Abstract
Histidine kinases (HKs) are major players in bacterial signaling. There has been an explosion of new HK crystal structures in the last five years. We globally analyze the structures of HKs to yield insights into the mechanisms by which signals are transmitted to and across protein structures in this family. We interpret known enzymological data in the context of new structural data to show how asymmetry across the dimer interface is a key feature of signal transduction in HKs, and discuss how different HK domains undergo asymmetric-to-symmetric transitions during signal transduction and catalysis. A thermodynamic framework for signaling that encompasses these various properties is presented and the consequences of weak thermodynamic coupling are discussed. The synthesis of observations from enzymology, structural biology, protein engineering and thermodynamics paves the way for a deeper molecular understanding of histidine kinase signal transduction.
Keywords: Histidine Kinases, Asymmetry, Signaling, Two-component systems, Bacteria
Introduction
Two-component systems represent a critical stimulus-response mechanism abundant in most bacteria. Stimulus is sensed by a histidine kinase (HK) and transmitted to a response regulator (RR), which in most cases binds to DNA and mediates a cellular response. A given bacterium can have tens to hundreds of different two-component systems (Ulrich and Zhulin, 2010) that allow it to sense and adapt to a variety of environmental signals such as osmotic changes (EnvZ/OmpR), temperature (DesK/C), small-molecules (NarX/Q, CitA/B) and antimicrobials (PhoQ/P) (Aguilar et al., 2001; Albanesi et al., 2009; Cheung and Hendrickson, 2010; Groisman, 2001; Kaspar et al., 1999; Miller et al., 1989; Rabin and Stewart, 1992; Russo and Silhavy, 1991; Tanaka et al., 1998). A molecular description of the structures and conformational dynamics of HKs is therefore fundamental to understanding bacterial signal transduction.
HKs catalyze the net transfer of a phosphoryl group from ATP to a histidine residue, and then to an aspartate of a response regulator resulting in signaling. Intriguingly, many HKs also function as phosphatases, which catalyze the hydrolytic cleavage of the phosphoryl group from the Asp residue of the response regulator. Sensory input in the form of ligand-binding or other environmental cues regulates the balance between these two opposing catalytic states.
The last five years have witnessed an explosion in high-resolution crystal structures of various domains of histidine kinases in various states (Albanesi et al., 2009; Diensthuber et al., 2013; Ferris et al., 2014, 2012; Marina et al., 2005; Mechaly et al., 2014; Wang et al., 2013). Although the structure of a full-length membrane-spanning HK has not yet been reported, low-resolution models based on disulfide cross-linking have been built (Molnar et al., 2014). Structural studies of intracellular and extracellular sensors have elucidated how these domains bind their ligands (Cheung and Hendrickson, 2010). The mechanisms by which HKs recognize their response regulators have been revealed through bioinformatics, protein engineering, and structural studies (Casino et al., 2010; Laub and Goulian, 2007; Podgornaia et al., 2013), and recently two structures of the ATP-bound Michaelis complex primed for autophosphorylation were reported (Casino et al., 2014; Mechaly et al., 2014). Despite this recent progress, many questions remain about the structural mechanism by which signals pass from the extracellular sensors to the cytoplasmic catalytic domain.
Here we compare and analyze the recent high-resolution structures of HK domains to glean insight into the structural ensembles and perturbations associated with signal transduction. Cryogenic crystal structures cannot tell us about the kinetics or molecular dynamics of signal transduction and catalysis, but they likely represent limiting states of conformational trajectories that can be used to inform various mechanistic models. Signaling can be understood in terms of thermodynamic coupling between distinct conformational states of individual protein domains. Transitions between symmetric and asymmetric states appear to be important in some domains. We explore how transitions within individual domains affect neighboring domains as signals are transmitted across the protein.
Domain architecture of histidine kinases
Histidine kinases are multi-domain proteins with considerable architectural variety across the family. The class of HKs discussed here are constitutive homodimers with extracellular sensors (Capra and Laub, 2012; Gao and Stock, 2009; Jung et al., 2012; Stock et al., 2000). Other HKs have transmembrane or intracellular sensors. CheA-linked chemo-attractant receptors are proteins that share some domains with HKs, but have distinct higher order structures yielding different biochemical properties (Hazelbauer et al., 2008).
Each subunit of a canonical HK (Fig. 1A) has an antiparallel pair of transmembrane (TM) helices near the N-terminus with the two helices separated by a periplasmic sensor domain. We use the term periplasmic (instead of “extracytoplasmic”) to refer to both Gram negative and Gram positive bacteria (Matias and Beveridge, 2006). A variety of different signal transducing elements connect the sensor/TM regions to the catalytic regions near the C-terminus of the protein.
Figure 1. Two-component system architecture.
(A) Schematic representation of a canonical TCS. Many of these domains are repeated in other protein classes. The DHp is the catalytic core of Histidine Kinases. (B) Variable domain structures from three sensor and signal transducing domains colored to highlight key features. The yellow helix (top) shows the dimeric interface. The white and black helices (bottom) show input and output helices. (C) Structures of conserved domains, DHp and catalytic, colored to highlight various features. DHp dimeric structure (top) has the N-terminal helix colored in orange and the C-terminal helix in blue. Critical regions for function are labeled and color-coded in the CA structure (bottom).
The periplasmic sensor domain is comprised of one or more diverse folds that include α/β PAS domains such as in CitA (Sevvana et al., 2008) and PhoQ (Cheung et al., 2008), or all α-helical structures like NarX (Cheung and Hendrickson, 2009) and TorS (Moore and Hendrickson, 2012) (Fig. 1B, top). The modes by which HK sensors respond to stimuli vary widely, as do the nature of the stimuli. Generally, stimulus sensing is transmitted through a key helix, which we define here as a periplasmic helix (p-helix) that connects the sensor to the transmembrane domain. Some HKs which respond to bilayer stress have only a small extracellular loop (Mascher, 2006).
The transmembrane domain in many HKs (PhoQ, CitA, LuxQ, EnvZ etc.) forms a 4-helical bundle in the membrane, with two TM helices from each monomer. In some cases like DesK and FixL each monomer has multiple helices, forming large transmembrane bundles.
Intracellular signal transducing domain(s) include a variety of domains like HAMP1, PAS2, GAF3 (Fig. 1B, bottom) and coiled-coils that reside in the cytoplasm just below the transmembrane domain. Transducing domains often exist as combinations or tandem-repeats and are generic signal transducers found in many other bacterial and mammalian proteins.
The dimerization and histidine phosphotransfer (DHp) domain is the site of the three catalytic reactions: Histidine phosphorylation, phosphotransfer to the response regulator, and, for bifunctional HKs, a phosphatase reaction (not a simple reversal of the phosphotransfer reaction). The DHp forms a homodimeric antiparallel 4-helical bundle with two helices connected by a hairpin loop (Fig. 1C, top). The catalytic histidine is located a few turns down the bundle on the solvent exposed side of helix 1. It begins a seven amino acid stretch of conserved sequence (Fig. 2A)
Residue # :1 2 3 4 5 6 7
AA sequence: H - (D/E) - (L/I) - (K/R) - (T/N) - P - L
The sidechain of the Asp or Glu (residue 2) is positioned to serve as a hydrogen bond acceptor or general acid/base while the Lys or Arg interact with acidic phosphoryl groups during phosphotransfer. The conserved (T/N)-P dipeptide (residues 5,6) is a locus for helix bending that allows the N-terminal end of helix 1 to adopt multiple conformations during catalysis (Fig. 2B). The bottom of the DHp bundle hosts the binding interface for the cognate response regulator as discussed previously (Capra and Laub, 2012; Podgornaia et al., 2013). The DHp is connected via a flexible loop to the catalytic domain.
