Abstract
The gene encoding PTPROt is methylated and suppressed in Chronic Lymphocytc Leukemia. PTPROt exhibits in vitro tumor suppressor characteristics through the regulation of B-cell receptor signaling. Here, we generated transgenic (Tg) mice with B-cell specific expression of PTPROt. While lymphocyte development is normal in these mice, crossing them with TCL1 Tg mouse model of CLL results in a survival advantage compared to the TCL1 Tg mice. Gene expression profiling of splenic B-lymphocytes before detectable signs of CLL followed by Ingenuity Pathway Analysis revealed that the most prominently regulated functions in TCL1 Tg vs non-transgenic (NTg) and TCL1 Tg vs PTPROt/TCL1 double Tg are the same and also biologically relevant to this study. Further, enhanced expression of the chemokine Ccl3, the oncogenic transcription factor Foxm1 and its targets in TCL1 Tg mice were significantly suppressed in the double Tg mice suggesting a protective function of PTPROt against leukemogenesis. This study also showed that PTPROt mediated regulation of Foxm1 involves activation of p53, a transcriptional repressor of Foxm1, which is facilitated through suppression of B-cell receptor signaling. These results establish the in vivo tumor suppressive function of PTPROt, and identify p53/Foxm1 axis as a key downstream effect of PTPROt-mediated suppression of BCR signaling.
INTRODUCTION
Protein tyrosine phosphatase receptor-type O (PTPRO) is a membrane anchored tyrosine phosphatase with varied functions in different tissues. It was originally cloned as glomerular epithelial protein 1 (GLEPP1) 1 with function in glomerular filtration and podocyte structure 2. This protein was also expressed at high level in the brain where it functions in axonogenesis and differentiation of neurons 3. A truncated isoform (PTPROt) identified in B-lymphoid cells was found to promote cell cycle arrest 4. A series of studies by our group and others have demonstrated its methylation and suppression in different types of cancers 5-10 and its in vitro and ex vivo growth suppressive characteristics 5, 7, 8, 11. In addition to understanding its functions, several studies including ours have identified its substrates in different cell types e.g. eph receptors in axons 12, SYK, Lyn and ZAP70 in lymphocytes 13, 14, BCR/ABL in myelogenous leukemia 11 and VCP in HCC 5. Recent studies using large number of human samples have demonstrated a prognostic function of PTPRO in breast cancer 15 and a biomarker function in esophageal squamous cell carcinoma 16. These studies have thus highlighted the physiological significance of PTPRO expression and its deregulation in diseased states.
Chronic Lymphocytic Leukemia (CLL) is the most common adult leukemia with 16,060 new cases in 2012 17. Despite advances made in treatment approaches and increase in 5-year relative survival rate over the past few decades, chronic lymphocytic leukemia (CLL) remains incurable. A role of aberrant protein tyrosine kinase activity (e.g. Lyn, SYK, ZAP70) and their downstream signaling supporting malignant proliferation and survival have been identified in CLL. Although the aberrant kinase activity is largely due to over-expression of tyrosine kinase genes, the lack of protein tyrosine phosphatase activity counterbalancing the kinase activity is also involved in the pathology of CLL. In this context, we have shown that PTPROt is significantly downregulated by transcriptional and epigenetic mechanisms in primary CLL 7 as well as in TCL1 Tg mouse model of CLL 18 relative to the respective normal B cells. Further, PTPROt plays an important role in B-cell receptor (BCR) signaling by dephosphorylating BCR signaling components Lyn kinase 14 and Syk 13. Additionally, ZAP70, a tyrosine kinase aberrantly expressed in B-CLL and predictive of worse outcome, is a bona fide substrate of PTPROt 14.
Despite all the indications of a critical role of PTPROt as a tumor suppressor in CLL, no studies have been performed to demonstrate its vivo functions in the context of CLL. Further, several mechanisms of CLL tumorigenesis have been identified based on studies conducted with human CLL samples and mouse models of CLL 19, 20. Among these mechanisms, aberrant expression of the TCL1 oncogene in CLL cells correlates with molecular subtypes and proliferation state 21. Importantly, ectopic expression of TCL1 in mouse B-lymphocytes causes a lymphoproliferative disorder on aging that mimics human CLL 22 and our previous studies have demonstrated suppression of PTPROt in this mouse model 18. These observations provide the rationale for exploring the role of PTPROt in leukemogenesis using the TCL1 Tg model of CLL and the mechanism associated with it. Here, we describe the generation of a transgenic mouse with PTPROt expression specifically in B-cells. These mice develop normally and live a normal life span. Further, they do not exhibit any defects in lymphocyte development. Crossing these mice with the TCL1 Tg mouse model of CLL alleviates the characteristics of CLL such as increased spleen weight and accumulation of leukemic CD5/CD19 cell population. Additionally, the double Tg mice exhibit an increased lifespan compared to the TCL1 Tg mice. While exploring the mechanism of this protective function of PTPROt we observed suppression of the chemokine Ccl3, a known marker of disease progression in CLL patients, and Foxm1 in the double Tg mice compared to the TCL1 Tg mice, implicating a direct role of Foxm1 in the pathogenesis of CLL. Further, we have demonstrated that inhibition of B-cell receptor signaling activates p53 to suppress FoxM1 expression in these mice.
MATERIALS AND METHODS
Antibodies
Antibodies used for Western blot: anti-HA (Covance, Princeton, NJ), anti-pY(416)Src, Src (Cell Signaling Technology, Danvers, MA), anti-pY 4G10 (Upstate), anti-pY20, anti-pY99 (Santa Cruz Biotechnology, Inc, Dallas, TX), anti-p53 (Santa Cruz Biotechnology, Inc, Dallas, TX), anti-GAPDH (Millipore, Billerica, MA), anti-Ku-70 (Neomarker, Fremont, CA). Following antibodies used for flow cytometry were from Beckton Dickinson and Company (San Jose, CA): B220, IgM, CD4, CD8, CD5, CD11b, GR-1.
