed with a prepared pCold-GST expression vector61 based plasmid, and grown at 37°C in M9 medium containing 100 μg ml−1 of ampicillin until reaching an optical density at 600 nm of 0.6–0.8. After the addition of 0.2 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 10 μM ZnCl2, the cells were further incubated at 16°C for overnight. The bacterial cells were harvested by centrifugation and resuspended with a lysis buffer [50 mM sodium phosphate, pH7.3, 150 mM NaCl, 0.005% Tween-20, and 0.5 mM tris(2-Carboxyethyl)phosphine (TCEP)]. The lysed cells were disrupted by sonication and then centrifuged. The supernatant was applied to glutathione-immobilized resin (GE Healthcare life sciences) and the resin was washed with the lysis buffer. The proteins bound to the resin was digested by HRV 3C protease (GE Healthcare life sciences) for overnight. The digested proteins were applied to Superdex-75 column (GE Heathcare life sciences) equilibrated with a buffer (10 mM sodium phosphate, pH7.3, 100 mM NaCl, 0.005% Tween-20, 0.5 mM TCEP, and 0.01% NaN3). The fractions including the protein of interest were collected and the protein was concentrated to 100–200 μM by using Amicon Ultra (Millipore). For NMR experiments, 0.5 M choline o-sulfate, 5% deuterium oxide, and 20 μM 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) were added to the protein solution. To prepare isotope-labeled proteins, M9 medium was prepared with 60% or 85% deuterium oxide with [15N]-ammonium chloride and [13C]- or [2H, 13C]-D-glucose. When the cells were grown in 85% deuterium oxide, 1g L−1 [2H, 13C, 15N]-algal amino acid mixture was added prior to the IPTG induction to enhance the protein harvest. To prepare amino acid selectively [13C]- or [15N]-labeled proteins, an amino acid uniformly labeled with 13C or 15N was added to the medium containing 85% deuterium oxide prior to the IPTG induction. Similarly, to prepare amino acid selectively protonated proteins, an amino acid without any isotope labeling was added to the medium containing 85% deuterium oxide prior to the IPTG induction.
NMR spectroscopy
NMR spectra were measured using Bruker AV700, AV850, and AV900 spectrometers equipped with cryogenic probes at 298K (MNMR center at University of MN). Sequential assignments of the backbone 1H, 13C, and 15N resonances of sNTD were achieved by triple-resonance NMR techniques combined with amino acid selective labeling41,62–65. Assignments of side chain 1H and 13C resonances were achieved by using a combination of 1H,13C-HSQC spectra of amino acid selectively [13C]-labeled sNTD (Supplementary Fig. 2) and 1H,1H-NOESY66 spectra of amino acid selectively protonated sNTD. 1H chemical shifts were directly referenced to DSS, while 13C and 15N chemical shifts were indirectly referenced by the absolute frequency ratios (13C/1H) = 0.251449530 and (15N/1H) = 0.101329118 with internal DSS67. All NMR spectra were processed and analyzed by using NMRPipe68 and Sparky69, respectively.
Structure calculations
The solution structures of sNTD were calculated with a distance geometry-simulated annealing protocol using X-PLOR-NIH70. The structure calculations and NOE peak assignments were performed in an iterative and manual manner. For distance restraints, the lower limitation was set to 1.8 Å and the upper limitation was set to 2.5, 3.0, 3.5, 4.0, 4.5, or 5.0 Å according to the NOE cross-peak intensity. Additionally, pseudo-atom corrections were applied66. In total, 1543 distance restraints were collected from 13C- and 15N-edited NOESY spectra and a series of 1H, 1H-NOESY spectra using amino acid selectively protonated sNTD. A total of 106 backbone dihedral angles (ϕ) were estimated using TALOS software based on 13Cα, 13Cβ, and 13C′ chemical shifts71. A total of 33 hydrogen bonds were identified from an H/D-exchanging experiment and the structures calculated in the early stage. The hydrogen bond restraints, 2.3–3.3 Å for N-O and 1.3–2.3 Å for HN-O, were introduced into the late stage of calculations. Additionally, Zn2+ ion and empirical restraints between Zn2+ ion and sNTD residues H65, C97, and C100 were introduced into the late stage of the calculations12. A total of 100 structures were calculated and the 10 lower-energy structures were selected. These 10 structures were used for the further calculation of an energy-minimized average structure. The quality of obtained structures was analyzed using MOLMOL72 and PROCHECK programs73. Structural figures were generated using MOLMOL and MacPyMOL softwares (Schrödinger).
Protein harvest assay
To monitor expression level and relative solubility of A3G NTD variants, the aforementioned protein expression and purification procedures were applied. The volumes of media (400 ml), lysis buffer (25 ml), and glutathion-immobilized resin (100 μl) were strictly fixed. The resin bound to the protein was resuspended with lysis buffer to adjust the total volume to 200 μl. 5 μl of the resuspension was applied to a sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) that was stained with Coomassie brilliant blue, and quantified for measured protein band density by Image J provided by US National Institute of Health. Each variant was independently cultured three times to calculate mean and s. d.
