Abstract
Reactivation of telomerase reverse transcriptase (TERT) expression enables cells to overcome replicative senescence and escape apoptosis, fundamental steps in the initiation of human cancer. Multiple cancer types, including up to 83% of glioblastomas (GBM), harbor highly recurrent TERT promoter mutations of unknown function but specific to two nucleotide positions. We identify the functional consequence of these mutations in GBM to be recruitment of the multimeric GABP transcription factor specifically to the mutant promoter. Allelic recruitment of GABP is consistently observed across four cancer types, highlighting a shared mechanism underlying TERT reactivation. Tandem flanking native ETS motifs critically cooperate with these mutations to activate TERT, likely by facilitating GABP heterotetramer binding. GABP thus directly links TERT promoter mutations to aberrant expression in multiple cancers.
The human telomerase is an enzyme critical for maintaining telomere length and chromosomal stability in stem cells(1, 2). The transcriptional regulation of the telomerase reverse transcriptase (TERT) gene, encoding the catalytic subunit of telomerase, is a rate-limiting step in modulating telomerase activity(3). Although normally silenced in somatic cells, TERT is aberrantly expressed in 90% of aggressive cancers, highlighting this event as a hallmark of tumorigenesis(4–6). Reactivating telomerase helps cells with finite lifespan to achieve limitless proliferative potential and bypass cellular senescence induced by DNA replication-associated telomere shortening. Understanding the mechanisms of aberrant TERT expression thus represents a crucial outstanding problem in cancer research.
Recently discovered non-coding mutations in the TERT promoter are among the most common genetic alterations observed across multiple cancer types, revealing a potentially causal biological mechanism driving increased telomerase activity in tumors (7–9). Specifically, one of two positions, G228A or G250A, is mutated in 20% of medulloblastomas(10), 44% of hepatocellular carcinomas (HCC)(11), 66% of urothelial carcinomas of the bladder(12), 71% of melanomas(7, 8), and 83% of primary glioblastomas (GBM)(9), making them the most recurrent single-nucleotide mutations observed in these cancer types. Both the G228A and G250A mutations are associated with increased TERT expression (fig. S1) and telomerase activity(13), and have prognostic power in bladder cancer and GBM(14–16). Both G>A transitions generate an identical 11bp sequence which is hypothesized to generate a de novo binding site for an ETS transcription factor(7). Despite these compelling findings and the central importance of TERT in human cancer, the precise function of the mutations has remained elusive since their initial discovery in melanoma patients.
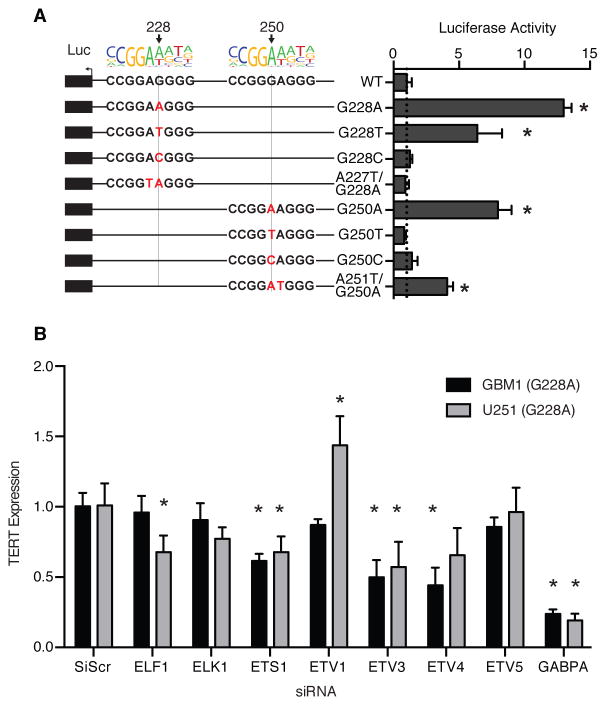
To determine whether the de novo ETS motif is necessary for mutant TERT activation, we performed site-directed mutagenesis of the core TERT promoter(17). The G228C, G250C, and G250T mutations did not increase promoter activity, highlighting the requirement for the G>A transition for TERT activation (Fig. 1A). Furthermore, removing the ETS motif while retaining the G228A mutation (A227T, G228A) resulted in a complete reduction of promoter activity to wild-type levels. Interestingly, the G228T mutation also partially increased promoter activity; this induction is consistent with the site being the second adenine position in an ETS motif, a position that is often degenerate for A/T(18). Mutating the second adenine position to thymine in the context of G250A (G250A, A251T) resulted in a similar intermediate level of promoter activity.
