Abstract
Mutations in Gap Junction Beta 2 (GJB2) have been reported to be a major cause of non-syndromic hearing loss in many populations worldwide. The spectrums and frequencies of GJB2 variants vary substantially among different ethnic groups, and the genotypes among these populations remain poorly understood. In the present study, we carried out a systematic and extended mutational screening of GJB2 gene in 1067 Han Chinese subjects with non-syndromic hearing loss, and the resultant GJB2 variants were evaluated by phylogenetic, structural and bioinformatic analysis. A total of 25 (23 known and 2 novel) GJB2 variants were identified, including 6 frameshift mutations, 1 nonsense mutation, 16 missense mutations and 2 silent mutations. In this cohort, c.235delC is the most frequently observed pathogenic mutation. The phylogenetic, structural and bioinformatic analysis showed that 2 novel variants c.127G>T (p.V43L), c.293G>C (p.R98P) and 2 known variants c. 107T>C (p.L36P) and c.187G>T (p.V63L) are localized at highly conserved amino acids. In addition, these 4 mutations are absent in 203 healthy individuals, therefore, they are probably the most likely candidate pathogenic mutations. In addition, 66 (24 novel and 42 known) genotypes were identified, including 6 homozygotes, 20 compound heterozygotes, 18 single heterozygotes, 21 genotypes harboring only polymorphism(s) and the wild type genotype. Among these, 153 (14.34%) subjects were homozygous for pathogenic mutations, 63 (5.91%) were compound heterozygotes, and 157 (14.71%) carried single heterozygous mutation. Furthermore, 65.28% (141/216) of these cases with two pathogenic mutations exhibited profound hearing loss. These data suggested that mutations in GJB2 gene are responsible for approximately 34.96% of non-syndromic hearing loss in Han Chinese population from Zhejiang Province in eastern China. In addition, our results also strongly supported the idea that other factors such as alterations in regulatory regions, additional genes, and environmental factors may contribute to the clinical manifestation of deafness.
Introduction
Hearing loss is one of the most common sensory defects, affecting one in 700–1000 newborns[1,2]. It can be classified as syndromic hearing loss (hearing loss with other symptoms such as diabetes), and non-syndromic hearing loss (hearing loss is the only obvious clinical phenotype). Approximately 50% of all cases of hearing loss have a genetic etiology or predisposition with autosomal dominant, autosomal recessive, X-linked or maternal patterns of inheritance[3,4]. To data, over 140 loci have been mapped for non-syndromic hearing loss, and 82 deafness-causing genes have been identified (The Hereditary Hearing loss Homepage. http://hereditaryhearingloss.org).
Mutations in the GJB2 gene, which encodes the connexin 26 (Cx26), are a major cause of non-syndromic hearing loss in many populations worldwide[5]. More than 150 different GJB2 variants have been identified, including missense, nonsense and frameshift mutation (The Connexin-deafness Homepage. http://davinci.crg.es/deafness). The spectrums of GJB2 mutations vary among different ethnic groups[5,6]. Of these variants, the c.35delG is the most prevalent GJB2-deafness-causing mutation in European populations, while c.235delC and c.167delT are the most frequent variants in Eastern Asian populations and Ashkenazi Jewish families, respectively[5,7–10]. In addition, the prevalence of GJB2 mutations differs among various populations, which varied from 7.9% to 44.1% among Caucasian cohorts and 4.5% to 34.7% in several Asian populations[5,6,11–17]. However, the spectrum and frequency of GJB2 mutations in the Chinese population are still poorly understood, and the prevalence of various GJB2 genotypes among different Chinese populations is less studied. In addition, it is anticipated that there are additional GJB2 mutation(s) associated with hearing loss among Chinese population.
In the present study, a systematic and extended mutational screening of GJB2 gene was carried out in a cohort of 1067 Han Chinese hearing-impaired subjects from Zhejiang Province, in Eastern China. We found 25 variants in this large cohort including missense mutation, nonsense mutation and frameshift mutation, which could be attributed to 66 genotypes. By phylogenetic analysis, structure-function relation and allelic frequency of these variants in the 203 Han Chinese healthy individuals from the same region, 4 putative pathogenic mutations were identified.
Subjects and Methods
Subjects and audiological examinations
A total of 1067 genetically unrelated hearing-impaired Chinese subjects were recruited from Zhejiang Province in Eastern China. The subjects were composed of 509 students from Schools for Deaf Children and Hearing and Speech Rehabilitation Centers, and 558 patients from the ENT Clinics of Zhejiang Province. A comprehensive history and physical examination were performed on these participants, so as to identify any syndromic findings, the history of the use of aminoglycosides, environmental and genetic triggers related to the hearing loss. Age-appropriate audiological evaluation was carried out as previous report [18]. Briefly, the pure-tone audiometry (PTA) was calculated from average of the audiometric thresholds at 500, 1000, 2000, 4000 and 8000 Hz. The severity of hearing loss was then classified into five grades: normal (<26 dB), mild (between 26 and 40 dB), moderate (between 41 and 70 dB), severe (between 71 and 90 dB), and profound (>90 dB). Auditory brainstem response (ABR), immittance testing and distortion product otoacoustic emissions (DPOAE) were also performed. The control DNA samples were obtained from 203 healthy Han Chinese subjects in the same region of China. This study has been approved by the Ethics Committees of both Zhejiang University and Wenzhou Medical University, and written informed consent was obtained from all participants.
