Abstract
Background
Several recent studies have identified that the TERT genetic polymorphism rs2853676 is associated with cancer risk, but presented inconsistent results. We investigated these inconclusive results by performing a meta-analysis to systematically evaluate the association.
Methods
We conducted a search in PubMed, Google Scholar and ISI Web of Science to select studies on the association between TERT rs2853676 and cancer risk. We conducted a stratified analysis using cancer type, ethnicity and source of controls. We calculated the odds ratios (OR) and 95% confidence intervals (CI). Article quality, heterogeneity, sensitivity, publication bias and statistical power were also assessed.
Results
26 articles covering 76 108 cases and 134 215 controls met our inclusion criteria. A significant association between TERT rs2853676 allele A and cancer susceptibility was demonstrated under a per-allele risk analysis (OR = 1.08, 95% CI = 1.04-1.13). Stratification analysis revealed an increased cancer risk in subgroups of glioma, lung cancer and ovarian cancer. No significant increase was found in melanoma, breast cancer, pancreatic cancer and colorectal cancer. In a subgroup analysis of lung cancer, a statistically significant increase was only observed in adenocarcinoma. Moreover, a stratified analysis performed for ethnic groups revealed that the significant increase was only observed in Caucasians, whereas a non-significant increase was found in Asians.
Conclusions
This meta-analysis suggests that the TERT genetic polymorphism rs2853676 is associated with increased risk of glioma, lung adenocarcinoma and ovarian cancer among Caucasians. Further functional studies are warranted to validate this association and investigate further.
Introduction
Cancer is a major global public health problem. Fourteen million people were diagnosed with cancer worldwide in 2012. By 2032, the global cancer incidence is predicted to reach to 25 million [1]. In the United States, cancer is the second leading cause of death following heart disease and the leading cause of death among adults aged between 40 and 79 years [2]. Although the causes of cancer are multi-factorial, genetic and environmental factors play an important role in cancer pathogenesis. Recent epidemiological studies have identified several genetic polymorphism loci on chromosome 5p15.33 that are associated with the risk of many types of cancer [3–5]. Chromosome 5p15.33 contains two key genes, cleft lip and palate transmembrane 1-like (CLPTM1L) and telomerase reverse transcriptase (TERT).
As the main catalytic subunit of telomerase, TERT is essential for the maintenance of telomere DNA length in chromosomes [6]. Telomerase is an RNA-dependent DNA polymerase that synthesizes repetitive DNA (TTAGGG repeats) sequences, which bind abundant specialized proteins onto the chromosome ends [7]. The telomeres prevent coding sequence erosion and protect chromosomes from rearrangements, fusion and genome instability by conducting chromosomal complete replication and regulating gene expression [8]. The expression of telomerase is extremely low in most normal human somatic cells, but is present in over 90% of human malignancies. In vitro immortalized cells and the stem cell lines of actively proliferating tissues show a high level of telomerase expression [9, 10]. The activation of telomerase is a vital step during cellular immortalization and the malignant transformation of human cells. This activation requires the TERT catalyst [11].
A series of important cancer-related polymorphisms have been reported within the TERT gene using a meta-analysis approach and have been identified as contributing to the risk of several cancers, such as the susceptibility to rs2736098 for lung and bladder cancer [12] and that to rs2736100 for lung cancer and glioma [13]. The rs2853676 polymorphism has been mapped to intron 2 of the TERT gene, which was implicated in an increased risk of glioma in 2009 [14]. Since then, several studies have assessed the association between the polymorphism and cancer risk, but have presented inconclusive results. We performed a meta-analysis to summarize the available evidence and more precisely characterize the relationship between the TERT rs2853676 polymorphism and cancer risk.
Material and Methods
Search strategy
According to the Meta-analysis of Observational Studies in Epidemiology guidelines [15], we conducted systematic searches in PubMed, Google Scholar and ISI Web of Science, up to September 20, 2014. We used the systemic literature search terms “TERT or rs2853676,” “polymorphism or variant” and “cancer or carcinoma or tumor or neoplasm.” All related reference articles and review articles were searched to identify additional relevant eligible publications. Unpublished data were also obtained from the authors by e-mail.
Inclusion and exclusion criteria
Identified studies meeting all of the following criteria were included: (1) articles about the TERT polymorphism rs2853676 and cancer risk that were published in English; (2) a case-control or case-cohort design addressing race and the numbers of affected and unaffected human control subjects; and (3) sufficient data to calculate an odds ratio (OR) with a 95% confidence interval (CI). The exclusion criteria were: (1) investigations in subjects with family cancer risk; (2) published as an abstract, summary, case report, comment letter, review or editorial; and (3) in overlapped case series, in which case all but the latest or largest study were excluded.
Data extraction
Data were extracted independently by two investigators, according to the inclusion and exclusion criteria listed above. Discrepancies were resolved by discussion and consensus. We extracted the first author, year of publication, cancer type, patient ethnicity, source of control group (population-based, hospital-based, multiple or nested-in-cohort controls), number of cases and controls, genotyping method, histological subtype, minor allele frequency, genotype and/or per-allele risk OR and 95% CI from each study. The data were extracted separately by population or cancer type, if these were explicitly given. The quality of each study was evaluated using previously published quality assessment criteria [16]. The quality scores of the studies ranged from 0 to 15. Scores ≤ 9 were considered to indicate low quality, while those > 10 were considered to indicate high quality.
