Abstract
Antibodies with protective activity are critical for vaccine efficacy. Affinity maturation increases antibody activity through multiple rounds of somatic hypermutation and selection in the germinal center. Identification of HIV-1 specific and influenza-specific antibody developmental pathways, as well as characterization of B cell and virus co-evolution in patients, has informed our understanding of antibody development. In order to counteract HIV-1 and Influenza viral diversity, broadly neutralizing antibodies precisely target specific sites of vulnerability and require high levels of affinity maturation. We present immunization strategies that attempt to recapitulate these natural processes and guide the affinity maturation process.
Keywords: B cell ontogeny, germinal center, initial recombinant, somatic hypermutation, broadly neutralizing antibodies, HIV-1, influenza, vaccine, HIV envelope glycoprotein, Influenza Hemagglutinin, immunization strategies, structure-based design, nanoparticles, viral evolution
Affinity maturation is the process by which antibodies gain increased affinity, avidity, and anti-pathogen activity and is the result of somatic hypermutation (SHM) of immunoglobulin genes in B cells, coupled to selection for antigen binding (Figure 1). This iterative process occurs in germinal centers (GCs), structures within secondary lymphoid tissues, and proceeds for weeks after acute infection or vaccination, or for many cycles during chronic infection [1]. The resulting antibodies can be highly mutated from their germline-encoded counterparts, with increases of several orders of magnitude in affinity for antigen compared to the corresponding naïve B cell receptors (BCRs)[2].
Figure 1. Overview of affinity maturation.
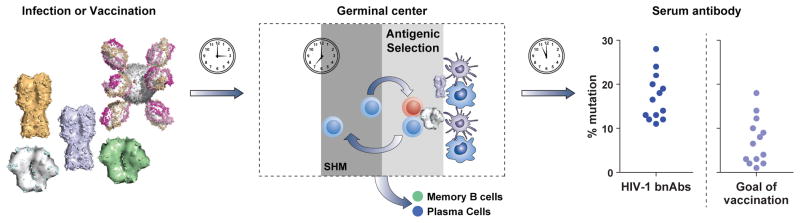
Left, Naïve or memory B cells are activated by exposure to viral antigens by infection or vaccination. Center, Activated naïve or memory B cells migrate to germinal centers within secondary lymphoid tissues such as lymph nodes [111, 112]. There, B cells cycle between a dark zone, where they undergo mutation and proliferate, and a light zone, where they undergo selection [1]. In the light zone, B cells compete for antigen on follicular dendritic cells, internalize the antigen, and present it to T follicular helper cells. The B cells with highest affinity internalize the most antigen, conferring an advantage in obtaining T cell help which in turn regulates survival, dwell time, and number of cycles of selection [105, 106]. Approximately 90% of selected cells return to the dark zone and repeat the cycle, while the remaining 10% exit to serve as memory cells or plasma cells [113]. Right, After sufficient time passes for multiple rounds of germinal center selection, the resulting antibodies may be highly mutated from their naïve precursors. While chronic infection may result in mutation levels upwards of 30% as seen in HIV-1 broadly neutralizing antibodies (bNAbs) [22], mutations of 10–20% may provide sufficient maturation to be effective [17, 18], and is more readily achieved by vaccination.
Why would affinity maturation need to be guided? In many cases, particularly for highly variable pathogens such as influenza and HIV-1, the antibodies typically elicited by vaccination or infection are poorly functional or insufficiently cross-reactive against multiple viral variants. Only a subset of antibodies that bind viral proteins can neutralize the virus, and an even smaller fraction is broadly neutralizing (cross-reactive). B cell selection is driven by affinity to the antigen that is presented in the germinal center, not by functionality that may be desirable in a vaccine context or measured in vitro, such as neutralization of heterologous viral strains [3, 4]. In many studies of HIV antibodies in which multiple variants of a neutralizing antibody lineage were identified [5–11], each lineage had members with broad cross-reactivity and others with poor activity, despite the antibodies containing similar levels of SHM. Thus, increasing SHM generated increasing functionality for some sub-lineages, but went “off track” for others (Penny Moore, personal communication and [11]) while the combined effects of broadly and poorly-neutralizing antibodies are only recently being appreciated [12]. Therefore, there is currently much discussion in the literature about how to guide affinity maturation.
On a guided journey, it is important to know where we want to go, start out headed in the right direction, not get lost along the way, and know when we have arrived at the desired endpoint. In this article, we will discuss recent findings regarding HIV-1 and influenza antibodies, new concepts for appropriate immunogen design and presentation, and strategies for priming and guiding the immune system along the maturation pathway.
Where do we want to go?
Antibodies can perform numerous antiviral functions, including neutralization of free virus, as well as Fc-requiring functions such as antibody-dependent cell-mediated cytotoxicity (ADCC). There are natural examples of differing ways to achieve potency and cross-reactivity: via a single antibody lineage that accounts for nearly all of the serum breadth and potency [9, 13], or by a collection of antibodies that collectively provide the observed breadth [14–16]. The required levels of SHM and affinity maturation may vary from target to target – for example, influenza neutralizing antibodies average 5–10% mutation from their germline genes [17, 18], while some classes of HIV-1 broadly neutralizing antibodies show mutation levels of 15–20% [6] and others show upwards of 30% mutation [13]. Even among the most highly mutated antibodies, not all of the mutations are required for full activity [18–20], and levels over 20% may be difficult to achieve by vaccination; therefore we suggest a goal of mutation levels closer to 5–20% for antibodies that target specific and multiple sites of vulnerability (Figure 1).
