Abstract
MicroRNAs (miRNAs) are involved in the progression and suppression of various diseases through translational inhibition of target mRNAs. Therefore, the alteration of miRNA biogenesis induces several diseases. The nuclear factor 90 (NF90)-NF45 complex is known as a negative regulator in miRNA biogenesis. Here, we showed that NF90-NF45 double-transgenic (dbTg) mice develop skeletal muscle atrophy and centronuclear muscle fibers in adulthood. Subsequently, we found that the levels of myogenic miRNAs, including miRNA 133a (miR-133a), which promote muscle maturation, were significantly decreased in the skeletal muscle of NF90-NF45 dbTg mice compared with those in wild-type mice. However, levels of primary transcripts of the miRNAs (pri-miRNAs) were clearly elevated in NF90-NF45 dbTg mice. This result indicated that the NF90-NF45 complex suppressed miRNA production through inhibition of pri-miRNA processing. This finding was supported by the fact that processing of pri-miRNA 133a-1 (pri-miR-133a-1) was inhibited via binding of NF90-NF45 to the pri-miRNA. Finally, the level of dynamin 2, a causative gene of centronuclear myopathy and concomitantly a target of miR-133a, was elevated in the skeletal muscle of NF90-NF45 dbTg mice. Taken together, we conclude that the NF90-NF45 complex induces centronuclear myopathy through increased dynamin 2 expression by an NF90-NF45-induced reduction of miR-133a expression in vivo.
INTRODUCTION
MicroRNAs (miRNAs) are functional small noncoding RNAs 21 to 23 nucleotides in length. miRNAs bind to 3′ untranslated regions (UTRs) of target mRNAs, leading to either mRNA degradation or translational inhibition. The functions of miRNAs influence biological phenomena and diseases such as development (1, 2), cell proliferation (3), differentiation (4), apoptosis (5), as well as tumorigenesis (6, 7).
The biogenesis of miRNAs involves several steps (8). First, miRNA genes are transcribed as primary miRNAs (pri-miRNAs) by RNA polymerase II (9). Subsequently, these pri-miRNAs are processed into precursor miRNAs (pre-miRNAs) by a microprocessor complex composed of Drosha and DGCR8 (10–14). Thereafter, the pre-miRNAs are transported from the nucleus to the cytoplasm (15–17) and processed into mature miRNA duplexes by the Dicer complex (18–20). The functional strand of the duplex (mature miRNA) is loaded into the RNA-induced silencing complex (RISC) (21–23). As part of the RISC, miRNA binds to target mRNAs and induces their translational inhibition or degradation (20, 22–24). Recently, it was reported that some RNA-binding proteins negatively regulate miRNA biogenesis. For example, Lin28A/B (25), the Musashi homolog 2/Hu antigen R complex (26), and the nuclear factor 90 (NF90; also referred to as interleukin enhancer binding factor 3 [ILF3], NFAR1, or DRBP76)-nuclear factor 45 (NF45) complex (27) suppress miRNA processing through binding to pri- or pre-miRNAs.
NF90 contains a functional nuclear localization signal, two double-stranded RNA (dsRNA)-binding motifs, a zinc finger nucleic acid-binding domain, and a glutamic acid-rich region. NF90 forms a complex with NF45, which carries out biological functions. We previously showed that the NF90-NF45 complex suppresses pri-miRNA cleavage and mature miRNA production through binding to pri-miRNAs in HEK293T cells (27). In addition, it has been reported that NF90 or the NF90-NF45 complex is involved in mRNA stabilization (28), translational repression (29, 30), replication of viral RNA (31), and transcription (32, 33) in vitro. However, the physiological functions of NF90 and/or NF45 remain largely unknown because NF90 or NF45 knockout (KO) mice exhibit perinatal lethality due to respiratory failure or embryonic lethality (32, 34). We have found that the overexpression of NF90 evokes heart failure and skeletal muscle atrophy through the repression of transcription factors, such as peroxisome proliferator-activated receptor γ coactivator 1 and nuclear respiratory factor 1, to regulate nuclear genes relevant to mitochondrial function in vivo (35). In contrast, the biological functions of the NF90-NF45 complex are uncertain in vivo. To address this issue, we generated and studied NF90-NF45 double-transgenic (dbTg) mice to uncover the functions of the NF90-NF45 complex in vivo. Here, we found that NF90-NF45 dbTg mice showed a decrease in muscular mass and that their skeletal muscles exhibited centronuclear muscle fibers. Moreover, the level of dynamin 2 (Dnm2), which is a known causative agent in centronuclear myopathy (CNM), was elevated in the skeletal muscle of NF90-NF45 dbTg mice, whereas the level of microRNA 133a (miR-133a), which translationally represses Dnm2 expression, was reduced in the same skeletal muscle. These observations revealed that the overexpression of NF90-NF45 leads to the development of centronuclear myopathy through an increase in the Dnm2 level caused by a downregulation of mature miR-133a.
MATERIALS AND METHODS
Generation of NF90-NF45 dbTg mice and genotyping.
NF90 transgenic (Tg) mice were generated as described previously (35). To generate NF45 Tg mice, mouse NF45 (mNF45) cDNA was amplified from the DC2.4 mouse dendritic cell line by reverse transcription-PCR (RT-PCR). The cDNA fragment was subcloned into the XhoI site of the pCAGGS vector provided by Jun-ichi Miyazaki, Osaka University (Osaka, Japan) (36). After digestion with SalI and HindIII, the fragment carrying the cytomegalovirus immediate early (CMV-IE) enhancer, the chicken β-actin promoter, mNF45 cDNA, and rabbit β-globin poly(A) was used for microinjection into fertilized eggs recovered from C57BL/6CrSlc females crossed with C57BL/6CrSlc males. NF90-NF45 dbTg mice were obtained by crossing NF90 Tg mice with NF45 Tg mice. Tg mice were identified by PCR analysis of genomic tail DNA using primers specific for mouse NF90 and NF45 cDNAs. All animal experiments were approved by the Division of Laboratory Animal Science, Science Research Center, Kochi University.
Western blot analysis.
Western blot analysis was performed as described previously (37).
Antibodies.
An anti-mouse NF90 antibody was produced as described previously (35). An anti-mouse NF45 antibody was produced by immunizing New Zealand White rabbits with a full-length recombinant His-mouse NF45 protein as described previously (38). Other antibodies used were anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Cell Signaling Technology, MA), anti-ILF3 (GeneTex, USA), and anti-Dnm2 (Abcam, United Kingdom).
X-ray computed tomographic analysis.
The muscle mass of the hindlimb was measured by Latheta X-ray computed tomography (Hitachi Aloca Medical, Ltd., Japan) according to the manufacturer's protocol.
Histological analysis.
The quadriceps of wild-type (WT) and NF90-NF45 dbTg mice was fixed with 10% phosphate-buffered formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E) and observed under a light microscope. Cross-sectional areas of myofibers were measured by using ImageJ software. More than 60 fibers were examined per section. To calculate the percentage of myofibers with centralized nuclei in WT and NF90-NF45 dbTg mice, we counted 100 fibers.
qRT-PCR.
Total RNA was isolated from the quadriceps of WT and NF90-NF45 dbTg mice by using TRIzol (Invitrogen, USA). Genomic DNA was removed by DNase-free treatment (Ambion, USA). cDNA was synthesized by using SuperScript III reverse transcriptase (Invitrogen, USA) and random hexamer primers according to the manufacturer's protocol. PCR was performed as described previously (35). Quantitative RT-PCR (qRT-PCR) for the detection of mature miRNAs was performed by using a TaqMan microRNA assay kit (Applied Biosystems, USA) according to the manufacturer's protocol. snoRNA202 was used as an internal control to normalize the RNA input.
Transmission electron microscopic analysis.
The hindlimbs of WT and NF90-NF45 dbTg mice were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3) for 2 h at 4°C. The tissues were then postfixed with 1% osmium tetroxide (or osmic acid) in 0.1 M phosphate buffer (pH 7.3) for 1 h at 4°C. After dehydration, the specimens were transferred to propylene oxide, embedded in Epon 812 (TAAB Laboratories Equipment, Reading, United Kingdom), and observed under an H-7100 electron microscope (Hitachi, Japan).