Figure 2. Catalytic core of Histidine Kinases.
A) Surface representation of the catalytic domain (green) and the response regulator (blue) are shown on their respective docking sites on the DHp bundle (left). Structural alignment of DHp bundles from various HKs (right) show that the core of the bundle is highly symmetric and conserved while the ends of the bundle vary. B) A sequence logo for the helices of DHp domains (based on Pfam 00512) with conserved positions. An acidic residue always follows the catalytic histidine. There is a loop of variable length between the two helices. C) A schematic showing how the handedness of the loop between the two DHp helices determines auto-phosphorylation geometry in the DHp bundle.
Examination of DHp domains from distantly related proteins reveal regions of high conformational conservation as well as high variability (Fig. 2B). These regions are also mirrored in comparisons of different catalytic states in a single HK. The top third of the bundle, above the conserved proline, is the docking site for the catalytic domain during autophosphorylation. It switches between symmetric and asymmetric conformations, which appear to correlate with phosphatase and kinase states of the enzyme, respectively. In contrast, the central core of the bundle is highly symmetric and conformationally homogeneous over a large number of HKs and catalytic states. Finally, the lowest part of the bundle is highly variable in length and handedness of the interhelical loop, fitting its role of providing sequence-specific interactions for cognate response regulator recognition.
The cis vs. trans autophosphorylation propensity of different histidine kinases stems from the intrinsic handedness of the hairpin loop between the two DHp helices (Ashenberg et al., 2013). If this loop turns right (Fig. 2C, right), then the catalytic domain of one monomer is closer to the catalytic histidine of the dimeric partner, and autophosphorylation proceeds in trans. In the alternate case (Fig. 2C, left), the loop turns left and the catalytic domain is closer to its own histidine and autophosphorylation occurs in cis. Altering the handedness of the loop between the helices can interconvert cis- and trans-kinases (Ashenberg et al., 2013; Casino et al., 2014).
The catalytic domain (CA), also known as the ATP binding domain, is a highly conserved α/β sandwich with three alpha helices packed against five anti-parallel beta strands (Fig. 1C, bottom). The nucleotide binds between two helices, and is held by a loop known as the ATP lid. Conserved nucleotide binding sequences that comprise the binding site are called the N, G1, G2 and F boxes named for critical residues within these sites (Kim and Forst, 2001).
We also define a “Gripper” helix in the CA domain, which works in conjunction with a Phe in the F-box to bind to the DHp domains in different manners as the protein transitions from one catalytic activity to the next (Fig. 4). It associates with the DHp using surface-exposed sidechains arranged along one face of the helix (at positions i, i+4, i+8, i+11), which differ from one kinase to another, but tend to be primarily hydrophobic. These sidechains are arranged like four sticky fingers, while the Phe serves as thumb. The length and orientation of the Gripper helix adjusts in response to the bound nucleotide.
Figure 4. Nucleotide dependent placement of the Gripper helix.
Structural representations of the DHp and CA domains are shown in ADP- (left) and ATP-(right) bound states show different associations with the Gripper. On the inactive side, Gripper residues interact with the DHp stem to form a 3 helix bundle, with the Phe (cyan) wedged into the dimeric interface. This conformation is also seen on both monomers in the ADP-bound state. On the active side, Gripper is released to facilitate autokinase activity. This conformation is only seen in one monomer of the ATP-bound state. DHp helices are colored as before. Gripper residues are conserved across several HKs.
The response regulator (RR) is typically a separate soluble protein with mixed α/β structure and a conserved aspartate that is phosphorylated during catalysis (Bourret, 2010). In some cases, a receiver domain can be fused directly to the HK.
Catalytic properties and enzyme mechanistic investigations of HKs
Autophosphorylation involves transferring the terminal phosphate from ATP to the Nε atom of the catalytic histidine to form a high-energy N-P bond. In vitro biochemical assays show that the KM for ATP for the autokinase reaction is ~10–200 µM across various HKs. This reaction is slow, with reported kcat values in the regime of 0.1–5 min−1 (Gutu et al., 2010; Jiang et al., 2000; Trajtenberg et al., 2010; Yeo et al., 2012). The binding constant (KD) for ATP is estimated to be tight, ~10 µM, from binding-studies of the non-hydrolyzable analogue ANP-PNP (Casino et al., 2014). The KD for ADP binding is weaker; ~80–100 µM based on isothermal calorimetry and photo-affinity labeling (Yeo et al., 2012). For reference, a typical bacterial cell has an ambient ATP concentration of ~1 mM (Buckstein et al., 2008); the ADP concentration is generally constant at about one tenth this value. Given these nucleotide affinities, it follows that at least one binding site of a typical HK is saturated with ATP/ADP at any given time in a cell.
Several HKs exhibit an intrinsic asymmetry across the dimer in the autokinase reaction. Early enzymological studies of NRII showed that in the presence of physiological concentrations of ATP and ADP, the protein is predominantly phosphorylated on only one of its two identical subunits. Both subunits are phosphorylated equally only when ADP is continuously recycled back to ATP (Jiang et al., 2000). Similar half-of-sites reactivity was reported for the kinase HK853, which showed fast accumulation of the hemi-phosphorylated form when treated with excess ATP (Casino et al., 2014). These observations propose that ADP inhibits the autokinase reaction by competing with ATP.
Further characterization of nucleotide-binding kinetics is required to understand how different nucleotides influence the kinase and phosphatase states. It has been known for a long time that HK phosphatase activity is stimulated by ADP as well as by ATP and non-hydrolyzable analogs (Igo et al., 1989). Recently, Groisman and coworkers have proposed a kinetic mechanism by which ADP inhibits kinase activity in PhoQ, analogous to end-product inhibition in enzymes. While they also propose that asymmetry is important, they predict that the release of ADP from the kinase is remarkably slow (t½ ~ 30 min) and limits turnover (Yeo et al., 2012). Indeed their model predicts that a single turnover occurs on a time scale slower than that of the doubling time for E. coli. These studies, however, were not performed under a constant ADP/ATP ratio, nor corrected for the build-up of ADP during the course of the reaction. Thus, it would be of interest to directly measure the off-rate of ADP following ATP hydrolysis for PhoQ.
His-to-RR phosphotransfer involves transfer of the phosphoryl group from phospho-His to an aspartate on the bound response regulator. The affinity (KD) between a HK and its cognate RR is of the order of 1 µM. Phosphotransfer is faster than the autophosphorylation, with reported kcat values on the order of 20–100 min−1 (Fisher et al., 1996; Yeo et al., 2012). Comparative studies show that non-cognate RRs have both a lowered affinity and slower phosphotransfer kinetics, which inhibits cross-talk between different two-component circuits in vivo (Fisher et al., 1996; Laub and Goulian, 2007). Unlike the autokinase reaction, enzymatic studies of phosphotransfer do not report any asymmetry across the dimer. RRs are typically present in greater abundance their cognate HKs (Li et al., 2014). For example it was estimated that in E. coli the steady state concentration of the kinase EnvZ is ~0.1 µM whereas its RR, OmpR, exists at 3.5 µM (Russo and Silhavy, 1991). Corresponding numbers for the kinase PhoQ and its response regulator PhoP are ~0.5 µM and 2.7 µM under non-inducing conditions (Yeo et al., 2012). Thus, since a given RR is not in large molar excess over its cognate HK, only a few turnovers are required to change the fractional population of phosphorylated response regulators, a fact consistent with relatively slow kinetic rates for in vitro autophosphorylation. Furthermore since many HKs exhibit both phosphatase and kinase activity, two hurdles are crossed with one leap and a single turnover affects both the production and the consumption of the phosphorylated response regulator.