Generation of PTPROt Tg mice and crossing with TCL1 Tg mice to obtain PTPROt/TCL1 double Tg mice
To express PTPROt specifically in the B-cell compartment of mice, human PTPROt cDNA was amplified using the primers 5’-CAGGATCCCAATGGTTACAGAGATGAATCC-3’ and 5’-CTACTCGAGAGCGTAATCTGGAACGTCGTATGGGTAGGACTTGCTAACATTCTCG-3’ and cloned in the BamHI and XhoI sites of pEmu plasmid (the same plasmid that was used to express TCL1 in the B-cells 23). The restriction sites are underlined and the sequence corresponding to the HA tag is depicted in bold. Microinjection of linearized transgenic construct into zygotes, implantation of microinjected zygotes into pseudo-pregnant recipient mice and weaning of founder mice was performed at the Genetically Engineered Mouse Modeling Shared Resource at The Ohio State University Comprehensive Cancer Center. Tail clips from founder mice were used for genotyping by Southern blot and PCR. Two founder lines were generated in C57BL6 background. Among these, founder line F1 had very low levels of PTPROt expression as detected by Western blot analysis whereas founder line F2 had easily detectable levels of PTPROt expression (Fig.1A). Preliminary characterization of lymphocyte development performed on the two founder lines yielded similar results (data not shown). Thus, data from the higher expressing founder line (F2) is presented here and the remaining studies have been performed with this founder. Characterization of the PTPROt Tg mice was performed in C57BL6 background. To cross them with TCL1 Tg mice they were first bred with C3H mice to obtain F1 generation in C57BL6/C3H background, the same background of TCL1 Tg mice. These mice were then crossed with homozygous TCL1 Tg mice to obtain both TCL1 Tg and PTPROt/TCL1 double Tg mice as littermates. One set of breeding was performed with hemizygous TCL1 Tg mice to obtain all four groups of mice (NTg, PTPROt Tg, TCL1 Tg and PTPROt/TCL1 double Tg) as littermates.
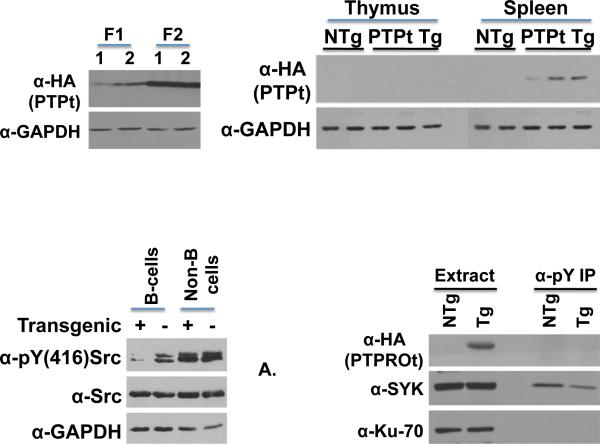
Figure 1. Characterization of PTPROt Tg mice.
Western blot analysis of whole cell extracts of total cells from the spleen of the two founders F1 and F2 (A) and from thymus and spleen of founder F2 (B) using α-HA (HA-tagged PTPROt) and α-GAPDH (normalizer). (C) Western blot analysis of whole cell extracts of CD19 sorted B-cells and non-B-cells from spleens using α-pY(416)Src (active Src/Lyn), α-Src (total Src) and GAPDH (normalizer). (D) Western blot analysis of whole cell extracts and phosphotyrosine enriched polypeptides from CD19 sorted spleen B-cells using α-HA (HA-tagged PTPROt), α-SYK (spleen tyrosine kinase) and α-Ku-70 (normalizer).
Lymphocyte development in PTPROt Tg mice
Lymph nodes, femur (for bone marrow), spleen and thymus were collected from seven week old NTg and PTPROt Tg mice (n=3). Lymphocytes were flushed out from the femur using PBS. Lymph nodes, thymus and spleen were gently crushed to release the lymphocytes. RBCs were removed from suspension of spleen cells using ficoll gradient. One million cells stained with the appropriate antibodies and respective isotype control antibodies were analyzed on FC500 flow cytometer (Beckman Coulter, Inc, Pasadena, CA).
Microarray analysis and Real-time RT-PCR
Spleen cells isolated from 4-month old NTg, TCL1 Tg and PTPROt/TCL1 double Tg mice were sorted for CD19+ cells using CD19 microbeads (Miltenyi). Purity of the sorted cells assessed by flow cytometry following staining with CD5/CD19 antibodies was found to be >90%. RNA was extracted from the cells using Trizol followed by further purification. The RNA was then used for microarray analysis on Affymetrix platform (moGene1.0 ST), performed at the Nucleic Acids Shared Resources at the Ohio State University Comprehensive Cancer Center. The data was normalized using RMA method 24. The microarray data were deposited in the GEO database (GSE60582). We used analysis of variance (ANOVA) to test differentially expressed genes using SAS software (SAS, Inc; Cary, NC). Biological functions and gene networks that were altered between the different groups of mice were identified using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc, Redwood City, CA). Expression of selected genes was validated by real-time RT-PCR using PrimeTime qPCR Assays (IDT).
Cell treatments
CD19 selected spleen lymphocytes isolated from TCL1 Tg mice were maintained in RPMI 1640 (Invitrogen) supplemented with 10% FBS (Invitrogen), 2 mmol/L of L-glutamine (Invitrogen) at 37°C in a humidified incubator with 5% CO2. The cells were treated with 2.5 μM SYK inhibitor R406 (Selleckchem, Houston, TX) and 2.5 μM BTK inhibitor PCI-32765 (Sellekchem, Houston, TX) for 48 hours.