Cell culture and transfection.
Mouse myoblast C2C12 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. For expression of mouse NF90 and NF45 proteins, a mouse NF90 cDNA fragment was subcloned into the XhoI site of the pEB-Multi vector (Wako, Japan), and a human NF45 cDNA fragment was inserted into the XhoI and KpnI sites of the pEB-Multi vector. The pEB-Multi-NF90 and -NF45 plasmids or the pEB-Multi control plasmid was transfected into C2C12 cells by using Lipofectamine LTX Plus reagent (Invitrogen) according to the manufacturer's instructions.
In vitro pri-miRNA processing assay.
An in vitro pri-miRNA processing assay was performed as described previously (27). Briefly, pri-miR-133a-1 and pri-miR-206 were amplified from cDNA prepared from the skeletal muscle of C57BL/6CrSlc mice by PCR with specific primers. The PCR products were subcloned into the pGEM-T-easy vector (Promega, WI, USA). The construction of a pGEM-T-easy-pri-miR-21 plasmid was described previously (27). The plasmids were linearized with SpeI and used for in vitro transcription to prepare a 32P-radiolabeled RNA probe. Whole-cell lysates (WCLs) were prepared from C2C12 cells, which were transfected with expression plasmids, by sonication in lysis buffer (20 mM HEPES-KOH [pH 8.0], 100 mM KCl, 0.2 mM EDTA, 5% glycerol, 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride), followed by centrifugation. Each 30-μl reaction mixture contained ∼25,000 cpm radiolabeled pri-miRNA probe, WCL from C2C12 cells, 1 U RNaseOUT (Invitrogen), and 6.4 mM MgCl2 in reaction buffer (20 mM HEPES-KOH [pH 8.0], 100 mM KCl, 0.2 mM EDTA, and 5% glycerol). After incubation of the mixture at 37°C within 90 min, processed RNA was purified by phenol-chloroform extraction and ethanol precipitation. The purified RNA probes were loaded onto a 12% denaturing polyacrylamide gel, and the intensities of the produced mature miRNAs were measured by using the BAS-2500 imaging system (Fuji Film, Japan).
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed with a 20-μl reaction mixture containing 50 mM Tris-HCl (pH 7.6), 100 mM NaCl, 5% glycerol, 2 mM MgCl2, 0.2% bovine serum albumin, 20 U RNaseOUT (Invitrogen), 1 μg Saccharomyces cerevisiae tRNA, and 25,000 cpm labeled RNA probe. The mixture was incubated at room temperature for 15 min and then electrophoresed on a 4% or 5% polyacrylamide gel containing 10% glycerol in 0.5× Tris-borate-EDTA. For supershift assays, recombinant proteins were preincubated with 0.5 or 1.0 μg, or 0.5 μl or 1.0 μl, of antibodies at room temperature for 15 min, followed by the addition of the labeled probe. Images were captured and the intensities of specific bands were measured by using the BAS-2500 imaging system.
Immunohistochemistry.
Immunohistochemistry was performed as described previously (35). The following antibodies were used: anti-mouse NF90 (3.6 ng/μl), anti-NF45 (3.6 ng/μl), anti-Dmn2 (2 ng/μl), and normal rabbit IgG (3.6 or 2 ng/μl).
Sequences of the PCR primers are available upon request.
miRNA microarray.
Total RNA was isolated from the quadriceps of WT and NF90-NF45 dbTg mice and labeled with Cy3 using an miRNA Complete labeling reagent and hybridization kit (Agilent Technologies) according to the manufacturer's instructions. The labeled RNA was hybridized on a SurePrint G3 mouse miRNA array kit (8×60K, release 16.0; Agilent Technologies). The array was scanned using a microarray scanner (Agilent Technologies) to measure the intensities of the microarray spots. The intensities of the spots were normalized by GeneSpring GX.
Whole-genome expression microarray.
Total RNA was isolated from the quadriceps of WT and NF90-NF45 dbTg mice. Single-stranded cDNA was generated from the amplified cRNA with the whole-transcriptome amplification kit module 1 (Affymetrix) and labeled biotin using the whole-transcriptome terminal labeling kit (Affymetrix). Following fragmentation, single-stranded cDNA was hybridized for 20 h at 48°C with a MoGene2.1ST array strip (Affymetrix) and a GeneChip expression 3′ amplification reagent hybridization control kit (Affymetrix). The array was scanned using an imaging station (Affymetrix) according to the manufacturer's protocol. Arrays were summarized and normalized with Expression Console using RMA-sketch.
Microarray data accession numbers.
All miRNA microarray data have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE61001. All whole-genome expression microarray data have been deposited in the GEO database under accession no. GSE67591.
RESULTS
The NF90-NF45 complex causes atrophy of skeletal muscle.
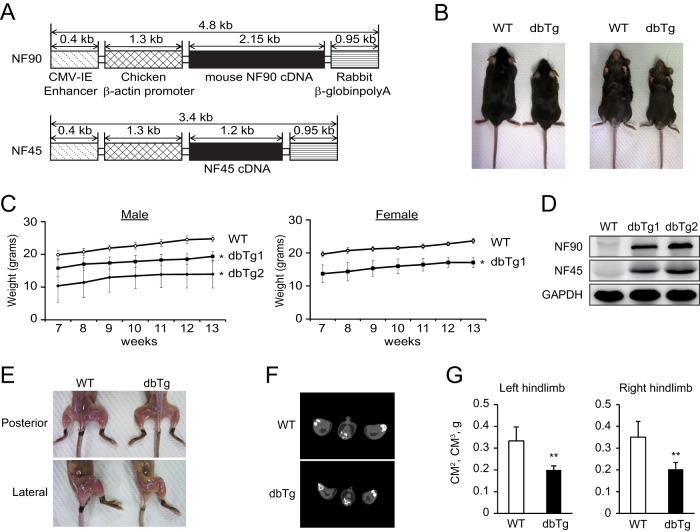
The complex of NF90 and NF45 is known to participate in viral RNA replication (31), transcription (32, 33), and miRNA biogenesis (27) in vitro. However, the physiological function of the NF90-NF45 complex is still unclear. To elucidate its functions, we generated NF90 Tg mice (35) and NF45 Tg mice. NF90-NF45 dbTg mice were obtained by crossing NF90 Tg mice with NF45 Tg mice, resulting in the establishment of two transgenic lines (dbTg1 and dbTg2). NF90 and NF45 transgene constructs were under the control of the chicken β-actin promoter and the cytomegalovirus (CMV) enhancer (Fig. 1A). Both lines of NF90-NF45 dbTg mice exhibited reductions in body size and weight compared to those of WT mice (Fig. 1B and C). To examine mRNA expression of NF90 and NF45 in various tissues of dbTg1 and dbTg2 mice, we performed semiquantitative RT-PCR analysis. Robust expression of NF90 and NF45 was detected in the eyes, heart, lung, and skeletal muscle of the two NF90-NF45 dbTg lines compared with that in WT mice (data not shown). Western blot analysis corroborated the dramatic elevation of NF90 and NF45 protein levels in the skeletal muscle of the two NF90-NF45 dbTg lines (Fig. 1D). Thereafter, we focused on the influence of NF90 and NF45 overexpression on the formation of skeletal muscle. NF90-NF45 dbTg mice showed a reduction of skeletal muscle by visual observation (Fig. 1E). In addition, we carried out X-ray computed tomography of the skeletal muscle of WT and NF90-NF45 dbTg mice. Gastrocnemius muscle volumes of the left and right hindlimbs of NF90-NF45 dbTg mice were significantly decreased compared to those of WT mice (Fig. 1F and G). These results showed that NF90-NF45 dbTg mice exhibited skeletal muscle atrophy.
FIG 1.