The lifetime of the phosphorylated response regulator is typically on the order of seconds to days (Bourret et al., 2010) and is controlled by both auto-dephosphorylation and intrinsic phosphatase activity from the HK. The phosphatase reaction is mechanistically distinct from the phosphotransfer step, and in some cases does not require the catalytic histidine. In cases where the catalytic histidine is required, it likely acts as a base to assist the attack of water or hydroxide on the phosphoryl-aspartyl group.
Is half-of-sites reactivity reflected in structures?
Individual domains of HKs adopt both symmetric and asymmetric conformations in different catalytic states, and there is now good reason to believe that some of these symmetry-asymmetry transitions are important for function.
Structural Analysis of Recent Structures of DHp and CA domains
The structures and symmetry of DHp and CA domains depends markedly on the catalytic act in which they have been crystallized. There are over 20 different structures of linked DHp-CA domains deposited in the PDB. In addition, there are several structures of DHp domains without the catalytic domains.
Structures with response regulator bound are believed to be in either the phosphotransfer or the phosphatase states. In both states, two RRs bind symmetrically to each dimeric DHp-CA unit and occupy the bottom half of the DHp bundle. The CA domain occupies the top half and also forms sparse contacts with the bound RR (Casino et al., 2009) (Fig. 2A). The phospho-accepting Asp group of the RR is bound to a structurally invariant and sequentially conserved region of the DHp near the catalytic His (Casino et al., 2010) and ADP nucleotides are bound to both catalytic subunits, consistent with the observation that ADP stimulates phosphatase activity. Overall, the structures with bound RRs are highly symmetric.
DHp-CA constructs crystallized in the absence of RR domains are much less symmetric (Casino et al., 2014; Diensthuber et al., 2013; Mechaly et al., 2014; Wang et al., 2013). We structurally aligned all the known structures using the middle of the DHp bundle as an anchor because this yielded the best overall alignment (see supplement for details). We then defined a coordinate system in which the dimeric axis of the DHp was coincident with the z-axis and the Cα atoms of the conserved proline were on the x-axis. Each structure was transformed into this coordinate system (see supplement for details).
Asymmetry in CA-DHp distances
We noticed marked asymmetry in our aligned ensemble. To distil a parameter that defines the structural state of each monomer, we measured the distance between the catalytic His and a conderved ATP-binding Asn in the CA domain, and constructed a correlation plot of this distance in chain A versus B (Fig. 3A). The autophosphorylation Michaelis complex is defined by a close approach between the CA and DHp domains; about 10–15 Å in the Michaelis complex versus 20–30 Å in other states, including the doubly ADP-bound states. The approach is asymmetric in that it occurs on only one side of the homodimer in all known structures. The other, inactive subunit shows an interaction more typical of the ADP-bound state. In contrast, structures representative of the phosphatase or phosphotransfer to the response regulator are much more symmetric.
Figure 3. Asymmetry in CA-DHp distances.
A) The distances between the catalytic His (on the DHp) and an ATP-binding Asn (on the CA) on each side of the dimer are correlated for all known HK structures. Structures representing the autophosphorylation Michaelis complex show significant asymmetry B) The DHp-CA distance is correlated with the angle between the Gripper helix and the DHp helix. The Gripper helix shows a very specific angular preference in in the autokinase Michaelis structures.
Gripper Helix mediates CA-DHp interactions
The Gripper helix, which is at the edge of the ATP lid near the G2 box (Fig. 4), adopts different orientations relative to the DHp in different catalytic states. The local structure of Gripper also varies with the bound nucleotide; when ATP is bound, Gripper is shortened by a turn, which allows the G2 box to form a P-loop like motif to bind the beta phosphate of ATP. Frequently, the hydrophobic residues on Gripper and/or the Phe from the F-box insert between DHp helices 1 and 2 (or 2’, depending on the bundle topology) wedging them apart.
We correlated the distance between the CA and DHp domains with the angle of the Gripper helix relative to the first helix of the DHp. For catalytically competent states the Gripper helix is close to the DHp helix (less than 15 Å) and oriented about 60 degrees from the DHp helix (Fig. 3B, red circle). This tight cluster is in direct contrast with the broad range of orientations adopted by the Gripper in the apo- and inactive states (Fig. 3B, blue circle).
Asymmetry of the DHp bundle
We see a large variation in the plasticity of the DHp bundle: the central core is highly invariant whereas the top and the bottom of the bundle vary significantly depending on the catalytic state.
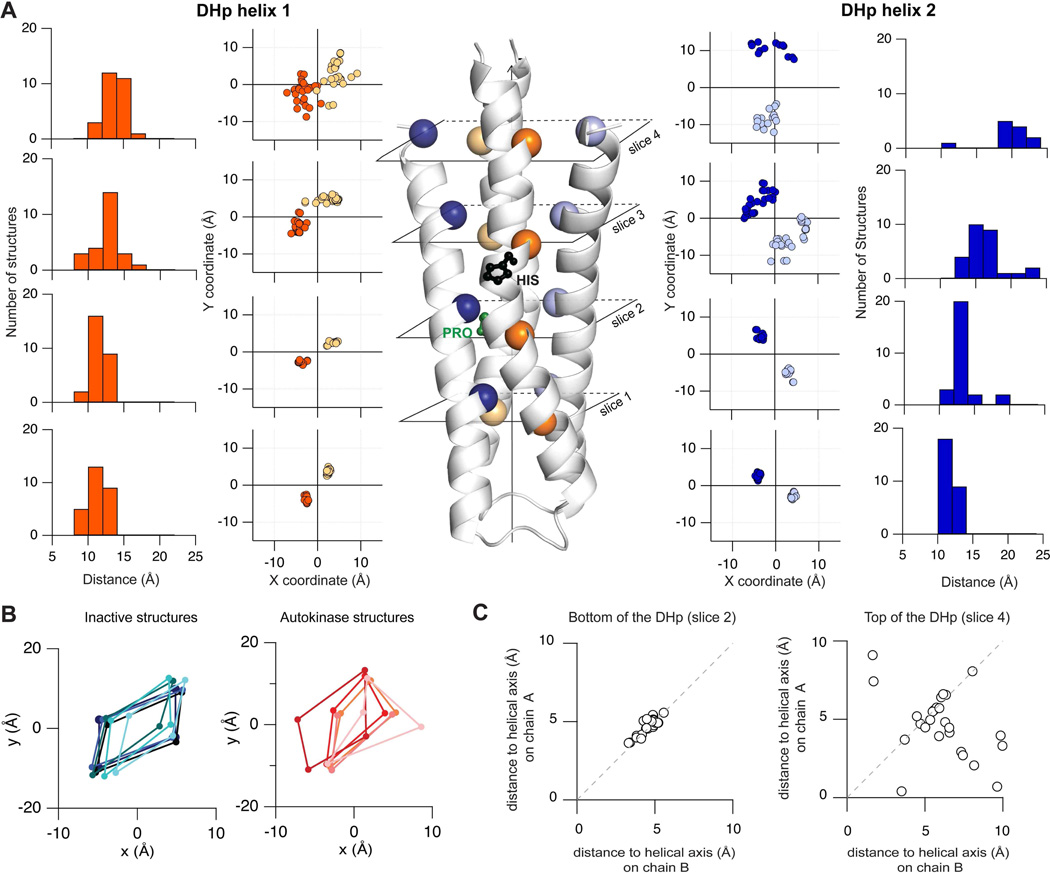
To quantify the variation we extracted Cα coordinates at four inward-facing positions along the bundle (Fig. 5A). Slice 1 corresponds to positions that had the closest distance between Cβ atoms across the dimer. At each slice, we computed the inter-monomer distance, and the distance to the bundle axis (z). In general, helix 1 is closer to its dimeric partner (8–16 Å) than helix 2 (10–25 Å). The variance of the distance distributions increased from 1.1 Å and 0.4 Å in the conserved core to 4.6 Å and 12.9 Å near the top of the bundle for helices 1 and 2 respectively. This is a remarkable change in variance across the bundle.