Starting in the right direction
The initial immune response is likely to be crucial in starting antibody lineages along the path to highly functional mature antibodies. The initial naïve B cell repertoire is highly diverse following VDJ recombination and selection against self-reactivity [2]. Naïve BCRs that target specific sites, or have certain characteristics such as utilizing a specific VH gene or displaying a long CDR H3, may be better suited than others to mature into highly functional antibodies [6, 21].
While most antibodies concentrate antigen-contacting amino acids in the CDR H3 (encoded by the VDJ junction), two groups of highly cross-reactive antibodies against influenza and HIV-1 bind primarily using the CDR H2, which is entirely encoded by the VH gene. Broadly neutralizing antibodies targeting the CD4-binding site (CD4bs) on the HIV-1 Envelope glycoprotein (Env) preferentially utilize the VH1-2*02 gene [8, 21, 22] or the VH1-46 gene [8] while those targeting the conserved influenza HA stem region utilize certain alleles of the VH1-69 gene [18, 23]. These genes contain critical binding motifs [18, 21, 23, 24] but also undergo SHM leading to increased affinity and neutralization breadth [18, 19, 21].
In addition, broadly cross-reactive antibodies that utilize the more typical CDR H3 recognition mode have been noted. In the case of HIV-1, many such antibodies display a characteristic elongated CDR H3 that is used to penetrate the extensive glycan shield found on the HIV-1 Env molecule [11, 25–27]. For influenza HA, the head region encodes significant diversity, yet a number of cross-reactive antibodies have been identified that target the receptor binding domain (RBD) within the head domain by using an aromatic residue located on the CDR H3 to mimic the HA-receptor interactions [28–31]. Importantly, antibodies with these modes of recognition have been isolated from multiple donors, indicating a common solution leading to broad neutralization.
This leads us to the hypothesis that immunogens that can bind to naive BCRs with favorable genetic properties can trigger the initial development of broadly neutralizing antibodies. In addition, there are other sites targeted by broadly neutralizing antibodies in multiple donors, such as the glycan-V3 or the membrane proximal external region of HIV-Env [9, 13, 32–36], that do not share genetic characteristics. Immunogens that bind to naive BCRs of multiple unrelated lineages may be more difficult to engineer, but would also have great potential as vaccines.
Guiding maturation
Following initial activation of B cells with preferred properties (such as BCR precursors of neutralizing antibodies or those that target known sites of vulnerability), further immunization will be required to drive the maturation process towards a refined set of broadly neutralizing antibodies. There have been a significant number of studies [18, 22, 37, 38] that have mapped the evolution of broadly neutralizing antibodies in both HIV-1 and Influenza. More recent studies have mapped the co-evolution of highly functional neutralizing antibodies and the viruses that elicit them. Applying this information to immunogen design as a way of re-eliciting these antibodies may be key to generating viable vaccine candidates. Two landmark studies of HIV-1 infection [6, 7], have followed antibody-virus co-evolution from the origin of the lineage through to maturation (Figure 2).
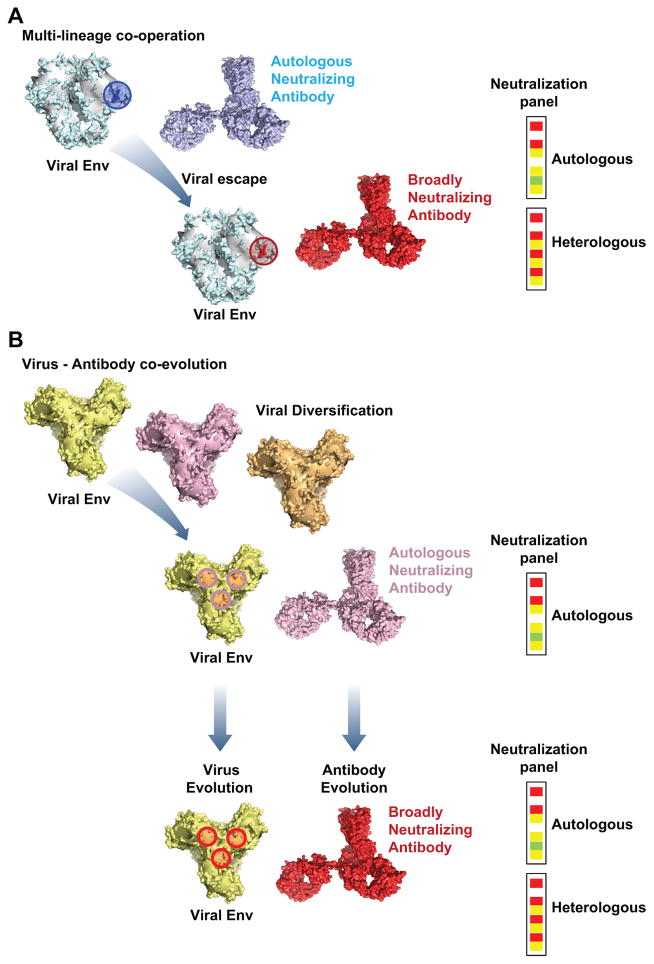
Figure 2. Development of broadly neutralizing antibodies following changes in the viral envelope HIV-1 gp140.