NF90-NF45 dbTg mice exhibit atrophy of skeletal muscle. (A) Transgene constructs of NF90 and NF45 used to generate dbTg mice. (B) Representative photographs of WT and dbTg1 mice at 15 weeks of age. (C) Growth curves of WT (n = 7), dbTg1 (n = 7), and dbTg2 (n = 3) mice (male) and those of WT and dbTg1 mice (female) (n = 7) at 7 to 13 weeks of age. Data are expressed as means ± standard deviations. *, P < 0.01 compared to the WT, determined by two-tailed Student's t test. (D) Western blot analysis of the levels of NF90 and NF45 in skeletal muscles of WT, dbTg1, and dbTg2 mice at 15 weeks of age. GAPDH was used as a loading control. (E) Visual observation of hindlimbs from WT and dbTg1 mice at 15 weeks of age. (F) Axial computed tomography images of the center of distal hindlimbs in WT and dbTg1 mice at 14 weeks of age. (G) X-ray computed tomographic analysis of the muscle masses of right and left hindlimbs in WT and dbTg1 mice at 14 weeks of age. Data are expressed as means ± standard deviations (n = 4). **, P < 0.01 compared to the WT, determined by two-tailed Student's t test.
Skeletal muscles of NF90-NF45 dbTg mice contain immature muscle fibers.
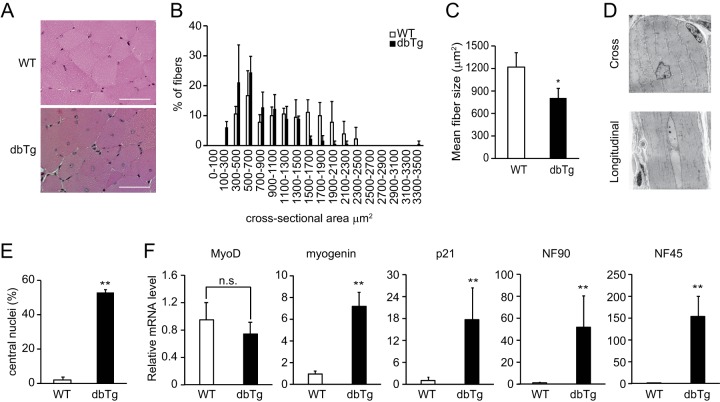
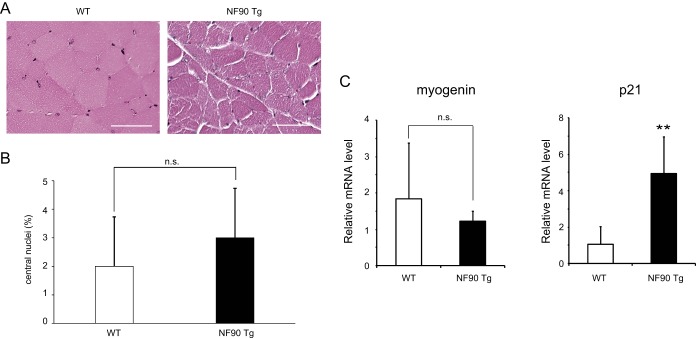
To address the cause of the reduction of skeletal muscle in NF90-NF45 dbTg mice, we performed histological analysis by H&E staining of the quadriceps in WT and NF90-NF45 dbTg mice at 15 weeks of age (Fig. 2A). The muscle fiber sizes of NF90-NF45 dbTg mice were obviously smaller than those of WT mice (Fig. 2B and C). Interestingly, we found that the nuclei were localized to the center of muscle fibers in NF90-NF45 dbTg mice (Fig. 2A). Transmission electron microscopic analysis also showed centronuclear muscle fibers in the skeletal muscle of NF90-NF45 dbTg mice (Fig. 2D). At 15 weeks of age, centronuclear muscle fibers were present in nearly 50% of the skeletal muscle fibers in NF90-NF45 dbTg mice (Fig. 2E) but were absent in the skeletal muscle of WT mice (Fig. 2E). Centronuclear muscle fibers are known as immature fibers (39). Myogenic markers such as MyoD, myogenin, and p21 are expressed in skeletal muscle at different developmental stages. Expression of MyoD is found in muscular tissues differentiated from myoblasts to myocytes, while myogenin and p21 are expressed during myogenesis from myocytes to myotubes (40–42). The levels of these markers are diminished in accordance with muscular maturation toward myofibers (43, 44). To examine the degree of skeletal muscle maturation in NF90-NF45 dbTg mice, we measured the mRNA expression of myogenic markers in the skeletal muscles of WT and NF90-NF45 dbTg mice by qRT-PCR. As expected, extremely high expression levels of NF90 and NF45 were observed in the skeletal muscle of NF90-NF45 dbTg mice (Fig. 2F). Notably, the expression levels of myogenin and p21 in NF90-NF45 dbTg mice were significantly higher than those in WT mice, while MyoD expression was unchanged (Fig. 2F). As mentioned above, the levels of these myogenic markers are largely decreased according to differentiation from myotubes to muscular maturation toward myofibers. Thus, these results supported the notion that the muscle maturation process was prevented by the overexpression of NF90 and NF45 in the skeletal muscle of NF90-NF45 dbTg mice at 15 weeks of age because the high levels of myogenin and p21 are maintained in the skeletal muscle of NF90-NF45 dbTg mice at the same age (Fig. 2F). NF90 is known to regulate the expression of myogenic regulatory factors, including MyoD, myogenin, and p21, at the mRNA and/or protein level (34). Therefore, it raises the possibility that high levels of myogenin and/or p21 directly cause the development of centronuclear myopathy in NF90-NF45 dbTg mice. On the other hand, we previously reported that NF90-alone Tg mice also developed skeletal muscle atrophy (35). However, muscular atrophy is accompanied by mitochondrial degeneration (35) but not by centralized nuclei (Fig. 3A and B). We also measured the expression levels of myogenin and p21 in the skeletal muscle of NF90 Tg mice. In consequence, the level of p21 alone was clearly elevated in NF90 Tg mice compared with that in WT mice (Fig. 3C), whereas there was no significantly difference in the expression levels of myogenin between WT and NF90 Tg mice, unlike between WT and NF90-NF45 dbTg mice (compare Fig. 2F and 3C). These results indicate that centronuclear myopathy caused by the overexpression of NF90-NF45 is accompanied by high levels of myogenin and p21, whereas NF90-alone-induced muscular atrophy bearing mitochondrial degeneration but not with centralized nuclei is related to the high expression level of p21 alone, suggesting that myogenin may directly participate in the development of centronuclear myopathy in NF90-NF45 dbTg mice. However, a previous report indicated that overexpression of myogenin leads to histologically normal muscular tissues in vivo (45). Therefore, it is thought that other factors besides myogenin cause skeletal muscle atrophy accompanying centronuclear muscle fibers in NF90-NF45 dbTg mice.
FIG 2.
Skeletal muscles of NF90-NF45 dbTg mice contain immature muscle fibers. (A) H&E staining of the cross-sectional area of quadriceps from WT and dbTg1 mice at 15 weeks of age. Bars, 50 μm. (B and C) Sizes of cross-sectional areas of quadriceps muscle fibers in WT and dbTg1 mice at 15 weeks of age were determined by using the ImageJ program. Sixty quadriceps fibers from each mouse were examined. The distribution of the cross-sectional areas (B) and the mean sizes of cross-sectional areas (C) are shown. Data shown in panels B and C are expressed as means ± standard deviations (n = 3). *, P < 0.05 compared to the WT, determined by two-tailed Student's t test. (D) Transmission electron microscopic analysis of quadriceps from dbTg1 mice at 15 weeks of age. Bars, 10 μm. (E) Number of centronuclear quadriceps fibers from WT and dbTg1 mice at 15 weeks of age. One hundred quadriceps fibers from each mouse were examined. Data are expressed as means ± standard deviations (n = 3). **, P < 0.01 compared to the WT, determined by two-tailed Student's t test. (F) Quantitative RT-PCR analysis of the expression levels of myogenic markers (MyoD, myogenin, and p21), NF90, and NF45 in quadriceps of WT and dbTg1 mice. Hypoxanthine phosphoribosyltransferase was used as an internal control. Data are expressed as means ± standard deviations (n = 4 to 5). **, P < 0.01 compared to the WT, determined by two-tailed Student's t test; n.s., not significant.