Other statistics
Data with continuous variable were analyzed using Analysis of variance (ANOVA). Mouse survival data were plotted (Kaplan-Meier survival curves) and analyzed using log-rank test. SAS software was used for data analysis.
RESULTS
Generation and preliminary characterization of PTPROt Tg mice
To evaluate the physiological significance of PTPROt in the context of CLL, we generated transgenic mice expressing HA-tagged PTPROt under the control of the VH promoter-Eμ enhancer, the same elements that drive TCL1 expression in TCL1 Tg mice 23. The expression of transgene was evaluated in whole cell extracts of spleen and thymus cells using antibody against HA-epitope. As anticipated, the transgene was specifically detected in the spleen but not in the thymus (Fig.1A). Next, the phosphorylation state of previously identified substrates of PTPROt (Lyn and SYK) was studied in B-cells purified from the spleen by positive selection. Immunoblotting with anti-pY(416)Src that detects activated Src/Lyn demonstrated markedly reduced phosphorylation in Tg mice compared to NTg counterpart (Fig.1B). In contrast, no alteration in phosphorylation of Src was observed in negatively selected cells (non-B-cells) used as controls (Fig.1B), reinforcing tissue-specific expression and biochemical effects in response to PTPROt overexpression. Because a specific antibody against phosphorylated SYK was unavailable, we immunoprecipitated phosho-proteins from whole cell extracts of spleen B-cells followed by immunoblotting with anti-SYK antibody to determine its phosphorylation state in the Tg mice. The data demonstrated diminished amount of SYK in the Tg sample following IP with anti-pY (Fig.1C), suggesting that it was hypophosphorylated in the spleen B-cells from Tg mice.
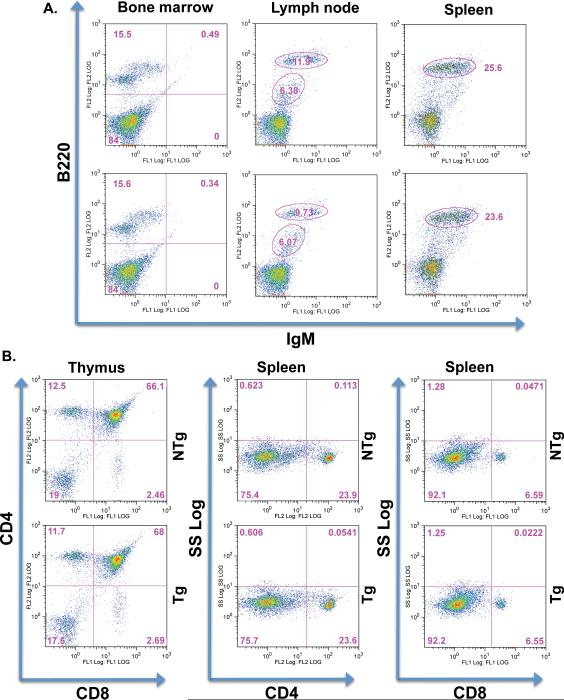
Next, flow cytometric analysis was performed on 7-week old mice to determine whether B-cell specific transgenic expression of PTPROt results in alterations of B-cell development in these mice. B220/IgM staining of cells from bone marrow, spleen and lymph nodes was used to study the pre-B to B-cell transition. No significant alterations in the B220+/IgM+ population of cells were observed in these lymphoid organs (Fig.2A). Additionally, T-cell development was also studied by flow cytometric analysis of CD4/CD8 cells in the thymus and spleen. This analysis also did not reveal any significant alterations in the CD4+/CD8+ population of cells in these lymphoid organs (Fig.2B). Similarly, both B- and T-cell development were indistinguishable in Tg and NTg mice at 16 months of age (Supplement, Fig.1). These data imply that any change in leukemic phenotype resulting from crossing PTPROt Tg mice with TCL1 Tg mice is not due to transgene effect on B-cell development.
Figure 2. Lymphocyte development in 7-week old PTPROt Tg mice.
(A) B-cell development in the bone marrow, lymph nodes and spleen studied by immunostaining for IgM and B220. (B) T-cell development in the thymus and spleen studied by immunostaining for CD4 and CD8.
