Abstract
Deregulation of cellular polarity proteins and their associated complexes leads to changes in cell migration and proliferation. The nitric oxide synthase 1 adaptor protein (NOS1AP) associates with the tumor suppressor protein Scribble to control cell migration and oncogenic transformation. However, how NOS1AP is linked to the cell signaling events that curb oncogenic progression has remained elusive. Here we identify several novel NOS1AP isoforms, NOS1APd, NOS1APe, and NOS1APf, with distinct cellular localizations. We show that isoforms with a membrane-interacting phosphotyrosine binding (PTB) domain can associate with Scribble and recognize acidic phospholipids. In a screen to identify novel binding proteins, we have discovered a complex consisting of NOS1AP and the transcriptional coactivator YAP linking NOS1AP to the Hippo signaling pathway. Silencing of NOS1AP reduces the phosphorylation of YAP and of the upstream kinase Lats1. Conversely, expression of NOS1AP promotes YAP and Lats1 phosphorylation, which correlates with reduced TEAD activity and restricted cell proliferation. Together, these data implicate a role for NOS1AP in the regulation of core Hippo signaling and are consistent with the idea that NOS1AP functions as a tumor suppressor.
INTRODUCTION
Cellular polarity is important for establishing and maintaining the shape and function of a cell. Loss of cellular polarity is associated with an epithelial-to-mesenchymal transition (EMT), where cells lose cell-cell adhesion and gain migratory and invasive properties that contribute to tumor progression. The Hippo signaling cascade, first identified in Drosophila melanogaster (1, 2), is highly conserved in mammals and is an important intracellular signaling pathway that controls tissue growth by modulating cell proliferation and cell differentiation (3). Deregulation of the Hippo signaling pathway connects the EMT to increased invasion, which has been associated with a number of different cancers (4). At the core of the Hippo complex are the serine threonine kinases MST1/2 and LATS1/2, which form a kinase cascade that controls the phosphorylation of the transcriptional coactivators YAP and TAZ (3, 5). Phosphorylation of YAP/TAZ on conserved serine residues restricts them to the cytosol, where they can associate with 14-3-3 proteins, be targeted for proteasomal degradation, or localize to regions of cell-cell contact, all of which prevent the nuclear localization and transcriptional activation of YAP/TAZ target genes (6). Recently, both genetic (7–9) and biochemical (9, 10) studies have revealed an association between the Hippo pathway and the polarity protein Scribble. Scribble is a large scaffolding protein that contains leucine-rich repeats (LRRs) at its N terminus and four PDZ domains that connect YAP/TAZ with MST1/2 and LATS1/2 (10), downstream of LKB1 and PAR-1 kinases (10). Together, these data suggest that Scribble regulates Hippo signaling and that the loss of Scribble is an important trigger to modify intracellular events leading to tumor development in a Hippo-dependent manner. However, the precise mechanism for the control of Hippo signaling by Scribble remains unknown.
We have previously shown that Scribble directly associates with nitric oxide synthase 1 adaptor protein (NOS1AP), also known as CAPON (carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase [nNOS] protein), and regulates dendritic spine development (11). NOS1AP has been implicated in a number of disorders, including schizophrenia (12), anxiety (13), QT syndrome (14), and, more recently, breast cancer (15). NOS1AP contains an amino-terminal phosphotyrosine binding (PTB) domain and a carboxyl-terminal PDZ binding motif that directly associates with the PDZ domain of nNOS (16, 17). The association of NOS1AP and nNOS has been shown to compete with the binding of nNOS and the postsynaptic density scaffolding protein PSD95, implicating NOS1AP in NMDA receptor signaling (17). The PTB domain of NOS1AP associates with (i) dexamethasone-induced Ras-related protein 1 (Dexras1), leading to a ternary complex between NOS1AP, Dexras1, and nNOS that is important for nNOS to activate Dexras1 (18); (ii) synapsins I, II, and III, establishing a complex that is important for the localization of nNOS to presynaptic terminals (16); and (iii) Scribble, allowing NOS1AP to bridge a complex between Scribble and Rho GTPase signaling through β-Pix and Git1 (11). In addition to a link with Rho GTPase, NOS1AP and Scribble associate with the planar cell polarity protein Vangl1 to regulate breast cancer development independent of β-Pix/Git1/Pak (15). Together, these results suggest that NOS1AP is an important adaptor protein controlling a number of different signaling complexes.
Whereas multiple isoforms of the NOS1AP gene have been annotated in the NCBI database, information regarding the functional significance of this diversity is lacking. As NOS1AP can exist in different complexes (11, 15), we suspect that these differences might be explained by NOS1AP isoforms with distinct biological functions. Here we report that NOS1AP is a highly spliced protein containing multiple isoforms. All NOS1AP isoforms containing the PTB domain associate with Scribble yet have unique tissue and subcellular localizations. The PTB domain is sufficient to recruit NOS1AP to the plasma membrane, where it associates with acidic phospholipids. Interestingly, NOS1AP and Scribble have been coupled to breast cancer regulation, while in separate studies, Scribble has been reported to scaffold Hippo components (9, 10, 15). Thus, we asked whether NOS1AP forms a complex with components of the Hippo pathway. Here we link NOS1AP variants with YAP and show that NOS1AP expression affects the nuclear accumulation of YAP and TEAD activation, both of which correlate with reduced proliferation. Thus, NOS1AP functions with Scribble to control Hippo pathway activity.
MATERIALS AND METHODS
Antibodies.
A NOS1APc rabbit polyclonal antibody was generated against a peptide corresponding to the rat NOS1APc sequence CAFPLLDPPPITRKRT (here pep-NOS1APc). An additional antibody was generated against the entire unique 30-kDa C-terminal extension region of NOS1APc. The following primers were used to amplify a unique region of the NOS1APc sequence: 5′-TTTTCGAATTCTATGTTTGAGAATTT and 3′-TTTGTCGACTGGTTACTACTCAAAGGACAG. The resulting fragment was cut with EcoR1 and Sal1 and subcloned into pGEXT3 to generate a purified glutathione S-transferase (GST) fusion protein that was used as an immunogen to generate GST-NOS1APc antibodies (here GST-NOS1APc). The pan-NOS1AP and Scribble antibodies were described previously (11); Scribble H-300, Yap, GST, c-Myc (9E10), and NOS1AP R-300 (here R-300) (catalog number sc-9138) were all obtained from Santa Cruz. nNOS was obtained from BD Transduction Laboratories. Green fluorescent protein (GFP) was obtained from Abcam. FLAG M2 was obtained from Sigma. Phospho-YAP(Ser127), Lats, and phospho-Lats (Ser909) were obtained from Cell Signaling Technologies.
Cell culture and transfections.
Human embryonic kidney HEK293T or HEK293A cells were grown at 37°C with 5% carbon dioxide in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. A rat primary epithelial cell line (IEC-18), a gift from Neale Ridgeway, Dalhousie University, was grown at 37°C with 5% carbon dioxide in DMEM supplemented with 5% heat-inactivated FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Immunoprecipitations and Western blotting.
Fresh or frozen rodent brains or cell lines were homogenized in NP-40 lysis buffer (10% glycerol, 1% NP-40, 20 mM Tris [pH 8.0], 37.5 mM NaCl) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, and10 μg/ml leupeptin. For lysis of rodent tissues, the NP-40 lysis buffer was combined with tissue at a 1/10 (wt/vol) ratio. Immunoprecipitation and Western blotting experiments were performed as previously reported (11). For analysis of Lats1, phospho-Lats1, and phospho-YAP levels, HEK293T cells were transfected with 1 to 10 μg of an expression plasmid encoding yellow fluorescent protein (YFP)-PTB, YFP-NOS1APa, or YFP-NOS1APc. The protein concentration in whole-cell lysates was quantified at 48 h posttransfection by using the Bradford assay (19), and lysates were prepared for Western blotting.
Immunocytochemistry.