FIG 3.
NF90 Tg mice do not display an accumulation of centronuclear myofibers. (A) H&E staining of the cross-sectional area of quadriceps from WT and NF90 Tg mice at 15 weeks of age. Bars, 50 μm. (B) Numbers of centronuclear quadriceps fibers in WT and NF90Tg mice at 15 weeks of age. One hundred quadriceps fibers from each mouse were examined. Data are expressed as means ± standard deviations (n = 3). Statistical significance was calculated by using two-tailed Student's t test. n.s., not significant. (C) Myogenin and p21 expression levels in the skeletal muscle of WT and NF90 Tg mice at 14 to 21 weeks of age were measured by qRT-PCR. Hypoxanthine phosphoribosyltransferase was used as an internal control. Data are expressed as means ± standard deviations (n = 4 to 6). Statistical significance was calculated by using two-tailed Student's t test. **, P < 0.01 compared to the WT.
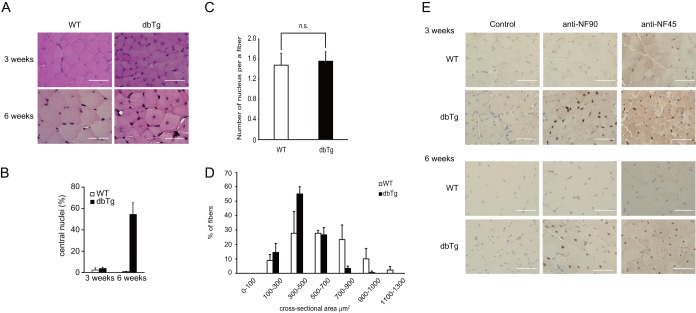
Next, we performed H&E staining of the skeletal muscles in WT and NF90-NF45 dbTg mice at 3 and 6 weeks of age. At 3 weeks of age, the skeletal muscle of NF90-NF45 dbTg mice did not give rise to centronuclear muscle fibers (Fig. 4A, top, and B). On the other hand, more nuclei seem to be present in the section from the skeletal muscle of NF90-NF45 dbTg mice than in that of WT mice (Fig. 4A, top). Therefore, we counted the number of nuclei in 100 skeletal muscle fibers of WT and NF90-NF45 dbTg mice at 3 weeks of age. In consequence, there was no difference in the numbers of nuclei per muscle fiber between WT and NF90-NF45 dbTg mice (Fig. 4C). Furthermore, the muscle fiber sizes of NF90-NF45 dbTg mice at 3 weeks of age were smaller than those of WT mice (Fig. 4D). Therefore, these results suggest that the larger number of nuclei present in the section from NF90-NF45 dbTg mice shown in Fig. 4A is caused by skeletal muscle fiber atrophy of NF90-NF45 dbTg mice. In contrast to the skeletal muscle of NF90-NF45 dbTg mice at 3 weeks of age, centronuclear muscle fibers were observed in the skeletal muscle of dbTg mice at 6 weeks of age (Fig. 4A, bottom, and B). We also measured the expressions of NF90 and NF45 in the skeletal muscle of WT and NF90-NF45 dbTg mice at 3 and 6 weeks of age by immunohistochemistry. Intriguingly, overexpression of NF90 and NF45 was observed in NF90-NF45 dbTg mice after 3 weeks of age (Fig. 4E). Collectively, these results indicated that the occurrence of centronuclear muscle fibers followed the overexpression of NF90 and NF45.
FIG 4.
Skeletal muscles of NF90-NF45 dbTg mice exhibit centronuclear muscle fibers at 3 to 6 weeks of age. Histological analysis of the quadriceps of WT and dbTg1 mice at 3 and 6 weeks of age was performed. (A) H&E staining. Bars, 50 μm. (B) Numbers of centronuclear quadriceps fibers in WT and dbTg1 mice at 3 and 6 weeks of age. One hundred quadriceps fibers from each mouse were examined. Data are expressed as means ± standard deviations (n = 3). **, P < 0.01 compared to the WT, determined by two-tailed Student's t test. (C) Numbers of nuclei in 100 skeletal muscle fibers of WT and NF90-NF45 dbTg mice at 3 weeks of age were counted. Data are expressed as means ± standard deviations (n = 3). (D) Sizes of cross-sectional areas of quadriceps muscle fibers in WT and dbTg1 mice at 3 weeks of age were determined by using the ImageJ program. Sixty quadriceps fibers from each mouse were examined. The distribution of the cross-sectional area is shown in panel A. Data are expressed as means ± standard deviations (n = 3). (E) Immunohistochemical analysis of NF90 and NF45. (Top) mice at 3 weeks of age; (bottom) mice at 6 weeks of age. Bars, 50 μm.
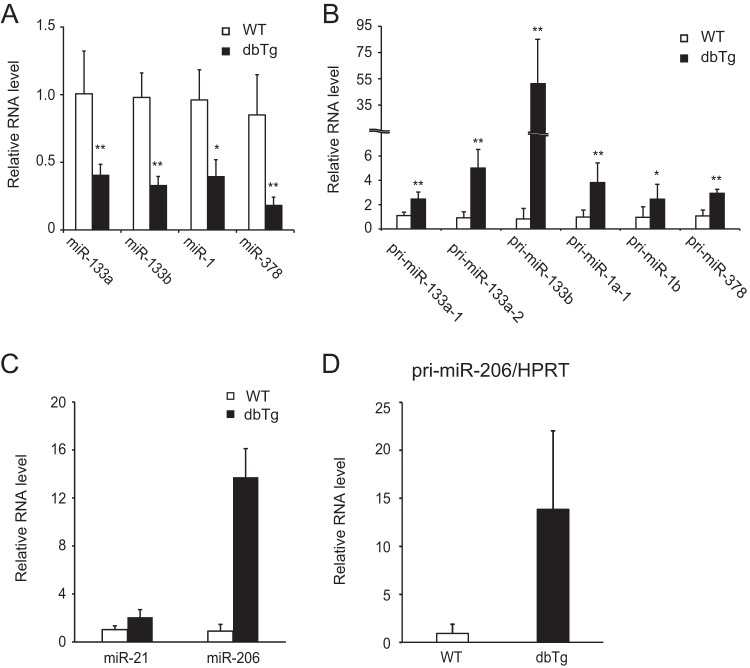
Myogenic miRNAs are attenuated in the skeletal muscle of NF90-NF45 dbTg mice.
In our previous study, we found that the NF90-NF45 complex functions as a negative regulator of miRNA biogenesis through inhibition of pri-miRNA processing (27). To examine alterations of miRNA expression in the skeletal muscle of NF90-NF45 dbTg mice, we performed comprehensive analyses of miRNA expression in the quadriceps of WT and NF90-NF45 dbTg mice. Compared with WT mice, microarray analysis demonstrated that the expression levels of 23 miRNAs were decreased by <0.5-fold in the quadriceps of NF90-NF45 dbTg mice (Table 1). Among the 23 miRNAs, we focused on miR-133a, miR-133b, miR-1, miR-378, and miR-206 because these miRNAs are reported to promote muscular maturation (46–48) and are referred to as myogenic miRNAs. qRT-PCR analysis showed that the levels of all of the examined myogenic miRNAs besides miR-206 were significantly decreased in the skeletal muscle of NF90-NF45 dbTg mice compared to those of WT mice (Fig. 5A). Unexpectedly, the expression level of miR-206 was dramatically elevated in the skeletal muscle of NF90-NF45 dbTg mice (Fig. 5C). To address the cause of the alteration in the levels of these miRNAs, we measured the levels of the corresponding pri-miRNAs in the skeletal muscles of WT and NF90-NF45 dbTg mice. The levels of all the pri-miRNAs examined were significantly increased in the skeletal muscle of NF90-NF45 dbTg mice compared to those of WT mice (Fig. 5B and D). Previously, we found that the binding of NF90-NF45 to pri-miRNAs impairs access of the microprocessor complex to the pri-miRNAs, resulting in a reduction of pre-miRNA and mature miRNA production (27). Therefore, these results together with those findings (27) raise the possibility that the reductions of miR-133a, miR-133b, miR-1, and miR-378 levels in the skeletal muscle of NF90-NF45 dbTg mice are due to a suppression of pri-miRNA processing by the overexpression of NF90 and NF45 through the association of these proteins with the pri-miRNAs. On the other hand, it was thought that the processing of pri-miR-206 would be unaffected by NF90-NF45-induced repression of pri-miRNA processing, probably because NF90-NF45 exhibits lower binding activity for pri-miR-206 than for other myogenic pri-miRNAs.