Distinct gene expression profiles between spleen B-cells from PTPROt/TCL1 double Tg and TCL1 Tg mice
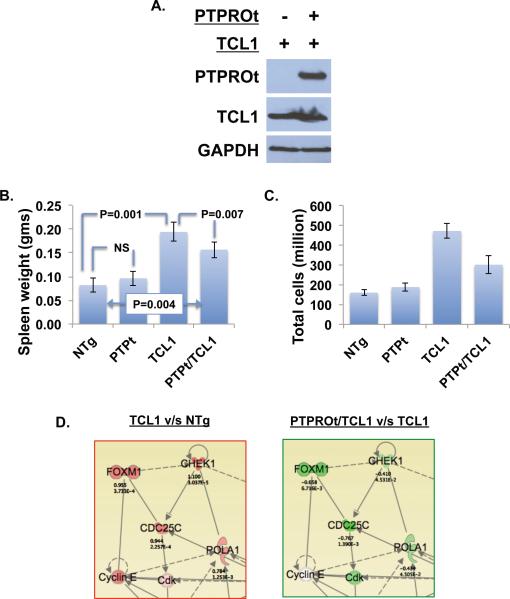
To evaluate the in vivo tumor suppressive function of PTPROt and understand the underlying mechanism, the PTPROt Tg mice were crossed to the TCL1 Tg mouse model of CLL. Western blot analysis was performed to check the expression of TCL1 and of both PTPROt and TCL1 in the spleen B-cells from TCL1 Tg and PTPROt/TCL1 double Tg mice, respectively (Fig.3A). Early changes in gene expression resulting from over-expression of PTPROt in TCL1 Tg background were assessed in 4-month old mice by microarray analysis. NTg and PTPROt Tg mice were also included in the analysis for comparison. At the time of sacrifice, a significant decrease in spleen weight (20%, P=0.007) as well as total number of cells (36%, P=0.007) was noted in the PTPROt/TCL1 double Tg group relative to the TCL1 Tg group (Fig.3B,C). It should be noted that there was no statistically significant difference in these parameters between NTg and PTPROt Tg mice. CD19+ B-cells from the spleens were used for microarray analysis on Affymetrix platform. Flow cytometry analysis confirmed >90% purity of the CD19 sorted B-cells (data not shown). We next sought to identify functions and networks associated with changes in gene expression between TCL1 Tg and NTg or PTPROt/TCL1. The Ingenuity Pathway Analysis (IPA) revealed that the most significantly regulated functions for both comparisons are the same and are also biologically relevant to these studies (Supplement, Fig.2). These observations suggest that PTPROt could reduce the tumorigenic properties of lymphocytes expressing TCL1. The gene networks with functions in Cell Cycle, DNA Replication, Recombination, Repair, and Cell Death were upregulated in TCL1 Tg mice (relative to NTg mice) but downregulated in the PTPROt/TCL1 double Tg mice (relative to TCL1 Tg mice) (Fig.3D). A functionally significant gene in this network is FoxM1, a member of the forkhead family of transcription factors that is emerging as a target for cancer diagnosis and therapies. FoxM1 has also recently been identified as a master regulator of proliferation of germinal centers 25. Validation of microarray data by real-time RTPCR indeed demonstrated increased expression of FoxM1 in the TCL1 Tg mice and relatively reduced expression in PTPROt/TCL1 double Tg mice (Fig.4A). Its upregulation in the spleen B-lymphocytes of TCL1 Tg mice could thus explain their increased proliferation 26 whereas its suppression in the double Tg mice suggests that PTPROt can reduce the proliferative potential of lymphocytes expressing TCL1. Interestingly, several other transcriptional targets of FoxM1 were also similarly upregulated in TCL1 Tg mice but not in the PTPROt/TCL1 double Tg mice relative to the NTg mice (Supplement, Fig.3 & Table 1). Among the cell cycle genes that are Foxm1 targets, Cdc20 and Cdc25c were also validated by real-time RT-PCR (Fig.4B,C). As observed with phenotypic data (Fig.3B,C), the expression of Foxm1 and Cdc20 was not significantly different between NTg and PTPROt Tg (Fig.4A,B). Although there was a small but significant increase in Cdc25c in the PTPROt Tg mice relative to the NTg mice, the increase observed in TCL1 Tg mice and the relative decrease in PTPTOt/TCL1 double Tg mice is much more dramatic and significant (Fig.4C). In addition to these cell cycle regulators, expression of Ccl3 (chemokine (C-C motif) ligand 3) was also enhanced in TCL1 Tg mice and inhibited in the double Tg mice. Expression of human CCL3, also known as Macrophage inflammatory protein-1a (MIP-1a), is induced in CLL cells by nurse-like cells through B-cell receptor signaling and, therefore, correlates with ZAP70 expression 27. Further, levels of CCL3 protein in the plasma of CLL patients correlate with risk of progression 28. Altered expression of Ccl3 in the TCL1 Tg and PTPROt/TCL1 double Tg mice was also validated by real-time RT-PCR (Fig.4D). Again, similar to Foxm1 and Cdc20, expression of Ccl3 was not significantly altered in the PTPROt Tg mice compared to the NTg mice (Fig.4D). Interestingly, CCL3 expression in CLL is enhanced in response to BCR activation 27. Since PTPROt is a negative regulator of BCR signaling 13, 14 it is logical to assume that altered expression of Ccl3 observed in the two groups of mice results from differential BCR signaling.
Figure 3. Effect of TCL1 expression on phenotype of PTPROt Tg mice.
(A) Western blot analysis of PTPROt and TCL1 in double Tg mice. (B) Spleen weight and (C) total number of spleen cells in NTg, PTPROt Tg, TCL1 Tg and double Tg mice. (D) Ingenuity pathway analysis of microarray data showing alteration in gene expression when comparing TCL1 Tg vs NTg and PTPROt/TCL1 Tg vs TCL1 Tg. Genes are represented as nodes using various shapes that represent the functional class of the encoded proteins. Solid/dash lines represent direct/indirect protein-protein interactions. Arrowheads represent activation. Red/green colors represent higher/lower expression.
Figure 4. Validation of microarray data.
Real-time RT-PCR for (A) FoxM1, (B) Cdc20, (C) Cdc25c and (D) Ccl3 in the CD19 sorted spleen B-cells of NTg, PTPROt Tg, TCL1 Tg and PTPROt/TCL1 double Tg mice. Statistical comparisons for fold change in expression were performed for PTPROt Tg, TCL1 Tg and PTPROt/TCL1 double Tg relative to NTg mice as well as between TCL1 Tg and PTPROt/TCL1 double Tg mice. Only statistically significant p-values (≤ 0.05) are depicted in the figure.