Cells were grown on 18-mm coverslips and transfected on 35-mm plates, as outlined previously (11). To visualize transfected cells, the coverslips were washed twice with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde (PFA) containing 4% sucrose in PBS for 10 min at room temperature. PFA was removed, followed by 3 washes in PBS. The coverslips were then incubated for 5 min at room temperature in BisBenzamide nuclear stain at a 1:2,000 dilution in PBS. Coverslips were then washed 2 times in PBS. Coverslips were then dipped into sterile double-deionized water and then mounted onto glass slides in 20 μl of mounting medium. All epifluorescent images were acquired on a 3i Marianas system (Intelligent Imaging Innovations) equipped with Slidebook 5.5. All post hoc imaging was done with Photoshop.
Cloning of NOS1AP isoforms.
The cerebellum was collected from an adult Wistar rat and flash frozen by using liquid nitrogen. The tissue was combined with 1.0 ml of TRIzol reagent (Invitrogen) per 50 mg of tissue, and total RNA was isolated according to the manufacturer's protocol. The RNA concentration was determined prior to RNA storage at −80°C until required. Isolated RNA was diluted to 0.5 μg in nuclease-free water for use in reverse transcription (RT) for cDNA synthesis. The diluted RNA was combined with oligo(dT) primers (250 ng/μl) heated to 65°C for 5 min and allowed to cool to 25°C for 10 min prior to the addition of RT master mix (RNase Out, 10 mM dithiothreitol [DTT], 1 mM deoxyribonucleotide triphosphate [dNTP], 10% Affinity Script buffer, 2% Affinity Script reverse transcriptase). The cDNA synthesis reaction was allowed to proceed for 1 h at 42°C, and the mixture was then incubated for a further 5 min at 95°C prior to snap-freezing. NOS1AP isoforms were amplified from rat brain cDNA by using the Expand High Fidelity PCR system (Roche) and the following full-length NOS1AP primers: 5′-TTTGCTTCGAATTCTGCAATGCCCAGCAAAACCAAGTAC and 3′-TTTAGCGTCGACTGCCTACTCAAAGGACAGCAG. Standard PCR cycling was performed on a Bio-Rad MyCycler as follows: 95°C for 3 min and 35 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 2.5 min, followed by 72°C for 10 min. Samples were stored at −20°C until use. The resulting PCR products were then cloned into the pGEM-T Easy vector (Pierce). This was used to transform DH5α cells, individual clones were selected, and plasmid DNA was isolated and sent for sequencing. Positive clones were then subcloned into YFP or myc vectors by using EcoRI and SalI restriction sites.
To identify the NOS1APf isoform, we utilized a 5′ rapid amplification of cDNA ends (RACE) kit (Ambion). The procedure was performed according to the manufacturer's instructions. Two sets of inner and outer 3′ primers were designed: (i) 5′-AAATGGTCCTCCTGGCTCTCC and 5′-AAAGCTGGATTGGAGCTTGG and (ii) 5′-AAATGGCTTCCGAGTTATGGG and 5′-AAATGGAGGATCCAGCAGAGG. Amplification products were cloned into pGEM-T Easy plasmid vectors according to the manufacturer's instructions (Promega). Conditions for the PCR were as follows: 95°C for 2 min; 95°C for 45 s, 56°C for 45 s, and 72°C for 1.5 min for 35 cycles; and 72°C for 10 min.
qPCR.
cDNA from different adult rat tissues was generated as outlined above. To determine the levels of expression, quantitative PCR (qPCR) conditions were developed according to the manufacturer's protocols for Brilliant II SYBR green qPCR master mix (Stratagene). The following primers were chosen: 5′-CTGGTGATGCAGGACCCTAG and 5′-CCCACTGTCCGTACGATTCT for the PTB domain, GCCTGGAGCTCATCAAGTTC and 5′-GGCGATCTCATCATCCAAAC for exon 10, 5′-GCAGACACTTCACGACCAGA and 5′-CTGTTCTCCCAGGACTCAGC for NOS1APc, 5′-AGAACCAGGGGTGATGGTG and 5′-AATGCTGGATTGGAGCTTT for NOS1APf, and 5′-GAGCTGTTTGCAGACAAAGTTC and 5′-CCCTGGCACATGAATCCTGG for cyclophilin A. To determine copy numbers, we used the YFP-NOS1APc or YFP-NOS1APf expression plasmid. Each of these constructs was serially diluted to generate a standard curve. This standard curve was used to determine the copy numbers of each construct in different tissue regions. A total of 3 independent runs were performed on each tissue, and the average and standard error of the mean (SEM) were calculated. To compare the ratios of the different isoforms, the threshold cycle (CT) values for cyclophilin A were used to normalize the expression level of each isoform by using the ΔΔCT method (20).
GST fusion proteins and in vitro binding experiments.
GST fusion proteins were produced and purified as described previously (11). Lipid spot blots were purchased from Echelon. Blots were reconstituted according to the manufacturer's instructions and then blocked in 3% bovine serum albumin (BSA) in Tris-buffered saline–Tween (TBST) for 1 h. A total of 0.5 μg/ml of either purified GST, GST-PTB, or GST-PTBinsert (PTB domain from NOS1APe) was added to 3% BSA in TBST. This mixture was incubated overnight at 4°C. Blots were then washed in TBST and incubated in anti-GST antibody at a 1:5,000 dilution. Blots were washed 3 times in TBST, incubated with goat anti-rabbit horseradish peroxidase (HRP) for 1 h, and then washed and incubated with ECL reagent (Pierce) to reveal positive reactions.
siRNA.
For knockdown of endogenous protein, we used a mix of four ON-TARGETplus rat NOS1AP Smart Pool small interfering RNAs (siRNAs) (product number L-099264-02-0005; Dharmacon) and a nontargeting siRNA control (Csi) nonhomologous to any known vertebrate sequence (product number SIC001-1NMOL; Sigma). For transfections, 20 μM siRNA (final concentration, 20 nM) was mixed with Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Complexes were added to IEC-18 cells (1 × 105 cells), incubated for 24 to 72 h, and prepared for Western blot analysis.
MCF7 stable lines.
MCF7 cells were grown to 60% confluence on 10-mm2 dishes prior to transfection. Ten micrograms of cDNA encoding YFP, YFP-NOS1APa, YFP-NOS1APc, YFP-NOS1APd, YFP-NOS1APe, or YFP-NOS1APf was transfected with Lipofectamine 2000 according to the manufacturer's protocols. Cells were maintained in serum-free DMEM overnight, washed, and returned to DMEM with 10% FBS. At 2 days posttransfection, 5 mg/ml Geneticin (G418) was added to the medium, and cells were maintained for 1 week in this medium. Fresh medium containing antibiotic was replaced every 3 days. Individual colonies that were fluorescent were selected and expanded. At least 3 independent lines for each construct were examined for localization and expression by Western blotting to ensure that the clones selected were expressing a protein of the expected size.
Live-cell imaging.
MCF7 cells stably expressing YFP, YFP-NOS1APa, YFP-NOS1APc, YFP-NOS1APd, YFP-NOS1APe, or YFP-NOS1APf were grown on 18-mm coverslips to 70 to 90% confluence. Cells were then washed in PBS, transferred into Hanks balanced salt solution (HBSS) supplemented with 10% FBS, and maintained at 37°C in a humidified heated chamber slide that was transferred to a 3i Marianas system with a Zeiss Axio observer. Confocal images were acquired on the 3i Marianas system equipped with a Yokogawa confocal scanner unit (CSU-X1) spinning-disk system. All images were captured with either a 515-nm or a 640-nm laser line. All post hoc imaging was done with Photoshop 6.0.
Thymidine incorporation studies.
A thymidine incorporation assay was used to analyze the serum-free or serum-induced proliferation of YFP- or YFP-NOS1APa-expressing cells as described previously (21, 22). The incorporation of 1 μCi/ml radioactive [methyl-3H]thymidine (Perkin-Elmer) into cellular DNA during a 15-min pulse time was quantified by liquid scintillation counting. The entire assay was repeated in two independent experiments, and the data are the averages of data from four independent replicates in each experiment.
TEAD transcriptional assays.