Figure 1.
The de novo ETS motif is critical for mutant TERT promoter activity in GBM. (A) TERT promoter-luciferase reporter assays for wild-type, G228A, G250A, or targeted mutation sequences. * P <0.05, Student’s t-test compared to wild-type (WT) (B) TERT expression relative to siScramble (siScr) 72 hours post ETS factor siRNA knockdown. * P <0.05, Student’s t-test compared to siScr. The results are an average of at least 3 independent experiments. Values are mean ± sd.
A siRNA screen of 13 ETS factors expressed in GBM revealed 5 ETS factors (ELF1, ETS1, ETV3, ETV4, GABPA) whose knock-down reduced TERT expression in at least one of two GBM cell lines harboring TERT promoter mutations (Fig. 1B, fig. S2, and fig. S3)(17). Only three factors (ETS1, ETV3, and GABPA) consistently reduced TERT expression in both lines. Of note, GABPA knockdown reduced TERT expression by as much as 50% within the first 24 hours, and sustained the largest effect on TERT expression amongst the ETS candidates throughout 72 hours (fig. S3). In contrast, knockdown of ETS1 and ELF1 resulted in a more modest reduction of TERT mRNA, and only reached statistical significance at 72 hours, suggesting their regulation of TERT is through indirect mechanisms. ETV3 is a transcriptional repressor in the ETS family and was thus not considered a candidate direct regulator of mutant TERT(19–21). Thus, the de novo ETS motif is critical for mutant TERT promoter activity in GBM, and one or more candidate ETS factors may regulate TERT expression directly through the G228A and G250A mutations.
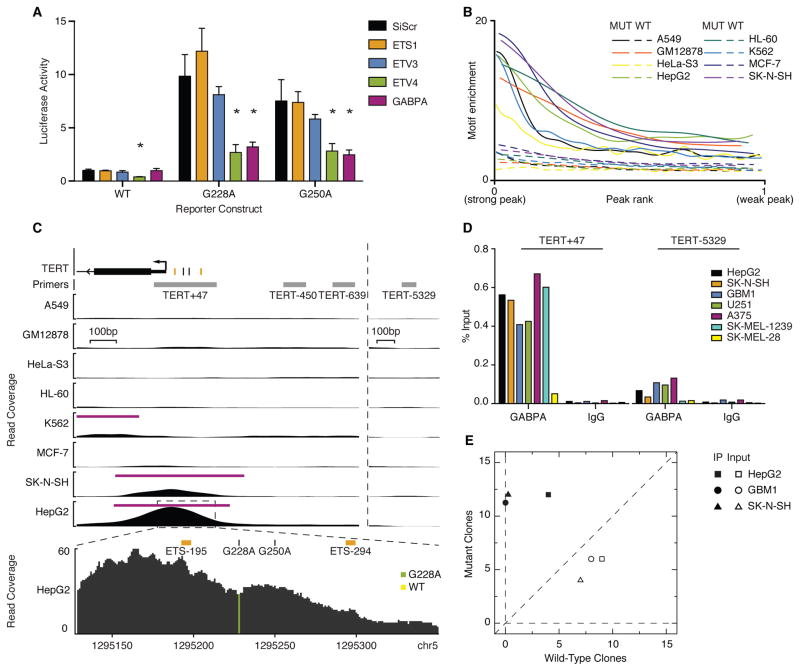
We next investigated whether regulation of TERT by ETS1, ETV3, ETV4, or GABPA depends upon the TERT promoter mutation status by testing the effect of siRNA knockdowns on activity of TERT promoter-driven luciferase reporters. Only GABPA knockdown significantly reduced mutant promoter activity without affecting wild-type promoter activity (Fig. 2A, fig. S4). While ETV4 knockdown reduced mutant promoter activity, it also significantly reduced the activity of the wild-type promoter, indicating the potential of ETV4 to bind and regulate the wild-type TERT promoter sequence in this assay. Knockdown of ETS1 and ETV3 did not significantly reduce promoter activity (Fig. 2A, fig. S4). GABPA was thus the only ETS factor that reproducibly affected TERT expression in a mutation-specific manner. Furthermore knockdown of GABPA did not significantly affect cell cycle or proliferation rate within this timeframe (fig. S5).