PTPROt expression in TCL1 Tg mice suppresses population of leukemic and inflammatory cells in the spleen
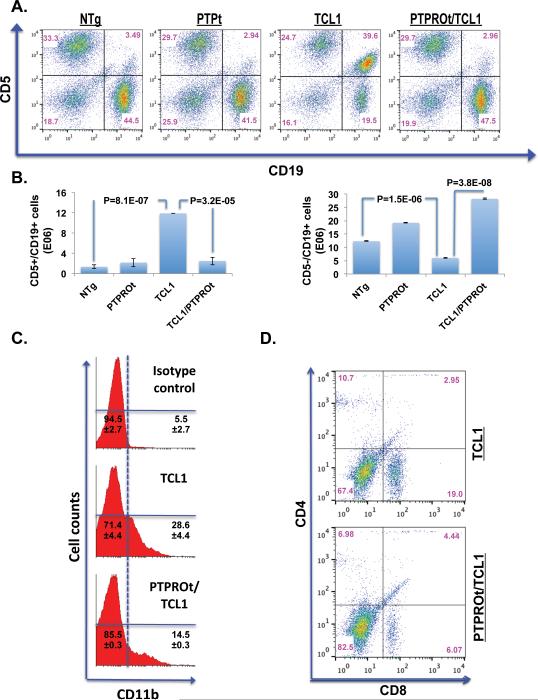
Accumulation of CD5+/CD19+ B-cells, a characteristic feature of CLL, is also observed in the TCL1 tg mice 23. To determine whether expression of PTPROt rescues this disease phenotype in double Tg mice, flow cytometry analysis was performed on spleen cells from the 8-month old mice. This time-point was selected because TCL1 Tg mice exhibit dramatic increase in the leukemic CD5+/CD19+ cell population at this time 23. The data indeed demonstrates a significant reduction in CD5+/CD19+ spleen cells in the double Tg mice relative to the TCL1 Tg mice (Fig.5A). Here again, no significant difference in the distribution of cell populations was observed between NTg and PTPROt Tg mice (Fig.5A). A plot of the absolute numbers of CD5+/CD19+ cells reflects the dramatic increase observed in TCL1 Tg mice (Fig.5B, left panel). Conversely, a plot of absolute numbers of CD5−/CD19+ cells shows a reduction of this population of cells in the TCL1 Tg mice (Fig5B, right panel). The upregulation of Ccl3, a chemo-attractant for monocytes, macrophages, dendritic cells as well as CD8+ T-cells prompted us to assess these cells in the spleens of TCL1 Tg and double Tg mice. As expected, a higher proportion of spleen cells from TCL1 Tg mice were positive for CD11b, a marker for monocytes and macrophages (Fig.5C). Similarly staining for T-cells with CD4 and CD8 showed larger population of CD8 cells in the TCL1 Tg mice compared to the double Tg mice (Fig.5D). This expansion in the population of CD8 cells in TCL1 Tg mice increased the CD8:CD4 ratio to ~2:1 from the normal 1:1 ratio observed in the PTPROt/TCL1 double Tg mice (Fig.5D) as well as in the NTg and PTPROt Tg mice (Supplement, Fig.1). These observations demonstrate that expression of PTPROt prevents infiltration of inflammatory cells into the spleen.
Figure 5. Cellular populations in the spleen of 8-month old TCL1 Tg and PTPROt/TCL1 double Tg mice.
(A) Flow cytometric analysis of spleen cells stained with CD5 and CD19 (CLL cells). Flow cytometry data for one representative sample out of three analyzed is presented. (B) Absolute numbers of CD5+/CD19+ (left panel) and CD5−/CD19+ (right panel) cells in the four groups of mice analyzed in (A). Data represents mean from 3 samples ± SD. (C) Flow cytometric analysis of spleen cells stained with CD11b (macrophages/monocytes). Flow cytometry data for one representative sample out of three analyzed is presented. Cell population percentages reflect mean of 3 samples ± SD. (D) Flow cytometric analysis of spleen cells stained with CD4 and CD8 (T-cells). Flow cytometry data for one representative sample out of three analyzed is presented.
Double Tg mice survive longer than the TCL1 Tg mice
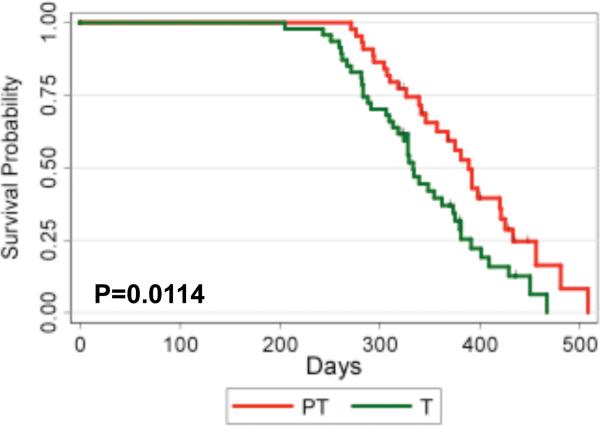
To determine whether expression of PTPROt confers a survival advantage to TCL1 Tg mice, a group of TCL1 Tg (n=47) and double Tg mice (n=44) was monitored for survival. The median survival for double Tg mice was 389 days (95% CI: 346, 421) whereas that for TCL1 Tg mice was 333 days (95% CI: 313, 373). The log-rank test revealed that the survival probabilities between the two groups were significantly different (p-value=0.0114) (Fig.6) with increased survival of the double Tg mice over that of TCL1 Tg mice. This observation reinforces the tumor suppressor characteristic of PTPROt.
Figure 6.
Kaplan Meier survival curve for TCL1 Tg (n=47) and PTPROt/TCL1 Tg (n=44) mice.