HEK293A cells were plated into 48-well tissue culture plates (17,500 cells/well) and allowed to adhere overnight. Cells were transfected with 75 ng of 8× GTIIC-luciferase (TEAD response element containing 8 TEAD binding sites) (a gift from Stefano Piccolo) (Addgene plasmid 34615), 75 ng of a β-galactosidase (β-Gal)-expressing control plasmid, and 50 ng of either the myc control, YFP-NOS1APa, or YFP-PTB with or without 20 ng YAP by using 0.4 μg polyethylenimine transfection reagent (Sigma-Aldrich) in Opti-MEM (Life Technologies). After 24 h of transfection, cells were lysed in 100 μl reporter lysis buffer (Promega), and β-Gal activity was determined by incubating a 1:1 mixture of the cell lysate with β-Gal assay buffer (200 mM sodium phosphate buffer, 2 mM MgCl2, 100 mM β-mercaptoethanol, 1.33 mg/ml ortho-nitrophenyl-β-galactoside) for 5 min prior to reading the absorbance at 420 nm. Luminescence was determined by mixing 10 μl of the cell lysate with 80 μl of luciferase assay substrate (Promega) in a white 96-well plate and immediately measuring total luminescence by using a FluoStar Omega plate reader. The fold change in luminescence was calculated by measuring the luciferase-to-β-Gal luminescence ratio for each sample and was expressed relative to the indicated control. All incubations were done under standard culture conditions (37°C in 95% air and 5% CO2).
Nucleotide sequence accession numbers.
All sequences have been deposited in the NCBI GenBank database with the following accession numbers: NOS1APd, KR558686; NOS1APe, KR558687; NOS1APf, KR558688.
RESULTS
Identification and characterization of multiple NOS1AP isoforms.
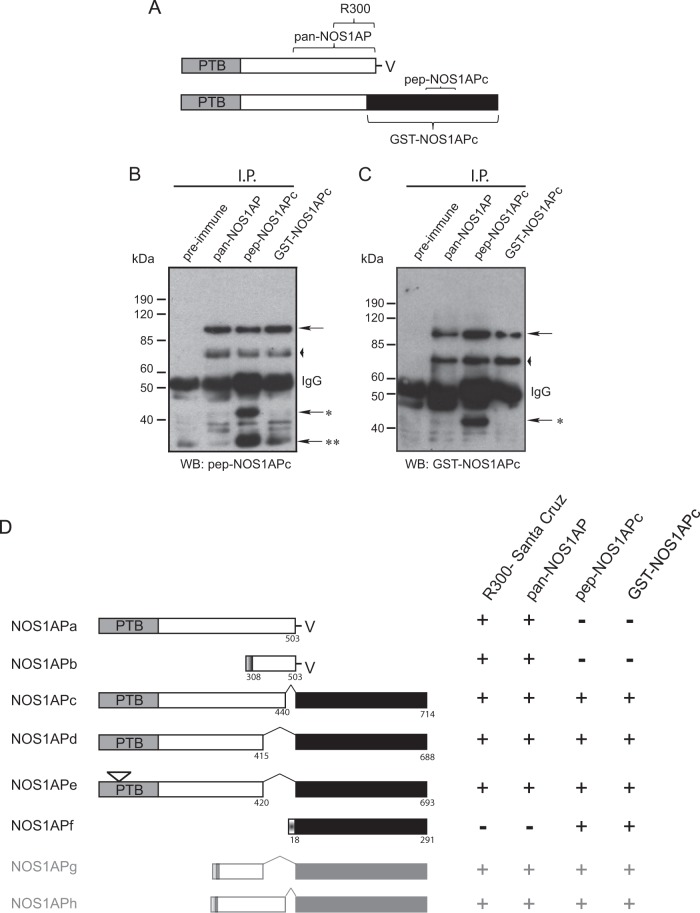
In a previous proteomic screen, we identified a splice variant of the nitric oxide synthase 1 adaptor protein (NOS1APa), which we termed NOS1APc (11). This isoform shares the phosphotyrosine binding (PTB) domain with NOS1APa but contains an ∼30-kDa C-terminal extension that is absent from NOS1APa (Fig. 1A). To better characterize the longer NOS1AP isoform, we generated two different NOS1APc isoform-specific antibodies, a peptide antibody raised against a unique peptide in the extended region of NOS1APc (CAFPLLDPPPITRKRT) (here pep-NOS1APc) and a GST fusion antibody generated against the entire unique 30-kDa C-terminal extension region of NOS1AP (here GST-NOS1APc) (see the schematic in Fig. 1A for regions against which the antibodies were raised). These antibodies, along with a pan-NOS1AP antibody (which detects multiple isoforms of NOS1AP, including NOS1APa) (11), were used to immunoprecipitate endogenous NOS1AP from adult brain lysates. The resulting Western blots were probed with either the pep-NOS1APc (Fig. 1B) or the GST-NOS1APc (Fig. 1C) antibodies. Both the pep-NOS1APc antibody and the GST-NOS1APc antibody revealed a band migrating at ∼95 kDa corresponding to NOS1APc, as previously reported (Fig. 1B and C, arrows). Interestingly, both NOS1APc isoform-specific antibodies detected a faster-migrating (70-kDa) band in the immunoprecipitates (Fig. 1B and C, arrowheads), indicating that there are uncharacterized isoforms with molecular masses similar to that of NOS1APa. Finally, two faster-migrating bands, at ∼40 kDa and 25 kDa, were detected in the pep-NOS1APc immunoprecipitates when probed with the pep-NOS1APc antibody, while only the 40-kDa band was detected by the GST-NOS1APc antibody (Fig. 1B and C, single [40 kDa] or and double [25 kDa] asterisks). These results suggest that mammals express multiple splice variants of NOS1AP in the rodent brain.
FIG 1.
Identification of multiple NOS1AP isoforms. (A) Schematic of NOS1APa and NOS1APc showing the regions of each protein that were used to generate the isoform-specific antibodies used in this study. (B and C) NOS1AP isoforms were precipitated (I.P.) from rat brain lysate with the indicated antibodies. Bound complexes were analyzed by immunoblotting using the antibodies indicated at the bottom. All NOS1AP antibodies tested precipitated and detected bands at 95 kDa (arrows) and at 70 kDa (arrowheads). Two faster-migrating bands at ∼40 kDa and 25 kDa were detected in both panels B and C when lysates were precipitated with pep-NOS1APC antibody (arrows with one or two asterisks). WB, Western blotting. (D, left) Schematic of NOS1AP isoforms (NOS1APb to -h) detected from the cerebellar cDNA screen and amino acids representing splice sites in NOS1APa. (Right) Summary of the antibodies used in this study and their relative affinities for the different NOS1AP isoforms. Shaded boxes in NOS1APb, -g, and -h represent the unique 5′ exon identified for NOS1APb. The small insert in the NOS1APe PTB domain represents the schematic where the LLLLQ insert is found.
To further characterize the nature of these other isoforms, we created a cDNA library from an adult rat cerebellum. Primers were chosen based on previously reported 5′ and 3′ sequences from rat NOS1APa and NOS1APc (11), which were used to amplify NOS1AP isoforms from a cerebellar cDNA library that we had generated. We initially identified and characterized NOS1APc through a bioinformatics approach (11); however, in this study, we cloned and sequenced NOS1APc from our cerebellar cDNA library. Sequence analysis of the resulting cDNAs revealed a different splice site than the one previously reported. While a number of the clones corresponded to NOS1APc (Fig. 1D), we identified other clones that contained a second unique splicing event in exon 10 of NOS1APa that was fused with the unique C-terminal region found in NOS1APc. We have termed this isoform NOS1APd (Fig. 1D). In addition, we identified a third NOS1AP isoform that was identical to NOS1APd, except for a unique 5-amino-acid insert (LLLLQ) in the PTB domain (NOS1APe) (Fig. 1D). Finally, using 5′ RACE, we identified another isoform identical to that previously annotated in the NCBI database (GenBank accession number NM_001190459.1), with a predicted molecular mass for the corresponding protein of ∼40 kDa; we have termed this isoform NOS1APf (Fig. 1D). None of the isoforms that we identified could explain the 70-kDa band observed on our Western blots (Fig. 1B and C, arrowheads). Whether these represent novel splicing of NOS1APb with the unique C-terminal region from NOS1APc to give rise to novel isoforms, which we have termed NOS1APg and -h (Fig. 1D), remains to be determined. Together, these results suggest that NOS1AP is a highly spliced protein and that multiple isoforms are expressed in different cell types.