Statistical analysis
Statistical analyses were performed using Stata 12.0 software (Stata Corporation, College Station, Texas). All of the tests were two-sided with a p-value. The Hardy-Weinberg equilibrium among the control subjects was assessed with a chi-square test, in which p < 0.05 suggested a significant deviation from equilibrium. The OR and 95% CI were calculated to assess the strength of the association between the rs2853676 polymorphism and cancer risk. The significance of the combined OR was determined with a Z test, in which p < 0.05 was considered statistically significant. Stratified analyses based on cancer type, ethnicity, histological subtype and source of controls were quantified with ORs and 95% CIs. Ethnicity data sets were categorized as Caucasian, Asian, African or multiple. If a cancer type contained only one data source, it was combined into the “other cancers” group.
The heterogeneity between the studies was calculated by Cochran’s Q-test, in which p < 0.10 indicated significant heterogeneity. If the heterogeneity was significant, the random-effects model (DerSimonian and Laird method) was applied [17], otherwise the fixed-effects model was used (Mantel-Haenszel method) [18]. The I 2 was calculated to quantitatively estimate the heterogeneity, with I 2 < 25%, I 2 = 25–75% and I 2 > 75% representing low, moderate and high heterogeneity, respectively [19].
Sensitivity analyses were performed by sequential removal of each study to assess the stability of the results. Begg’s funnel plots and Egger’s linear regression tests were used to examine the publication bias, in which p < 0.05 indicated statistical significance [20]. Moreover, we estimated the statistical power of each subgroup analysis. Power analyses of the meta-analyses were all conducted using PS (Power and Sample Size Calculations) software version 3.0.5. To guard against Type I errors, α was typically set at 0.05, while β was set at 0.20 to guard against Type II errors, thus a sufficient power of the statistical test would be greater than 80% [21].
Results
Eligible studies
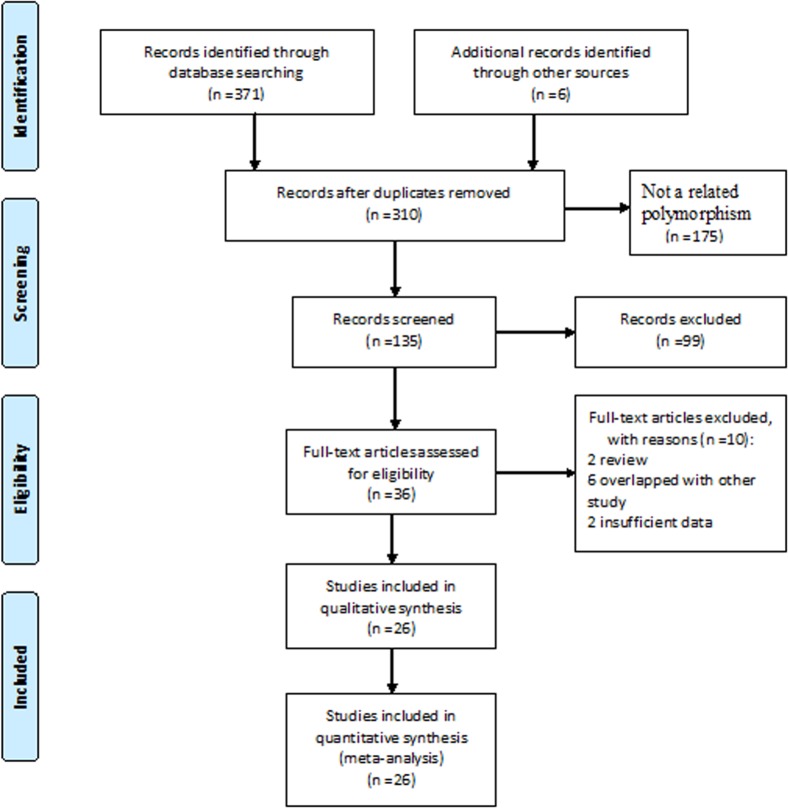
After a comprehensive search, 310 relevant articles were retrieved. Screening of the titles and abstracts excluded 175 articles. Following a full text review and detailed evaluations, 26 articles covering 32 case-control studies with 76 108 cases and 134 215 controls met our inclusion criteria (Fig 1) [4, 14, 22–45]. Among the 32 studies, nine focused on glioma [14, 31, 32, 37, 40], three each on lung cancer [4, 26, 45], breast cancer [22, 39, 43] and melanoma [24, 33, 35], two each on pancreatic cancer [23, 27], ovarian cancer [31, 38] and colorectal cancer [36, 42], and one each on nasopharyngeal cancer [25], endometrial cancer [28], neuroblastoma [34], prostate cancer [41], testicular germ cell cancer [29], acute lymphoblastic leukemia [44], skin squamous cell carcinoma and Basal cell carcinoma [33].
Fig 1. Flow chart of the literature search and selection procedures.
Six studies focused on Asians [25, 26, 31, 37, 44, 45], twenty-four on Caucasians [14, 22–24, 27–30, 32–36, 38, 40–43] and one each on Africans [39] and multiple populations [4]. Ten studies used population-based controls [14, 24, 32, 36, 42, 43, 45], eight used hospital-based controls [14, 25, 31, 33, 35, 37, 40, 44], five used nested-in-cohort controls [22, 28, 33] and nine used multiple controls [4, 23, 26, 27, 29, 30, 38, 39, 41]. The studies used genotyping methods such as Illumina, iPLEX and TaqMan (Table 1). The minor allele frequencies of the control subjects were 25.31% in Caucasians, 17.10% in Asians and 26.4% in Africans.