Foxm1 is regulated by p53 through the B-cell receptor (BCR) signaling pathway
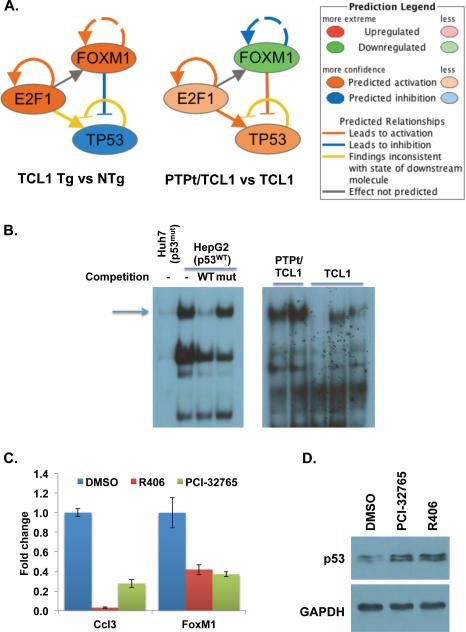
To determine the mechanism of Foxm1 upregulation in the TCL1 Tg mice and its suppression by PTPROt in the double Tg mice, we used Ingenuity Pathway Analysis (IPA) to determine the most significantly altered transcription factors in the TCL1 Tg mice relative to the NTg mice and the double Tg mice. TP53 was the top ranked transcription factor that was predicted to be inhibited in the TCL1 Tg mice in both comparisons. Conversely, TP53 was predicted as activated in the double Tg mice relative to the TCL1 Tg mice (Fig.7A). Interestingly, TP53 can negatively regulate FOXM1 through its inhibition of E2F1 29, 30. Analysis of DNA binding activity of p53 by EMSA indeed demonstrated that it was higher in the double Tg mice compared to the TCL1 Tg mice (Fig.7B). Since p53 itself is regulated by serine phosphorylation 31, it is likely that altered activity of p53 in the Tg mice was a result of signaling molecules regulated by tyrosine phosphorylation. Because PTPROt inhibits the B-cell receptor pathway by targeting Lyn kinase 14 and SYK 13 we used a clinically active inhibitor against SYK (R406) to determine whether Foxm1 expression is regulated by PTPROt through this pathway. We also tested a new inhibitor, PCI-32765/ibrutinib that targets BTK in the B-cell receptor signaling pathway and has been recently approved by FDA for use in the treatment of CLL, for its effect on Foxm1 expression. Real-time RT-PCR demonstrated that treatment of spleen B-cells isolated from TCL1 Tg mice with the inhibitors resulted in suppression of Foxm1 (Fig.7C). As expected, the expression of Ccl3 was also suppressed following inhibition of BCR signaling (Fig7C). We also observed that expression of p53 was higher in the inhibitor treated cells (Fig. 7D). These observations suggest that inhibition of BCR signaling by PTPROt induces p53 that in turn represses Foxm1.
Figure 7. Mechanism of Foxm1 regulation.
(A) IPA predicted interaction among TP53, E2F1 and FOXM1 in TCL1 Tg mice and PTPROt/TCL1 double Tg mice. (B) p53 DNA binding activity in TCL1 Tg and PTPROt/TCL1 double Tg mice. HepG2 (p53WT) and Huh7 (p53mut) were used as positive and negative controls, respectively. (C) Fold change in expression of Ccl3 and Foxm1 as measured by real-time RT-PCR in CD19+ cells isolated from TCL1 Tg mice treated with inhibitors as indicated. (D) Immunoblot analysis of p53 in treated cells. Data is normalized to GAPDH.
DISCUSSION
In this study we have investigated, for the first time, the in vivo function of PTPROt in CLL through the generation of a Tg mouse with B-cell specific expression of PTPROt. We have demonstrated that these mice develop normally and have a normal life span. Further, phosphorylation of previously identified substrates of PTPROt, Lnk and SYK kinase, was suppressed in the B-lymphocytes but not the T-lymphocytes isolated from the spleens of the Tg mice. Importantly, these mice did not exhibit any significant changes in lymphocyte development. This observation indicates that any phenotypic and pathological alterations observed in the double Tg mice obtained by crossing the PTPROt Tg mice with the TCL1 Tg mice, a mouse model of CLL, is not a result of altered lymphocyte development due to exogenous expression of PTPROt.
Assessment of the pathology of double Tg mice demonstrated a decrease in spleen weight and the total number of spleen cells compared to the TCL1 Tg mice suggesting that expression of PTPROt has a growth inhibitory effect in the background of TCL1 expression. Microarray analysis performed to explore the mechanism of PTPROt function showed that the gene expression profile of the TCL1 Tg mice exhibited oncogenic properties when compared to the non-transgenic mice and the double Tg mice. This suggests that the phenotype of the double Tg mice is similar to the NTg mice.
A striking observation is the significant upregulation of Ccl3 (macrophage inflammatory protein 1a), a marker of CLL disease progression 28, in the TCL1 Tg mice and its suppression in the double Tg mice. Consistent with its chemo-attractant function 28, 32, the population of CD11b+ and CD8+ cells was higher in the TCL1 Tg mice but lower in the double Tg mice. As a consequence, the percent of CD19+ B-cells was reduced in the TCL1 Tg mice compared to the double Tg mice. These results demonstrate that PTPROt can inhibit the infiltration of monocytes and T-lymphocytes that support the growth of CLL cells 33.
Another novel observation is the suppression of the oncogenic transcription factor Foxm1 and its targets in the double Tg mice. Foxm1, a proliferation-associated transcription factor belonging to the FOX family of transcription factor, is involved in the regulation of cell cycle genes and is an important target for cancer diagnosis and therapy. Although CLL has been traditionally known as a disease of decreased apoptosis rather than increased proliferation, it is now recognized that CLL cells proliferate and such proliferation has also been observed in the TCL1 Tg mice 26. Upregulation of FOXM1 has not been previously reported in primary human CLL probably due to the use of peripheral blood lymphocytes instead of spleen cells. It is conceivable that lymphocytes are proliferating in the spleen and become dormant once in circulation. It thus becomes important to target the expression of FOXM1 in the spleen lymphocytes to inhibit their proliferation and the increase in peripheral white blood cell counts.