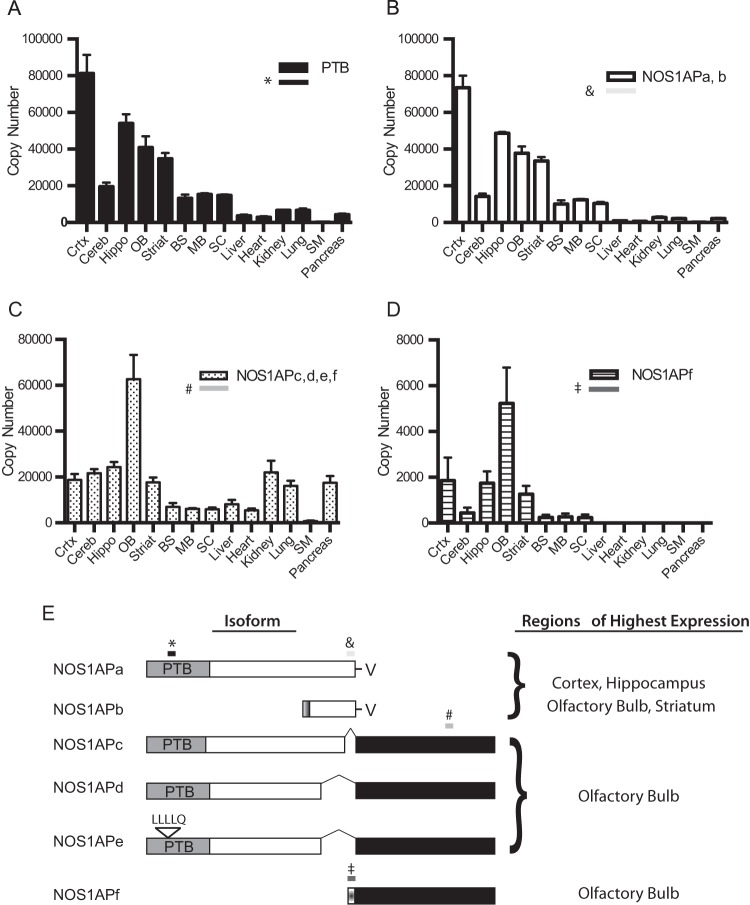
Since we had isolated a number of different NOS1AP cDNAs, we next wanted to determine the relative expression of each isoform in several adult tissues. PCR primers were chosen to flank unique regions in each NOS1AP isoform for quantitative PCR analysis (Fig. 2A to E). Since most of the isoforms identified to date contained the PTB domain, we first quantified the number of mRNA copies of NOS1AP isoforms containing the sequence corresponding to this region (Fig. 2A). High levels of PTB-containing NOS1AP isoforms were found in the cortex, hippocampus, olfactory bulb, and striatum (Fig. 2A). Next, a primer set was chosen to amplify sequences in the PDZ binding motif common to both NOS1APa and NOS1APshort (23, 24) (here NOS1APb). Although the distribution of the NOS1APa/b isoforms was similar to that of NOS1AP isoforms containing the PTB domain (Fig. 2B), the expression levels were lower, suggesting the presence of isoforms containing the PTB domain that do not contain the PDZ binding motif (compare Fig. 2A and B). Next, we selected primers that would amplify the unique C-terminal regions of NOS1APc, -d, -e, and -f and determined their expression patterns in different tissues. The highest copy numbers of mRNA for NOS1APc, -d, -e, and -f were found in the olfactory bulb, with similar levels being found in the cortex, hippocampus, cerebellum, striatum, kidney, and pancreas (Fig. 2C). Lower levels were found in the brainstem, midbrain, spinal cord, and heart and skeletal muscle. Since NOS1APf contained a unique 5′ region, we designed primers to amplify this region to determine mRNA levels of NOS1APf. The overall levels were lower than those of NOS1APc, -d, and -e, suggesting that NOS1APf was a subset of NOS1APc, -d, and -e. The highest levels of NOS1APf expression were seen in the olfactory bulb, with lower levels in the cortex, hippocampus, and striatum; very little expression was found in nonneuronal tissues (Fig. 2D). Taken together, NOS1APa is expressed at higher levels throughout the central nervous system (CNS), while the expressions of NOS1APc, -d, and -e are more evenly distributed across multiple tissues, with the exception of higher levels in the olfactory bulb. The functional significance of these isoform differences remains to be determined.
FIG 2.
qPCR analysis of different NOS1AP transcripts. (A) mRNA copy numbers of NOS1AP isoforms that contain the PTB domain from different tissues. (B) mRNA copy numbers of NOS1AP isoforms that contain the C-terminal region of exon 10 of rat NOS1APa. (C) Copy numbers of NOS1AP isoforms that contain the unique C-terminal region found in NOS1APc, -d, -e, and -f. (D) Copy numbers of NOS1AP constructs that contain the unique 5′ region of NOS1APf. (E, left) Schematic of the regions (shaded boxes with symbols) in each isoform (*, NOS1APa; &, NOS1APa/b; #, NOS1APc, -d, -e, and -f; ‡, NOS1APf) that were analyzed by quantitative PCR in panels A to D. (Right) Tissues with the highest mRNA expression levels for the given NOS1AP isoforms. Abbreviations: Crtx, cortex; Cereb, cerebellum; Hippo, hippocampus; OB, olfactory bulb; Striat, striatum; BS, brain stem; MB, midbrain; SC, spinal cord; SM, skeletal muscle.
NOS1AP isoforms containing the PTB domain are directed to membranes.
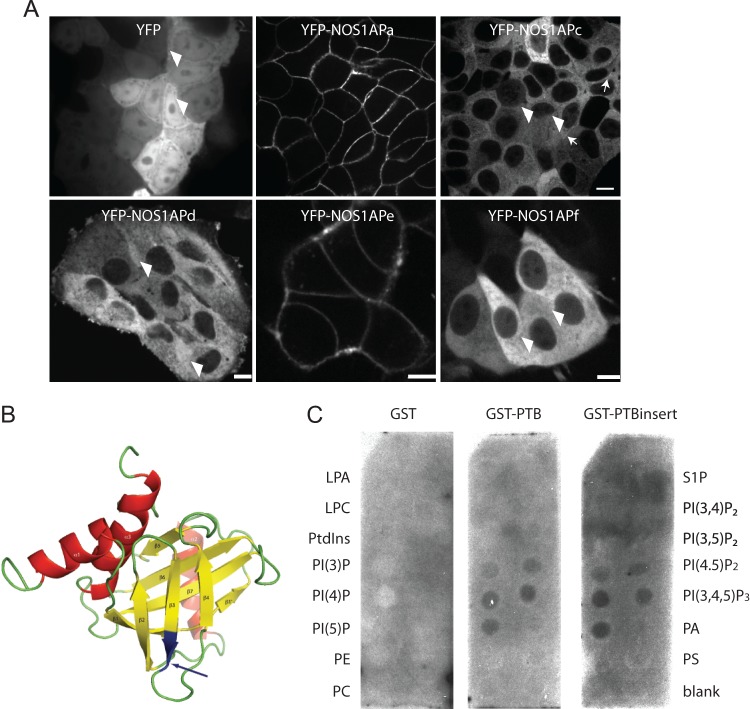
Since NOS1AP has been shown to associate with Scribble (11, 15), and Scribble localizes to the basolateral membranes of MDCK cells, we had anticipated that NOS1AP would localize to basolateral membranes along with Scribble in polarized MDCK cells; however, we saw a diffuse staining pattern using several different NOS1AP antibodies (data not shown). One possibility is that cells contain multiple NOS1AP isoforms that may have different subcellular localizations within the cell, and since our NOS1AP antibodies recognize multiple isoforms, this may manifest as diffuse staining within the cell. Therefore, to better characterize the subcellular localization of the different isoforms, we generated stable cell lines expressing each of the different NOS1AP isoforms as YFP fusions in both the HEK293T and MCF7 cell lines. While similar results were obtained from the HEK293 cell lines, we report here only the MCF7 stable line expression pattern. YFP-NOS1APa and YFP-NOS1APe are enriched at cell-cell contacts (Fig. 3A, middle). Both YFP-NOS1APc and YFP-NOS1APd remained cytoplasmic, with the exception of a weak localization observed for NOS1APc at cell-cell contacts (Fig. 3A, top right, arrows). Interestingly, YFP-NOS1APd, which differed from NOS1APe by 5 amino acids in the PTB domain, was enriched in the cytoplasm (Fig. 3A, bottom left). Finally, YFP-NOS1APf was diffusely distributed in the cytoplasm (Fig. 3A, bottom right). We noted that weak nuclear localization was also detected for YFP and the YFP-NOS1APc, -d, and -f isoforms (Fig. 3A, arrowheads). These results demonstrate that the NOS1AP variants localize differently when expressed in epithelial cells.