Table 1. Study characteristics of the association between the rs2853676 polymorphism and cancer risk in this meta-analysis.
| Author(year) | Ethnicity | Cancer type | Source | Method | Cases/Controls | MAF | OR (95%CI) | Score |
|---|---|---|---|---|---|---|---|---|
| Hunter et al.(2007) | Caucasian | Breast | Nested in cohort | Illumina | 1145/1142 | 23.8 | 0.97(0.81–1.15) | 12 |
| Amundadottir et al. (2009) a | Caucasian | Pancreas | Multiple | Illumina | 1896/1939 | 23.8 | 0.90(0.81–1.00) | 11 |
| Falchi et al. (2009) | Caucasian | Melanoma | PB | Illumina | 3131/3702 | 23.8 | 0.98(0.89–1.07) | 13 |
| Shete et al. (French 2009) | Caucasian | Glioma | PB | Illumina | 1361/1490 | 27.0 | 1.29(1.15–1.44) | 14 |
| Shete et al. (German2009) | Caucasian | Glioma | PB | Illumina | 498/565 | 27.0 | 1.32(1.10–1.59) | 14 |
| Shete et al. (Sweden 2009) | Caucasian | Glioma | PB | Illumina | 639/760 | 27.0 | 1.30(1.10–1.54) | 14 |
| Shete et al. (England 2009) | Caucasian | Glioma | PB | Illumina | 631/1433 | 27.0 | 1.14(0.99–1.32) | 14 |
| Shete et al. (America 2009) | Caucasian | Glioma | HB | Illumina | 1247/2234 | 27.0 | 1.26(1.14–1.41) | 12 |
| Bei et al. (2010) b | Asian | Nasopharynx | HB | Illumia | 1582/1894 | 16.3 | 0.97(0.85–1.11) | 12 |
| Hsiung et al. (2010) | Asian | Lung | Multiple | Illumia | 2539/2535 | 17.3 | 1.08(0.98–1.20) | 10 |
| Petersen et al. (2010) c | Caucasian | Pancreas | Multiple | Illumia | 1955/1995 | 23.8 | 0.96(0.86–1.07) | 11 |
| Prescott et al. (2010) d | Caucasian | Endometrium | Nested in cohort | Taqman | 651/1605 | 26.1 | 1.08(0.94–1.25) | 12 |
| Turnbull et al. (2010) | Caucasian | Testicular germ cell cancer | Multiple | Illumia | 979/4947 | 27.7 | 0.75(0.67–0.84) | 9 |
| Beesley et al. (2011)e | Caucasian | Ovarian | Multiple | iPLEX | 990/3687 | 26.0 | 1.05(0.93–1.18) | 10 |
| Chen et al. (2011) | Asian | Glioma | HB | iPLEX | 948/1041 | 16.0 | 1.27(1.08–1.49) | 11 |
| Egan et al. (2011) | Caucasian | Glioma | PB | Illumia | 639/649 | 28.7 | 1.22(1.03–1.44) | 13 |
| Nan 1 et al. (2011) | Caucasian | Melanoma | Nested in cohort | Taqman | 218/840 | 25.2 | 1.42(1.13–1.78) | 13 |
| Nan 2 et al. (2011) | Caucasian | SCC(skin) | Nested in cohort | Taqman | 281/840 | 25.2 | 1.10(0.88–1.36) | 13 |
| Nan 3 et al. (2011) | Caucasian | BCC(skin) | Nested in cohort | Taqman | 284/840 | 25.2 | 1.03(0.83–1.29) | 13 |
| Wang et al. (2011) | Caucasian | Neuroblastoma | HB | Illumia | 2251/6097 | 27.3 | 1.03(0.95–1.11) | 11 |
| Bodelon et al. (2012) f | Caucasian | Melanoma | HB | Illumia | 796/770 | 30.0 | 1.17(0.84–1.63) | 12 |
| Hofer et al. (2012) | Caucasian | Colorectal | PB | Taqman | 137/1705 | 25.0 | 1.20(0.91–1.58) | 14 |
| Liu et al. (2012) | Asian | Glioma | HB | iPLEX | 312/311 | 23.9 | 0.89(0.69–1.15) | 11 |
| Terry et al. (2012) g | Caucasian | Ovarian | Multiple | TaqMan | 2112/2456 | 20.0 | 1.13(1.02–1.25) | 10 |
| Zheng et al. (2012) | African | Breast | Multiple | Illumia | 1508/1383 | 22.8 | 1.02(0.91–1.16) | 12 |
| Jin et al. (2013) | Caucasian | Glioma | HB | iPLEX | 433/463 | 15.9 | 1.56(1.23–1.98) | 11 |
| Kote-Jarai et al. (2013) | Caucasian | Prostate | Multiple | Illumia or iPLEX | 22301/22320 | 25.8 | 1.09(1.05–1.12) | 11 |
| Pellatt 1 et al. (2013) | Caucasian | Colorectal | PB | TaqMan | 2308/2914 | 26.0 | 1.04(0.94–1.14) | 13 |
| Pellatt 2 et al. (2013) h | Caucasian | Breast | PB | Illumia | 3533/4102 | 19.0 | 1.23(1.00–1.51) | 13 |
| Sheng et al. (2013) | Asian | ALL | HB | Taqman | 567/669 | 16.1 | 1.19(0.97–1.47) | 12 |
| Zhao et al. (2013) | Asian | Lung | PB | SNPscan | 782/778 | 18.0 | 0.98(0.82–1.18) | 14 |
| Park et al. (2014) | Multiple | Lung | Multiple | Illumia | 17454/57789 | 26.4 | 1.05(1.02–1.09) | 11 |
ALL: Acute lymphoblastic leukemia; SCC: Squamous cell carcinoma; BCC: Basal cell carcinoma; PB: Population based; HB: Hospital based; MAF: Minor allele frequency in control subjects;
a,c Adjusted for study, sex, ancestry and five principal components of population stratification;
b,f Adjusted for age and gender;
d,e Adjust for age and study;
g Adjusted for age (continuous), center, oral contraceptive use, parity, family history of breast or ovarian cancer, and tubal ligation;
h Adjusted for age, study, BMI, vigorous activity in referent year, parity, age at first birth, alcohol consumption, and genetic admixture.