(A) Multi-lineage cooperation. Autologous neutralizing antibodies develop early in infection and can neutralize many autologous viruses but do not neutralize heterologous Tier 2 viral strains. Mutations that confer viral escape from these early, autologous neutralizing antibodies create the epitope for later, broadly cross-reactive antibodies of the same [6] or different [40] lineages that can neutralize both the autologous strains and also many heterologous viral strains. (B) Virus-antibody co-evolution. Viral diversification through mechanisms such as viral mutation or super-infection leads to the development of an epitope on the virus which selects for a specific B cell that can target a known site of vulnerability and gives autologous neutralization. Following selection of this initial B cells, ongoing virus evolution drives the maturation of the B cell which results in antibodies capable of heterologous neutralization increases in neutralization breadth and potency [6, 7].
In each study, mature HIV-1-neutralizing antibodies were isolated from the donor, followed by next-generation “deep” sequencing (NGS) of B cells from multiple earlier time points, and the resulting sequences were used to infer the unmutated common ancestor (UCA) and intermediate antibodies along the developmental pathway of these broadly neutralizing antibodies.
In the first study, the UCA of the CD4bs-directed CH103 lineage bound to but did not neutralize the initial transmitted/founder virus, while intermediate antibodies and later isolates bound to and neutralized the virus with increasing potency, in tandem with increases in the antibody SHM. The mature antibodies also showed neutralization activity against later viral isolates from the donor [7]. The maturation pathway included structural changes necessary to ensure focused binding that was minimally affected by viral mutation [39]. In addition, the authors identified a second lineage of antibodies that arose earlier in infection that also targets the CD4bs, and exerted selective pressure on the viral Env leading to escape mutations that improved the epitope for the CH103 lineage [40]. A similar phenomenon has been observed in studies of escape from plasma (polyclonal) neutralization [41–43], and may be a common occurrence in HIV infection.
The second study described the V1V2-directed CAP256-VRC26 lineage [6]. Like other broadly neutralizing V1V2 antibodies, the heavy chain contains a very long and anionic CDR H3 [25]. The earliest sequences of the lineage, and a UCA derived from them, already contained the long CDR H3. Thus, the signature CDR H3 loop was derived from the original VDJ recombination event, rather than increasing in size via SHM and insertions. This UCA bound and neutralized an early viral isolate, emphasizing the importance of the naive B cell in the development of functional antibodies. However, incremental increases in SHM, much of it focused on the CDR H3, did lead to incrementally improved affinity, breadth and potency.
In both lineage studies [6, 7], viral sequence diversification was immediately followed by the development of neutralization breadth. The relevance of this association is unclear; it is possible that a particular viral sequence is required to activate the ‘correct’ naïve BCR and also facilitate development of that antibody lineage, or that sampling of multiple variations on the epitope encoded by viral variants allows for selection of multiple related BCRs as well as iterative antibody refinement over time. The role of multiple viruses in the latter concept is supported by the analysis of viral escape mutations: early escape mutations in the viruses that conferred resistance to the earliest antibodies were tolerated by some of the later, more broadly cross-reactive antibodies [6]. The inference is that adaptation to the early viral escape mutations allowed for tolerance of variation within the target epitope, fortuitously conferring the ability to neutralize heterologous strains.
Immunogen choices
In addition to careful sequence selection and design, the form of the immunogen [44] can be critical (Figure 3). Effective immunogens need to present sites of vulnerability, while in some way minimizing the presence of non-neutralizing epitopes [4, 45]. Many viral immunogens undergo significant structural rearrangements; thus, structural stabilization can dramatically improve immunogenicity. This was demonstrated in a recent vaccination study against respiratory syncytial virus (RSV), in which mutations that locked the RSV fusion glycoprotein molecule in the preferred prefusion form generated ~50-fold higher levels of neutralizing antibodies compared to the postfusion form [46].
Figure 3. Structural definition of protein immunogens.