Figure 2.
GABPA selectively regulates and binds the mutant TERT promoter across multiple cancer types. (A) Wild-type, G228A, or G250A luciferase activity 72 hours post ETS siRNA knockdown in GBM1 cultured cells, scaled to WT-siScramble (siScr). The results are an average of at least 3 independent experiments. Values are mean ± sd * P <0.05, Student’s t-test compared to siScr. (B) Enrichment of mutant (CCGGAA) or wild-type (CCGGAG) hexamer sequences in ENCODE GABPA ChIP-seq peaks relative to flanking regions. (C) ENCODE GABPA ChIP-seq data at the proximal TERT promoter and around distal qPCR primers. Native ETS motifs and mutation positions are annotated by orange and black tick marks respectively. Inset shows allelic read coverage at G228A in HepG2 cells. (D) GABPA ChIP-qPCR for the TERT promoter and a nearby control locus in seven cancer cell lines. Values represent mean % input based on triplicate qPCR measurments. N=1 for each cell line. (E) Allelic variant frequency in GABPA (IP) or input control DNA.
To determine the in vivo binding specificity to the mutant TERT promoter sequence (‘CCGGAA’) relative to the wild-type sequence (‘CCGGAG’) amongst the candidate ETS factors, we analyzed publicly available ChIP-seq data for GABPA, ELF1, ETS1, and ETV4(22, 23). While all factors display significant enrichment of the sequence found in the mutant TERT promoter relative to the wild-type sequence, we found that GABPA peaks contained significantly greater enrichment of the mutant motif compared to ETS1 or ETV4 peaks (P-value = 5.1×10−8 for ETS1 and 1.8×10−8 for ETV4, Wilcoxon rank-sum test)(Fig. 2B, fig. S6). This genome-wide analysis supports the binding specificity for the motif created by the TERT promoter mutations, and suggests that GABPA binding may be more sensitive to these promoter mutations. Furthermore, this enrichment is not observed in DNase I hypersensitivity peaks in the same cells, demonstrating that the motif enrichment does not represent sequence biases in areas of open chromatin (fig. S6). Among the eight ENCODE cell lines with GABPA ChIP-seq, only HepG2 hepatocellular carcinoma cells and SK-N-SH neuroblastoma cells, both of which harbor heterozygous G228A mutations, displayed significant GABPA binding at the TERT promoter (Fig. 2C). In contrast, none of the TERT mutant cell lines showed ELF1 binding at the TERT promoter (fig. S7). Likewise, ChIP of ETS1 and ETV4 did not show binding at the mutant TERT promoter in vivo (fig. S7). An in vitro single-molecule protein binding assay further confirmed that ETV4 does not stably bind the mutated sequence (fig. S8)(17). These results are consistent with the fact that only GABPA knockdown shows immediate reduction on TERT expression (fig. S3) and implicate GABPA to be the only ETS factor among the candidates to directly bind the mutant TERT promoter. All of the cell lines that did not show GABPA binding (K562, GM12878, A549, Hela, MCF-7, HL-60) were derived from cancers in which TERT promoter mutations are absent or uncommon(9). Strikingly, 100% of the GABPA ChIP-seq reads covering the mutated site within the TERT promoter contain G228A, suggesting that GABPA selectively binds the mutant allele in vivo and that it cannot recognize and bind the wild-type sequence (Fig. 2C). Recruitment of GABP to the G250A mutant sequence was confirmed in vitro using a single-molecule protein binding assay. In contrast, no binding event of GABP was detected for the wild-type TERT sequence (fig. S8). Mutant allele-specific DNase I hypersensitivity and Pol II recruitment was also observed in these lines (fig. S9).
To confirm that GABPA is specifically recruited to the mutant allele, we performed GABPA ChIP in HepG2, SK-N-SH, two GBM lines, and three melanoma lines (Table S1)(17). All cell lines harboring either the G228A or G250A mutation showed significant GABPA binding in the TERT core promoter (P-value = 0.016, Wilcoxon Rank Sum Test, Fig. 2D). In contrast, the TERT wild-type melanoma line SK-MEL-28 showed no GABPA binding at the TERT promoter compared to the other lines (P-value = 0.007, Weisberg t-test for outliers). Consistent with our findings of specificity for the mutant allele in the ENCODE ChIP-seq data, the GABPA-immunoprecipitated DNA from the heterozygous mutant cell lines HepG2, SK-N-SH, and GBM1 all showed significant bias towards the mutant allele compared to input control DNA (P-value = 1.264×10−5, Fisher’s Exact Test, Fig. 2E). Furthermore, we confirmed that both heterozygous mutations in the TERT promoter resulted in allelic deposition of H3K4me3 and allele-specific expression (fig. S10). Nucleosome positioning analysis from micrococcal nuclease-digested H3K4me3 ChIP-seq(24) data revealed that both mutation positions lie within a nucleosome free region, with the upstream nucleosomes containing the H3K4me3 modifications (fig. S10). These data demonstrate that GABPA is selectively recruited to the mutant TERT allele across multiple cancer types and results in allele-specific activation of TERT.