TABLE 1.
Downregulated miRNAs (<0.5-fold change) in skeletal muscles of a dbTg1 mouse (n = 1) compared with those in a WT mouse (n = 1)
| miRNAb | Fold change | Tissue distribution | Target(s) in skeletal musclea | Reference(s) |
|---|---|---|---|---|
| mmu-miR-29c* | 0.21 | Unclear | Unclear | |
| mmu-miR-193 | 0.28 | Brown adipose tissue | Cdon, Igfbp5 | 58 |
| mmu-miR-193b | 0.37 | |||
| mmu-miR-378 | 0.32 | Muscle | MyoR, IGF1R | 47, 59 |
| mmu-miR-29c | 0.32 | Ubiquitous | YY1 | 60, 61 |
| mmu-miR-3107 | 0.32 | Unclear | Unclear | |
| mmu-miR-185 | 0.33 | Unclear | Unclear | |
| mmu-miR-365 | 0.33 | Brown adipose tissue | Unclear | 58 |
| mmu-miR-136 | 0.40 | Unclear | Unclear | |
| mmu-miR-30a* | 0.42 | Unclear | Unclear | |
| mmu-miR-133a* | 0.42 | Unclear | Unclear | |
| mmu-miR-133a | 0.43 | Muscle | SRF, nPTB, UCP2 | 46, 62, 63 |
| mmu-miR-133b | 0.47 | |||
| mmu-miR-196a | 0.44 | Unclear | Unclear | |
| mmu-miR-30a | 0.44 | Ubiquitous | Snai1 | 64–66 |
| mmu-miR-30c | 0.45 | |||
| mmu-miR-345-5p | 0.45 | Unclear | Unclear | |
| mmu-miR-30e* | 0.46 | Unclear | Unclear | |
| mmu-miR-151-5p | 0.47 | Unclear | PLM | 67 |
| mmu-miR-101b | 0.48 | Unclear | Unclear | |
| mmu-miR-378* | 0.48 | Unclear | Unclear | |
| mmu-miR-1 | 0.49 | Muscle | HDAC4, Cx43, Pax7, c-Met, G6PD | 46, 52, 68–70 |
| mmu-miR-361 | 0.49 | Unclear | Unclear |
IGF1R, insulin-like growth factor 1 receptor; SRF, serum response factor; HDAC4, histone deacetylase 4; G6PD, glucose-6-phosphate dehydrogenase.
Asterisks indicate the antisense form of miRNA.
FIG 5.
Myogenic miRNA levels are altered in the skeletal muscles of NF90-NF45 dbTg mice. (A and B) Measurements of the levels of myogenic mature miRNAs and pri-miRNAs in quadriceps of WT and dbTg1 mice. RNAs isolated from the quadriceps of WT and dbTg1 mice were analyzed by qRT-PCR with specific primers. (A) Expression levels of mature miR-1, miR-133a, miR-133b, and miR-378 were measured by TaqMan microRNA assays. snoRNA202 was used as an internal control. Data are expressed as means ± standard deviations. (B) Expression levels of pri-miR-133a-1, pri-miR-133a-2, pri-miR-133b, pri-miR-1a-1, pri-miR-1b, and pri-miR-378 were measured by qRT-PCR. Hypoxanthine phosphoribosyltransferase was used as an internal control. Data shown in panels A and B are expressed as means ± standard deviations (n = 4 to 9). *, P < 0.05; **, P < 0.01 (compared to the WT, determined by two-tailed Student's t test). (C) Expression levels of mature miR-21 and miR-206 were measured by TaqMan microRNA assays. snoRNA202 was used as an internal control. Data are expressed as means ± standard deviations (n = 5 to 11). (D) The expression level of pri-miR-206 was measured by qRT-PCR. Hypoxanthine phosphoribosyltransferase (HPRT) was used as an internal control. Data are expressed as means ± standard deviations (n = 5).
The NF90-NF45 complex suppresses the processing of pri-miR-133a.
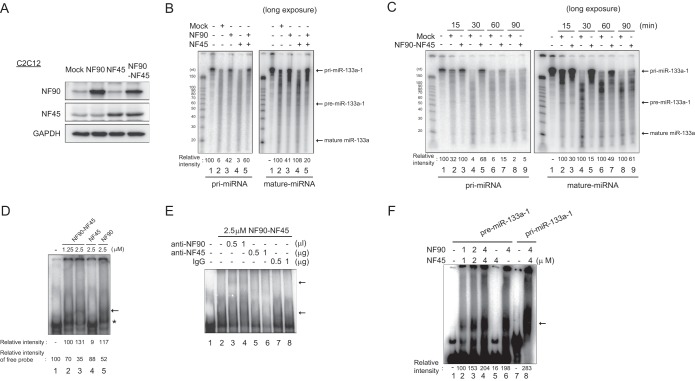
It has been reported that depletion of miR-133, the expression of which was significantly reduced in NF90-NF45 dbTg mice (Fig. 5A), evokes centronuclear myopathy in vivo (49). As shown in Fig. 2A to E, overexpression of NF90-NF45 induced skeletal muscle atrophy accompanied by centronuclear muscle fibers. These findings suggest that the miR-133 pathway regulated by NF90-NF45 causes muscular atrophy with centralized nuclei in the skeletal muscle of NF90-NF45 dbTg mice. Accordingly, we focused on the regulation of miR-133 biogenesis by the NF90-NF45 complex. To clarify whether the NF90-NF45 complex participates in the processing of pri-miR-133, we performed an in vitro pri-miRNA processing assay. We prepared whole-cell lysates of murine C2C12 cells transfected with expression plasmids for NF90 and/or NF45 (Fig. 6A). A radiolabeled pri-miR-133a-1 probe was incubated with the cell lysates, and the processed RNA was then purified by phenol-chloroform extraction and ethanol precipitation. The processed RNA probes were loaded onto a denaturing gel and visualized by autoradiography. Compared with the mock-transfected control, the generation of mature miR-133a was partially inhibited by the overexpression of NF90 alone (Fig. 6B, lanes 2 and 3). On the other hand, cleavage of pri-miR-133a-1 was not affected by elevated levels of NF45 alone (Fig. 6B, lanes 2 and 4). Notably, the levels of mature miR-133a were remarkably decreased by the overexpression of both NF90 and NF45 (Fig. 6B, right, lanes 2 and 5). Furthermore, we performed a time course experiment on the processing assay to validate the results shown in Fig. 6B. The almost all-radiolabeled pri-miRNA probe was processed to pre-miRNA and mature miRNA after a >30-min reaction time in the processing assay using the lysates from the mock-transfected control (Fig. 6C, lane 4), whereas the overexpression of NF90-NF45 caused the residue of the pri-miRNA probe at the 30-min reaction time in the processing assay (Fig. 6C, lane 5), confirming that the overexpression of NF90-NF45 depresses the processing of pri-miRNA to mature miRNA in skeletal muscle tissue. Subsequently, to examine whether the NF90-NF45 complex binds to pri-miR-133a-1, we prepared recombinant murine NF90 and NF45 fusion proteins in Escherichia coli and applied these proteins to an electrophoretic mobility shift assay (EMSA) with the pri-miR-133a-1 probe. The EMSA probed with pri-miR-133a-1 clearly showed that NF90-NF45 was bound to pri-miR-133a-1 in vitro (Fig. 6D, arrow, compare lane 1 to lanes 2 and 3). On the other hand, the binding activity of NF90 alone was weaker than that of NF90-NF45 (Fig. 6D, arrow and asterisk, compare lanes 2 and 3 to lane 5). NF45 alone did not exhibit binding to pri-miR-133a-1 (Fig. 6D, lane 4). To confirm whether the major band indicated by the arrow in Fig. 6D was due to the NF90-NF45 complex binding to pri-miRNA, we carried out a supershift assay using polyclonal anti-NF90 and -NF45 antibodies. The major protein-RNA complex indicated by the bottom arrow was supershifted by the anti-NF90 antibody (Fig. 6E, top arrow, compare lane 2 to lanes 3 and 4), while the addition of the anti-NF45 antibody resulted in the absence of the major band (Fig. 6E, compare lane 2 to lanes 5 and 6), indicating that the major band is the complex of NF90-NF45 and pri-miRNA. We also performed an EMSA with pri-miR-133a-1 or pre-miR-133a-1 to examine whether the NF90-NF45 complex discriminates between pri- and pre-miR-133a-1. As shown in Fig. 6F, NF90-NF45 or NF90 bound to pre-miR-133a-1 (lanes 2 to 4 and 6), but NF45 alone did not exhibit binding to the pre-miRNA (lane 5). Unlike pri-miRNA, there was no difference in binding activity with pre-miRNA between NF90-NF45 and NF90 (Fig. 6F, compare lanes 4 and 6). On the other hand, the binding activity of NF90-NF45 with pri-miRNA was obviously greater than that of this complex with pre-miRNA (Fig. 6F, compare lanes 4 and 8). These results suggest that although NF90-NF45 or NF90 binds to both the overhanging region and the stem-loop region of pri-miR-133a-1, NF90-NF45 prefers to bind to the overhanging region of pri-miR-133a-1, which is not a common secondary structure of pri- and pre-miRNAs.