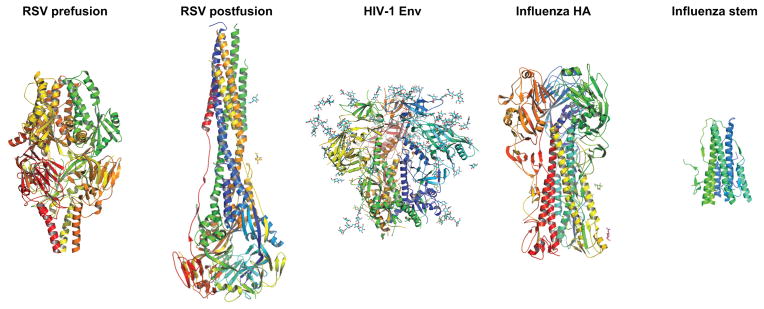
Left, Respiratory Syncytial Virus fusion glycoprotein exists in two forms: a metastable, neutralization-sensitive, prefusion form [46] (PDB ID:4MMT) and a stable, postfusion form [114] (PDB ID:3RRR). Center, HIV-1 BG505 SOSIP.664 trimer structure displays the near-native viral prefusion form [26] (PDB ID:4TVP). Right, Full-length Influenza Hemagglutinin H1 trimer [115] (PDB ID:1RUZ) and a model of the designed H1 stem immunogen [74] based on PDB ID:1RU7. All molecules are shown in ribbon representation with glycans shown in stick representation.
While HIV-1 Env gp120 molecules bear many neutralizing epitopes, they also present many non-neutralizing epitopes that are absent from functional trimers on the viral surface [47]. Most HIV Env-based vaccines tested in animals or humans have elicited antibodies that neutralize laboratory-adapted strains, but not primary isolates. These responses have been mapped to non-neutralizing epitopes, most often the Env V3 loop, which can be immunodominant on soluble Env proteins but are shielded on the native Env spike [48]. Several studies have shown that it is possible to change the focus of the immune response by mutating sequences, introducing masking glycans or deleting residues which leads to increased immune responses against previously sub-dominant epitopes [49–52]. In addition, a recent study illustrated how B cell progenitors of non-neutralizing antibodies had a competitive advantage over B cell progenitors of broadly neutralizing antibodies, explaining to a certain extent the empirical responses seen in many immunogenicity studies over the years [4].
HIV-1 immunogen design has recently experienced a number of promising advances. In the past, HIV-1 Env protein immunogens have been monomeric [53] or trimers held together with immunogenic non-HIV domains [54, 55]. The proteins presented V3 and other suboptimal epitopes, and human clinical trials all reported low neutralization levels [53, 56]. Thus, the development by Moore and colleagues [57–59] of BG505 SOSIP.664 Env trimers that closely mimic the prefusion closed form of the viral spike is a major breakthrough. These molecules bind to broadly neutralizing antibodies, including those that target recently discovered quaternary epitopes [6, 25, 60, 61], but show little or no binding to non-neutralizing antibodies such as those targeting the V3 loop [58]. Structural characterization of these trimers [26, 62, 63] can allow further design of stabilized immunogens from additional strains [57], which may translate to improved immunogenic responses. Meanwhile, careful engineering and screening has allowed identification of a number of Env gp120 molecules capable of binding to the germline versions of the VRC01-class of antibodies [4, 64, 65]. Such designs may be further improved by a new understanding of in vivo viral escape pathways, as described in studies using humanized mice infused with VRC01 [66] and also from studies of virus mutations in the donor from which VRC01 was isolated [67, 68].
In the case of Influenza, stable HA trimers have been available for many decades but the development of a universal vaccine is still awaited [69]. During typical immunization schemes, the HA head region is antigenically dominant [70, 71]. However the head region also varies the most between immunogens typically resulting in limited neutralization responses that do not result in cross-reactive antibodies. Recent work has served to provide an understanding for this limited neutralization alongside the antigenic drift of influenza while also proposing the development of preemptive vaccine strategies to improve vaccine efficacy [72, 73]. Multiple design efforts have also focused on creating a HA stem-region focused immunogen, leading to the elicitation of cross-reactive antibodies in preclinical studies [74–76].
These exciting developments in understanding the role of protein stabilization as well as immunogen selection allow the use of creative and logical strategies that are aimed at eliciting affinity matured neutralizing antibodies of either specific lineages or towards specific targets.
Proposed immunization strategies
Based on the studies described above and other related research efforts, several groups [6, 43, 64, 77, 78] have suggested the following general vaccine concepts for induction of cross-reactive antibodies. (i) Prime with modified designed viral proteins that engage the reverted germline versions of known mature antibodies [64, 65], then boost with mutants thereof that introduce glycans and/or sequence variation that are not recognized by the germline antibodies, but are neutralized by intermediate or mature antibodies [64, 65, 78]. These priming molecules could also include designs generated from the early viruses identified in donors such as NIH donor 45 [68], CHAVI donor 505 [7] or CAPRISA donor 256 [6] that show affinity to UCA or germline antibody sequences. Given the highly glycosylated nature of HIV-1 immunogens, the initial immunogen could be tailored to “open” up the area of interest by removing proximal glycans to first generate a broad response to the area of focus followed by immunizations with more “closed” immunogens that would force mutations in antibodies that are still targeting the area of interest (Figure 4A).