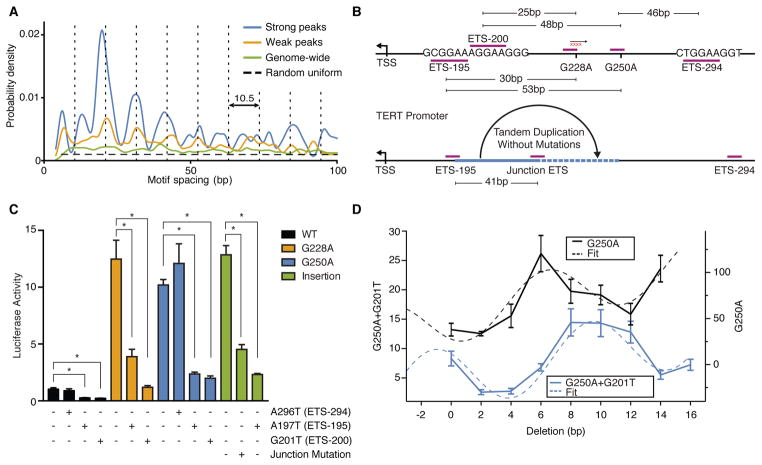
GABPA is unique among the large ETS transcription factor family as it is the only obligate multimeric factor(25, 26). GABPA dimerizes with GABPB, and the resulting heterodimer (GABP) forms a fully functional transcription factor that can both bind DNA and activate transcription(27). GABPA has a single transcript isoform that is widely expressed across tissue types, while GABPB is encoded by either the GABPB1 or GABPB2 gene and GABPB1 contains multiple isoforms(28, 29). A subset of GABPB isoforms contain leucine zipper-like domains which allow two GABP heterodimers to form a heterotetramer complex capable of binding two GABPA motifs (core consensus ‘CCGGAA’) in proximity to each other, and further stimulating transcription(30). Consistent with this fact, genome-wide analysis of ENCODE GABPA ChIP-seq data showed that peaks containing two GABPA motifs have significantly higher binding enrichment scores compared to peaks with just one or zero motifs (P-value =1.6×10−157, Wilcoxon rank-sum test, fig. S11, fig. S12). Analysis of GABPA motif spacing within peaks containing 2 motifs revealed that strong peaks are more likely to have a separation distance shorter than 50bp compared to weak peaks (Fig. 3A, fig. S11). Moreover, this increase in likelihood occurred at discrete spacing that aligned well with the 10.5bp periodicity of relaxed B-DNA, highlighting the importance of having two GABPA binding sites in phase and separated by full helical turns of double stranded DNA. This periodicity was unique to GABPA and is not observed in ELF1 or ETS1 ChIP-seq data (fig. S11). The Fourier spectrum of the enrichment also spiked around the helical frequency in strong GABPA peaks, but not in weak peaks or the genomic background (fig. S13). This analysis suggested that two proximal motifs in helical phase act synergistically to recruit a GABP heterotetramer complex.
Figure 3.
G228A and G250A cooperate with the native ETS sites ETS-195 and ETS-200 and fall within spacing for GABP heterotetramer recruitment. (A) Distribution of motif separation in weak and strong GABP peaks. Vertical dotted lines denote periodicity of 10.5bp. Horizontal dashed line indicates the theoretical null distribution. (B) Native and de novo putative ETS-binding sites in the core TERT promoter. (C) Site-directed mutagenesis of the GABP heterotetramer motifs in the wild-type, G228A, G250A, or insertion TERT reporter constructs. Mutation of the ETS-195, ETS-294, or junction motif are indicated by ‘+’. The results are an average of at least 3 independent experiments. Values are mean ± sd * P <0.05, Student’s t-test. (D) Site-directed mutagenesis deleting between 2 to 16 base pairs at the G228A site. Deletions were tested for promoter activity in a G250A or G250A+G201T background. The sinusoidal fits were obtained by using the model a sin(2π(x − b)/10.5) + cx + d. The results are an average of at least 3 independent experiments. Values are mean ± sd.