FIG 6.
The NF90-NF45 complex binds to pri-miR-133a-1 and suppresses miR-133a processing. (A) Western blot analysis of NF90 and NF45 in whole-cell extracts of C2C12 cells transfected with expression plasmids. Anti-GAPDH was used as a loading control. (B and C) In vitro pri-miR-133a-1 processing assay. A radiolabeled pri-miR-133a-1 probe was incubated with whole-cell extracts of C2C12 cells transfected with expression plasmids. Processed RNA was separated on a 12% polyacrylamide-urea gel and visualized by autoradiography. The intensities of bands corresponding to pri-miR-133a-1 and miR-133a were measured with a densitometer and are presented as relative band intensities. (D) EMSA performed with the pri-miR-133a-1 probe and both recombinant murine NF90 and NF45 proteins (lanes 2 and 3), NF45 alone (lane 4), or NF90 alone (lane 5) at the indicated protein concentrations. The intensities of bands corresponding to the specific RNA-protein complex and free probe, indicated by an arrow and an asterisk, respectively, were measured with a densitometer and are presented as relative band intensities. (E) An EMSA was performed with pri-miR-133a-1 and 2.5 μM NF90-NF45. The indicated amounts of anti-NF90 antibody (lanes 3 and 4), anti-NF45 antibody (lanes 5 and 6), or control normal rabbit IgG (lanes 7 and 8) were incubated with the recombinant proteins in binding buffer prior to probe addition. The positions of the RNA-protein complex and the supershifted complex are indicated by arrows. (F) EMSA performed with pre-miR-133a-1 and pri-miR-133a-1 probes and both recombinant NF90 and NF45 proteins (lanes 2 to 4 and 8), NF45 alone (lane 5), or NF90 alone (lane 6) at the indicated protein concentrations. The intensities of bands corresponding to the specific RNA-protein complex indicated by an arrow were measured with a densitometer and are presented as relative band intensities.
Taken together, these findings shown in Fig. 6 suggest that the NF90-NF45 complex suppresses pri-miR-133-1 processing through the preferential binding of NF90-NF45 to the overhanging region of pri-miR-133a-1.
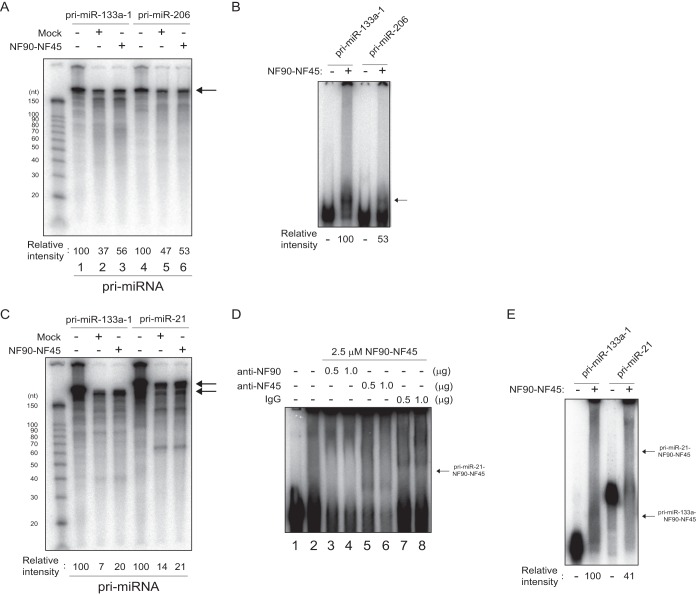
Regarding miR-206, the level of the mature miRNA was strikingly increased by the overexpression of NF90-NF45 (Fig. 5C). To examine the cause of this phenomenon, we performed a processing assay and EMSA. The processing assay showed that there is no large difference in the extent of pri-miR-206 processing between mock-transfected control cells and NF90-NF45-overexpressing cells relative to the extent of pri-miR-133 processing inhibition (Fig. 7A, compare lane 5 to lane 6 and lane 2 to lane 3). Furthermore, the binding activity of NF90-NF45 was ∼0.5-fold lower for pri-miR-206 than for pri-miR-133a-1 (Fig. 7B). As mentioned above several times, the high binding activity of NF90-NF45 with pri-miRNA is necessary for the repression of pri-miRNA processing by these proteins (27). Therefore, the rise in the level of miR-206 in the skeletal muscle of NF90-NF45 dbTg mice would be due to the facilitation of pri-miR-206 processing caused by the low binding activity of NF90-NF45 with pri-miR-206.
FIG 7.
Facilitation of pri-miR-206 and pri-miR-21 processing is caused by the low level of association of NF90-NF45 with pri-miRNAs. (A) Radiolabeled pri-miR-133a-1 and pri-miR-206 probes were incubated with whole-cell extracts of C2C12 cells transfected with expression plasmids. Processed RNA was separated on a 12% polyacrylamide-urea gel and visualized by autoradiography. The intensities of bands corresponding to pri-miR-133a-1 and pri-miR-206 were measured with a densitometer and are presented as relative band intensities. (B) EMSA performed with pri-miR-133a-1 or pri-miR-206 and recombinant murine NF90 and NF45. The intensities of bands corresponding to the specific RNA-protein complex indicated by an arrow were measured with a densitometer and are presented as relative band intensities. (C) Radiolabeled pri-miR-133a-1 and pri-miR-21 probes were incubated with whole-cell extracts of C2C12 cells transfected with expression plasmids. Processed RNA was separated on a 12% polyacrylamide-urea gel and visualized by autoradiography. The intensities of bands corresponding to pri-miR-133a-1 and pri-miR-21 were measured with a densitometer and are presented as relative band intensities. (D) An EMSA was performed with pri-miR-133a-1 and 2.5 μM NF90-NF45. The indicated amounts of anti-NF90 antibody (lanes 3 and 4), anti-NF45 antibody (lanes 5 and 6), or control normal rabbit IgG (lanes 7 and 8) were incubated with the recombinant proteins in binding buffer prior to probe addition. The position of the RNA-protein complex is indicated by an arrow. (E) EMSA performed with pri-miR-133a-1 or pri-miR-21 and recombinant murine NF90 and NF45. The intensities of bands corresponding to the specific RNA-protein complex indicated by arrows were measured with a densitometer and are presented as relative band intensities.