Figure 5. Asymmetry in the DHp bundle.
A) 23 structures of DHp domains were structurally aligned such that the z-axis coincided with the dimer axis (see supplement). X,Y coordinates extracted at 4 different slices across the bundle show that the core of the bundle is highly invariant whereas the top of the bundle varies significantly. Inter-monomer Cα-Cα distance distributions are also shown. B) Structures that represent ADP-bound, Apo and inactive states show a symmetric diamond like geometry (blue) in the XY coordinates at slice 4, whereas ATP-bound Michaelis complex structures show a distorted kite-like geometry (red). C) A third correlation plot of distances from the central helical axis to chain A vs. chain B shows that slice 2 is highly symmetric whereas slice 4 is asymmetric.
The large variance at the top of the bundle is presumably because of conformational transitions associated with docking the CA domain in different catalytic states. Projections in the x and y plane of coordinates at the top of the DHp (slice 4) reveal that in apo and ADP-bound states, the bundle forms a symmetric diamond-like shape whereas in ATP- and ATP-analogue bound structures, the bundle is more distorted (Fig. 5B). In some extreme cases like DesK, the helical crossing angles at the top of the DHp switch from being left-handed (in kinase-competent states) to right-handed (in phosphatase-competent states).
We and others (Mechaly et al., 2014; Wang et al., 2013) propose that the signal triggers catalysis by inducing an asymmetric bend in the DHp stem that promotes strong interactions between the Gripper and the DHp bundle on the inactive side of the dimer while concomitantly releasing the Gripper from the active side of the dimer. In general, the consequence of ligand-induced activation involves remodeling the energetic landscape of the DHp stem so that it preferentially populates an asymmetric structure. Thus structural asymmetry is directly mirrored in functional asymmetry (half-of-sites reactivity discussed above).
Signal transducing elements: HAMP, PAS and polar linkers
Stimulus-dependent remodeling of the DHp bundle relies on the extracellular signal reaching the top of the DHp bundle. Commonly the signal must pass through a HAMP1, PAS2 or GAF3 domain, and exit C-terminal helices that encode the output signal.
Structural topology of a HAMP domain
HAMP domains are parallel 4-helical bundles with two helices from each monomer. A highly conserved glycine marks the end of the first helix and the beginning of a loop that wraps around the structure (Fig. 6A). A conserved glutamate marks the beginning of the second HAMP helix. When viewed from the top, the bundle has two interfaces: a close inter-subunit interface (Fig. 6E, Cα-Cα distance 4.0–5.5 Å) and a far intra-subunit interface (Cα-Cα distance 7.0–8.5 Å).
Figure 6. HAMP Domains.
A) Sequence alignment of six HAMP domains of known structure show a conserved Gly and a semi-conserved Glu. B) Three different mechanisms proposed for HAMP signaling are described in cartoon form. C) An aligned ensemble of 26 HAMP domains was used to extract XY coordinates and distance distributions at 2 locations as in Fig. 5. The distribution for helix 1 (orange) is skewed outwards at the bottom of the bundle whereas helix 2 is skewed inwards (blue). D) The negative slope of the correlation between intermonomer Cα-Cα distances show that displacements in helix 1 and 2 are anti-correlated. E) The two overlaid structures, from Af1503 (2LFR,dark) and Aer2 (1I44, light) show a diagonal displacement of helices.
Proposed models for signal transduction via HAMPs
Different mechanisms have been proposed to describe HAMP mediated signaling; these are not always mutually exclusive (Fig. 6B).
The gear-box mechanism suggests that the HAMP is a bi-stable structure with two nearly iso-energetic states that interconvert via helical rotations. This model was proposed because the original structure of the Af1503 HAMP showed knobs-to-knobs coiled-coil packing that differed from what is typically seen in four-fold symmetric homotetrameric coiled-coils, so the authors postulated that a second state with a more canonical knobs-to-holes packing must also exist (Hulko et al., 2006). More recent studies provided some support for this hypothesis, but also showed that the structures deviate significantly from ideal coiled-coils (as reflected in deviations in Crick angles). Thus, the precise formalism of coiled coil registry change envisioned in the gearbox mechanism is restrictive, and might be expanded to include additional changes in helical packing and dynamics.
The diagonal scissoring mechanism proposes that the HAMP alternates between one state in which the helices are tightly packed, and a second in which they splay out at the C-terminal and form a loose bundle. These states have been crystallized in structures of tandem HAMP repeats (Fig. 6E) (Airola et al., 2010). Further evidence comes from disulfide cross-linking studies of Tar, which show that interactions across the close interface are critical for function. Stapling these helices together yielded the kinase-on or kinase-off state of the protein depending on the position of the staple (Swain and Falke, 2007; Swain et al., 2009).
The dynamic bundle model proposes that one signaling state of the HAMP has significantly more disorder than the other (Parkinson, 2010; Stewart, 2014). In its most extreme form, controlled local unfolding of the HAMP has been proposed as the final signal transduction event (Schultz and Natarajan, 2013). Specifically, a folded and tightly packed HAMP domain restrains the downstream domain in an inactive state, whereas an unfolded, loosely packed HAMP allows the subsequent domain to access all the necessary signaling states. The rationale for this model comes from HAMP-adenylate cyclase proteins, which show localized unfolding of a few helical turns between the HAMP and the enzyme in structures of the active state (Tews et al., 2005). There is no direct evidence for HAMPs unfolding in HKs, although a number of mutational studies on NarX (Appleman et al., 2003), Af1503 (Ferris et al., 2012, 2011), Tsr (Ames et al., 2008) and Tar (Swain and Falke, 2007) reveal that adding large sidechains to the core of the HAMP has a striking effect on kinase activity.
Recent structures of CpxA suggest that the bottom of the HAMP might also be involved in a tertiary interaction with the catalytic domain on the inactive side of the asymmetric autokinase state. The generality of this interaction remains to be seen. Consensus in the field is that HAMPs are highly malleable structures that occupy a relatively shallow energy landscape allowing them to shuttle between different conformations very easily. This marginal stability is a key feature of signal transduction.
Symmetric, anti-correlated helical displacement in HAMPs
Although the HAMP often connects directly to the highly asymmetric stem of the DHp bundle, all known structures of HAMP domains are relatively symmetric homodimers: the Cα RMSD between monomeric chains is < 1Å on average. In order to quantify the symmetry, we generated an aligned ensemble of 26 reported HAMP structures in a manner analogous to the DHp and extracted the X and Y coordinates for inward facing Cα atoms at the top and the bottom of the bundle (see supplement for technical details).