Investigation of the DNA sequence flanking the mutation sites revealed three native ETS binding motifs (ETS-195, ETS-200, and ETS-294) (Fig. 3B). To determine whether these flanking ETS motifs are required for mutant TERT activation, we performed site-directed mutagenesis of the flanking ETS sites with or without the G228A or G250A mutation. Mutating ETS-195 or ETS-200 alone reduced promoter activity from the relatively low level of the wild-type promoter, and also significantly reduced activity in the context of G228A or G250A. In contrast, mutating ETS-294 had no effect on promoter activity in the context of G250A despite being closer than ETS-195 or ETS-200 (Fig. 3C). These data demonstrate that both ETS-195 and ETS-200 are required for aberrant activity of the mutant TERT promoter. The GABPB1 isoforms required for GABP heterotetramer formation are the predominant isoforms expressed in GBM melanoma, hepatocellular carcinoma and bladder urothelial carcinoma, all tumor types prone to TERT promoter mutations (fig. S14).
To test whether ETS motif spacing is essential for mutant TERT promoter activation, we performed a series of deletions in 2bp increments between the native ETS site and the G250A mutation, effectively bringing G250A out of phase and back into phase with the native ETS motifs. While the wild-type reporter construct displays only noise level fluctuations in activity, we observed clear periodic behavior in the G250A reporter, suggesting the recruitment of a GABP heterotetramer (Fig. 3D, fig. S11). However, G250A promoter activity peaked after deleting six base pairs, which brought the G250A site in phase with the ETS-200 site by perfect 4 helical turns. Mutating ETS-195, although reducing the TERT activation level (Fig. 3C), did not change the periodic pattern, implying a preferential interaction of GABP with ETS-200 instead of ETS-195 (fig. S11). Repeating the experiment with a mutated ETS-200, however, led to a translation in 10.5bp periodicity, which was now consistent with pairing between G250A and ETS-195 (Fig. 3D). These results strongly suggest that GABP may be able to bind and switch between both native ETS motifs in the context of G250A, consistent with the fact that both native ETS motifs are essential for robust TERT activation (Fig. 3C).
The critical role of two adjacent ETS motifs in aberrant TERT activation was further strengthened by our analysis of an oligodendroglioma tumor containing a unique, heterozygous 41bp tandem duplication within the core TERT promoter. While this sample was wild-type at G228A and G250A, we found that the junction of the duplication event generated a de novo ETS motif that is 41bp away from the native downstream ETS-195 motif (Fig. 3B). The promoter sequence containing this duplication induces elevated promoter activity similar to the G228A and G250A mutant sequences, despite its wild-type status at these positions (Fig. 3C). Mutagenesis of either the native ETS-195 site or the de novo junction ETS site significantly reduced promoter activity, once again demonstrating that this duplication satisfies the prerequisite for GABP heterotetramer recruitment (Fig. 3C).
We have thus identified GABP as the critical ETS transcription factor activating TERT expression in the context of highly recurrent promoter mutations. Although many ETS transcription factors can bind similar DNA sequence motifs, GABP is unique in that it can bind neighboring ETS motifs as a heterotetrameric complex. We show that strong GABPA ChIP-seq peaks contain a periodicity of approximately 10.5bp between neighboring ETS motifs, consistent with the binding of a GABP complex at two locations separated by full helical turns of DNA. This genome-wide pattern is reproduced in the context of TERT promoter mutations, where both G228A and G250A are separated from two tandem proximal native ETS motifs by 2.9/2.4 (ETS-195/ETS-200) and 5.0/4.6 (ETS-195/ETS-200) helical turns respectively. We propose that TERT promoter mutations cooperate with both of these native ETS sites to recruit GABP. Further work is necessary to elucidate which other transcription factors are interacting with GABP at the mutant TERT promoter in order to drive aberrant transcription. Additionally, both TERT promoter mutations fall within a GC-rich repeat sequence with potential to form a G-quadruplex, DNA secondary structure which can regulate gene expression(32, 33). A potential impact of TERT promoter mutations on this predicted secondary structure and on the complex relationship between secondary structure and GABP recruitment may also play a role in deregulating TERT expression. The cancer-specific interaction of GABP with the TERT core promoter mutations highlights a common mechanism utilized by many cancers to overcome replicative senescence.