Meta-analysis results
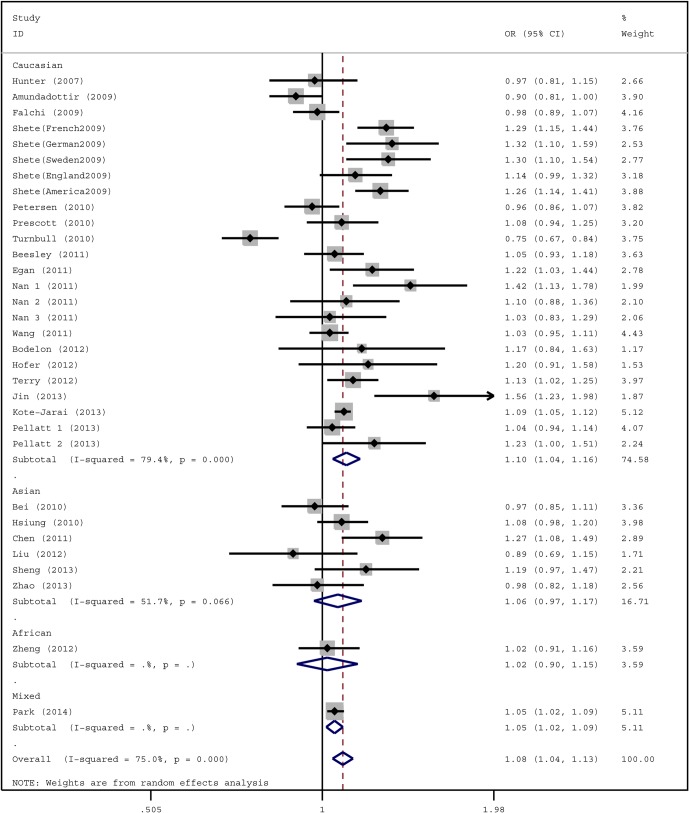
Eight studies were based on adjustment data [23, 25, 27, 28, 30, 35, 38, 43]. These studies had a small effect on the synthesis and did not significantly alter the OR, which agreed with previous results [46, 47]. Based on the data from all 32 studies, we found a significant increased cancer risk for the TERT rs2853676 A allele under a per-allele risk analysis (OR = 1.08, 95% CI = 1.04–1.13, p < 0.001), with a statistical power of 100%. The results from a random effect model showed significant heterogeneity (p heterogeneity < 0.001, I 2 = 75.0%) (Fig 2).
Fig 2. Forest plot of the ORs for the overall cancer risk associated with the rs2853676 polymorphism.
Stratification analysis identified increased cancer risk in subgroups of glioma (per-allele OR = 1.25, 95% CI = 1.19–1.32, p heterogeneity = 0.123, I 2 = 36.9%), lung cancer (per-allele OR = 1.05, 95% CI = 1.02–1.08, p heterogeneity = 0.654, I 2 = 0.0%), ovarian cancer (per-allele OR = 1.10, 95% CI = 1.01–1.18, p heterogeneity = 0.358, I 2 = 0.0%). No significant increase in risk was found in melanoma, breast cancer, pancreatic cancer and colorectal cancer (Table 2). In a subgroup analysis of lung cancer, a statistically significant increase was observed in adenocarcinoma (OR = 1.14, 95% CI = 1.09–1.19, p heterogeneity = 0.616, I 2 = 0.0%) (Table 3). A non-significant difference was found in squamous cell carcinoma (OR = 0.98, 95% CI = 0.92–1.05, p heterogeneity = 0.762, I 2 = 0.0%) and small cell lung carcinoma (OR = 1.05, 95% CI = 0.97–1.15, p heterogeneity = 0.826, I 2 = 0.0%) (data not shown). Moreover, a stratified analysis performed on the ethnicity of the groups revealed that the significant risk was only observed in Caucasians (per-allele OR = 1.10, 95% CI = 1.04–1.16, p heterogeneity < 0.001, I 2 = 79.4%), but a non-significant risk was found in Asians (per-allele OR = 1.06, 95% CI = 0.97–1.17, p heterogeneity = 0.066, I 2 = 51.7%). (Fig 2 and Table 2).
Table 2. Stratified analyses of the rs2853676 polymorphism and cancer risk.