Unlike the DHp (Fig. 5A), the HAMP (Fig. 6C) does not have distinct regions of structural variability; instead the coordinates suggest smaller rigid body shifts that are concerted over the whole bundle. Earlier analysis showed that helical rotation is a small component of the observed variance across structures (Molnar et al., 2014). Remarkably, displacements in the two HAMP helices are anti-correlated: in structures where helix 1 tilts outwards, helix 2 moves inwards (Fig. 6D) suggesting that helical tilts are a large component of the input-output signaling through HAMPs. Distance distributions show a variance of ~2 Å at both the top of helix 1 and the bottom of helix 2, consistent with these regions being important for signal input and output.
The variance in Cα coordinates from the aligned HAMPs is much smaller than the variance at the DHp stem, so some sort of signal amplification must occur between the HAMP and the DHp. Furthermore since the HAMP conformations are all symmetric, whereas the autokinase states of the DHp and CA are decidedly asymmetric, a symmetry-asymmetry transition must also occur between the HAMP and the DHp.
Structure and signal transduction through PAS domains
The canonical PAS domain is a mixed α/β structure containing a central antiparallel 5-stranded beta sheet (Fig. 1C) surrounded by several helices. This central beta sheet is curved like a baseball glove and the two faces make contacts critical in signaling. The inner surface, or palm of the glove, is responsible for binding cofactors or ligands (e.g. heme, tricarboxylic acids) while the outer surface makes contacts between dimers or with flanking output helices (via a conserved Asp-Ile-Thr (DIT) motif). The PAS architecture allows ligand-binding to alter the packing and dynamics of the flanking alpha helices that transmit the signal. PAS domains are found as linkers as well as periplasmic sensors; they are the dominant sensor domains for HKs in B.subtilis (Chang et al., 2010; Cheung and Hendrickson, 2010).
Structural alignments of all known PAS domains, and details of their signaling mechanisms have been reviewed extensively elsewhere (Henry and Crosson, 2011; Möglich et al., 2009b), so we highlight only a few results from the laboratory of Möglich and Moffat. These coworkers have shown that the length of the helical linker between the PAS domain and the catalytic histidine is tightly controlled and varies by multiples of seven across the family, indicating that a heptad register is involved in transmitting signals downstream of the PAS (Möglich et al., 2009a). Using these principles, they engineered a blue light sensitive HK by adding a light sensing PAS domain above the DHp of FixL (Diensthuber et al., 2013). Recently, the structure of the CovS histidine kinase has provided the first structural view of a natural HK connected to a PAS domain (Wang et al., 2013). The structure is remarkably asymmetric, especially in the PAS output helices that connect it to the DHp.
Symmetry-Asymmetry transitions through polar linkers
The short helical linkers between the HAMP/PAS and the DHp have special properties that facilitate asymmetric signaling. The HAMP and DHp domains are typical helical bundles with hydrophobic interiors and polar exteriors yielding a regular pattern of polar and apolar residues in their sequences. This pattern is interrupted in the short linker, often by insertion of a single residue such that an ideal coiled-coil cannot smoothly connect the two domains. Biochemical studies on several HKs show that systematically manipulating the helical registry in these linkers can completely alter signaling (Möglich et al., 2009a; Stewart and Chen, 2010; Winkler et al., 2012). Furthermore, these linkers are often rich in polar residues (Fig. 7A), which must be buried (at the thermodynamic cost of dehydration) at the dimer interface. The net result is conformational strain such that no single symmetric state presents a deep energy well, and the linker helices are free to bend asymmetrically as seen in structures of DesK (Fig. 7B) and the AF1503-EnvZ chimera (Fig. 7C). Thus the symmetric conformational of the HAMP is converted into an asymmetric conformation in the DHp.
Figure 7. Symmetry-asymmetry transitions through polar linkers.
A) Polar clusters located a few residues above the catalytic histidine are present in several HKs. Alternate packing arrangements of the polar clusters result in asymmetric bending of the linker as seen in structures of the temperature-sensitive DesK (B, left) and Af1503-EnvZ chimera (C, left). The asymmetry profiles (B,C right) plot the Cα RMSD of computed over a 7-residue moving window for chains A and B superimposed on chains B and A respectively, and the Cα-Cα distance for each residue across the dimer. There is striking asymmetry in the linker in many HK structures.
Sensory elements: Periplasmic and Transmembrane domains
Sensor domains are the most diverse part of histidine kinases. They can be extracellular, transmembrane or intracellular and they differ in their structures and ligand binding modes (Cheung and Hendrickson, 2010; Mascher et al., 2006).
The structurally elusive transmembrane domains
There is no crystal structure for any TM in this family. A set of NMR structures for the monomeric transmembrane units from ArcB, QseC and KdpD (Maslennikov et al., 2010) have been solved in detergent micelles at high temperature, however these structures have limited utility because the dimer interface is not obvious. Instead, the structure of a phototaxis sensory rhodopsin II- transducer complex (HtrII) is often used as a model for the HK transmembrane four-helical bundle (Moukhametzianov et al., 2006).
Site-directed mutagenesis, disulfide cross-linking and molecular dynamics simulations have been used to characterize structure and helical periodicity in the TM domain of PhoQ (Goldberg et al., 2010; Lemmin et al., 2013). These studies show that a polar residue, which forms a small water pocket in the core of the TM domain, is critical for signal transduction. Remarkably, this residue can be moved up or down a turn or even moved to a neighboring TM helix without significant perturbations to the function. Replacements with other polar residues (e.g. Asn, Gln, Arg, His) at that position maintain the phenotype, while a hydrophobic mutation switches the protein to a phosphatase state. Thus, the helical bend induced at the water pocket allows the TM helices to switch between left- and right-handed crossing angles in the two signaling states.
Recent data on PhoQ in which ligand-dependent disulfide cross-linking experiments were interpreted using a Bayesian framework, showed that two different structural states were required to adequately fit the data (Molnar et al., 2014). The main difference between these states is a diagonal displacement of the TM helices (Fig. 8A).
Figure 8. The TM and periplasmic sensory elements.
A) Two structural states of the transmembrane bundle of PhoQ inferred using Bayesian modeling of disulfide crosslinking data. The states differ in their helical packing by a diagonal or reciprocal displacement of the helices. B) A simplified cartoon of three proposed models for conformational changes in the sensor domain. C) The magnitude of the displacement in the z and xy coordinates of the p-helices of Tar (left) and DctB (right) upon ligand binding. A small z-shift (pistoning) and a larger xy-shift (scissoring) are seen in both cases as the helix enters the membrane.
Diversity in Extracellular Sensory mechanisms
Sensory mechanisms are diverse across the HK family. Many sensors bind to a ligand directly, but the binding modes vary. For example nitrate binds in a cavity at the dimer interface of NarX (Cheung and Hendrickson, 2009); magnesium ions bind to an acidic patch at the periphery between the PhoQ sensor and the lipid bilayer (Cheung et al., 2008), and in CitA and DctB, the ligands (citrate and malonate) bind inside the PAS domain (Fig. 1B, top) (Sevvana et al., 2008; Zhou et al., 2008). Other HK sensors sense the state of a secondary protein that binds to the ligand; for example the sensor domain of LuxQ interacts with LuxP, which binds directly to the ligand AI-2 (Neiditch et al., 2006).