Mutational analysis of GJB2 gene
Genomic DNAs were isolated from the blood samples of all participants using Universal Genomic DNA Extraction Kit Ver.3.0 (TaKaRa, Dalian, China). The DNA fragments spanning the entire coding region of GJB2 gene were amplified by PCR using the oligodeoxynucleotides: forward-5’TATGACACTCCCCAGCACAG3’ and reverse-5’GGGCAATGCTTAAACTGGC3’. Then the purified GJB2 fragments were analyzed by direct sequencing as detailed previously[19]. Subsequently, the mutations were identified by comparing the resultant sequence with wild type GJB2 sequence (GenBank accession No: NM_004004.5). The sequence of each amplicon was confirmed by sequencing in both directions.
Phylogenetic analysis
A total of 23 species Cx26 amino acid sequences were used in the inter-species analysis (S2 Table). The conservation index (CI) was defined as the percentage of species with amino acid residues identical to that of human at that position. Alignments and analysis were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Bioinformatic analysis
In order to predict the effect of the putative GJB2 mutation on protein function, the pathogenic potential of these missense mutations was evaluated by PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) and SIFT (http://sift.jcvi.org/) programs.
Results
Study samples
The study samples were Han Chinese recruited from Schools of Deaf Children and ENT clinics of Zhejiang Province. Subjects who had a history of exposure to aminoglycoside antibiotics, or other clearly identifiable cause(s) of hearing loss (such as bacterial meningitis, labyrinthitis, temporal bone fracture involving the cochlea, active chronic otitis media, cholesteatoma), or syndromic deafness were excluded from this study. A total of 1067 subjects (609 males and 458 females), who exhibited sensorineural hearing loss as the sole symptom, were analyzed. The age of subjects ranged from 1 year old to 41 years old, with a median age of 17 years. The age-at-onset of the hearing loss in these subjects ranged from congenital to 41 years old, with a median age of 8 years. Audiological studies showed that there was a wide range of different degrees of hearing loss in these subjects: 485 subjects exhibited profound hearing loss, 403 patients had severe hearing loss, 145 individuals suffered moderate hearing loss, and 34 cases with mild hearing loss. In addition, 203 Han Chinese subjects (112 males and 91 females) recruited from the same region, exhibited normal hearing and didn’t have a family history of hearing loss. The age of these participants ranged from 8 years old to 27 years old, with a median age of 16 years (S1 Table).
Mutation analysis of GJB2 gene
DNA fragments spanning the entire coding region of GJB2 gene were PCR-amplified from genomic DNA samples of 1067 hearing-impaired and 203 control subjects, and each fragment was sequenced. As shown in Table 1, 25 (2 novel and 23 known) nucleotide changes in GJB2 gene were identified when comparing the resultant sequence with the wild type sequence (GenBank accession NM_004004.5). Among these mutations, 2 novel variants (p.V43L and p.R98P) were identified in heterozygous state. When looking at the allelic frequency of GJB2 gene in healthy individuals, we found 3 variants (p.V27I, p.V37I and p.E114G) in 203 Chinese controls with the frequencies of 22.91%, 4.93% and 15.02%, respectively. Furthermore, the missense mutations were evaluated with phylogenetic analysis by comparing the human GJB2-encoding connexin-26 (Cx26) amino acid residues with other 22 species in data bank. As shown in Table 1, all the 16 missense mutations observed in our study changed amino acids which are well conserved in evolution with the conservation index (CI) ranged from 39.13% to 100%. In particular, CI of 13 variants were >90%, CI of other 2 variants including p.G4D and p.R98P were between 80% and 50%, and CI for p.T123N variant was 39.13%. In addition, localization of these mutations in the secondary structure of Cx26 protein was exhibited in S1 Fig The variants p.V43L, p.V63L and p.G160S reside at the extracellular loops of Cx26, the variants p.V27I, p.I30V, p.L36P, p.V37I, p.R143W, p.E147K and p.I203T occur in transmembrane domains, and the variants p.G4D, p.G12V, p.V95M, p.R98P, p.E114G and p.T123N are located at amino termini and intracellular loop of the protein. Moreover, both the PolyPhen-2 and SIFT programs showed that 9 of these missense mutations were damaging (Table 1).
Table 1. Variants in the GJB2 gene among 1067 Han Chinese subjects with hearing loss.