| Category | No. of | Random effect model | Fixed effect model | I 2 (%) | P heterogeneity | P egger | Power (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| data sets | Cases/Controls | OR(95%CI) | P | OR(95%CI) | P | |||||
| Total | 32 | 76108/134215 | 1.08(1.04–1.13) | <0.001 | - | - | 75.0 | <0.001 | 0.369 | 100.0 |
| Cancer type | ||||||||||
| Glioma | 9 | 6708/8946 | - | - | 1.25(1.19–1.32) | <0.001 | 36.9 | 0.123 | 0.687 | 100.0 |
| Lung cancer | 3 | 20075/61102 | - | - | 1.05(1.02–1.08) | 0.002 | 0.0 | 0.654 | 0.821 | 96.2 |
| Breast cancer | 3 | 6186/6627 | - | - | 1.04(0.95–1.14) | 0.357 | 38.4 | 0.197 | 0.618 | 24.8 |
| Melanoma | 3 | 4145/5312 | 1.16(0.89–1.50) | 0.265 | - | - | 78.5 | 0.010 | 0.399 | 99.4 |
| Pancreas | 2 | 3851/3934 | - | - | 0.93(0.86–1.00) | 0.055 | 0.0 | 0.405 | - | 48.0 |
| Ovarian cancer | 2 | 3102/6143 | - | - | 1.10(1.01–1.18) | 0.021 | 0.0 | 0.358 | - | 75.0 |
| Colorectal | 2 | 2445/4619 | - | - | 1.06(0.96–1.16) | 0.238 | 0.0 | 0.337 | - | 30.7 |
| Other cancer | 8 | 28896/38372 | 1.01(0.92–1.11) | 0.794 | - | - | 83.5 | <0.001 | 0.383 | 12.4 |
| Ethnicity | ||||||||||
| Caucasian | 24 | 50416/67815 | 1.10(1.04–1.16) | 0.001 | - | - | 79.4 | <0.001 | 0.410 | 100.0 |
| Glioma | 7 | 5448/7594 | - | - | 1.27(1.20–134) | <0.001 | 0.0 | 0.471 | 0.314 | 100.0 |
| Breast cancer | 2 | 4678/5244 | 1.09(0.86–1.37) | 0.490 | - | - | 66.2 | 0.085 | - | 69.1 |
| Asian | 6 | 6730/7228 | 1.06(0.97–1.17) | 0.203 | - | - | 51.7 | 0.066 | 0.841 | 45.6 |
| Glioma | 2 | 1260/1352 | 1.08(0.76–1.53) | 0.669 | - | - | 81.2 | 0.021 | - | 19.0 |
| Lung cancer | 2 | 3321/3313 | - | - | 1.06(0.97–1.15) | 0.232 | 0.0 | 0.360 | - | 45.2 |
| African | 1 | 1508/1383 | 1.02(0.90–1.15) | 0.749 | - | - | - | - | - | 6.1 |
| Multiple | 1 | 17454/57789 | 1.05(1.02–1.09) | 0.004 | - | - | - | - | - | 94.3 |
| Source of control | ||||||||||
| Population based | 10 | 13659/18098 | 1.15(1.06–1.25) | 0.001 | - | - | 66.2 | 0.002 | 0.133 | 100.0 |
| Hospital based | 8 | 8136/13479 | 1.14(1.03–1.28) | 0.016 | - | - | 73.9 | <0.001 | 0.471 | 100.0 |
| Nested in cohort | 5 | 2579/3587 | - | - | 1.09(1.00–1.18) | 0.045 | 44.3 | 0.127 | 0.487 | 54.5 |
| Multiple | 9 | 51734/99051 | 1.00(0.94–1.07) | 0.917 | - | - | 84.8 | <0.001 | 0.148 | 5.0 |
Random effects model was applied when P value for heterogeneity test<0.10, otherwise, fixed effect model was used;
Power calculations assume a = 0.05.
Table 3. Stratified analyses of the rs2853676 polymorphism in lung adenocarcinoma.
| Author(year) | Ethnicity | Cancer type | Source | Method | Cases/Controls | OR (95%CI) |
|---|---|---|---|---|---|---|
| Hsiung et al. (2010) | Asian | Adenocarcinoma | Multiple | Illumia | 1930/2535 | 1.06(0.95–1.19) |
| Zhao et al. (2013) | Asian | Adenocarcinoma | PB | SNPscan | 359/778 | 1.04(0.83–1.31) |
| Park et al. (MEC 2014) | Multiple | Adenocarcinoma | Multiple | Illumia | 252/9587 | 1.19(0.96–1.47) |
| Park et al. (WHI 2014) | Multiple | Adenocarcinoma | Multiple | Illumia | 760/5825 | 1.11(0.98–1.25) |
| Park et al. (DeCode Genetics 2014) | Caucasian | Adenocarcinoma | PB | Illumia | 346/11225 | 1.03(0.86–1.22) |
| Park et al. (Harvard 2014) | Caucasian | Adenocarcinoma | PB | Illumia | 488/970 | 1.26(1.06–1.51) |
| Park et al. (HGF Germany 2014) | Caucasian | Adenocarcinoma | PB | Illumia | 188/479 | 1.18(0.84–1.67) |
| Park et al. (IARC GWAS 2014) | Multiple | Adenocarcinoma | Multiple | Illumia | 586/3174 | 1.25(1.08–1.44) |
| Park et al. (MDACC 2014) | Caucasian | Adenocarcinoma | Multiple | Illumia | 619/1133 | 1.09(0.93–1.28) |
| Park et al. (NCI GWAS 2014) | Multiple | Adenocarcinoma | Multiple | Illumia | 1836/5686 | 1.19(1.09–1.29) |
| Park et al. (SLRI/ Toronto 2014) | Caucasian | Adenocarcinoma | PB | Illumia | 89/488 | 1.07(0.76–1.51) |
PB: Population based; HB: Hospital based;
The overall OR = 1.14, 95% CI: 1.09–1.19, P <0.001; P heterogeneity = 0.616, I 2 = 0.0%.
A stratified analysis by source of controls indicated a significantly increased risk associated with population-based, hospital-based, and nested-in-cohort controls, with ORs of 1.15 (95% CI = 1.06–1.25), 1.14 (95% CI = 1.03–1.28), and 1.09 (95% CI = 1.00–1.18), respectively. No significant increase was found in multiple-source controls. A stratified analysis by cancer type was also performed in Caucasians and Asians. The results for glioma and breast cancer were the same in Caucasians as in the overall population, but a non-significant increase in risk was found for glioma and lung cancer in Asians (Table 2).
Heterogeneity test and sensitivity analyses
Significant heterogeneity existed, mainly in all cancer and the subgroups of ethnicity, population-based controls, hospital-based controls and multiple-source controls. However, most of the heterogeneity disappeared, except melanoma and “other cancers,” in the analysis of cancer type subgroups (Table 2).