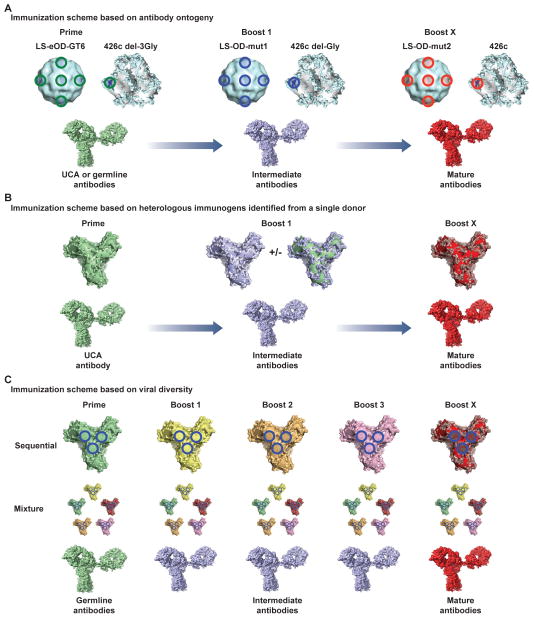
Figure 4. Immunization strategies to target specific sites and generate affinity matured cross-specific immune responses.
(A) Strategy based on antibody ontogeny as defined from mature antibody identification and subsequent deep-sequencing data [13, 22, 37]. Initial priming immunogens such as Lumazine synthase-eOD-GT6 (LS-eOD-GT6) or HIV-1 gp140 clade C strain 426cΔ3-Gly are selected and designed based on their ability to bind to specific UCA or germline antibodies in vitro [64, 65]. These priming molecules can be used to boost the immune response or modified to ensure binding to only intermediate or mature antibodies. (B) Strategy based on antibody-viral co-evolution study information. This strategy would mimic natural infection and antibody evolution where all immunogens are designed based on viral sequences identified in a donor. Given that the viral population is transient and not uniform at any given time, immunogens based on a number of sequences may be used to enable development of the desired immune response. (C) Strategy based on viral diversity. Either through a sequential or mixture type of immunization strategy, the immune system would be inundated with many epitopes of interest. The common sites of vulnerability on the viral target would inherently be the only conserved regions between the diverse molecules and thus over time would lead to a targeted and affinity matured response aimed at these sites.
(ii) Boost with variants so as to mimic the natural antibody - virus co-evolution pathway so as to recapitulate viral evolution in a single donor [6, 43, 77–79], for example as seen in the CH103 and VRC26 studies [6, 7]. The boosting immunogens would be designed based on early escape variants, and multiple later variants that escape from the immune response. These variants would bind to intermediates along the pathway to the mature antibodies, with either single variants from each time point assessed or a combination of variants [79]. A related strategy would include immunogens that bind to an earlier “helper” lineage, as defined in [40]. Using a template that is representative of antibody binding modalities that are shared among multiple donors would seem to give a higher chance of success [6, 15, 26] (Figure 4B). Early assessments of this concept by Haigwood and colleagues [79–81] have shown modest improvement over single-immunogen regimens. The strength of this approach is that the template viral Env molecules have elicited broadly neutralizing antibodies in humans with one defined path for antibody maturation already mapped out.
(iii) Heterologous immunizations of well characterized molecules in either a mixture format or via sequential immunizations over time may generate somatic hypermutation focused on specific sites. The immunogens could include variants identified from well-characterized donors, or currently circulating viral strains. An advantage over strategy (ii) is the increased viral antigenic variation compared to using sequences from a single donor (Figure 4C). Each new immunization would elicit immune responses against the whole variable antigen but the conserved sites of vulnerability inherent to the viral protein are unchanged and this allows the boosting effect to specifically target these areas [24, 82, 83]. This can be tailored to specific viral subgroups or epitopes but in fact, the empirical process may define the viral sites of vulnerability that are most easily targeted by the immune system [84–87].
Many vaccine concepts, including trivalent influenza vaccine, include multiple viral sequences; the key to making this more effective may be selection of variants with specific properties such as binding to the UCAs of a favored class of antibodies. The choice between these strategies may depend on the epitope of interest. Virus-antibody co-evolution data is only available for a few epitopes; and a recent modeling study predicted that sequential immunogens would be more effective than mixed immunogens to target the HIV-1 Env CD4bs epitope [88].
Help along the way
As the lessons of the recent studies are applied to immunogen design, it will be crucial to use vaccine platforms of sufficient immunogenicity. Duration of antigen presentation is likely to be one central factor in generating high levels of immunity. For example, a simply longer interval between influenza vaccine doses resulted in higher levels of neutralizing antibodies [89]. Studies of several cohorts of HIV-1 infected individuals found that the potency and cross-reactivity of HIV neutralizing antibodies in serum correlated with both level of viremia [90–92] and duration of infection [92]. The presence of antigen for extended periods of time is thus likely to be important in any vaccine regimen. Yet, most preclinical vaccine studies, and indeed all HIV-1 human clinical trials to date, have relied on non-replicating vectors or protein subunits. The goal of extended antigen presence may be achieved with replication-competent vectors such as adenovirus-4, or non-replicating systems that have slow release or long residency in secondary lymphoid tissue [93]. Nanoparticles [94] such as ferritin displaying viral proteins [95] are preferentially taken up by dendritic cells [96], with the added benefit of improved B cell receptor clustering and activation and increased immunogenicity [97–99].