Despite this diversity, all sensors couple ligand-binding/stimulus-sensing to a conformational change that propagates across the receptor. The p-helix (defined earlier) is often found at the dimer interface of the sensor. Many of the known ligand binding sites are proximal to the p-helix and trigger structural rearrangements in it that propagate to the TM domains.
Asymmetry in sensor domains
Asymmetry is often a critical feature of HK stimulus sensing, but the details of the asymmetry vary. LuxQ, for example, forms an asymmetric 2:2 complex upon binding to its cognate periplasmic protein, LuxP (Neiditch et al., 2006). TorS begins in an asymmetric 2:2 complex with its periplasmic partner, TorT, in the apo-state, and the activated complex becomes symmetric upon TMAO binding (Moore and Hendrickson, 2012). The homodimeric aspartate receptor, closely related to sensor HKs, binds two Asp ligands with negative cooperativity, and signals from the asymmetric, singly-bound state (Chervitz and Falke, 1996; Yu and Koshland, 2001). Even in cryogenic crystal structures of dimeric sensor domains, large deviations from 2-fold rotational symmetry exist. For example, the PhoQ sensor domain forms a number of asymmetric interactions across the dimer interface, which suggests they are important for signaling and cannot be maintained in a two-fold symmetrical structure (Cheung et al., 2008).
Proposed models for stimulus-structure coupling
Given the diversity in stimulus-sensing mechanisms across different HKs, a number of models have been proposed for ligand-induced structural perturbations.
The piston model (Fig. 8B, left), which was based on the chemoreceptor Tar (Chervitz and Falke, 1996), proposes that ligand binding generates an ~1 Å piston-like downward displacement in the p-helix that propagates into the TM bundle along an axis normal to the bilayer plane. Pistoning has generally been discussed within a rigid-coupling framework, with many papers suggesting that the 1Å displacement propagates to the TM helices in an all-or-nothing fashion (Chervitz and Falke, 1996; Falke and Erbse, 2009). The scissor-blade model (Fig. 8B, middle), was also initially proposed for Tar based on crystallography (Milburn et al., 1991), but fell out of favor after disulfide crosslinking supported pistoning. Here, the ligand/signal induces a scissor-blade like diagonal displacement of the p-helices relative to the membrane normal, such that they splay out as they enter the membrane. Recently there has been renewed interest in scissoring since the sensor domains of the heparin binding hybrid kinase BT4663 (Lowe et al., 2012) and the magnesium sensitive PhoQ (Molnar et al., 2014) were both proposed to scissor. A third model suggests that helical rotations are the primary conformational motion required for signaling (Fig. 8B, right). This is based on the LuxP-LuxQ complex in which the asymmetric ligand-bound state requires rotation of the transmembrane helices (Neiditch et al., 2006).
Pistoning and Scissoring in Tar and DctB
We analyzed the sensors of the aspartate receptor, Tar and the dicarboxylate receptor DctB which have physiologically relevant dimeric structures in both apo-and ligand-bound states. We aligned all structures to a z-axis, normal to the membrane, and measured changes in the x,y and z coordinates of Cα atoms to evaluate ligand-induced pistoning and scissoring (details in supplement). Structural alignments between two distinct conformations of other sensors have been analyzed in detail elsewhere (Cheung and Hendrickson, 2009; Molnar et al., 2014).
Two general results emerged. First, diagonal displacement explains a large component of the variance between apo- and ligand-bound structures. While Tar shows a greater vertical displacement compared to DctB, consistent with the long-standing idea that ligand-binding induces a piston-like vertical displacement in Tar, for both DctB and Tar the lateral xy displacement is much more pronounced than the vertical displacement and increases as the p-helix approaches the membrane (Fig. 8C). There is no appreciable change in helical phase or rotation. Pistoning and scissoring cannot be completely decoupled in a simple x-y-z coordinate system, but this simple analysis qualitatively recapitulates results from a full parameterization using an orthogonal basis set (Molnar et al., 2014). Additionally, the conformational changes are highly asymmetric across the dimer interface; in both Tar and DctB, the magnitude of the shifts was generally larger on chain A of the dimer, than on chain B (Fig. 8C).
Many more structures of sensors are required to assess if there is any generality in sensory mechanisms across the HK family. It is especially useful to think of the sensor-TM as a thermodynamically coupled equilibrium because it is the energetic effect of the ligand-induced structural change rather than the mechanical details that dictates how the signal is transduced.
Statistical-Thermodynamic Model and Coupling of Conformational Transitions
Given the multi-domain architecture of histidine kinases, and the relatively independent nature of each domain, it is helpful to think of signal transduction as a series of equilibria between thermodynamically linked protein domains. These equilibria could be tightly coupled leading to a switchable cooperative two-state system, or they can be weakly coupled, leading to the population of many intermediates.
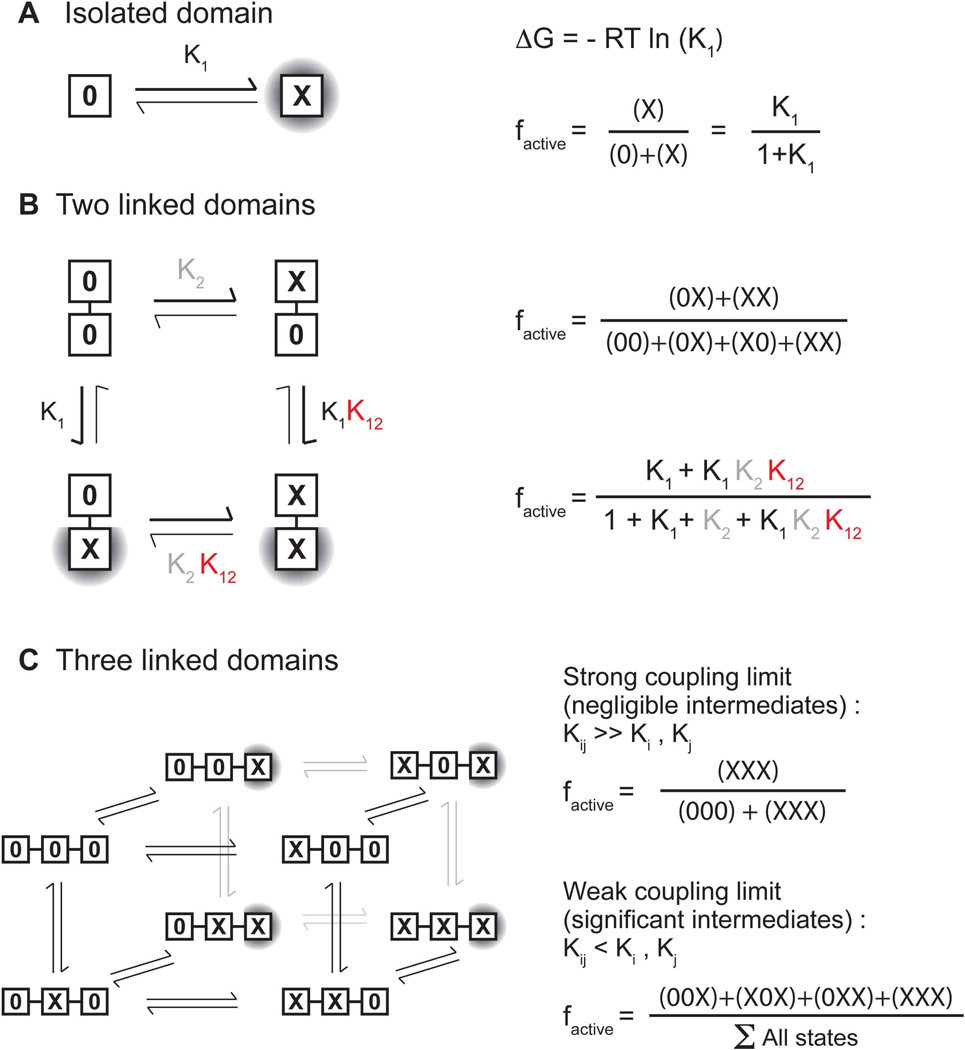
To illustrate this conceptually, consider an isolated catalytic domain that can sample two states, an active state (X) or an inactive state (0) (Fig. 9A). The fractional population of the active state dictates the maximal response of the protein to a stimulus and is dependent on the free energy difference (ΔG1) between the two states and temperature. If this catalytic domain is linked to a transmembrane domain, which can also be either active (X) or inactive (0) (Fig. 9B), the system has 2^2 = 4 states and the population of the active catalytic unit is governed not only by it’s own energy gap (ΔG1), but also by the energy gap of the linked domain (ΔG2) and the strength of the coupling (ΔG12). Three such domains yield 2^3 = 8 states and so on. Formally, the fractional population of a given state is the statistical weight of that state divided by the sum of the statistical weights for all the states, or the partition function. Each additional linked domain contributes an intrinsic energy gap, and a coupling energy to the partition function. These additional energetic terms can alter the basal population of the active kinase, thereby modulating the inducible response (Grigoryan et al., 2011).