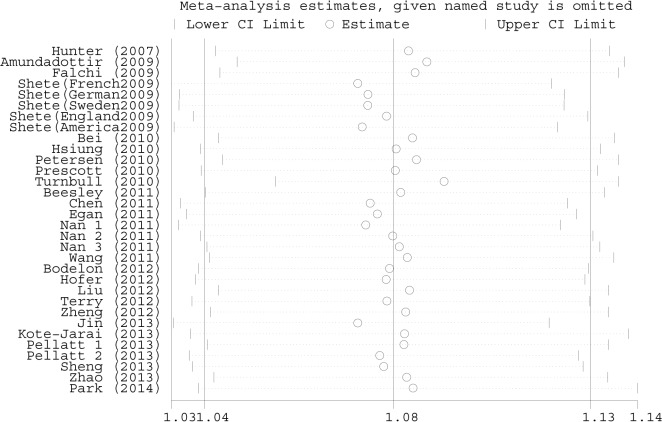
A sensitivity analysis was conducted to assess the influence of each study, by sequential omission of each eligible study. The results showed that the significance of the OR was not affected by any single study (Fig 3). In addition, after the removal of a study that resulted in a departure from the Hardy-Weinberg equilibrium, no significant alteration was found in the OR.
Fig 3. Sensitivity analysis of the overall ORs.
The results were calculated by omitting each eligible study. Meta-analysis random-effects estimates (exponential form) were used.
Publication bias
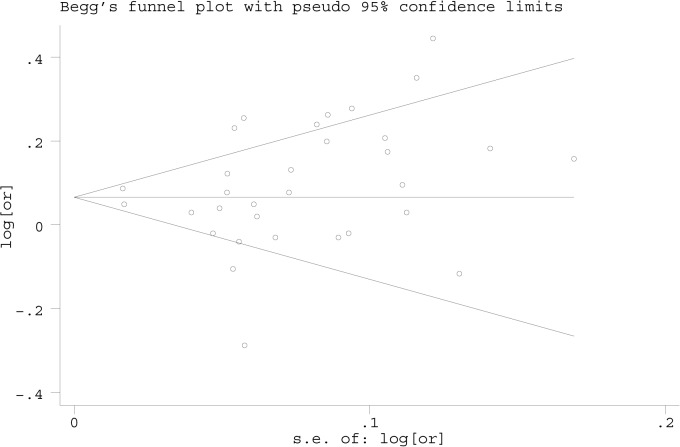
Publication bias was assessed with Begg’s funnel plots and Egger’s test. The shapes of the funnel plots did not show any evidence of publication bias (Fig 4). No significant publication bias was found by Egger’s test in the overall or subgroup analyses (Table 2).
Fig 4. Funnel plot analysis to detect publication bias for the rs2853676 polymorphism in the 32 data sets.
Discussion
The TERT gene is the main catalytic subunit of telomerase, which is encoded by a single-copy gene, mapped on chromosome 5p15.33 and contains 16 exons and 15 introns spanning about 35 kb [48]. The gene consists of three distinct structural domains: an RNA-binding domain, a reverse transcriptase domain and a carboxy- terminal extension, which is thought to represent the putative thumb domain of TERT [49]. A high level of TERT expression is involved in a variety of human malignancies. TERT may play an important role in the pathogenesis of cancer [6, 9, 50]. The TERT gene sequence has been proposed as a general mechanism affecting individual susceptibility to cancer risk [4, 5, 47]. A growing number of epidemiological studies have been conducted in response, which have provided evidence that TERT polymorphisms contribute to cancer development [4, 5].
The polymorphism rs2853676 is located in intron 2 of the TERT gene. The association between this polymorphism and cancer risk has been assessed in several studies, which showed inconclusive results. Only one meta-analysis demonstrated strong evidence that rs2853676 increased the risk of central nervous system tumors, but the evidence for the risk of lung cancer was weak [51]. A recent study from the Population Architecture using the Genomics and Epidemiology and Transdisciplinary Research in Cancer of the Lung consortia identified that rs2853676 was associated with an increased risk of lung adenocarcinoma [4]. Our meta-analysis suggested that the TERT genetic polymorphism rs2853676 allele A increased several cancer risk, based on 76 108 cases and 134 215 controls. The association mainly existed in the Caucasian population, especially for glioma, lung cancer and ovarian cancer. No significant association was found in melanoma, breast cancer, pancreatic cancer or colorectal cancer. In a subgroup analysis of lung cancer, a statistically significant association was only observed in adenocarcinoma. Interestingly, the Asian population showed no significant result in any type of cancer. Notably, one study described an increased risk for prostate cancer [41], whereas another described a reduced risk for testicular germ cell cancer [29]. Further studies are required to validate these associations in urogenital system tumors.
The heterogeneity between our studies was significantly reduced in the analysis of the cancer type subgroups, indicating that the effect of TERT polymorphisms may be modified by tumor origin. The effect may be cancer-type specific and play a different role in the etiology of different tumors [47]. However, the exact functional mechanisms underlying the association between the rs2853676 polymorphism and cancer remains unclear. Several studies have suggested that telomere length alters cancer risk [5, 33, 52]. However, this alteration has not been observed in rs2853676 [28, 38, 43], except by Melin et al., who detected potential relevance at higher ages in a small sample [53]. The other plausible mechanisms underlying the association between the rs2853676 polymorphism and cancer risk may be attributable to environmental risk factors or genetic background. The modification of the TERT function is likely to also play an important role. A high linkage disequilibrium with other nearby biologically potential functional polymorphisms or disease-causing mutations may also exist. In National Cancer Institute controls, rs2853676 was in modest linkage disequilibrium (r2 = 0.25, D’ = 0.82) with rs2736100 and showed a similar pattern of association with lung adenocarcinoma (OR = 1.16, p = 3.44×10–4) [54]. A recent Japanese study also identified that TERT rs2853677 (European ancestry: r2 = 0.59) was associated with lung adenocarcinoma (p = 3.1×10–40) [55]. Park and collaborators speculated that the association between rs2853676 and adenocarcinoma may be influenced by rs2736100 and rs2853677 [4].