| Nucleotide Change | Effect on Protein | dbSNP ID | Conservation Index (%) a | Allele Frequency in Affected Subjects (%, 2134 allele) | Allele Frequency in Controls (%, 406 allele) | PolyPhen-2 | SIFT | Characterization of Variant |
|---|---|---|---|---|---|---|---|---|
| Frameshift mutations (Deletions and insertions) | ||||||||
| c.35insG | G12GfsX36 | - | 0.09 | 0 | Pathogenic | |||
| c.35delG | G12VfsX2 | rs80338939 | 0.09 | 0 | Pathogenic | |||
| c.176_191del16 | G59AfsX18 | - | 0.75 | 0 | Pathogenic | |||
| c.235delC | L79CfsX3 | rs80338943 | - | 13.96 | 0 | Pathogenic | ||
| c.299_300delAT | H100RfsX14 | rs111033204 | - | 2.25 | 0 | Pathogenic | ||
| c.512_513insAACG | A172EfsX39 | - | 0.52 | 0 | Pathogenic | |||
| Nonsense mutation | ||||||||
| c.139G>T | p.E47X | rs104894398 | 100 | 0.09 | 0 | Pathogenic | ||
| Missense mutations | ||||||||
| c.11G>A | p.G4D | rs111033222 | 56.52 | 0.33 | 0 | Benign | Tolerated | Polymorphism |
| c.35G>T | p.G12V | rs1801002 | 100 | 0.05 | 0 | Damaging | Damaging | Pathogenic |
| c.79G>A | p.V27I | rs2274084 | 100 | 25.21 | 22.91 | Damaging | Tolerated | Polymorphism |
| c.88A>G | p.I30V | rs374625633 | 100 | 0.14 | 0 | Benign | Tolerated | Polymorphism |
| c.107T>C | p.L36P | 100 | 0.05 | 0 | Damaging | Damaging | Putative pathogenic | |
| c.109G>A | p.V37I | rs72474224 | 100 | 8.86 | 4.93 | Damaging | Tolerated | Pathogenic |
| c.127G>T b | p.V43L | 100 | 0.05 | 0 | Damaging | Damaging | Putative pathogenic | |
| c.187G>T | p.V63L | 100 | 0.05 | 0 | Damaging | Damaging | Putative pathogenic | |
| c.283G>A | p.V95M | rs111033299 | 91.30 | 0.09 | 0 | Damaging | Damaging | Pathogenic |
| c.293G>Cb | p.R98P | 78.26 | 0.05 | 0 | Damaging | Damaging | Putative pathogenic | |
| c.341A>G | p.E114G | rs2274083 | 91.30 | 19.07 | 15.02 | Benign | Tolerated | Polymorphism |
| c.368C>A | p.T123N | rs111033188 | 39.13 | 0.23 | 0 | Benign | Tolerated | Polymorphism |
| c.427C>T | p.R143W | rs80338948 | 100 | 0.09 | 0 | Damaging | Damaging | Pathogenic |
| c.439G>A | p.E147K | 100 | 0.09 | 0 | Damaging | Damaging | Pathogenic | |
| c.478G>A | p.G160S | rs34988750 | 100 | 0.33 | 0 | Damaging | Tolerated | Polymorphism |
| c.608T>C | p.I203T | rs76838169 | 95.65 | 0.14 | 0 | Damaging | Damaging | Polymorphism |
| Silent variants | ||||||||
| c.81C>T | p.V27 | 100 | 0.05 | 0 | Polymorphism | |||
| c.444C>T | p.A148 | 100 | 0.05 | 0 | Polymorphism | |||
a The conservation index (CI) was calculated by comparing the human amino acid variants with other 22 species (See S1 Table). The CI was then defined as the percentage of species from the list of 23 different species that have the wild-type nucleotide at that position.
b The novel variants.
The p.G4D, p.V27I, p.E114G and p.T123N variants were considered as polymorphisms to comply with the standard of whose frequency >5% in controls or CIs<60%. In addition, the p.I30V, p.G160S and p.I203T variants were also classified as benign since they were reported in controls in other populations[6,13,20,21]. On the other hand, the frequency of p.V37I variant in hearing-impaired subjects was much higher than in our control cohort. Furthermore, this variant has been reported to be associated with mild hearing loss among Chinese and Japanese populations[22]. Taken together, we identified 9 GJB2 variants in this large cohort of Han Chinese population to be pathogenic, including the 5 known deafness-associated mutations (p.G12V, p.V37I, p.V95M, p.R143W and p.E147K). The allelic frequency of pathogenic mutations in GJB2 gene among this Chinese cohort was 27.13%. In particular, c.235delC was the most frequent mutation with the allelic frequency of 13.96%, followed by c.299_300delAT (2.25%).
Genotypes and phenotypes of GJB2
As shown in S3 Table, a total of 66 (24 novel and 42 known) genotypes of GJB2 was observed in the studied cohort of 1067 hearing-impaired subjects, including 44 (16 novel and 28 known) genotypes carrying at least one of pathogenic mutation(s), 21 (8 novel and 13 known) genotypes harboring only polymorphism(s) and a genotype lacking any copy of variants. In this study, we found 34.96% (373/1067) of the hearing-impaired cases carrying at least one pathogenic mutation, among which 153 (14.34%) were homozygous for pathogenic mutations and 63 (5.91%) were compound heterozygotes, while the other 157 (14.71%) subjects only had one heterozygous mutation. In particular, c.235delC homozygote was the most common pathogenic genotype, accounting for 70.59% (108/153) of the homozygotes group, and 10.12% of the entire subjects. Furthermore, the [c.235delC]/[c.299_300delAT] was found to be the most frequent genotype with a frequency of 30.16% (19/63) in compound homozygotes group. Among the 216 subjects with biallelic pathogenic mutations (Table 2), 65.28% (141/164) of these cases exhibited profound hearing loss.