Figure 9. A thermodynamic framework for signal transduction.
A) shows a single domain that has two states, an active state (X) or an inactive state (0). The population of the active state is directly related to the energy difference between these two states i.e. the equilibrium constant. B) shows two thermodynamically coupled domains. The fraction of the active state depends on two equilibrium constants and a coupling energy. C) extends this formalism to three linked domains. In the limit of rigid, all-or-nothing coupling between the domains, the system reduces to a 2 state system but in the limit of weak coupling, multiple intermediates contribute to the active population.
Why is a thermodynamic coupling framework useful for HKs?
Thermodynamic linkage is particularly appropriate for multi-domain proteins like HKs because it allows each domain to access independent conformational states and dynamics. The entire protein need not utilize the same mode of motion in order to transmit a signal. As long as each domain has an energetic input and output, it will sense the state of a neighboring domain and populate conformational states accordingly.
This energetic effect of one domain on its neighbors is called a coupling energy. The molecular origin of coupling can be enthalpic (via the formation of new bonds or the removal of steric hindrance) or entropic (via increased dynamics or release of solvent). Indeed, it is not always necessary for a large ΔGcoupling to yield a large structural change or vice versa. In the strong coupling limit, the coupling interaction is larger than the intrinsic energy gap of a given domain, intermediates are sparsely populated and a multi-state system is effectively reduced to a 2-state (on-off) system (Fig. 9C). Conversely, in the non-rigid or weak coupling limit, the effect of turning “on” one domain is to alter the equilibrium constant between the different states of the coupled domain. Weak thermodynamic coupling allows for the accrual of multiple domains capable of binding different ligands and populating intermediates along the way. For example VirA binds sugars via the periplasmic-binding protein, ChvE, as well as phenolics. Moreover, both VirA (Lin et al., 2014) and ChvE (Hu et al., 2013) are regulated by pH. PhoQ responds to divalent cations, protons and antimicrobials. In both cases each ligand can alter the signaling response (Bader et al., 2005; Vescovi et al., 1996).
The same framework also explains why domains from different HKs can be combined to form functional chimeric proteins when the energetic input and output for each domain is preserved (Utsumi et al., 1989). It can easily be extended to include additional domains, asymmetric conformations of individual domains, and ligand binding (Grigoryan et al., 2011). The challenge is to molecularly understand how proteins achieve weak-thermodynamic coupling and whether allosteric networks and mechanisms are conserved across the family (Kuriyan and Eisenberg, 2007).
Universal Themes in HK Signal Transduction
Recent structures of HKs yielded many general insights into bacterial signal transduction. Symmetric to asymmetric transitions within the homodimer, appear to be ubiquitous in many HK domains and also in their enzymological function. A similar connection between half-of-sites reactivity and asymmetry is seen in homodimeric receptor tyrosine kinases like EGFR (Leahy, 2010) and in chemoreceptors (Li and Hazelbauer, 2014), suggesting that this might be a general strategy for signal transduction. In HKs, asymmetric transitions are often encoded into structures via the fundamental properties of helical bundles such as their hydrophobic registry and side-chain packing propensities. Diagonal displacement of 4-helix bundles where pairs of helices slip in opposing directions is a general mode of helical-bundle malleability. This allows HKs to have shallow but a finely tuned energy landscapes such that small changes induced by stimuli, can propagate via thermodynamic coupling to yield a significant physiological response. The controlled reshuffling of the populations of different conformers as a consequence of thermodynamic coupling is at the heart of transmembrane signal transduction.
Supplementary Material
Acknowledgements
We thank Bruk Mensa and Nathan Schmidt for helpful discussions, and Thomas Lemmin for carefully reading the manuscript and providing feedback. The DeGrado lab acknowledges support from NIH GM54616 and AI074866, as well as support from the MRSEC program of NSF (DMR-1120901). M.P.B. was supported by the Jane Coffin Childs foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
HAMP domains are named for being present in Histidine kinases, Adenylate cyclases, Methyltransferases and Phosphodiesterases.
PAS domains are named for being first discovered in Periodic circardian proteins, Aryl hydrocarbon nuclear translocator protein and Single-minded protein.
GAF domains are named after being present in cGMP-specific phosphodiesterases, Adenylyl cylases and Formate hydrogenlases.