Several limitations in this meta-analysis should be addressed. First, our meta-analysis only presented limited studies that were available to adjust the estimates, and more individual data would be required to draw a more precise conclusion. Second, gene-gene and gene-environment interactions may have influenced our results, as cancer is mainly caused by genetic and environmental factors. However, no appropriate information was available to test this. Third, not all of the authors of the included studies agreed to provide their data and exact genotype data were reported in a minority of the studies. The analysis was therefore only conducted with an additive model (per-allele risk analysis). Fourth, in this meta-analysis, the power of several subgroup results was < 80%, indicating that additional high-level studies are still needed.
In conclusion, this meta-analysis suggests that the TERT genetic polymorphism rs2853676 is associated with an increased risk of glioma, lung adenocarcinoma and ovarian cancer among Caucasians, suggesting that the association may be cancer-type and ethnically specific. To validate this association and investigate our findings further, functional studies are warranted.
Supporting Information
A list of full-text excluded articles.
(DOC)
Meta-analysis on Genetic Association Studies Checklist.
(DOCX)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (NSFC 31170720). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bender E. Developing world: Global warning. Nature 2014;509:S64–65. 10.1038/509S64a [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Ma J, Zou Z, Jemal A. Cancer Statistics, 2014. CA Cancer J Clin. 2014; 64:9–29. 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 3. Freier K, Pungs S, Flechtenmacher C, Bosch FX, Lichter P, Joos S, et al. Frequent high telomerase reverse transcriptase expression in primary oral squamous cell carcinoma. J Oral Pathol Med. 2007;36:267–272. [DOI] [PubMed] [Google Scholar]
- 4. Park SL, Fesinmeyer MD, Timofeeva M, Caberto CP, Kocarnik JM, Han Y, et al. Pleiotropic associations of risk variants identified for other cancers with lung cancer risk: The PAGE and TRICL consortia. J Natl Cancer Inst. 2014;106:dju061 10.1093/jnci/dju061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. 10.1038/ng.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Young NS. Telomere biology and telomere diseases: implications for practice and research. Hematology Am Soc Hematol Educ Program. 2010;2010:30–35. 10.1182/asheducation-2010.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66:407–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pooley KA, Sandhu MS, Tyrer J, Shah M, Driver KE, Luben RN, et al. Telomere length in prospective and retrospective cancer case-control studies. Cancer Res. 2010;70:3170–3176. 10.1158/0008-5472.CAN-09-4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science 1994;266:2011–2015. [DOI] [PubMed] [Google Scholar]
- 10. Wyatt HD, West SC, Beattie TL. In TERT preting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. 10.1093/nar/gkq370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang A, Zheng C, Lindvall C, Hou M, Ekedahl J, Lewensohn R, et al. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000;60:6230–6235. [PubMed] [Google Scholar]
- 12. Wu H, Qiao N, Wang Y, Jiang M, Wang S, Wang C, et al. Association between the Telomerase Reverse Transcriptase (TERT) rs2736098 polymorphism and cancer risk: evidence from a case-control study of non-small-cell lung cancer and a meta-analysis. PLoS One. 2013; 8:e76372 10.1371/journal.pone.0076372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou P, Gu A, Ji G, Zhao L, Zhao P, Lu A. The TERT rs2736100 polymorphism and cancer risk: a meta-analysis based on 25 case-control studies. BMC Cancer. 2012;12:7 10.1186/1471-2407-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. 10.1038/ng.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology. J Amer Med Assoc. 2000;283:2008–2012. [DOI] [PubMed] [Google Scholar]
- 16. He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, et al. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: Evidence from a meta-analysis. Sci Rep. 2014;4:6159 10.1038/srep06159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. [DOI] [PubMed] [Google Scholar]
- 18. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 19. Higgins J, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. [DOI] [PubMed] [Google Scholar]
- 20. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muncer S, Taylor S, Craigie M. Power dressing and meta-analysis: incorporating power analysis into meta-analysis. J Adv Nurs. 2002; 38:274–280. [DOI] [PubMed] [Google Scholar]
- 22. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. 10.1038/ng.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JA, Pastinen T, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41:915–919. 10.1038/ng.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42:599–603. 10.1038/ng.601 [DOI] [PubMed] [Google Scholar]
- 26. Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6:e1001051 10.1371/journal.pgen.1001051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. 10.1038/ng.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prescott J, McGrath M, Lee IM, Buring JE, De Vivo I. Telomere length and genetic analyses in population-based studies of endometrial cancer risk. Cancer 2010;116:4275–4282. 10.1002/cncr.25328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. 10.1038/ng.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beesley J, Pickett HA, Johnatty SE, Dunning AM, Chen X, Li J, et al. Functional polymorphisms in the TERT promoter are associated with risk of serious epithelial ovarian and breast cancers. PLoS one 2011;6:e24987 10.1371/journal.pone.0024987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen H, Chen Y, Zhao Y, Fan W, Zhou K, Liu Y, et al. Association of Sequence Variants on Chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) With Glioma Susceptibility in a Chinese Population. Am J Epidemiol. 2011;173:915–922. 10.1093/aje/kwq457 [DOI] [PubMed] [Google Scholar]