FIG 3.
NOS1AP isoforms with different subcellular localizations. (A) Spinning-disk confocal images of MCF7 cells stably expressing YFP, YFP-NOS1APa, NOS1APc, NOS1APd, NOS1APe, and NOS1APf. Note a faint enrichment of YFP-NOS1APc at cell-cell contacts (arrows) and low levels of nuclear localization in YFP, YFP-NOS1APc, NOS1APd, and NOS1APf (arrowheads). Bar = 20 μm. (B) NMR structure model of the Drosophila NUMB PTB domain (42). The arrows point to the insert in NOS1APe positioned in the predicted surface-exposed loop in the protein. (C) A membrane covered with different phospholipids was probed with purified GST, GST-PTB, or GST-PTBinsert (the NOS1APe PTB domain containing the LLLLQ insert). LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PtdIns, phosphatidylinositol.
PTB domains of multiple proteins, including Dab1 (25) and SHC (26), have been shown to associate with the plasma membrane, while an 11-amino-acid insert in the NUMB PTB domain has been reported to confer phospholipid recognition of NUMB (27, 28). The membrane localization of NOS1APa suggests that the PTB domain in NOS1AP isoforms may be critical for membrane localization. Furthermore, the lack of membrane enrichment of NOS1APc or NOS1APd to membranes may be a function of an inter- or intramolecular interaction created by the extended C-terminal region that blocks the association of PTB domains with membranes. We noted that an alignment of the NOS1AP sequence with the Drosophila NUMB PTB nuclear magnetic resonance (NMR) structure revealed that the insert would be positioned in a surface-exposed loop in the protein (Fig. 3B, arrow). It is unlikely that this would disrupt the structure of the PTB domain, yet in the context of NOS1APe, the LLLLQ insert may either induce a conformational change or relieve an interaction allowing the PTB domain to access membranes. To test for the possibility that the PTB domain directs NOS1APa and NOS1APe to the membrane, we performed a lipid strip assay. Purified GST-PTB domains or GST alone was incubated with different types of lipids found in cell membranes that had been spotted onto hydrophobic membranes. After washing, the PTB domains were detected with an anti-GST antibody. We found that the PTB domains from all NOS1AP isoforms associated primarily with phosphoinositides (Fig. 3C, middle). Specifically, the PTB domain bound with the highest affinity to phosphatidylinositol 4-phosphate [PtdIns(4)P], PtdIns(5)P, and phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3]. However, the 5-amino-acid insert did not provide any additional selectivity in lipid recognition to NOS1APe, while GST alone did not bind any of the phospholipids (Fig. 3C, middle and left). Together, these results are consistent with the idea that the PTB domain recruits NOS1APa and NOS1APe to the plasma membrane. These results also indicate that acidic phospholipids are the primary targets for these isoforms.
The identification of novel NOS1AP isoforms and the differences in their localization within the cell next led us to determine if there were distinct protein-protein complexes forming with the different NOS1AP isoforms. Since we have previously shown that NOS1AP associates with Scribble through a PTB-PDZ interaction (11), we first tested whether the novel NOS1AP isoforms containing the PTB domain could associate with Scribble. Additionally, as a positive control for binding, the lysates were also probed with nNOS, as it is precipitated in Scribble and NOS1AP complexes. First, we precipitated different NOS1AP isoforms or Scribble from adult rat brain lysates using our pan-NOS1AP, pep-NOS1APc, or Scribble antibodies and probed for the presence of bound proteins. As anticipated, the pan-NOS1AP, pep-NOS1APc, and Scribble antibodies all precipitated complexes that contained Scribble (Fig. 4A, top) and NOS1APc (Fig. 4A, middle). Interestingly, reprobing the membranes with anti-nNOS revealed that only the pan-NOS1AP and Scribble antibodies precipitated complexes containing nNOS (Fig. 4A, bottom). This suggests that NOS1APa is the primary isoform that binds nNOS. To more directly test if NOS1AP isoforms containing the PTB domain could interact with Scribble, we cotransfected HEK293T cells with cDNA encoding YFP-NOS1APa, -c, -d, -e, or -f together with myc-Scribble. The YFP-tagged NOS1AP isoforms were immunoprecipitated, and the membranes were probed for the presence of myc-Scribble. Scribble associated with NOS1APa, -c, -d, and -e but, as expected, not with NOS1APf, since this isoform lacks the PTB domain (Fig. 4B and C). Thus, the association of NOS1APd and NOS1APe with Scribble likely involves the PTB domain, as demonstrated previously for NOS1APa and NOS1APc.
FIG 4.
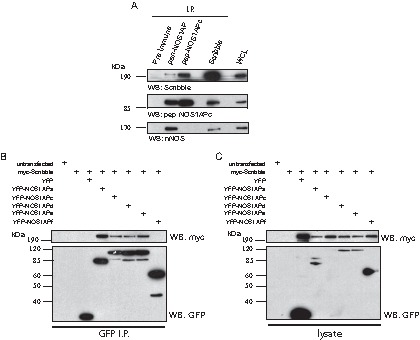
NOS1AP isoforms containing the PTB domain interact with Scribble. (A) Endogenous NOS1AP isoforms were precipitated (I.P.) from rat brain lysate with the indicated antibodies. Bound complexes were analyzed by immunoblotting using anti-Scribble, anti-nNOS, and pep-NOS1APc antibodies, as indicated at the bottom. (B and C) cDNA constructs encoding the indicated proteins were transfected into HEK293T cells. (B) Bound complexes were analyzed with anti-myc and anti-GFP antibodies. (C) Whole-cell lysates with input levels.
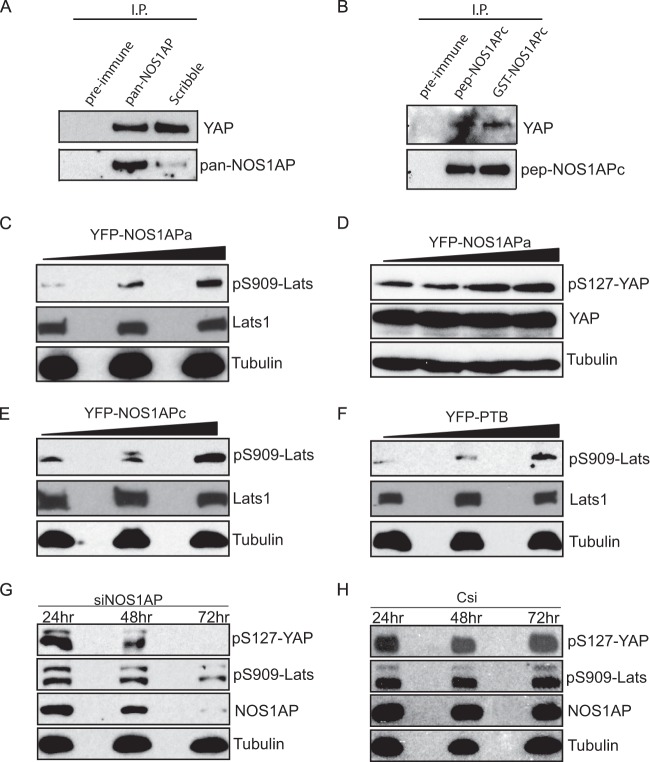
Scribble is a regulator of cell polarity that functions to sort proteins into defined membrane domains (29). A recent study linked NOS1AP to Scribble tumor-suppressive activity through the regulation of anchorage-independent cell growth (15). Importantly, the Hippo pathway effectors YAP and TAZ can induce anchorage-independent growth, and Scribble has been reported to control the assembly of these components (10). This gave rise to the question of whether NOS1AP is part of a Scribble/YAP/TAZ complex. To determine if NOS1AP is associated with the Hippo pathway, we precipitated endogenous NOS1AP or Scribble from distinct cell lines using pan-NOS1AP or Scribble antibodies and probed the precipitates for YAP. Consistent with data from a previous report (7), Scribble coprecipitated with YAP (Fig. 5A). Notably, we also detected multiple NOS1AP isoforms that were able to interact with YAP (Fig. 5A and B), suggesting that NOS1AP is in the same complex as Scribble and YAP. These data demonstrate that multiple NOS1AP isoforms may have a function in Hippo signaling.