Adjuvants are likely to be crucial in promoting affinity maturation. The licensed adjuvant, alum, likely provides a “depot effect” that increases the persistence of antigen, in addition to effects on immune cells [100]. New generations of adjuvants [101] are taking advantage of the molecular mechanisms of B cell maturation. For example, a cholera toxin-derived adjuvant increased SHM and GC size via interactions with follicular dendritic cells and complement [102, 103]. B cell factors have also been directly incorporated into vaccine constructs: chimeras with APRIL, CD40L, or BAFF increased B cell expression of activation-induced cytidine deaminase (AID), a crucial enzyme for SHM, in vitro and some led to improved antibodies in vivo [93, 104]. Augmentation of T follicular helper cell interactions [105, 106] is also likely to improve vaccine responses and needs to be further developed within vaccine regimens [107].
Determining our “arrival”
Testing vaccine concepts and choosing those likely to succeed in human clinical trials requires thoughtful measurements of immunity. Studies that carefully assess immunogen(s), adjuvants, immunization schemes, animal models and the resulting immune responses in conjunction with protection against challenge are critical. In the search for a universal influenza vaccine or a HIV-1 vaccine, the development of somatic hypermutation and broadly neutralizing or cross-reactive antibodies is a priority. Thus, sera neutralization assays and ADCC assays can be combined with assessment of SHM levels by cloning antigen-specific B cells [17, 108] or by high throughput next-generation sequencing [109, 110].
Conclusions
Based on the lessons learned from human infections, an effective immunization scheme would utilize carefully characterized and stable immunogens that preferentially contain broadly neutralizing epitopes over non-neutralizing epitopes. Initial priming immunogens would be capable of binding to unmutated ancestors of families of broadly neutralizing antibodies. It would allow stimulation of the immune system to generate a sustained response over time and should maximize appropriate T helper function. In guiding the appropriate immune response, thoughtful choices of the initial steps, the boosting immunogens, adjuvants, and duration of the regimen, coupled with well-defined and carefully measured goals, may greatly improve the chances of successful vaccination.
Highlights.
Broadly neutralizing HIV-1 and Influenza antibodies possess significant levels of affinity maturation.
Choice of initial immunogens may be critical to select naïve B cells that can mature appropriately.
Viral diversity leads to development of broadly neutralizing antibodies.
Recent HIV-1 and Influenza immunogen design breakthroughs are reviewed.
Antibody-virus co-evolution studies in HIV-1 and Influenza infections suggest immunization strategies.
Acknowledgments
We thank Rebecca M. Lynch and Evan M. Cale for careful reading of the manuscript, and Masaru Kanekiyo, Nancy S. Longo, Penny L. Moore and Tongqing Zhou for helpful discussions. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–39. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Chan TD, Brink R. Affinity-based selection and the germinal center response. Immunol Rev. 2012;247(1):11–23. doi: 10.1111/j.1600-065X.2012.01118.x. [DOI] [PubMed] [Google Scholar]
- 3.McGuire AT, et al. Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D. J Virol. 2014;88(5):2645–57. doi: 10.1128/JVI.03228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuire AT, et al. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346(6215):1380–3. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Doria-Rose NA, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. One of two landmark studies following HIV-antibody coevolution in a single donor from the time of infection through development of broad neutralizing antibodies. Naive BCR with long CDRH3 engaged initial virus; subsequent SHM led to improved potency, affinity, and breadth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–76. doi: 10.1038/nature12053. One of two landmark studies following HIV-antibody coevolution in a single donor from the time of infection through development of broad neutralizing antibodies. Virus diversification immediately preceded breadth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–7. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109(47):E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sok D, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A. 2014;111(49):17624–9. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein F, et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med. 2014;211(12):2361–72. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonsignori M, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86(8):4688–92. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 16.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Pappas L, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516(7531):418–22. doi: 10.1038/nature13764. Demonstrated that neutralizing antibodies directed to the stem region of influenza Hemagglutinin develop in a similar manner in multiple donors. Such antibodies arise from particular VH gene alleles, bear short CDHR3s, and gain activity by one of two single mutations. [DOI] [PubMed] [Google Scholar]
- 19.Zhou T, et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgiev IS, et al. Antibodies VRC01 and 10E8 neutralize HIV-1 with high breadth and potency even with Ig-framework regions substantially reverted to germline. J Immunol. 2014;192(3):1100–6. doi: 10.4049/jimmunol.1302515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West AP, Jr, et al. Structural Basis for Germline Usage of a Potent Class of HIV-1 Antibodies Targeting the CD4 Binding Site. Proc Natl Acad Sci U S A. 2012;109(30):E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–70. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittle JR, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol. 2014;88(8):4047–57. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Pancera M, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514(7523):455–61. doi: 10.1038/nature13808. Structural characterization of stable and soluble “SOSIP” HIV Env trimer molecule in complex with antibodies PGT122 and 35O22 at 3.5 Å resolution allowing definition of the gp41 and gp120 interacting regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108(34):14216–21. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489(7417):526–32. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R, et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20(3):363–70. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PS, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A. 2012;109(42):17040–5. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ofek G, et al. Structural basis for HIV-1 neutralization by 2F5-like antibodies m66 and m66.6. J Virol. 