DNA deamination assay
Synthesized DNA fragment d(5′-ATTCCCAATT-3′)12 was dissolved in a buffer (10 mM sodium phosphate, pH7.3, 100 mM NaCl, 0.005% Tween-20, and 0.5 mM TCEP) at a concentration of 100 μM. Purified A3G CTD-2K3A or sNTD was added to the DNA solution at a final concentration of 20 μM. The mixture was incubated at 25°C. One-dimensional 1H spectra were measured 0.5, 3, and 24 hours after the addition of protein.
E. coli co-expression and GST-pull down assay
BL21 (DE3) cells were transformed with a pCold-GST-NTD variant expression plasmid, pRSFDuet-Vif-176-CBF-β, and pACYCDuet-EloBC and grown at 37°C in 400 ml of Luria-Bertani medium containing 100 μg ml−1 of ampicillin, 25 μg ml−1 of kanamycin, and 30 μg ml−1 of chloramphenicol until reaching an optical density at 600 nm of 0.6–0.8. After the addition of 1 mM IPTG, the cells were further incubated at 16°C for overnight. The harvested cells were applied to the protein purification procedure described above. The proteins were collected using 100 μl volume of glutathione-immobilized resin. The resin bound to the protein was resuspended with lysis buffer to adjust the total volume to 200 μl. 5 μl of the resuspension separated by SDS-PAGE, which was stained with Coomassie brilliant blue, and quantified for protein band density. For each variant, the assay was repeated in three independent cell culture.
Degradation assay using human cells
Human embryonic kidney (HEK293T) cells in 12-well plates were co-transfected with pcDNA3.1-based A3G or sNTD expression vector (0.66 μg) and pcDNA-HVif, pcDNA-HVif(H43A Y44A), or empty pcDNA3.1 vector (1.32 μg) by using FuGENE HD (Promega). After the incubation for 24 hours, 2 μM MG132 or dimethylsulfoxide was added to the media and the cells were further incubated for 24 hours. Resulting cell lysates were separated by SDS-PAGE and immunoprobed by using mouse monoclonal anti-c-myc antibody (9E10) (Sigma-Aldrich). The protein bands were visualized and scanned with an Odyssey imaging system (LI-COR).
HIV single-cycle assays
Single-cycle infectivity experiments were conducted in triplicate as described35,74. 293T cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 0.5% pen/strep. Cells were plated 250,000 cells per well of a 6-well plate and 24 hrs later transfected (TransIT-LT1; Mirus Bio) with 1μg of previously reported Vif-proficient or Vif-deficient HIV-1 IIIB proviral expression constructs75 and 0ng, 25ng, 50ng, or 100ng of A3G-myc expression constructs. DNA quantities were equalized with empty pcDNA3.1 (Life Technologies). 48hrs post-transfection, the producer cells and the virus containing-supernatants were harvested. A portion of the virus containing-supernatants was used to determine infectivity by incubating with CEM-GFP reporter cells plated at 25,000 cells per well in a 96 well plate following an additional 48 hrs by GFP flow cytometry. The remainder of the virus containing-supernatants was centrifuged through a 20% sucrose cushion to partially purify viral particles for analysis by immunoblotting. Cell and viral pellets were lysed in 2.5X Laemmeli reducing sample buffer for immunoblotting. Expression of A3G-myc in cell and viral lysates were detected by use of an anti-c-Myc antibody (Sigma-Aldrich) and expression of Vif in cell lysates was detected by an anti-Vif antibody (NIH AIDS Reagent Program). Tubulin and p24 served as loading controls for cell and viral lysates, respectively, and detected with an anti-tubulin antibody (Covance) and an anti-p24 antibody (in-house).
Surface charge and pocket volume analysis
The sNTD NMR structure (Model 1) was used as the template to generate the A3G-NTD homology model, using PRIME (Schrödinger, LLC), including energy minimization to resolve atomic clashes. Surface electrostatics were calculated in Maestro (Schrödinger, LLC). Pocket volume calculations were performed using SiteMap (Schrödinger, LLC).
Supplementary Material
Acknowledgments
This work was supported by grants from the US National Institute of Health (AI073167 to H.M, GM091743 to R.S.H.). Salary support for T.K. was provided in part by a Toyobo Biotechnology Foundation Fellowship. The University of Minnesota Supercomputing and NMR center (NSF BIR-961477) provided NMR instrumentation. We thank C. Kojima (Institute for Protein Research, Osaka University, Japan) for the pCold vector, K. Strebel (US National Institute of Health) for the pcDNA-hVif plasmid, M. Katahira for advice on structure calculations, Y. Iwatani for advice on degradation assays, Y. Xia for advice on NMR experiments, Y. Hong for supporting preparation of expression vectors, F. Liu for advice on human cell experiments, and K. Walters for editing the manuscript.