FIG 5.
NOS1AP binds to and modulates the phosphorylation of YAP. (A) Rat brain lysate was precipitated with pan-NOS1AP or Scribble antibodies. Coprecipitated complexes were analyzed by immunoblotting with YAP antibodies (top) and pan-NOS1AP (bottom). (B) HEK293T cell lysate was precipitated with pep-NOS1APc or GST-NOS1APc antibodies. The resulting precipitated complexes were probed with YAP (top) or pep-NOS1APc (bottom) antibodies. (C to F) HEK293T cells were transfected with increasing amounts of a plasmid encoding YFP-NOS1APa (C and D), YFP-NOS1APc (E), or the YFP-PTB domain alone (F). The lysates were analyzed for endogenous Lats1 and YAP and with phospho-Lats1 (detecting S909) and phospho-YAP (detecting S127) antibodies. (G and H) IEC-18 cells transfected with NOS1AP-targeting siRNA (siNOS1AP) (G) or control siRNA (Csi) (H) were lysed at the indicated time points. Equal amounts of cell lysate were prepared for Western blotting and analyzed with the indicated antibodies.
The MST1/2-Sav1 complex activates the downstream Lats kinases, which in turn phosphorylate YAP, leading to its nuclear exclusion (3). To examine whether YAP phosphorylation is influenced by NOS1AP, we transfected HEK293T cells with increasing amounts of cDNA encoding YFP-NOS1APa or YFP-NOS1APc and probed the lysates with phosphospecific antibodies against Lats1 and YAP. Increasing amounts of NOS1APa promoted the kinase activity of endogenous Lats1 by increasing phosphorylation in the activation loop at S909 (Fig. 5C). This increase in Lats1 phosphorylation was accompanied by an increased phosphorylation of YAP at S127 (Fig. 5D), suggesting that NOS1AP regulates YAP phosphorylation through Lats1 activity. Similar results were seen for NOS1APc in the activation of Lats1 (Fig. 5E). We next tested whether increased expression of the NOS1AP PTB domain alone affects the levels of Lats1. Indeed, PTB domain expression induced Lats1 phosphorylation at S909 (Fig. 5F), indicating that this effect is PTB domain dependent.
To examine whether the loss of NOS1AP has the opposite effect of overexpression on Lats1/YAP phosphorylation, we knocked down endogenous NOS1AP expression by siRNA and monitored the levels of pLats1/pYAP. A pool of siRNAs effectively reduced NOS1AP levels over 72 h compared to the control, which correlated with decreased phosphorylation of Lats1 and YAP (Fig. 5G and H). We next examined the localization of YAP in epithelial MCF7 cells stably expressing YFP or YFP-NOS1APa and stained for endogenous YAP. Unlike cells expressing YPF alone (Fig. 6B), where YAP was enriched in the nucleus (Fig. 6C), cells plated at a similar density expressing YFP-NOS1APa (Fig. 6E) revealed a more diffuse distribution of YAP throughout the cell (Fig. 6F). Taken together, these data demonstrate that NOS1AP expression levels affect both the subcellular localization and activity of Lats1 and YAP.
FIG 6.
NOS1APa reduces YAP nuclear accumulation. MCF7 cells stably expressing YFP or YFP-NOS1APa were fixed and costained with anti-GFP and anti-YAP antibodies. Hoechst stains cell nuclei. Bar = 20 μm.
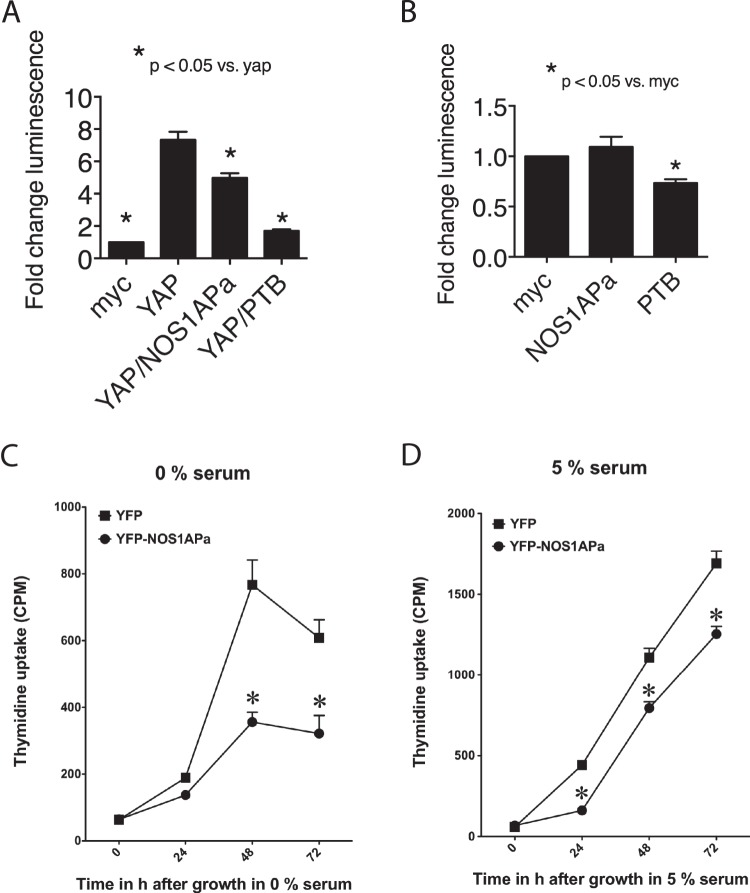
The TEAD family of transcription factors binds to and is activated by YAP (30). We next tested whether NOS1APa affected the ability of YAP to activate TEAD transcription by utilizing a previously described TEAD-luciferase reporter system (31, 32). Here we coexpressed myc-NOS1APa and FLAG-YAP in the presence of the TEAD reporter and found that NOS1APa suppressed the ability of YAP to activate this reporter (Fig. 7A), consistent with the redistribution of YAP staining to the cytoplasm (Fig. 6). In addition, since we had shown that the PTB domain was sufficient to regulate Lats1 activity, we tested if the PTB domain influenced YAP-mediated TEAD activation. Overexpression of the myc-PTB domain in the presence of FLAG-tagged YAP was able to suppress TEAD in the reporter assay to a greater extent than NOS1APa (Fig. 7A). Finally, we examined whether NOS1AP or the PTB domain was able to suppress TEAD activation in the absence of YAP. Reporter activity was significantly suppressed in the presence of the PTB domain (Fig. 7B), further implicating the NOS1AP PTB domain as the functional region in NOS1AP-mediated Hippo component regulation. These results show that NOS1APa has the ability to control YAP-dependent TEAD activity and possibly also target genes involved in proliferation and apoptosis.
FIG 7.
The PTB domain is sufficient to reduce YAP-mediated TEAD activity. (A and B) HEK293T cells transfected with TEAD-luciferase together with the plasmids indicated and lysates were prepared for luciferase measurements after 24 h. Data show luciferase activity normalized against β-galactosidase activity. Histograms show means ± SEM from 3 independent experiments. Differences were considered significant by Student's t test at a P value of <0.05. (C and D) MCF7 cells stably expressing YFP-NOS1APa incorporate less radiolabeled thymidine over a 72-h period without (C) or with (D) 5% serum than do MCF7 cells stably expressing YFP. Differences were considered significant by Student's t test at a P value of <0.05 (asterisk).