2014;88(5):2426–41. doi: 10.1128/JVI.02837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–37. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardoso RM, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22(2):163–73. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Morris L, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One. 2011;6(9):e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39(2):245–58. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doores KJ, et al. Two Classes of Broadly Neutralizing Antibodies within a Single Lineage Directed to the High-Mannose Patch of HIV Envelope. J Virol. 2015;89(2):1105–18. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fera D, et al. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc Natl Acad Sci U S A. 2014;111(28):10275–80. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Gao F, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158(3):481–91. doi: 10.1016/j.cell.2014.06.022. Demonstrated, in an HIV-infected donor, that viral mutations conferring escape from an early antibody lineage created the epitope for a second, more cross-reactive lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Moore PL, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18(11):1688–92. doi: 10.1038/nm.2985. Demonstrated, in two HIV-infected donors, that viral mutations conferring escape from early antibodies created the epitope for later, more cross-reactive antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy MK, et al. Viral escape from neutralizing antibodies in early subtype A HIV-1 infection drives an increase in autologous neutralization breadth. PLoS Pathog. 2013;9(2):e1003173. doi: 10.1371/journal.ppat.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wibmer CK, et al. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 2013;9(10):e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guttman M, et al. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce MG, et al. Outer domain of HIV-1 gp120: antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. J Virol. 2013;87(4):2294–306. doi: 10.1128/JVI.02717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342(6158):592–8. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong T, et al. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol. 2012;86(7):3574–87. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254(1):225–44. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobin GJ, et al. Deceptive imprinting and immune refocusing in vaccine design. Vaccine. 2008;26(49):6189–99. doi: 10.1016/j.vaccine.2008.09.080. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava IK, et al. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J Virol. 2003;77(4):2310–20. doi: 10.1128/JVI.77.4.2310-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeffs SA, et al. Truncated gp120 envelope glycoprotein of human immunodeficiency virus 1 elicits a broadly reactive neutralizing immune response. J Gen Virol. 2002;83(Pt 11):2723–32. doi: 10.1099/0022-1317-83-11-2723. [DOI] [PubMed] [Google Scholar]
- 52.Crooks ET, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366(2):245–62. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–44. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 54.Yang X, et al. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sliepen K, et al. Immunosilencing a highly immunogenic protein trimerization domain. J Biol Chem. 2015;290(12):7436–42. doi: 10.1074/jbc.M114.620534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitisuttithum P. HIV-1 prophylactic vaccine trials in Thailand. Curr HIV Res. 2005;3(1):17–30. doi: 10.2174/1570162052772933. [DOI] [PubMed] [Google Scholar]
- 57.Pugach P, et al. A native-like SOSIP.664 trimer based on a HIV-1 subtype B env gene. J Virol. 2015;89(6):3380–95. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. Demonstration that an engineered stable and soluble “SOSIP” HIV Env trimer is an antigenic mimic of intact Env on the viral surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ringe RP, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A. 2013;110(45):18256–61. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blattner C, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5):669–80. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515(7525):138–42. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Lyumkis D, et al. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342(6165):1484–90. doi: 10.1126/science.1245627. Structural characterization of the stable and soluble BG505 SOSIP.664 HIV Env trimer molecule in complex with VRC-PG04 using cryo-electron microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Julien JP, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342(6165):1477–83. doi: 10.1126/science.1245625. Structural characterization of stable and soluble BG505 SOSIP.664 HIV Env trimer molecule in complex with PGT122 using X-ray crystallography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Jardine J, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340(6133):711–6. doi: 10.1126/science.1234150. One of two studies describing the design of immunogens that bind V-gene reverted forms of broadly neutralizing HIV antibodies of the VRC01 class, with the goal of eliciting such antibodies. Tested ability of immunogens to engage and activate naive B cells in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.McGuire AT, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210(4):655–63. doi: 10.1084/jem.20122824. One of two studies describing the design of immunogens that bind V-gene reverted forms of broadly neutralizing HIV antibodies of the VRC01 class, with the goal of eliciting such antibodies. Tested ability of immunogens to engage and activate naive B cells in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horwitz JA, et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A. 2013;110(41):16538–43. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67*.Lynch RM, et al. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol. 2015;89(8):4201–13. doi: 10.1128/JVI.03608-14. A description of the HIV viral escape pathway in the donor of a broadly neutralizing antibody. Several escape mutants also affect other members of this class of antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu X, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol. 2012;86(10):5844–56. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 70.Caton AJ, et al. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31(2 Pt 1):417–27. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 71.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–71. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 72.Fonville JM, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346(6212):996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koel BF, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342(6161):976–9. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 74.Mallajosyula VV, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A. 2014;111(25):E2514–23. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol. 2013;3(5):521–30. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krammer F, et al. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87(12):6542–50. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Derdeyn CA, Moore PL, Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr Opin HIV AIDS. 2014;9(3):210–6. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haynes BF, et al. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature biotechnology. 2012;30(5):423–33. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79*.Malherbe DC, et al. Envelope Variants Circulating as Initial Neutralization Breadth Developed in Two HIV-Infected Subjects Stimulate Multiclade Neutralizing Antibodies in Rabbits. J Virol. 2014;88(22):12949–67. doi: 10.1128/JVI.01812-14. Early test of the hypothesis that B cells develop broad antibodies by exposure to the evolving viral envelope population. Used immunogens based on HIV sequences isolated at multiple time points during the development of neutralization breadth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malherbe DC, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85(11):5262–74. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pissani F, et al. Motif-optimized subtype A HIV envelope-based DNA vaccines rapidly elicit neutralizing antibodies when delivered sequentially. Vaccine. 2012;30(37):5519–26. doi: 10.1016/j.vaccine.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120(5):1663–73. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A. 2012;109(23):9047–52. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim JH, Excler JL, Michael NL. Lessons from the RV144 Thai Phase III HIV-1 Vaccine Trial and the Search for Correlates of Protection. Annu Rev Med. 2015;66:423–37. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 85.Zolla-Pazner S, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS One. 2014;9(2):e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–86. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fouda GG, et al. Infant HIV Type 1 gp120 Vaccination Elicits Robust and Durable Anti-V1V2 Immunoglobulin G Responses and Only Rare Envelope-Specific Immunoglobulin A Responses. J Infect Dis. 2015;211(4):508–17. doi: 10.1093/infdis/jiu444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S, Mata-Fink, Kreigsman B. Manipulating the Selection Forces during Affinity Maturation to Generate Cross-Reactive HIV Antibodies. Cell. 2015:785–97. doi: 10.1016/j.cell.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belshe RB, et al. Safety and immunogenicity of influenza A H5 subunit vaccines: effect of vaccine schedule and antigenic variant. J Infect Dis. 2011;203(5):666–73. doi: 10.1093/infdis/jiq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doria-Rose NA, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84(3):1631–6. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piantadosi A, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol. 2009;83(19):10269–74. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83(2):757–69. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwa S, et al. CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge. J Virol. 2014;88(17):9579–89. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee LA, Wang Q. Adaptations of nanoscale viruses and other protein cages for medical applications. Nanomedicine. 2006;2(3):137–49. doi: 10.1016/j.nano.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 95.Kanekiyo M, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499(7456):102–6. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiang SD, et al. Pathogen recognition and development of particulate vaccines: does size matter? Methods. 2006;40(1):1–9. doi: 10.1016/j.ymeth.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 97.Temchura VV, et al. Targeting and activation of antigen-specific B-cells by calcium phosphate nanoparticles loaded with protein antigen. Biomaterials. 2014;35(23):6098–105. doi: 10.1016/j.biomaterials.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 98.Bachmann MF, et al. The influence of antigen organization on B cell responsiveness. Science. 1993;262(5138):1448–51. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 99.Yin Z, et al. Boosting immunity to small tumor-associated carbohydrates with bacteriophage qbeta capsids. ACS Chem Biol. 2013;8(6):1253–62. doi: 10.1021/cb400060x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lambrecht BN, et al. Mechanism of action of clinically approved adjuvants. Curr Opin Immunol. 2009;21(1):23–9. doi: 10.1016/j.coi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 101.Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med. 2013;19(12):1597–608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 102.Mattsson J, et al. Complement activation and complement receptors on follicular dendritic cells are critical for the function of a targeted adjuvant. J Immunol. 2011;187(7):3641–52. doi: 10.4049/jimmunol.1101107. [DOI] [PubMed] [Google Scholar]
- 103.Bemark M, et al. A unique role of the cholera toxin A1-DD adjuvant for long-term plasma and memory B cell development. J Immunol. 2011;186(3):1399–410. doi: 10.4049/jimmunol.1002881. [DOI] [PubMed] [Google Scholar]
- 104.Melchers M, et al. Targeting HIV-1 envelope glycoprotein trimers to B cells by using APRIL improves antibody responses. J Virol. 2012;86(5):2488–500. doi: 10.1128/JVI.06259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509(7502):637–40. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kulkarni RR, et al. Activation of the RIG-I pathway during influenza vaccination enhances the germinal center reaction, promotes T follicular helper cell induction, and provides a dose-sparing effect and protective immunity. J Virol. 2014;88(24):13990–4001. doi: 10.1128/JVI.02273-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208(1):181–93. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He L, et al. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sundling C, et al. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol. 2014;192(8):3637–44. doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Apel M, Berek C. Somatic mutations in antibodies expressed by germinal centre B cells early after primary immunization. Int Immunol. 1990;2(9):813–9. doi: 10.1093/intimm/2.9.813. [DOI] [PubMed] [Google Scholar]
- 112.Jacob J, et al. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354(6352):389–92. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 113.Oprea M, Perelson AS. Somatic mutation leads to efficient affinity maturation when centrocytes recycle back to centroblasts. J Immunol. 1997;158(11):5155–62. [PubMed] [Google Scholar]
- 114.McLellan JS, et al. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85(15):7788–96. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gamblin SJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303(5665):1838–42. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]