Table 2. Genotypes and phenotypes of GJB2 in the 216 hearing-impaired subjects with two pathogenic mutations.
| Genotypes | Severity of Hearing Loss (PTA0.5-8k Hz) a | Number | |||
|---|---|---|---|---|---|
| Mild | Moderate | Severe | Profound | ||
| [c.235delC] b / [c.235delC] | 1 | 2 | 23 | 82 | 108 |
| [c.235delC; c.478G>A] / [c.235delC] | 0 | 0 | 0 | 1 | 1 |
| [c.235delC; c.478G>A] / [c.235delC; c.478G>A] | 0 | 0 | 0 | 1 | 1 |
| [c.299_300delAT] / [c.299_300delAT] | 0 | 1 | 0 | 5 | 6 |
| [c.109G>A] / [c.109G>A] | 5 | 13 | 9 | 7 | 34 |
| [c.79G>A;c.109G>A;c.341A>G] / [c.109G>A] | 0 | 2 | 0 | 1 | 3 |
| [c.235delC] / [c.35insG] | 0 | 0 | 0 | 2 | 2 |
| [c.235delC] / [c.35delG] | 0 | 0 | 0 | 1 | 1 |
| [c.235delC] / [c.35G>T;c.79G>A; c.341A>G] | 0 | 0 | 1 | 0 | 1 |
| [c.235delC] / [c.127G>T;c.79G>A; c.341A>G] | 0 | 0 | 0 | 1 | 1 |
| [c.235delC] / [c.139G>T] | 0 | 0 | 0 | 1 | 1 |
| [c.235delC] / [c.176_191del16] | 0 | 1 | 2 | 4 | 7 |
| [c.235delC] / [c.299_300delAT] | 0 | 0 | 4 | 15 | 19 |
| [c.235delC;79G>A] / [c.299_300delAT] | 0 | 0 | 0 | 1 | 1 |
| [c.235delC;79G>A] / [c.439G>A] | 0 | 1 | 0 | 1 | 2 |
| [c.235delC] / [c.512_513insAACG] | 0 | 2 | 1 | 3 | 6 |
| [c.299_300delAT] / [c.139G>T] | 0 | 0 | 0 | 1 | 1 |
| [c.299_300delAT] / [c.176_191del16] | 0 | 0 | 0 | 5 | 5 |
| [c.299_300delAT] / [c.512_513insAACG] | 0 | 0 | 0 | 1 | 1 |
| [c.235delC] / [c.109G>A] | 2 | 1 | 1 | 1 | 5 |
| [c.176_191del16] / [c.109G>A] | 0 | 0 | 0 | 2 | 2 |
| [c.283G>A] / [c.109G>A] | 0 | 0 | 0 | 1 | 1 |
| [c.283G>A; c.79G>A; c.341A>G] / [c.109G>A] | 0 | 0 | 1 | 0 | 1 |
| [c.293G>C] / [c.109G>A] | 0 | 0 | 0 | 1 | 1 |
| [c.299_300delAT] / [c.109G>A] | 0 | 1 | 0 | 2 | 3 |
| [c.427C>T] / [c.109G>A] | 0 | 0 | 1 | 1 | 2 |
| Total | 8 | 24 | 43 | 141 | 216 |
a PTA pure-tone audiometry.
b The pathogenic mutations were in bold.
Among the 18 genotypes of single heterozygotes, 16 (4 novel and 12 known) genotypes carried pathogenic mutations (c.35delG, c.176_191del16, c.235delC, c.299_300delAT, c.512_513insAACG, p.V37I, p.V95M and p.R143W) and 2 novel genotypes harbored the putative deafness-associated mutations (p.L36P and p.V63L). In addition to the genotypes contained pathogenic mutation(s) mentioned above, there were 22 genotypes did not carry any deafness-associated mutation(s), involving 65.04% (694/1067) cases of this cohort. Of these nonpathogenic genotypes, 21 genotypes were found only harboring polymorphisms including 2 genotypes with silent variants (c.81C>T, p.V27 and c.444C>T, p.A148) and 19 genotypes carrying missense variants (p.G4D, p.V27I, p.I30V, p.E114G, p.T123N, p.G160S and p.I203T).
Clinical and genetic characterization of hearing-impaired subjects carrying the 4 putative GJB2 mutations
A genotype-phenotype correlation analysis was performed among 4 probands carrying the 4 putative deafness-associated GJB2 mutations. The comprehensive history and physical examination including audiological assessment indicated that hearing impairment was the sole clinical phenotype in these subjects, and no other clinical abnormalities was observed in members of their families such as diabetes, muscular diseases, visual loss and neurological disorders. All the 4 subjects did not have a history of exposure to neither aminoglycosides nor other ototoxic drugs. Moreover, there was no evidence that the 4 cases had encountered any other external cause to be responsible for hearing loss. Audiological assessments indicated that they all exhibited profound hearing loss at ages younger than 2 years old, with flat-shaped pattern of audiometric configuration.
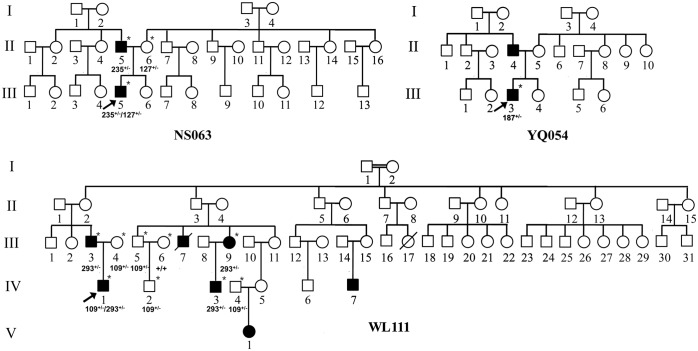
As illustrated in Table 3 and Fig 1, 2 of the 4 subjects had a family history of hearing loss. Proband NS063-III-5, whose genotype was in a heterozygous state of c.127G>T (p.V43L) and c.235delC mutations, suffered profound congenital hearing loss. Among the other family members, only his father exhibited moderate hearing defect (45 dB at right ear, 42 dB at left ear; sloping-shaped pattern). In addition, the proband WL111-IV-1, bearing the [c.293G>C]/[c.109G>A] genotype, showed profound congenital hearing loss. Among this pedigree, the c.293G>C (p.R98P) variant was also detected in III-3, III-9 and IV-3 in heterozygous state. Audiological examinations showed mild hearing loss in III-3 with sloping-shaped pattern, and the III-9 and IV-3 exhibited profound congenital hearing loss.