Finally, the mechanism of Foxm1 regulation deserves comment. Previous studies have shown that B-cell receptor pathway regulates the expression of Ccl3 27. Since PTPROt is a negative regulator of kinases in this pathway it is conceivable that suppression of Ccl3 in the double Tg mice is caused by inhibition of BCR signaling. However, no such regulation of Foxm1 has been studied. Interestingly, IPA of microarray data showed that TP53, a negative regulator of FOXM1 29, 30, is inhibited in the TCL1 Tg mice compared to both NTg mice and the double Tg mice. To determine whether TP53, and consequently FOXM1, are also regulated by the BCR signaling pathway we used inhibitors against specific kinases in the pathway. This study indeed showed that Foxm1 expression is also under the control of BCR signaling in the B-lymphocytes. Importantly, this study has delineated a relationship between important tumor suppressors PTPROt and p53 and their role in the regulation of the oncogene Foxm1. In this context, it is noteworthy that loss of p53 associated with deletion of chromosome 17p in CLL patients is considered to cause aggressive phenotype 34, 35.
Supplementary Material
Supplement, Figure 1. Lymphocyte development in 16-month old PTPROt Tg mice. B-cell development in the bone marrow and spleen (B220 and IgM staining) and T-cell development in the spleen (CD4 and CD8 staining).
Supplement, Figure 2. Gene expression changes in TCL1 Tg mice and PTPROt/TCL1 double Tg mice. Ingenuity pathway analysis of microarray data showing changes in significantly altered biological functions when compared between TCL1 Tg vs NTg and TCL1 Tg vs PTPROt/TCL1 double Tg, p-value represents the likelihood of the function being regulated due to random chance
Supplement, Figure 3. Expression of the transcriptional targets of Foxm1 in the TCL1 Tg and PTPROt/TCL1 double Tg mice relative to the NTg mice.
ACKNOWLEDGEMENTS
We thank the Ohio State University Comprehensive Cancer Center Microarray, Analytical Cytometry, and Genetically Engineered Mouse Modeling shared resources for technical assistance. We thank Kalpana Ghoshal, Sarmila Majumder, and Jharna Datta for valuable discussions; Satavisha Roy for experimental assistance; and Alexey Efanov for help with flow cytometry. This work was supported, in part, by the NIH grant CA101956 (STJ).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interests.
SUPPLEMENTARY MATERIAL
Supplementary information is available at Leukemia's website.
REFERENCES
- 1.Wiggins RC, Wiggins JE, Goyal M, Wharram BL, Thomas PE. Molecular cloning of cDNAs encoding human GLEPP1, a membrane protein tyrosine phosphatase: characterization of the GLEPP1 protein distribution in human kidney and assignment of the GLEPP1 gene to human chromosome 12p12-p13. Genomics. 1995 May 1;27(1):174–181. doi: 10.1006/geno.1995.1021. [DOI] [PubMed] [Google Scholar]
- 2.Wharram BL, Goyal M, Gillespie PJ, Wiggins JE, Kershaw DB, Holzman LB, et al. Altered podocyte structure in GLEPP1 (Ptpro)-deficient mice associated with hypertension and low glomerular filtration rate. The Journal of clinical investigation. 2000 Nov;106(10):1281–1290. doi: 10.1172/JCI7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltran PJ, Bixby JL, Masters BA. Expression of PTPRO during mouse development suggests involvement in axonogenesis and differentiation of NT-3 and NGF-dependent neurons. The Journal of comparative neurology. 2003 Feb 17;456(4):384–395. doi: 10.1002/cne.10532. [DOI] [PubMed] [Google Scholar]
- 4.Aguiar RC, Yakushijin Y, Kharbanda S, Tiwari S, Freeman GJ, Shipp MA. PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood. 1999 Oct 1;94(7):2403–2413. [PubMed] [Google Scholar]
- 5.Hsu SH, Motiwala T, Roy S, Claus R, Mustafa M, Plass C, et al. Methylation of the PTPRO gene in human hepatocellular carcinoma and identification of VCP as its substrate. Journal of cellular biochemistry. 2013 Aug;114(8):1810–1818. doi: 10.1002/jcb.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramaswamy B, Majumder S, Roy S, Ghoshal K, Kutay H, Datta J, et al. Estrogen-mediated suppression of the gene encoding protein tyrosine phosphatase PTPRO in human breast cancer: mechanism and role in tamoxifen sensitivity. Mol Endocrinol. 2009 Feb;23(2):176–187. doi: 10.1210/me.2008-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motiwala T, Majumder S, Kutay H, Smith DS, Neuberg DS, Lucas DM, et al. Methylation and silencing of protein tyrosine phosphatase receptor type O in chronic lymphocytic leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007 Jun 1;13(11):3174–3181. doi: 10.1158/1078-0432.CCR-06-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motiwala T, Kutay H, Ghoshal K, Bai S, Seimiya H, Tsuruo T, et al. Protein tyrosine phosphatase receptor-type O (PTPRO) exhibits characteristics of a candidate tumor suppressor in human lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004 Sep 21;101(38):13844–13849. doi: 10.1073/pnas.0405451101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Motiwala T, Ghoshal K, Das A, Majumder S, Weichenhan D, Wu YZ, et al. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene. 2003 Sep 25;22(41):6319–6331. doi: 10.1038/sj.onc.1206750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob ST, Motiwala T. Epigenetic regulation of protein tyrosine phosphatases: potential molecular targets for cancer therapy. Cancer gene therapy. 2005 Aug;12(8):665–672. doi: 10.1038/sj.cgt.7700828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motiwala T, Majumder S, Ghoshal K, Kutay H, Datta J, Roy S, et al. PTPROt inactivates the oncogenic fusion protein BCR/ABL and suppresses transformation of K562 cells. The Journal of biological chemistry. 2009 Jan 2;284(1):455–464. doi: 10.1074/jbc.M802840200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Shintani T, Ihara M, Sakuta H, Takahashi H, Watakabe I, Noda M. Eph receptors are negatively controlled by protein tyrosine phosphatase r eceptor type O. Nature neuroscience. 2006 Jun;9(6):761–769. doi: 10.1038/nn1697. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Juszczynski P, Takeyama K, Aguiar RC, Shipp MA. Protein tyrosine phosphatase receptor-type O truncated (PTPROt) regulates SYK phosphorylation, proximal B-cell-receptor signaling, and cellular proliferation. Blood. 2006 Nov 15;108(10):3428–3433. doi: 10.1182/blood-2006-03-013821. [DOI] [PubMed] [Google Scholar]