Furthermore, we utilized pri-miR-21 as a negative control for the pri-miRNA processing assay and EMSA, because there was no marked change in the level of miR-21 between the skeletal muscle of WT and that of NF90 Tg mice by our miRNA microarray analysis (GEO database accession no. GSE61001) and qRT-PCR analysis (Fig. 5C). As shown in Fig. 7C, the reduction in the processing of pri-miR-21 by the overexpression of NF90-NF45 was not prominently occurring compared with the extent of pri-miR-133a processing inhibition by these proteins. On the other hand, the binding activity of NF90-NF45 with pri-miR-21 was ∼0.6-fold lower than that of NF90-NF45 with pri-miR-133a (Fig. 7E). The complex of NF90-NF45 and pri-miR-21 was identified by a supershift assay using anti-NF90 and -NF45 antibodies (Fig. 7D, arrow). Therefore, like the processing of pri-miR-206 in NF90-NF45-overexpressing cells, these results suggest that the low affinity of NF90-NF45 for pri-miR-21 would cause a reduction in pri-miR-21 processing inhibition, leading to the prevention of the NF90-NF45-induced negative regulation of miR-21 production.
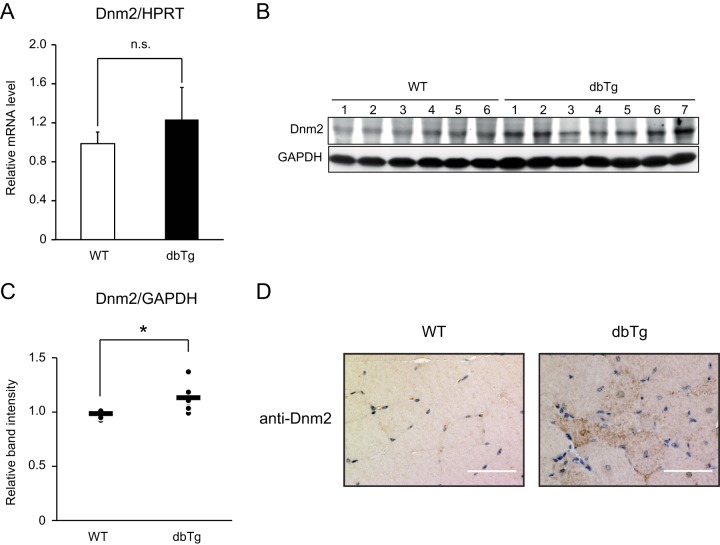
The level of Dnm2 is elevated in the skeletal muscle of NF90-NF45 dbTg mice.
It has been reported that a reduction of miR-133a causes centronuclear muscle fibers following an elevation of the expression level of Dnm2, which is a known causative gene of centronuclear myopathy (49) and a target of miR-133a. Therefore, we measured the mRNA and protein expression levels of Dnm2 in the skeletal muscles of WT and NF90-NF45 dbTg mice. As shown in Fig. 8A to C, the mRNA levels of Dnm2 were similar in WT and NF90-NF45 dbTg mice, while the protein level of Dnm2 was prominently elevated in the skeletal muscle of NF90-NF45 dbTg mice. To confirm the rise in Dnm2 protein levels in NF90-NF45 dbTg mice, we performed immunohistochemistry using a Dnm2 antibody. This analysis revealed that NF90-NF45 dbTg mice exhibit an accumulation of Dnm2 inside myofibers, while intracellular accumulation was not observed in the skeletal muscle of WT mice (Fig. 8D). These results confirm the elevation of Dnm2 protein levels in the skeletal muscle of NF90-NF45 dbTg mice. These findings, together with the data shown in Fig. 2A, D, and E and 4, suggest that the centronuclear muscle fibers in the skeletal muscle of NF90-NF45 dbTg mice are caused by an increase in the Dnm2 level through a decrease in miR-133a-mediated translational repression.
FIG 8.
Dnm2 protein levels are elevated in the skeletal muscles of NF90-NF45 dbTg mice. (A) Dnm2 mRNA expression in the skeletal muscles of WT (n = 7) and dbTg1 (n = 7) mice was analyzed by qRT-PCR. Hypoxanthine phosphoribosyltransferase (HPRT) was used as an internal control. Data are expressed as means ± standard deviations. Statistical significance was calculated by using two-tailed Student's t test. n.s., not significant. (B) Western blot analysis of Dnm2 levels in tissue extracts of skeletal muscles from WT and dbTg1 mice. GAPDH was used as a loading control. (C) Intensities of specific bands in the Western blot analysis shown in panel B were measured with a densitometer and are presented as a graph. Each dot and bar indicate one experiment and an average, respectively. *, P < 0.05 compared to the WT, determined by two-tailed Student's t test. (D) Immunohistochemical analysis of Dnm2 in quadriceps from WT and dbTg1 mice at 15 weeks of age. Bars, 50 μm.
DISCUSSION
Human centronuclear myopathies (CNMs) are characterized by muscle weakness and centronuclear muscle fibers in skeletal muscle. CNMs are categorized as three main forms: the X-linked recessive form (XLCNM), with a neonatal phenotype caused by MTM1 gene mutations; the classical autosomal dominant forms, with mild, moderate, or severe phenotypes caused by Dnm2 gene mutations; and an autosomal recessive form, with severe and moderate phenotypes caused by BIN1 gene mutations. It has been reported that the Dnm2 level is increased in the skeletal muscle of XLCNM patients and in a mouse model of XLCNM (50), indicating that an elevation of the Dnm2 level induces muscular atrophy and centronuclear muscle fibers. Indeed, the overexpression of Dnm2 in the skeletal muscle of Tg mice causes an accumulation of centronuclear myofibers in skeletal muscle (49). In this study, we found that NF90-NF45 dbTg mice exhibited centronuclear muscle fibers in their skeletal muscles (Fig. 2A, D, and E and 4) and that the protein level of Dnm2 was remarkably increased in the skeletal muscle of NF90-NF45 dbTg mice (Fig. 8). The skeletal muscle abnormalities in NF90-NF45 dbTg mice were very similar to those in CNMs, suggesting that the expression of the NF90-NF45 complex may influence the occurrence of this disease.
NF90 KO mice exhibited perinatal lethality associated with muscle weakness and respiratory failure (34). In addition, it has been proposed that the muscular abnormality in NF90 KO mice is due to defects in skeletal myocyte differentiation arising from insufficient expression of myogenic regulatory factors such as MyoD, myogenin, and p21 (34). Furthermore, NF90 has been reported to enhance the posttranscriptional stabilization of MyoD and p21 mRNAs through the association of NF90 with 3′ UTRs of the RNAs (34). In this report, we found that there was no significant difference in the expression of MyoD mRNA between WT and NF90-NF45 dbTg mice (Fig. 2F). On the other hand, NF90 KO mice exhibited a reduction in the expression of MyoD in the skeletal muscle (34). Moreover, it has been reported that NF90 stabilizes posttranscriptional mRNA of MyoD through the binding of NF90 to the MyoD 3′-UTR RNA (34). MyoD promotes differentiation from myoblasts to myocytes in the early stages of myogenesis (43). Following the transition to late differentiation from myocytes to myotubes, the expression of this factor is downregulated (43). We utilized the RNA extracted from the skeletal muscle of WT and NF90-NF45 dbTg mice at 15 weeks of age to measure the level of MyoD shown in Fig. 2F. The status of the centronuclear myopathy observed in the skeletal muscle of NF90-NF45 dbTg mice at 15 weeks of age seems to be similar to that of myotubes in myogenesis. As mentioned above, the expression of MyoD is largely reduced in late differentiation, including in myotubes and myofibers (43). In general, the skeletal muscle of NF90-NF45 dbTg mice and WT mice at 15 weeks of age differentiates into myotubes and myofibers. Therefore, the negative regulatory machinery for the expression of MyoD would be more activated in the skeletal muscle of WT and NF90-NF45 dbTg mice at 15 weeks of age than in mouse embryos and newborn mice. Thus, the level of MyoD may not be elevated in the skeletal muscle of NF90-NF45 dbTg mice at 15 weeks of age (Fig. 2F). On the other hand, the expression levels of myogenin and p21 were prominently increased in the skeletal muscle of NF90-NF45 dbTg mice compared to those of WT mice (Fig. 2F), while the overexpression of NF90 alone significantly enhanced the expression of p21 (Fig. 3C). Therefore, these results, together with previously reported findings (34), indicate that NF90-NF45 or NF90 elevates the level of p21 through the posttranscriptional stabilization of p21 mRNA in the skeletal muscle of NF90-NF45 dbTg and NF90 Tg mice. Unlike p21, the rise in the level of myogenin was observed in the skeletal muscle of NF90-NF45 dbTg mice alone (Fig. 2F and 3C). Accordingly, the expression of myogenin would be positively regulated by NF90-NF45 through transcriptional control, mRNA stabilization, or miRNA-mediated translational repression. Taken together, the high levels of p21 and myogenin induced by the overexpression of NF90-NF45 may cause the inhibition of muscle maturation, thereby leading to muscular atrophy. However, muscle atrophy with centralized nuclei does not occur by the overexpression of p21 or myogenin in mice (35, 45). Thus, it is thought that other factors besides p21 and myogenin cause the occurrence of skeletal muscle atrophy accompanying centronuclear muscle fibers in NF90-NF45 dbTg mice.