Table 3. Summary of clinical and molecular data for 4 Han Chinese subjects carrying the putative deafness-associated mutations in GJB2 gene.
| Genotype | Proband | Gender a | Audiometric Configuration | Age-at-onset (Years) | PTA b (dB) c Right Ear | PTA(dB) Left Ear | Level of Hearing Loss | Family History of Hearing Loss |
|---|---|---|---|---|---|---|---|---|
| [c.107T>C] d / [c.79G>A; c.341A>G] | NS110 | F | Flat | <1 | 114 | 108 | Profound | No |
| [c.235delC;c.79G>A; c.341A>G] / [c.127G>T] | NS063 | M | Flat | <1 | 115 | 113 | Profound | Yes |
| [c.187G>T] / [c.79G>A; c.341A>G] | YQ054 | F | Flat | 2 | 109 | 110 | Profound | No |
| [c.293G>C] / [c.109G>A] | WL111 | F | Flat | <1 | 108 | 112 | Profound | Yes |
a F female, M male.
b PTA pure-tone audiometry.
c dB decibel.
d The (putative) pathogenic mutations were in bold.
Fig 1. Three Han Chinese pedigrees with hearing loss carrying the GJB2 putative mutations.
Affected individuals are indicated by filled symbols. An arrow denotes probands. The interviewed and sequenced individuals are marked by asterisks. +/- denotes heterozygote; +/+ denotes wild type.
Discussion
GJB2 is the most common causative gene for hereditary hearing loss in many populations worldwide, and most of the GJB2 sequence variations described to date were localized in the coding region (The Connexin-deafness Homepage. http://davinci.crg.es/deafness). In fact, the GJB2 mutation spectrums and frequencies vary among different ethnic population[5,6]. In the present study, we analyzed the spectrum and frequency of GJB2 variants in 1067 Chinese non-syndromic hearing loss subjects from Zhejiang Province in eastern China, and investigated the clinical and genetic characterization of patients with putative GJB2 mutations. A total of 25 different nucleotide changes in the GJB2 gene were identified in this cohort, 34.96% of subjects in the cohort harbored GJB2 pathogenic mutations, which can be classified into 44 genotypes, including homozygotes, compound heterozygotes and single heterozygotes (only one mutant allele). These results indicate that GJB2 mutations are the important causes of hearing loss in hearing-impairment Han Chinese population.
In the present study, we revealed a high prevalence (27.13%) of GJB2 mutations in Chinese hearing loss patients, which is compatible with other reports[13,23]. Among these variants, c.235delC mutation was observed with a frequency of 13.96% in our cohort. The c.235delC mutation in the GJB2 gene predominantly occurs in people of East Asian, the incidences of the mutation ranged from 6.7% to 22.5% in several other Chinese deaf cohorts according to previous studies, which were 5.3% and 10.9% in two reported Japanese hearing loss cohorts, 1.5% in a cohort from Mongolia, 5.1% and 6.9% in two cohorts of Korean deafness patients[11,14,24–28]. The high frequency of c.235delC mutation in multiple East Asian populations raises the possibility of a common ancestral founder event. Interestingly, c.235delC mutation has not been reported in south Asian populations such as Indian, Pakistanis, and Bangladeshis[5]. Instead, p.W24X, p.W77X and p.Q124X are the prevalent GJB2 mutations in these populations[5,12,29,30]. Interestingly, the observed racial divergence of South and East Asian is consisted with the studies of mtDNA haplogroup analysis in that predicting country of ancestral origin (http://www.mitomap.org, 2015). It has been well accepted that haplogroup L3 gave rise to two lineages: M and N. In addition, the major haplogroups B and F in East Asian were rising from macrohaplogroup N, and the mitochondrial haplogroups in South Asian were macrohaplogroup M and its derivatives. These results indicate that c.235delC mutation might have arisen after the racial divergence of South and East Asian occurred. It would be necessary to systematically investigate the GJB2 mutations of remaining Asian countries to precisely elucidate the origin of c.235delC mutation[31]. Of the other GJB2 deletion/frameshift mutations associated with hearing loss, c.176_191del16 and c.299_300delAT were previously reported in Korean and Japanese hearing loss cohorts[14,25], and c.512_513insAACG mutation has been also detected in Chinese and Japanese deaf patients but in a much lower frequency[13,14].
The c.35delG mutation of GJB2 was previously reported to be the most prevalent mutation (21%) in a cohort of north Indian population with hearing loss[32]. In particular, among these populations with European ancestry, the c.35delG mutation causes severe to profound hearing loss, accounted for approximately 70% of all recessive mutations of the gene[7]. In fact, the mutations at position 35 are found to be exceptionally low in eastern Asian. Dai et al reported in a large cohort that 12 Chinese patients carried c.35delG mutation; however, the majority of these patients was Uigur from Xinjiang area[13]. In this cohort of Han people from Zhejiang Province in eastern China, the deletion, insertion and G to T mutation at position 35 were respectively detected in 5 unrelated subjects, with all the 3 mutations found in compound heterozygous state. Of the 5 cases, 4 subjects also carried c.235delC mutation, and the other one patient had c.79G>A polymorphism.