Overexpression of NOS1AP leads to a decrease in the amount of YAP localized to the nucleus and a reduction in the ability of YAP to activate TEAD. Therefore, we tested whether NOS1AP influences proliferation. To address this, we used a thymidine incorporation assay to measure the proliferation rate of MCF7 cells stably expressing NOS1APa compared to a line expressing YFP alone. In the NOS1APa-expressing cells, we saw a reduction in proliferation in the absence and presence of serum (Fig. 7C and D). These data indicate that NOS1AP functions though YAP to influence both basal and stimulated cell growth.
DISCUSSION
In this study, we have identified several novel NOS1AP isoforms by using antibodies against the C-terminal region of NOS1APc, whose sequence we previously identified through bioinformatics strategies (11). All NOS1AP isoforms detected are abundantly distributed in the brain, with lower levels being found in nonneuronal tissue. While some PTB domain-containing NOS1AP isoforms are targeted to the plasma membrane, where they can associate with inositol phospholipids, others that bind Scribble do not share this localization, indicating that different NOS1AP complexes exist in mammalian cells. This idea is consistent with our results that link NOS1AP to proteins in the Hippo pathway by demonstrating that the signaling activities of the transcriptional coactivator YAP and its upstream kinase Lats1 are controlled by NOS1AP. Thus, our data are consistent with NOS1AP functioning as an important regulator of the Hippo pathway to control cellular proliferation.
To determine whether the NOS1AP gene produces splice variants, we sequenced a number of clones obtained from a cerebellar cDNA library. This led to the identification of two new isoforms that are variants of NOS1APc, which we have named NOS1APd and NOS1APe. The NOS1APc isoform, which migrates at 95 kDa, results from a splicing event that fuses amino acid 440 of Rattus norvegicus NOS1APa with the unique C-terminal region found in NOS1APc. The splice variants NOS1APd and NOS1APe result from a splicing event that fuses amino acid 415 of Rattus NOS1APa with the unique C-terminal region that we have identified in NOS1APc (11), with NOS1APe having a further 5-amino-acid insert in its PTB domain (Fig. 1D). We also identified a shorter isoform that contains a unique 17-amino-acid sequence 5′ to the C-terminal region common to the NOS1APc, -d, and -e isoforms. This isoform, which we named NOS1APf, has previously been annotated in the NCBI database (GenBank accession number NM_001190459). Intriguingly, we identified a protein that migrated at ∼70 kDa, which precipitated with and was recognized by all antibodies used in our study. We surmise that this protein cannot be NOS1APa, as two of the antibodies that were used to detect this isoform recognize epitopes unique to NOS1APc. However, to date, we have been unsuccessful in identifying the nature of this isoform. We speculate that part of the protein is common to regions in NOS1APa, while it also contains the C-terminal extension identified in NOS1APc. One interesting possibility is that this isoform is a fusion between a previously reported NOS1APshort isoform (NOS1APb) that is upregulated in patients with bipolar disorders and schizophrenia (23) and the C-terminal extension found in NOS1APc (NOS1APg and -h) (Fig. 1E). Taken together, these results demonstrate a diversity of NOS1AP splice variants that may have different functions and targets in cell lines and tissues.
NOS1APe contains a small (LLLLQ) insert in its PTB domain, which is important for its membrane localization, as NOS1APd lacking this motif has a cytoplasmic localization (Fig. 3C). Since NOS1APe, which contains the insert, and NOS1APa, which lacks this insert, exclusively localize to the plasma membrane, this suggests that the C-terminal extensions found in NOS1APc (and NOS1APd) likely form an intra- or intermolecular interaction that restricts the PTB domain from recognizing membranes. In the context of NOS1APe, the LLLLQ insert may relieve this restriction, allowing binding of NOS1APe to the membrane. Consistent with this view, PTB domains have a preference for membrane recognition (33), and Numb, a protein involved in Drosophila neurogenesis, expresses a splice form that contains an 11-amino-acid insert in the α2 helix and β2 strand in its PTB domain, which is critical for phospholipid association (28). Additionally, the insert in NOS1APe flanks regions rich in basic residues. Stretches of basic residues in the PTB domains have been suggested to be essential for PTB domain-mediated membrane recognition (33). Thus, although we do not know the precise mechanism by which the NOS1AP PTB domain targets the membrane, we find that the short NOS1APe insert is found in a stretch of lysine residues, creating a basic region in the domain. Whether the 5-amino-acid insert exposes this basic stretch allowing for electrostatic interactions with membranes or disrupts a protein-protein interaction permitting the PTB domain to bind phospholipids remains to be tested. Nonetheless, our results show that similar to PTB domains contained in other proteins, the NOS1AP PTB domain can direct the protein to phospholipid-rich regions.
The PTB domain is also essential for the association with Scribble, as NOS1APf, which lacks this domain, fails to interact. However, the abundant expression of NOS1APf mRNA in the CNS suggests that this isoform may have a distinct role in the brain that remains to be established. Scribble has been shown to associate with membranes through its LRR while the PDZ domains act as scaffolds for various ligands and are critical for the functions of Scribble in controlling cellular proliferation and migration (34–36). We have previously reported that the NOS1AP PTB domain can interact with PDZ domains 3 and 4 in Scribble. Our current findings show that isoforms that are not preferentially enriched at the membrane can still associate with Scribble. Thus, it seems likely that NOS1APa and -e link Scribble to its function at the membrane, while NOS1APc and -d connect Scribble to spatially distinct events. This idea would also be consistent with data from a recent report suggesting the existence of different NOS1AP-Scribble complexes (15).
Our findings demonstrate that NOS1AP forms a complex with the transcriptional coactivator YAP (Fig. 5A to C) as well as with Scribble. Furthermore, the expression of increasing amounts of NOS1AP induces the phosphorylation of YAP at S127, while NOS1AP silencing abolishes this effect. Likewise, the expression levels of NOS1AP alter the phosphorylation state of Lats1, indicating that NOS1AP acts upstream of both Lats1 and YAP. Similar results on phosphorylation were obtained with other isoforms containing the PTB domain as well as with the expression of the PTB domain alone. We also found that NOS1AP induces the redistribution of YAP to the cytoplasm in stable MCF7 cells that express NOS1APa. This effect coincided with reduced cell proliferation as well as with decreased transcriptional activity of the YAP target TEAD. It is unlikely that NOS1AP proteins directly associate with YAP, since we were unable to detect binding between overexpressed NOS1AP and YAP. This suggests that other proteins are necessary to bridge the interaction, and these proteins are likely important in influencing the role of NOS1APa in TEAD activity, as full-length NOS1APa was less effective than the PTB domain alone in suppressing YAP-dependent TEAD activity. Nonetheless, given that the PTB domain of NOS1AP is sufficient to influence the activity of YAP/Lats1, we speculate that NOS1AP, through the PTB domain, exerts its effect on Hippo signaling functions through Scribble, which is required for proper Hippo pathway activity (9, 10). It will be of interest to further define the role of the PTB domain in the context of Hippo signaling and the roles that the different NOS1AP isoforms play, as they all contain the PTB domain but have differential subcellular localizations within the cell (Fig. 3). Finally, YAP and TAZ function as transcriptional coactivators, and TAZ has been shown to associate with Scribble and members of the Hippo core complex (9); thus, whether TAZ functionally connects with a Scribble-NOS1AP complex to regulate Hippo-dependent signaling remains to be tested. Nonetheless, our data demonstrate that NOS1AP plays an important role in regulating Hippo signaling to control cellular proliferation. This is important in light of NOS1AP's function as a tumor suppressor in breast cancer (15).
In summary, we have shown that NOS1AP is a highly spliced protein, with the different isoforms having differential tissue expression and subcellular localizations and forming different signaling complexes. Given that NOS1AP has been implicated in a number of disorders, including bipolar disorder (23, 37), schizophrenia (38), QT syndrome (14, 39, 40), anxiety (13), and post-traumatic stress disorder (PTSD) (41), it will be important to determine if any of the isoforms identified here play a role in modifying or antagonizing any of these disorders. Further characterization of the nature of splicing in different tissues will help dissect the role of NOS1AP in disease processes.