REFERENCES
- Aguilar PS, Hernandez-Arriaga aM, Cybulski LE, Erazo aC, de Mendoza D. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001;20:1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola MV, Watts KJ, Bilwes AM, Crane BR. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure. 2010;18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi D, Martín M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleman JA, Chen L-L, Stewart V. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 2003;185:4872–4882. doi: 10.1128/JB.185.16.4872-4882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenberg O, Keating AE, Laub MT. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J. Mol. Biol. 2013;425:1198–1209. doi: 10.1016/j.jmb.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Thomas Sa, Page SC, Creager-Allen RL, Moore AM, Silversmith RE. Measurement of response regulator autodephosphorylation rates spanning six orders of magnitude. Methods Enzymol. 2010;471:89–114. doi: 10.1016/S0076-6879(10)71006-5. [DOI] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Miguel-Romero L, Marina A. Visualizing autophosphorylation in histidine kinases. Nat. Commun. 2014;5:3258. doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Chang C, Tesar C, Gu M, Babnigg G, Joachimiak A, Raj Pokkuluri P, Szurmant H, Schiffer M. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J. Bacteriol. 2010;192:1156–1159. doi: 10.1128/JB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz Sa, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal structure of a functional dimer of the PhoQ sensor domain. J. Biol. Chem. 2008;283:13762–13770. doi: 10.1074/jbc.M710592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Hendrickson Wa. Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 2010;13:116–123. doi: 10.1016/j.mib.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Hendrickson Wa. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–1151. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Coles M, Lupas AN, Hartmann MD. Crystallographic snapshot of the Escherichia coli EnvZ histidine kinase in an active conformation. J. Struct. Biol. 2014:1–4. doi: 10.1016/j.jsb.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Dunin-Horkawicz S, Hornig N, Hulko M, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. Mechanism of regulation of receptor histidine kinases. Structure. 2012;20:56–66. doi: 10.1016/j.str.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Dunin-Horkawicz S, Mondéjar LG, Hulko M, Hantke K, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19:378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Fisher SL, Kim SK, Wanner BL, Walsh CT. Kinetic comparison of the specificity of the vancomycin resistance VanSfor two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SD, Clinthorne GD, Goulian M, DeGrado WF. Transmembrane polar interactions are required for signaling in the Escherichia coli sensor kinase PhoQ. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8141–8146. doi: 10.1073/pnas.1003166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Moore DT, DeGrado WF. Transmembrane communication: general principles and lessons from the structure and function of the M2 proton channel, K+ channels, and integrin receptors. Annu. Rev. Biochem. 2011;80:211–237. doi: 10.1146/annurev-biochem-091008-152423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA. The Pleiotropic Two-Component Regulatory System PhoP-PhoQ. 2001;183:1835. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu AD, Wayne KJ, Sham L-T, Winkler ME. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 2010;192:2346–2358. doi: 10.1128/JB.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JT, Crosson S. Ligand-Binding PAS Domains in a Genomic, Cellular, and Structural Context. Annu. Rev. Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhao J, Degrado WF, Binns AN. Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. 2013;110 doi: 10.1073/pnas.1215033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Igo MM, Ninfa aJ, Stock JB, Silhavy TJ. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Jiang P, Peliska Ja, Ninfa aJ. Asymmetry in the autophosphorylation of the two-component regulatory system transmitter protein nitrogen regulator II of Escherichia coli. Biochemistry. 2000;39:5057–5065. doi: 10.1021/bi992921w. [DOI] [PubMed] [Google Scholar]
- Jung K, Fried L, Behr S, Heermann R. Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 2012;15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Kaspar S, Perozzo R, Reinelt S, Meyer M, Pfister K, Scapozza L, Bott M. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 1999;33:858–872. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- Kim D, Forst S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology. 2001;147:1197–1212. doi: 10.1099/00221287-147-5-1197. [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Leahy DJ. The ins and outs of EGFR asymmetry. Cell. 2010;142:513–515. doi: 10.1016/j.cell.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Lemmin T, Soto CS, Clinthorne G, DeGrado WF, Dal Peraro M. Assembly of the transmembrane domain of E. coli PhoQ histidine kinase: implications for signal transduction from molecular simulations. PLoS Comput. Biol. 2013;9:e1002878. doi: 10.1371/journal.pcbi.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Selective allosteric coupling in core chemotaxis signaling complexes. Proc. Natl. Acad. Sci. U. S. A. 2014;2014:1–6. doi: 10.1073/pnas.1415184111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-H, Pierce BD, Fang F, Wise A, Binns AN, Lynn DG. Role of the VirA histidine autokinase of Agrobacterium tumefaciens in the initial steps of pathogenesis. Front. Plant Sci. 2014;5:195. doi: 10.3389/fpls.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe EC, Baslé A, Czjzek M, Firbank SJ, Bolam DN. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. 2012 doi: 10.1073/pnas.1200479109. doi:10.1073/pnas.1200479109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina A, Waldburger CD, Hendrickson Wa. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 2006;264:133–144. doi: 10.1111/j.1574-6968.2006.00444.x. [DOI] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslennikov I, Klammt C, Hwang E, Kefala G, Okamura M, Esquivies L. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. 2010 doi: 10.1073/pnas.1001656107. doi:10.1073/pnas.1001656107/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias VRF, Beveridge TJ. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 2006;188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechaly AE, Sassoon N, Betton J-M, Alzari PM. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 2014;12:e1001776. doi: 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim S-H. Three-Dimensional Structures of the Ligand-Binding Domain of the Bacterial Aspartate Receptor. Science. 1991;254:1342–1346. doi: 10.1126/science.1660187. (80-.). [DOI] [PubMed] [Google Scholar]
- Miller SI, Kukral aM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers Ra, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J. Mol. Biol. 2009a;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers Ra, Moffat K. Structure and Signaling Mechanism of Per-ARNT-Sim Domains. Structure. 2009b;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar KS, Bonomi M, Pellarin R, Clinthorne GD, Gonzalez G, Goldberg SD, Goulian M, Sali A, DeGrado WF. Cys-Scanning Disulfide Crosslinking and Bayesian Modeling Probe the Transmembrane Signaling Mechanism of the Histidine Kinase, PhoQ. Structure. 2014;22:1239–1251. doi: 10.1016/j.str.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JO, Hendrickson Wa. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure. 2012;20:729–741. doi: 10.1016/j.str.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukhametzianov R, Klare JP, Efremov R, Baeken C, Göppner A, Labahn J, Engelhard M, Büldt G, Gordeliy VI. Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature. 2006;440:115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- Podgornaia AI, Casino P, Marina A, Laub MT. Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling. Structure. 2013;21:1636–1647. doi: 10.1016/j.str.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin RS, Stewart V. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8419–8423. doi: 10.1073/pnas.89.18.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo D, Silhavy J. EnvZ Controls the Concentration of Phosphorylated OmpR to Mediate Osmoregulation of the Porin Genes medium. J. Mol. Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- Stewart V. The HAMP signal-conversion domain: static two-state or dynamic three-state? Mol. Microbiol. 2014;91:853–857. doi: 10.1111/mmi.12516. [DOI] [PubMed] [Google Scholar]
- Stewart V, Chen L-L. The S helix mediates signal transmission as a HAMP domain coiled-coil extension in the NarX nitrate sensor from Escherichia coli K-12. J. Bacteriol. 2010;192:734–745. doi: 10.1128/JB.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock M, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Saha S, Tomomori C, Ishima R, Liu D, Tong KI, Park H, Dutta R, Qin L, Swindells M, Yamazaki T, Ono AM, Kainosho M, Inouye M, Ikura M. NMR structure of the histidine kinase domain of E. coli osmosensor EnvZ. Nat. Biotechnol. 1998;396:88–92. doi: 10.1038/23968. [DOI] [PubMed] [Google Scholar]
- Tews I, Findeisen F, Sinning I, Schultz A, Schultz JE, Linder JU. The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science. 2005;308:1020–1023. doi: 10.1126/science.1107642. [DOI] [PubMed] [Google Scholar]
- Trajtenberg F, Graña M, Ruétalo N, Botti H, Buschiazzo A. Structural and enzymatic insights into the ATP binding and autophosphorylation mechanism of a sensor histidine kinase. J. Biol. Chem. 2010;285:24892–24903. doi: 10.1074/jbc.M110.147843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- Vescovi EG, Soncini FC, Groisman EA. Mg2+ as an Extracellular Signal : Environmental Regulation of Salmonella Virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, Senadheera DB, Cvitkovitch DG, Chen L, Goodman SD, Han A. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler K, Schultz A, Schultz JE. The S-helix determines the signal in a Tsr receptor/adenylyl cyclase reporter. J. Biol. Chem. 2012;287:15479–15488. doi: 10.1074/jbc.M112.348409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo W-S, Zwir I, Huang HV, Shin D, Kato A, Groisman Ea. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol. Cell. 2012;45:409–421. doi: 10.1016/j.molcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EW, Koshland DE. Propagating conformational changes over long (and short) distances in proteins. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y-F, Nan B, Nan J, Ma Q, Panjikar S, Liang Y-H, Wang Y, Su X-D. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.