Fifteen additional missense mutations (13 known and 2 novel) were detected among these hearing loss patients, including 4 pathogenic mutations, 7 polymorphisms and 4 unknown variants. To identify putative pathogenic mutation, the unknown variants were further evaluated using following criteria: (1) conservation index greater than 75%, (2) absent in the 203 controls, (3) potential structural and functional alterations, predicted by their locations, PolyPhen-2 and SIFT programs, and (4) pedigree analysis if possible. The p.L36P, p.V43L, V63L and p.R98P variants, which occurred at evolutionarily conserved amino acid residues and absent from the control group, were predicted as pathogenic mutations by PolyPhen-2 and SIFT programs. The p.L36P mutation was first described in trans with 35delG in a hearing loss patient of African descent[33]. This putative disease-causing mutation locates at the transmembrane domain of Cx26 protein, which is involved in the oligomerization of hexameric connexon hemichannels. The resultant changed protein may lead to the reduction of junctional permeability in cells that is essential for ions and small metabolites exchange. Variants p.V43L and V63L, which reside at the first extracellular loop of Cx26, may result in gap-junction channels dysfunction among adjacent cells. The Val at 43 position changed to Met in Cx26 was previously observed in a hearing loss cohort from Taiwan and considered to be associated with the disease[34]. Here, we report the novel p.V43L variant as a potential deafness-associated mutation in Han Chinese population from Zhejiang province. The p.V63L mutation was first reported in a Taiwan cohort, and then it was also found in other Chinese population with hearing loss[13,34]. However, the pathogenic potential of this variant is difficult to define since it has been reported only in the heterozygous state without accompanying other mutations. Another novel GJB2 variant we found in this study was the p.R98P mutation which occurred in amino termini and intracellular loop, it is proposed that this mutation may impair the gap junctional intercellular communication in cochlea and finally lead to hair cell dysfunction. More interestingly, we found that the p.R98P variant co-segregates with the phenotype of hearing loss in the WL111 pedigree (Fig 1). But based on the current data, it is not possible to determine whether this variant is a dominant or a recessive mutation. Taking together, these 4 amino acid changes are very likely to be the possible causes of hearing loss. However, the functional impact of these putative causal variants remains speculative, the establishment of recombinant expression systems and animal models would be necessary for further investigation.
The p.V37I variant was originally reported as a polymorphism[35]. Subsequently, a series of studies reported that this variant exists in deafness patients in homozygous or compound heterozygous state among different populations[5,36]. However, its pathogenicity is still controversial. Ethnic differences were observed in the allele frequency of p.V37I variant, as it was not detected in the control subjects from Italy, Spain, Germany, Greece, Israel, Ghana, and Austria in spite of a high prevalence of the mutation in Eastern Asian population such as Chinese and Japanese descent[22]. In addition, the p.V37I mutant protein has normal subcellular localization and membrane expression as the wild type protein[37]. In this study, we found that the V37I allele was identified in 8.86% and 4.93% of the Chinese patient and control alleles, respectively. The p.V37I variant is located at transmembrane domain of Cx26, and the Valine residue at position 37 is highly conserved among different species. Moreover, functional analysis has proved that the coexpression at equimolar levels of wild type and mutant Cx26 proteins (p.V37I) in Xenopus oocytes resulted in a dominant-negative effect of the mutation[38]. In general, this variant results in a rather mild phenotype, which is consistent with previous reports that p.V37I missense mutation leads to a less severe hearing loss phenotype than frameshift mutations such as c.35delG and c.235delC mutations[39,40]. Taking into consideration the analysis of clinical, genetical and functional data regarding p.V37I available in the publications, this variant could also be a primary cause that leads to predisposition for hearing loss depending on genetic background and/or environmental factors.
In this report, we found 66 genotypes of GJB2 in the large cohort of 1067 hearing-impaired subjects including 24 novel genotypes. Totally, 20.25% of the subjects harbored two GJB2 pathogenic alleles, either homozygotes or compound heterozygotes, and there was a fairly high frequency (14.71%) of single heterozygosity in this cohort. More interestingly, patients with homozygous or compound heterozygous for the same pathogenic mutation exhibit a wide variation in the phenotypic expression of hearing loss. These data strongly supported the hypothesis that other factors may contribute to the clinical manifestation of GJB2-related deafness, such as alterations in regulatory region, additional mutations in other genes such as GJB6 and GJB3, and environmental factors[6,41,42]. Accordingly, identification of GJB2 mutations can yield considerable helpful information to those hearing impaired population and their families. Understanding of the cause of hearing loss will effectively promote genetic counseling and implementation of various intervention strategies such as early rehabilitation and fitting of hearing aids, which is particularly important for infants in affected families. In addition, further studies emphasize on the disease associated factors are very important that would enable us to gain a deeper understanding of the complex molecular pathogenic mechanisms of hearing loss.
Supporting Information
The Cx26 protein has four transmembrane domains (M1–M4), connected by two extracellular loops (E1 andE2) and a cytoplasmic loop (CL). The NT and CT denote the N- and C-termini of the protein.