ACKNOWLEDGMENTS
This work was supported by grants from the NSERC and the EJLB Foundation (J.P.F.) and the CIHR (C.J.S.). L.C. was funded in part by the CRTP, Dalhousie University. J.P.F. is a Tier 2 Canadian Research Chair in Molecular Mechanisms of Brain Repair.
REFERENCES
- 1.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 2.Harvey KF, Pfleger CM, Hariharan IK. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114:457–467. doi: 10.1016/S0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 3.Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 4.Gumbiner BM, Kim NG. 2014. The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci 127:709–717. doi: 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enderle L, McNeill H. 2013. Hippo gains weight: added insights and complexity to pathway control. Sci Signal 6:re7. doi: 10.1126/scisignal.2004208. [DOI] [PubMed] [Google Scholar]
- 6.Barry ER, Camargo FD. 2013. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol 25:247–253. doi: 10.1016/j.ceb.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Verghese S, Waghmare I, Kwon H, Hanes K, Kango-Singh M. 2012. Scribble acts in the Drosophila fat-hippo pathway to regulate warts activity. PLoS One 7:e47173. doi: 10.1371/journal.pone.0047173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig F, Schafer T, Ramachandran H, Mlodzik M, Huber TB, Kuehn EW, Kim E, Kramer-Zucker A, Walz G. 2009. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci U S A 106:8579–8584. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. 2011. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 10.Mohseni M, Sun J, Lau A, Curtis S, Goldsmith J, Fox VL, Wei C, Frazier M, Samson O, Wong KK, Kim C, Camargo FD. 2014. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol 16:108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richier L, Williton K, Clattenburg L, Colwill K, O'Brien M, Tsang C, Kolar A, Zinck N, Metalnikov P, Trimble WS, Krueger SR, Pawson T, Fawcett JP. 2010. NOS1AP associates with Scribble and regulates dendritic spine development. J Neurosci 30:4796–4805. doi: 10.1523/JNEUROSCI.3726-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. 2004. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet 74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu LJ, Li TY, Luo CX, Jiang N, Chang L, Lin YH, Zhou HH, Chen C, Zhang Y, Lu W, Gao LY, Ma Y, Zhou QG, Hu Q, Hu XL, Zhang J, Wu HY, Zhu DY. 2014. CAPON-nNOS coupling can serve as a target for developing new anxiolytics. Nat Med 20:1050–1054. doi: 10.1038/nm.3644. [DOI] [PubMed] [Google Scholar]
- 14.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O'Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A. 2006. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet 38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 15.Anastas JN, Biechele TL, Robitaille M, Muster J, Allison KH, Angers S, Moon RT. 2012. A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene 31:3696–3708. doi: 10.1038/onc.2011.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffrey SR, Benfenati F, Snowman AM, Czernik AJ, Snyder SH. 2002. Neuronal nitric-oxide synthase localization mediated by a ternary complex with synapsin and CAPON. Proc Natl Acad Sci U S A 99:3199–3204. doi: 10.1073/pnas.261705799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. 1998. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron 20:115–124. doi: 10.1016/S0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 18.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. 2000. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron 28:183–193. doi: 10.1016/S0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Muruganandan S, Roman AA, Sinal CJ. 2010. Role of chemerin/CMKLR1 signaling in adipogenesis and osteoblastogenesis of bone marrow stem cells. J Bone Miner Res 25:222–234. doi: 10.1359/jbmr.091106. [DOI] [PubMed] [Google Scholar]
- 22.Muruganandan S, Parlee SD, Rourke JL, Ernst MC, Goralski KB, Sinal CJ. 2011. Chemerin, a novel PPARgamma target gene that promotes mesenchymal stem cell adipogenesis. J Biol Chem 286:23982–23995. doi: 10.1074/jbc.M111.220491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu B, Wratten N, Charych EI, Buyske S, Firestein BL, Brzustowicz LM. 2005. Increased expression in dorsolateral prefrontal cortex of CAPON in schizophrenia and bipolar disorder. PLoS Med 2:e263. doi: 10.1371/journal.pmed.0020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrel D, Du Y, Komlos D, Hadzimichalis NM, Kwon M, Wang B, Brzustowicz LM, Firestein BL. 2009. NOS1AP regulates dendrite patterning of hippocampal neurons through a carboxypeptidase E-mediated pathway. J Neurosci 29:8248–8258. doi: 10.1523/JNEUROSCI.5287-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolt PC, Chen Y, Liu P, Bock HH, Blacklow SC, Herz J. 2005. Phosphoinositide binding by the disabled-1 PTB domain is necessary for membrane localization and Reelin signal transduction. J Biol Chem 280:9671–9677. doi: 10.1074/jbc.M413356200. [DOI] [PubMed] [Google Scholar]
- 26.Ravichandran KS, Zhou MM, Pratt JC, Harlan JE, Walk SF, Fesik SW, Burakoff SJ. 1997. Evidence for a requirement for both phospholipid and phosphotyrosine binding via the Shc phosphotyrosine-binding domain in vivo. Mol Cell Biol 17:5540–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie J, Li SS, McGlade CJ. 2004. A novel PTB-PDZ domain interaction mediates isoform-specific ubiquitylation of mammalian Numb. J Biol Chem 279:20807–20815. doi: 10.1074/jbc.M311396200. [DOI] [PubMed] [Google Scholar]
- 28.Dho SE, French MB, Woods SA, McGlade CJ. 1999. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem 274:33097–33104. [DOI] [PubMed] [Google Scholar]
- 29.Albertson R, Chabu C, Sheehan A, Doe CQ. 2004. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J Cell Sci 117:6061–6070. doi: 10.1242/jcs.01525. [DOI] [PubMed] [Google Scholar]
- 30.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 33.DiNitto JP, Lambright DG. 2006. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta 1761:850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Zeitler J, Hsu CP, Dionne H, Bilder D. 2004. Domains controlling cell polarity and proliferation in the Drosophila tumor suppressor Scribble. J Cell Biol 167:1137–1146. doi: 10.1083/jcb.200407158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. 2008. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murdoch JN, Henderson DJ, Doudney K, Gaston-Massuet C, Phillips HM, Paternotte C, Arkell R, Stanier P, Copp AJ. 2003. Disruption of scribble (Scrb1) causes severe neural tube defects in the circletail mouse. Hum Mol Genet 12:87–98. doi: 10.1093/hmg/ddg014. [DOI] [PubMed] [Google Scholar]
- 37.Berrettini W. 2003. Bipolar disorder and schizophrenia: not so distant relatives? World Psychiatry 2:68–72. [PMC free article] [PubMed] [Google Scholar]
- 38.Brzustowicz LM. 2008. NOS1AP in schizophrenia. Curr Psychiatry Rep 10:158–163. doi: 10.1007/s11920-008-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamshidi Y, Nolte IM, Dalageorgou C, Zheng D, Johnson T, Bastiaenen R, Ruddy S, Talbott D, Norris KJ, Snieder H, George AL, Marshall V, Shakir S, Kannankeril PJ, Munroe PB, Camm AJ, Jeffery S, Roden DM, Behr ER. 2012. Common variation in the NOS1AP gene is associated with drug-induced QT prolongation and ventricular arrhythmia. J Am Coll Cardiol 60:841–850. doi: 10.1016/j.jacc.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raitakari OT, Blom-Nyholm J, Koskinen TA, Kahonen M, Viikari JS, Lehtimaki T. 2009. Common variation in NOS1AP and KCNH2 genes and QT interval duration in young adults. The Cardiovascular Risk in Young Finns Study. Ann Med 41:144–151. doi: 10.1080/07853890802392529. [DOI] [PubMed] [Google Scholar]
- 41.Lawford BR, Morris CP, Swagell CD, Hughes IP, Young RM, Voisey J. 2013. NOS1AP is associated with increased severity of PTSD and depression in untreated combat veterans. J Affect Disord 147:87–93. doi: 10.1016/j.jad.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Li SC, Zwahlen C, Vincent SJ, McGlade CJ, Kay LE, Pawson T, Forman-Kay JD. 1998. Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat Struct Biol 5:1075–1083. doi: 10.1038/4185. [DOI] [PubMed] [Google Scholar]