- 14.Motiwala T, Datta J, Kutay H, Roy S, Jacob ST. Lyn kinase and ZAP70 are substrates of PTPROt in B-cells: Lyn inactivation by PTPROt sensitizes leukemia cells to VEGF-R inhibitor pazopanib. Journal of cellular biochemistry. 2010 Jul 1;110(4):846–856. doi: 10.1002/jcb.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YT, Li FF, Ke C, Li Z, Li ZT, Zou XF, et al. PTPRO promoter methylation is predictive of poorer outcome for HER2-positive breast cancer: indication for personalized therapy. Journal of translational medicine. 2013;11:245. doi: 10.1186/1479-5876-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You YJ, Chen YP, Zheng XX, Meltzer SJ, Zhang H. Aberrant methylation of the PTPRO gene in peripheral blood as a potential biomarker in esophageal squamous cell carcinoma patients. Cancer letters. 2012 Feb 28;315(2):138–144. doi: 10.1016/j.canlet.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.American Cancer Society . Cancer Facts and Figures. American Cancer Society; 2012. [Google Scholar]
- 18.Motiwala T, Zanesi N, Datta J, Roy S, Kutay H, Checovich AM, et al. AP-1 elements and TCL1 protein regulate expression of the gene encoding protein tyrosine phosphatase PTPROt in leukemia. Blood. 2011 Dec 1;118(23):6132–6140. doi: 10.1182/blood-2011-01-323147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pekarsky Y, Zanesi N, Croce CM. Molecular basis of CLL. Seminars in cancer biology. 2010 Dec;20(6):370–376. doi: 10.1016/j.semcancer.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanesi N, Balatti V, Bottoni A, Croce CM, Pekarsky Y. Novel insights in molecular mechanisms of CLL. Current pharmaceutical design. 2012;18(23):3363–3372. doi: 10.2174/138161212801227104. [DOI] [PubMed] [Google Scholar]
- 21.Herling M, Patel KA, Khalili J, Schlette E, Kobayashi R, Medeiros LJ, et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia. 2006 Feb;20(2):280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 22.Zanesi N, Aqeilan R, Drusco A, Kaou M, Sevignani C, Costinean S, et al. Effect of rapamycin on mouse chronic lymphocytic leukemia and the development of nonhematopoietic malignancies in Emu-TCL1 transgenic mice. Cancer research. 2006 Jan 15;66(2):915–920. doi: 10.1158/0008-5472.CAN-05-3426. [DOI] [PubMed] [Google Scholar]
- 23.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proceedings of the National Academy of Sciences of the United States of America. 2002 May 14;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003 Apr;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Molecular systems biology. 2010 Jun 8;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enzler T, Kater AP, Zhang W, Widhopf GF, 2nd, Chuang HY, Lee J, et al. Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression. Blood. 2009 Nov 12;114(20):4469–4476. doi: 10.1182/blood-2009-06-230169. [DOI] [PubMed] [Google Scholar]
- 27.Burger JA, Quiroga MP, Hartmann E, Burkle A, Wierda WG, Keating MJ, et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009 Mar 26;113(13):3050–3058. doi: 10.1182/blood-2008-07-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivina M, Hartmann E, Kipps TJ, Rassenti L, Krupnik D, Lerner S, et al. CCL3 (MIP-1alpha) plasma levels and the risk for disease progression in chronic lymphocytic leukemia. Blood. 2011 Feb 3;117(5):1662–1669. doi: 10.1182/blood-2010-09-307249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009 Dec 3;28(48):4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R, et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Molecular cancer therapeutics. 2011 Jun;10(6):1046–1058. doi: 10.1158/1535-7163.MCT-11-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends in molecular medicine. 2010 Nov;16(11):528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schall TJ, Bacon K, Camp RD, Kaspari JW, Goeddel DV. Human macrophage inflammatory protein alpha (MIP-1 alpha) and MIP-1 beta chemokines attract distinct populations of lymphocytes. The Journal of experimental medicine. 1993 Jun 1;177(6):1821–1826. doi: 10.1084/jem.177.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R, et al. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood. 2011 May 19;117(20):5463–5472. doi: 10.1182/blood-2010-12-324210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Chen G, Feng L, Zhang W, Pelicano H, Wang F, et al. Loss of p53 and altered miR15-a/16-1short right arrowMCL-1 pathway in CLL: insights from TCL1-Tg:p53(−/−) mouse model and primary human leukemia cells. Leukemia. 2014 Jan;28(1):118–128. doi: 10.1038/leu.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008 Oct 15;112(8):3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement, Figure 1. Lymphocyte development in 16-month old PTPROt Tg mice. B-cell development in the bone marrow and spleen (B220 and IgM staining) and T-cell development in the spleen (CD4 and CD8 staining).
Supplement, Figure 2. Gene expression changes in TCL1 Tg mice and PTPROt/TCL1 double Tg mice. Ingenuity pathway analysis of microarray data showing changes in significantly altered biological functions when compared between TCL1 Tg vs NTg and TCL1 Tg vs PTPROt/TCL1 double Tg, p-value represents the likelihood of the function being regulated due to random chance
Supplement, Figure 3. Expression of the transcriptional targets of Foxm1 in the TCL1 Tg and PTPROt/TCL1 double Tg mice relative to the NTg mice.