(TIF)
(DOC)
(DOC)
(DOCX)
Acknowledgments
We thank the patients and their families for the participation in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the 973 Program (2014CB541704) from the Ministry of Science and Technology of China (MXG), projects (81470685 and 81330024) from National Natural Science Foundation of China (YC and MXG), and China Postdoctoral Science Foundation (2013M531742 to JZ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mehl AL, Thomson V (2002) The Colorado newborn hearing screening project, 1992–1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 109: E7 [DOI] [PubMed] [Google Scholar]
- 2. Morton CC, Nance WE (2006) Newborn hearing screening—a silent revolution. N Engl J Med 354: 2151–2164. [DOI] [PubMed] [Google Scholar]
- 3. Hilgert N, Smith RJ, Van Camp G (2009) Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res 681: 189–196. 10.1016/j.mrrev.2008.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dror AA, Avraham KB (2010) Hearing impairment: a panoply of genes and functions. Neuron 68: 293–308. 10.1016/j.neuron.2010.10.011 [DOI] [PubMed] [Google Scholar]
- 5. Chan DK, Chang KW (2014) GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope 124: E34–53. 10.1002/lary.24332 [DOI] [PubMed] [Google Scholar]
- 6. Kenneson A, Van Naarden Braun K, Boyle C (2002) GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med 4: 258–274. [DOI] [PubMed] [Google Scholar]
- 7. Lucotte G, Diéterlen F (2005) The 35delG mutation in the connexin 26 gene (GJB2) associated with congenital deafness: European carrier frequencies and evidence for its origin in ancient Greece. Genet Test 9: 20–25. [DOI] [PubMed] [Google Scholar]
- 8. Yan D, Park HJ, Ouyang XM, Pandya A, Doi K, Erdenetungalag R, et al. (2003) Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in east Asians. Hum Genet 114: 44–50. [DOI] [PubMed] [Google Scholar]
- 9. Yao J, Lu Y, Wei Q, Cao X, Xing G (2012) A systematic review and meta-analysis of 235delC mutation of GJB2 gene. J Transl Med 10: 136 10.1186/1479-5876-10-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, et al. (1998) Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med 339: 1500–1505. [DOI] [PubMed] [Google Scholar]
- 11. Tekin M, Xia XJ, Erdenetungalag R, Cengiz FB, White TW, Radnaabazar J, et al. (2010) GJB2 mutations in Mongolia: complex alleles, low frequency, and reduced fitness of the deaf. Ann Hum Genet 74: 155–164. 10.1111/j.1469-1809.2010.00564.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. RamShankar M, Girirajan S, Dagan O, Ravi Shankar HM, Jalvi R, Rangasayee R, et al. (2003) Contribution of connexin26 (GJB2) mutations and founder effect to non-syndromic hearing loss in India. J Med Genet 40: e68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dai P, Yu F, Han B, Liu X, Wang G, Li Q, et al. (2009) GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med 7: 26 10.1186/1479-5876-7-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsukada K, Nishio S, Usami S (2010) A large cohort study of GJB2 mutations in Japanese hearing loss patients. Clin Genet 78: 464–470. 10.1111/j.1399-0004.2010.01407.x [DOI] [PubMed] [Google Scholar]
- 15. Liu XZ, Xia XJ, Ke XM, Ouyang XM, Du LL, Liu YH, et al. (2002) The prevalence of connexin 26 (GJB2) mutations in the Chinese population. Hum Genet 111: 394–397. [DOI] [PubMed] [Google Scholar]
- 16. Estivill X, Fortina P, Surrey S, Rabionet R, Melchionda S, D'Agruma L, et al. (1998) Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet 351: 394–398. [DOI] [PubMed] [Google Scholar]
- 17. Mueller RF, Nehammer A, Middleton A, Houseman M, Taylor GR, Bitner-Glindzciz M, et al. (1999) Congenital non-syndromal sensorineural hearing impairment due to connexin 26 gene mutations—molecular and audiological findings. Int J Pediatr Otorhinolaryngol 50: 3–13. [DOI] [PubMed] [Google Scholar]
- 18. Shen Z, Zheng J, Chen B, Peng G, Zhang T, Gong S, et al. (2011) Frequency and spectrum of mitochondrial 12S rRNA variants in 440 Han Chinese hearing impaired pediatric subjects from two otology clinics. J Transl Med 9: 4 10.1186/1479-5876-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li R, Greinwald JH Jr., Yang L, Choo DI, Wenstrup RJ, Guan MX. (2004) Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J Med Genet 41: 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han SH, Park HJ, Kang EJ, Ryu JS, Lee A, Yang YH, et al. (2008) Carrier frequency of GJB2 (connexin-26) mutations causing inherited deafness in the Korean population. J Hum Genet 53: 1022–1028. 10.1007/s10038-008-0342-7 [DOI] [PubMed] [Google Scholar]
- 21. Yilmaz A (2014) Bioinformatic Analysis of GJB2 Gene Missense Mutations. Cell Biochem Biophys [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 22. Bason L, Dudley T, Lewis K, Shah U, Potsic W, Ferraris A, et al. (2002) Homozygosity for the V37I Connexin 26 mutation in three unrelated children with sensorineural hearing loss. Clinical genetics 61: 459–464. [DOI] [PubMed] [Google Scholar]