Supplementary Material
Acknowledgments
This work was generously supported by NCI R01CA163336 (JSS), the Sontag Foundation Distinguished Scientist Award (JSS), NCI P50CA097257 and P01CA118816-06 (MSB, SMC, JFC); NCI R01CA169316-01 (JFC), the Grove Foundation, the Karen Osney Brownstein Endowed Chair (JFC), the Anne and Jason Farber Foundation, and a gift from the Dabbiere family. Additional support was provided by NCI R25CA112355 (KMW), R01CA52689 (KMW, JKW, MRW), P50CA097257 (KMW, MRW), the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, the Founder Professorship from the Grainger Engineering Breakthroughs Initiative (JSS), and donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. Provisional patent application (application number: 62/145,579) has been filed by University of California, San Francisco and University of Illinois. All transcriptome sequencing data have been deposited in the European Genome-phenome Archive under accession number EGAS00001001242.
Footnotes
References and Notes
- 1.Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 3.Weinrich SLS, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 4.Phd PCB, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncology. 2013;14:534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 5.Kim NW, et al. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 7.Horn S, et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science. 2013 doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 8.Huang FW, et al. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science. 2013 doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remke M, et al. TERT promoter mutations are highly recurrent in SHH subgroup medulloblastoma. Acta Neuropathol. 2013;126:917–929. doi: 10.1007/s00401-013-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quaas A, et al. Frequency of TERT promoter mutations in primary tumors of the liver. Virchows Arch. 2014 doi: 10.1007/s00428-014-1658-7. [DOI] [PubMed] [Google Scholar]
- 12.Papadopoulos N, et al. TERT Promoter Mutations Occur Early in Urothelial Neoplasia and are Biomarkers of Early Disease and Disease Recurrence in Urine. Cancer Res. 2013:1–18. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinagre JAO, et al. Frequency of TERT promoter mutations in human cancers. Nature Communications. 2013;4:1–6. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 14.Rachakonda PS, et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. PNAS. 2013;110:17426–17431. doi: 10.1073/pnas.1310522110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon M, et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-Oncology. 2014;0:1–8. doi: 10.1093/neuonc/nou158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiegl-Kreinecker S, et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro-Oncology. 2015 doi: 10.1093/neuonc/nov010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See supplementary materials on Science Online.
- 18.Wei GH, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. The EMBO Journal. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klappacher GW, et al. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. 2002;109:169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 20.El Kasmi KC, et al. Cutting edge: A transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. 2007;179:7215–7219. doi: 10.4049/jimmunol.179.11.7215. [DOI] [PubMed] [Google Scholar]
- 21.Sawka-Verhelle D, et al. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. 2004;279:17772–17784. doi: 10.1074/jbc.M311991200. [DOI] [PubMed] [Google Scholar]
- 22.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenhorst PC, et al. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genome Res. 2011;25:2147–2157. doi: 10.1101/gad.17546311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagarajan RP, et al. Recurrent epimutations activate gene body promoters in primary glioblastoma. Genome Res. 2014;24:761–774. doi: 10.1101/gr.164707.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson CC, Brown TA, Mcknight SL. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- 26.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 27.LaMarco K, Thompson CC, Byers BP, Walton EM, Mcknight SL. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- 28.Gugneja S, Virbasius JV, Scarpulla RC. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol Cell Biol. 1995;15:102–111. doi: 10.1128/mcb.15.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De La Brousse FC, Birkenmeier EH, King DS, Rowe LB, Mcknight SL. Molecular and genetic characterization of GABP beta. Genes Dev. 1994;8:1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]
- 30.Sawada J, Goto M, Sawa C, Watanabe H, Handa H. Transcriptional activation through the tetrameric complex formation of E4TF1 subunits. EMBO J. 1994;13:1396–1402. doi: 10.1002/j.1460-2075.1994.tb06393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arita H, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 32.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim KW, et al. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J Am Chem Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 34.Brennan CW, et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamim S, et al. Genomic analyses reveal broad impact of miR-137 on genes associated with malignant transformation and neuronal differentiation in glioblastoma cells. 2014;9:e85591–e85591. doi: 10.1371/journal.pone.0085591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlson SM, et al. Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. 2011;4:rs11–rs11. doi: 10.1126/scisignal.2002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead Ben, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.