Footnotes
ACCESSION CODES
Atomic coordinates: Protein Data Bank (PDB) code 2MZZ
NMR constraints: Biological Magnetic Resonance Bank (BMRB) number 25509
AUTHOR CONTRIBUTIONS
T.K. designed the strategy for searching soluble A3G NTD mutants and all of A3G NTD mutants, performed harvest assay, Vif-binding assay in E. coli, Vif-dependent degradation assay, and deamination assay, prepared NMR samples, determined the solution structure; E.M.L. performed single-cycle infectivity assays; M.S. optimized human cell experiments; S.S. analyzed surface charge distribution and binding pocket volumes of the NTD structure; J.Z. supported harvest assays; L.C. and M.H. supported preparation of expression plasmids and the Vif-binding assay in E. coli. T.K., E.M.L., S.S., C.A.S., R.S.H. and H. M. contributed to writing and editing the manuscript.
References
- 1.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–98. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–83. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Refsland EW, et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–84. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooms M, et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–21. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Sato K, et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10:e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–4. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 9.Navarro F, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–86. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Rathore A, et al. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J Mol Biol. 2013;425:4442–54. doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harjes S, et al. Impact of H216 on the DNA binding and catalytic activities of the HIV restriction factor APOBEC3G. J Virol. 2013;87:7008–14. doi: 10.1128/JVI.03173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. Embo J. 2009;28:440–51. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan MA, et al. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J Virol. 2007;81:5000–13. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svarovskaia ES, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–8. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 16.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–61. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douaisi M, et al. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem Biophys Res Commun. 2004;321:566–73. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Desimmie BA, et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–45. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Baig TT, Love RP, Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol. 2014;5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager S, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–5. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–9. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 22.Kao S, et al. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–7. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 25.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–13. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 26.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–10. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pery E, Rajendran KS, Brazier AJ, Gabuzda D. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83:2374–81. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G, He Z, Wang T, Xu R, Yu XF. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J Virol. 2009;83:8674–82. doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–52. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–33. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 31.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–9. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 32.Harjes E, et al. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–32. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008 doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shandilya SM, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohn MF, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–50. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siu KK, Sultana A, Azimi FC, Lee JE. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byeon IJ, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura S, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–10. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 39.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–7. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–90. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman EN, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–70. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 43.Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–4. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–3. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 46.Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A. 2004;101:3927–32. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101:5652–7. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita T, Kamada K, Hatcho K, Adachi A, Nomaguchi M. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 2008;10:1142–9. doi: 10.1016/j.micinf.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Lavens D, et al. Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic Acids Res. 2010;38:1902–12. doi: 10.1093/nar/gkp1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shandilya SM, Bohn MF, Schiffer CA. A computational analysis of the structural determinants of APOBEC3’s catalytic activity and vulnerability to HIV-1 Vif. Virology. 2014;471–473C:105–116. doi: 10.1016/j.virol.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlow DC, Short SA, Wolfenden R. Role of glutamate-104 in generating a transition state analogue inhibitor at the active site of cytidine deaminase. Biochemistry. 1996;35:948–54. doi: 10.1021/bi951498y. [DOI] [PubMed] [Google Scholar]
- 52.Chen KM, et al. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bach D, et al. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogerd HP, Cullen BR. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA. 2008;14:1228–36. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang T, et al. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–24. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L, et al. Function analysis of sequences in human APOBEC3G involved in Vif-mediated degradation. Virology. 2008;370:113–21. doi: 10.1016/j.virol.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 57.Reingewertz TH, et al. Mapping the Vif-A3G interaction using peptide arrays: a basis for anti-HIV lead peptides. Bioorg Med Chem. 2013;21:3523–32. doi: 10.1016/j.bmc.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–15. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albin JS, et al. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J Biol Chem. 2010;285:40785–92. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi K, Kojima C. pCold-GST vector: a novel cold-shock vector containing GST tag for soluble protein production. Protein Expr Purif. 2008;62:120–7. doi: 10.1016/j.pep.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–67. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 63.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–90. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A Suite of Triple Resonance NMR Experiments for the Backbone Assignment of 15N, 13C, 2H Labeled Proteins with High Sensitivity. J Am Chem Soc. 1994;116:11655–11666. [Google Scholar]
- 65.Jaipuria G, Krishnarjuna B, Mondal S, Dubey A, Atreya HS. Amino acid selective labeling and unlabeling for protein resonance assignments. Adv Exp Med Biol. 2012;992:95–118. doi: 10.1007/978-94-007-4954-2_6. [DOI] [PubMed] [Google Scholar]
- 66.Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 67.Wishart DS, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–40. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 68.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 69.Goddard TDK, DG . Sparky. Vol. 3. University of California; San Francisco: [Google Scholar]
- 70.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 71.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 72.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 73.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 74.Albin JS, Hache G, Hultquist JF, Brown WL, Harris RS. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J Virol. 2010;84:10209–19. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hache G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–24. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.