- 32. Egan KM, Thompson RC, Nabors LB, Olson JJ, Brat DJ, Larocca RV, et al. Cancer susceptibility variants and the risk of adult glioma in a US case-control study. J Neurooncol. 2011;104:535–542. 10.1007/s11060-010-0506-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nan H, Qureshi AA, Prescott J, De Vivo I, Han J. Genetic variants in telomere-maintaining genes and skin cancer risk. Hum Genet. 2011;129:247–253. 10.1007/s00439-010-0921-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature 2011;469:216–220. 10.1038/nature09609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bodelon C, Pfeiffer RM, Bollati V, Debbache J, Calista D, Ghiorzo P, et al. On the interplay of telomeres, nevi and the risk of melanoma. PLoS one 2012;7:e52466 10.1371/journal.pone.0052466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hofer P, Baierl A, Bernhart K, Leeb G, Mach K, Micksche M, et al. Association of genetic variants of human telomerase with colorectal polyps and colorectal cancer risk. Mol Carcinog. 2012;51: E176–182. 10.1002/mc.21911 [DOI] [PubMed] [Google Scholar]
- 37. Liu HB, Peng YP, Dou CW, Su XL, Gao NK, Tian FM, et al. Comprehensive study on associations between nine SNPs and glioma risk. Asian Pac J Cancer Prev. 2012;13:4905–4908. [DOI] [PubMed] [Google Scholar]
- 38. Terry KL, Tworoger SS, Vitonis AF, Wong J, Titus-Ernstoff L, De Vivo I, et al. Telomere length and genetic variation in telomere maintenance genes in relation to ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2012;21:504–512. 10.1158/1055-9965.EPI-11-0867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng Y, Ogundiran TO, Adebamowo C, Nathanson KL, Domchek SM, Rebbeck TR, et al. Lack of association between common single nucleotide polymorphisms in the TERT-CLPTM1L locus and breast cancer in women of African ancestry. Breast Cancer Res Treat. 2012;132:341–345. 10.1007/s10549-011-1890-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin TB, Zhang JY, Li G, Du SL, Geng TT, Gao J, et al. RTEL1 and TERT polymorphisms are associated with astrocytoma risk in the Chinese Han population. Tumor Biol. 2013; 34:3659–3666. 10.1007/s13277-013-0947-0 [DOI] [PubMed] [Google Scholar]
- 41. Kote-Jarai Z, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Dadaev T, Jugurnauth-Little S, et al. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet. 2013;22:2520–2528. 10.1093/hmg/ddt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pellatt AJ, Wolff RK, Herrick J, Lundgreen A, Slattery ML. TERT’s role in colorectal carcinogenesis. Mol Carcinog. 2013;52:507–513. 10.1002/mc.21885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pellatt AJ, Wolff RK, Torres-Mejia G, John EM, Herrick JS, Lundgreen A, et al. Telomere length, telomere-related genes, and breast cancer risk: The breast cancer health disparities study. Genes Chromosomes Cancer 2013;52:595–609. 10.1002/gcc.22056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheng X, Tong N, Tao G, Luo D, Wang M, Fang Y, et al. TERT polymorphisms modify the risk of acute lymphoblastic leukemia in Chinese children. Carcinogenesis 2013;34:228–235. 10.1093/carcin/bgs325 [DOI] [PubMed] [Google Scholar]
- 45. Zhao Z, Li C, Yang L, Zhang X, Zhao X, Song X, et al. Significant association of 5p15.33 (TERT–CLPTM1L genes) with lung cancer in Chinese Han population. Exp Lung Res. 2013;39:91–98. 10.3109/01902148.2012.762436 [DOI] [PubMed] [Google Scholar]
- 46. Voils CI, Crandell JL, Chang Y, Leeman J, Sandelowski M. Combining adjusted and unadjusted findings in mixed research synthesis. J Eval Clin Pract. 2011;17:429–434. 10.1111/j.1365-2753.2010.01444.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yin J, Li Y, Yin M, Sun J, Liu L, Qin Q, et al. TERT-CLPTM1L polymorphism rs401681 contributes to cancers risk: evidence from a meta-analysis based on 29 publications. PLoS one 2012;7:e50650 10.1371/journal.pone.0050650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 1999;232:97–106. [DOI] [PubMed] [Google Scholar]
- 49. Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 2008;455:633–637. 10.1038/nature07283 [DOI] [PubMed] [Google Scholar]
- 50. Zachos I, Konstantinopoulos PA, Vandoros GP, Karamouzis MV, Papatsoris AG, Podimatas T, et al. Predictive value of telomerase reverse transcriptase expression in patients with high risk superficial bladder cancer treated with adjuvant BCG immunotherapy. J Cancer Res Clin Oncol. 2009;135:1169–1175. 10.1007/s00432-009-0557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mocellin S, Verdi D, Pooley KA, Landi MT, Egan KM, Baird DM, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104:840–854. 10.1093/jnci/djs222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Campa D, Martino A, Varkonyi J, Lesueur F, Jamroziak K, Landi S, et al. Risk of multiple myeloma is associated with polymorphisms within telomerase genes and telomere length. Int J Cancer 2015;136: E351–358. 10.1002/ijc.29101 [DOI] [PubMed] [Google Scholar]
- 53. Melin BS, Nordfjäll K, Andersson U, Roos G. hTERT cancer risk genotypes are associated with telomere length. Genet Epidemiol. 2012;36:368–372. 10.1002/gepi.21630 [DOI] [PubMed] [Google Scholar]
- 54. Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. 10.1016/j.ajhg.2009.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–903. 10.1038/ng.2353 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A list of full-text excluded articles.
(DOC)
Meta-analysis on Genetic Association Studies Checklist.
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.