- 23. Yuan Y, You Y, Huang D, Cui J, Wang Y, Wang Q, et al. (2009) Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in China. J Transl Med 7: 79 10.1186/1479-5876-7-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Usami S, Nishio SY, Nagano M, Abe S, Yamaguchi T (2012) Simultaneous screening of multiple mutations by invader assay improves molecular diagnosis of hereditary hearing loss: a multicenter study. PLoS One 7: e31276 10.1371/journal.pone.0031276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park HJ, Hahn SH, Chun YM, Park K, Kim HN (2000) Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope 110: 1535–1538. [DOI] [PubMed] [Google Scholar]
- 26. Lee KY, Choi SY, Bae JW, Kim S, Chung KW, Drayna D, et al. (2008) Molecular analysis of the GJB2, GJB6 and SLC26A4 genes in Korean deafness patients. Int J Pediatr Otorhinolaryngol 72: 1301–1309. 10.1016/j.ijporl.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang YC, Kung CY, Su MC, Su CC, Hsu HM, Tsai CC, et al. (2002) Mutations of Cx26 gene (GJB2) for prelingual deafness in Taiwan. Eur J Hum Genet 10: 495–498. [DOI] [PubMed] [Google Scholar]
- 28. Shi GZ, Gong LX, Xu XH, Nie WY, Lin Q, Qi YS. (2004) GJB2 gene mutations in newborns with non-syndromic hearing impairment in Northern China. Hear Res 197: 19–23. [DOI] [PubMed] [Google Scholar]
- 29. Santos RL, Wajid M, Pham TL, Hussan J, Ali G, Ahmad W, et al. (2005) Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet 67: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bajaj Y, Sirimanna T, Albert DM, Qadir P, Jenkins L, Bitner-Glindzicz M. (2008) Spectrum of GJB2 mutations causing deafness in the British Bangladeshi population. Clin Otolaryngol 33: 313–318. 10.1111/j.1749-4486.2008.01754.x [DOI] [PubMed] [Google Scholar]
- 31. Dzhemileva LU, Barashkov NA, Posukh OL, Khusainova RI, Akhmetova VL, Kutuev IA, et al. (2010) Carrier frequency of GJB2 gene mutations c.35delG, c.235delC and c.167delT among the populations of Eurasia. J Hum Genet 55: 749–754. 10.1038/jhg.2010.101 [DOI] [PubMed] [Google Scholar]
- 32. Khandelwal G, Bhalla S, Khullar M, Panda NK (2009) High frequency of heterozygosity in GJB2 mutations among patients with non-syndromic hearing loss. J Laryngol Otol 123: 273–277. 10.1017/S0022215108002892 [DOI] [PubMed] [Google Scholar]
- 33. Propst EJ, Stockley TL, Gordon KA, Harrison RV, Papsin BC (2006) Ethnicity and mutations in GJB2 (connexin 26) and GJB6 (connexin 30) in a multi-cultural Canadian paediatric Cochlear Implant Program. Int J Pediatr Otorhinolaryngol 70: 435–444. [DOI] [PubMed] [Google Scholar]
- 34. Hwa HL, Ko TM, Hsu CJ, Huang CH, Chiang YL, Oong JL, et al. (2003) Mutation spectrum of the connexin 26 (GJB2) gene in Taiwanese patients with prelingual deafness. Genet Med 5: 161–165. [DOI] [PubMed] [Google Scholar]
- 35. Kelley PM, Harris DJ, Comer BC, Askew JW, Fowler T, Smith SD, et al. (1998) Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am J Hum Genet 62: 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim SY, Park G, Han KH, Kim A, Koo JW, Chang SO, et al. (2013) Prevalence of p.V37I variant of GJB2 in mild or moderate hearing loss in a pediatric population and the interpretation of its pathogenicity. PLoS One 8: e61592 10.1371/journal.pone.0061592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oguchi T, Ohtsuka A, Hashimoto S, Oshima A, Abe S, Kobayashi Y, et al. (2005) Clinical features of patients with GJB2 (connexin 26) mutations: severity of hearing loss is correlated with genotypes and protein expression patterns. Journal of Human Genetics 50: 76–83. [DOI] [PubMed] [Google Scholar]
- 38. Palmada M, Schmalisch K, Bohmer C, Schug N, Pfister M, Lang F, et al. (2006) Loss of function mutations of the GJB2 gene detected in patients with DFNB1-associated hearing impairment. Neurobiol Dis 22: 112–118. [DOI] [PubMed] [Google Scholar]
- 39. Cryns K, Orzan E, Murgia A, Huygen PL, Moreno F, del Castillo I, et al. (2004) A genotype-phenotype correlation for GJB2 (connexin 26) deafness. J Med Genet 41: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao FF, Ji YB, Wang DY, Lan L, Han MK, Li Q, et al. (2011) Phenotype–Genotype Correlation in 295 Chinese Deaf Subjects with Biallelic Causative Mutations in the GJB2 Gene. Genet Test Mol Biomarkers 15: 619–625. 10.1089/gtmb.2010.0192 [DOI] [PubMed] [Google Scholar]
- 41. del Castillo I, Villamar M, Moreno-Pelayo MA, del Castillo FJ, Alvarez A, Telleria D, et al. (2002) A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med 346: 243–249. [DOI] [PubMed] [Google Scholar]
- 42. Ortolano S, Di Pasquale G, Crispino G, Anselmi F, Mammano F, Chiorini JA. (2008) Coordinated control of connexin 26 and connexin 30 at the regulatory and functional level in the inner ear. Proc Natl Acad Sci U S A 105: 18776–18781. 10.1073/pnas.0800831105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Cx26 protein has four transmembrane domains (M1–M4), connected by two extracellular loops (E1 andE2) and a cytoplasmic loop (CL). The NT and CT denote the N- and C-termini of the protein.
(TIF)
(DOC)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.