Previous studies have shown that alterations in miRNA expression are involved in muscular maturation. Dicer is a positive regulator of the miRNA processing pathway, and Dicer knockout mice exhibit skeletal muscle atrophy and apoptosis through reductions in the levels of miR-1, miR-133, and miR-206 (51). miR-1 targets histone deacetylase 4 (HDAC4), a transcriptional repressor of muscle gene expression (46). Moreover, miR-1 and miR-206 translationally repress Pax7, a member of the paired-box family of transcription factors that play a crucial role in regulating the developmental program of embryonic myoblasts (52). miR-133 targets serum response factor (SRF), which enhances myoblast proliferation and inhibits myogenic differentiation and Dnm2, a known causative agent in CNMs (46, 49). Besides miR-1, miR-133, and miR-206, miR-378 suppresses MyoR, a repressor of MyoD transcriptional activity (47). In addition, we performed whole-genome expression microarray analysis of the skeletal muscles of WT and NF90-NF45 dbTg mice to explore other candidate factors, which are regulated by the NF90-NF45-myogenic miRNA pathway, for the development of muscle atrophy accompanying centronuclear muscle fibers. In consequence, we found that the expression levels of 725 genes were increased by >2-fold in the skeletal muscles of NF90-NF45 dbTg mice compared to those of WT mice (GEO database accession no. GSE67591). Among the 725 genes, we attempted to explore predicted targets of miR-133, miR-1, and miR-378, the expression levels of which were significantly decreased in the skeletal muscle of NF90-NF45 dbTg mice (Fig. 5A), using TargetScan, a computational tool for miRNA target prediction. Finally, we found several predicted targets of miR-133, miR-1, and miR-378. The predicted targets of each miRNA were as follows: the predicted miR-133 targets were Aif1l, Atp6ap2, Col8a1, Srgap3, and Vat1; the predicted miR-1 targets were Anxa2, Anxa4, Atp6ap2, Atp6v1a, Azin1, Coro1b, G6pdx, H3f3b, Serp1, Sh3bgrl3, and Zfp36l2; and the predicted miR-378 targets were H3f3b and Vat1. Subsequently, we performed gene ontology (GO) analysis of the above-mentioned targets. This analysis indicated the possibility that Aif1l, Coro1b, G6pdx, Serp1, and Sh3bgrl3 are involved in the regulation of actin and microtubule cytoskeletons (see Table S1 in the supplemental material). These observations suggest that the various targets of myogenic miRNAs, the processing of which is regulated by NF90-NF45, participate in the development of the muscle abnormalities of NF90-NF45 dbTg mice.
It was recently reported that the suppression of the miRNA processing pathway induces various human diseases. Lin28A/B enhances the growth of breast and colon tumors via inhibition of let-7 miRNA biogenesis (25). Fragile X-associated tremor/ataxia syndrome, a neurodegenerative disorder, is caused by the expansion of 55 to 200 CGG repeats in the 5′ untranslated region of FMR1 that inhibits Drosha-mediated pri-miRNA processing through binding of the Drosha-DGCR8 complex to CGG repeats (53). Cardiac dysfunction in type I myotonic dystrophy is implicated in the reduction of miR-1 processing caused by the fall in the activity of MBNL1 as a positive regulator of processing from pre-miR-1 to miR-1 (54). These findings indicate that negative regulators in the miRNA biogenesis pathway are deeply involved in human disease development. The NF90-NF45 complex also acts as a negative regulator in miRNA biogenesis (27). In this study, we found that NF90-NF45 induces centronuclear muscle fibers through suppression of pri-miR-133a processing by binding of NF90-NF45 to pri-miR-133a.
It has also been reported that the level of NF90 is elevated in non-small-cell lung, epithelial ovarian, and breast cancers (55–57). We also observed a significant increase in the NF90-NF45 expression level in hepatocellular carcinoma compared to that in adjacent normal tissues (T. Higuchi and S. Sakamoto, unpublished data). Further analysis indicated that NF90-NF45 plays a role in the promotion of tumorigenesis through inhibition of tumor suppressor miRNA biogenesis (Higuchi and Sakamoto, unpublished). These findings raise the possibility that NF90-NF45-mediated inhibition of miRNA biogenesis is involved in the development of various diseases, including myopathy and cancer. In the future, we will explore the mechanisms of various human diseases via the inhibition of miRNA processing by the NF90-NF45 complex, which may provide new insights into the development of various diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Japan Society for the Promotion of Science (JSPS) grant-in-aid for JSPS fellows (grant no. 12J10665) and a JSPS grant-in-aid for scientific research (C) (grant no. 25460371). This work was also supported by a grant from the Nakatomi Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01297-14.
REFERENCES
- 1.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen CZ, Li L, Lodish HF, Bartel DP. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 5.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. 2005. A microRNA polycistron as a potential human oncogene. Nature 435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. 2004. Processing of primary microRNAs by the microprocessor complex. Nature 432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 13.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 14.Landthaler M, Yalcin A, Tuschl T. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohnsack MT, Czaplinski K, Gorlich D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. 2004. Nuclear export of microRNA precursors. Science 303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 18.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 20.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 21.Hammond SM, Bernstein E, Beach D, Hannon GJ. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 22.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563–574. doi: 10.1016/S0092-8674(02)00908-X. [DOI] [PubMed] [Google Scholar]
- 23.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Olsen PH, Ambros V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol 216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 25.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. 2011. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhury NR, de Lima Alves F, de Andres-Aguayo L, Graf T, Caceres JF, Rappsilber J, Michlewski G. 2013. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev 27:24–38. doi: 10.1101/gad.199190.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y. 2009. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol 29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim J, Lim H, Yates JR III, Karin M. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell 10:1331–1344. doi: 10.1016/S1097-2765(02)00730-X. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, Fontoura BM, Martin WD, Barber GN. 2008. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci U S A 105:4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tominaga-Yamanaka K, Abdelmohsen K, Martindale JL, Yang X, Taub DD, Gorospe M. 2012. NF90 coordinately represses the senescence-associated secretory phenotype. Aging (Albany NY) 4:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin HJ, Kim SS, Cho YH, Lee SG, Rho HM. 2002. Host cell proteins binding to the encapsidation signal epsilon in hepatitis B virus RNA. Arch Virol 147:471–491. doi: 10.1007/s007050200001. [DOI] [PubMed] [Google Scholar]
- 32.Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D. 2010. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J Biol Chem 285:8256–8267. doi: 10.1074/jbc.M109.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichman TW, Muniz LC, Mathews MB. 2002. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol Cell Biol 22:343–356. doi: 10.1128/MCB.22.1.343-356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. 2005. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem 280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 35.Higuchi T, Sakamoto S, Kakinuma Y, Kai S, Yagyu K, Todaka H, Chi E, Okada S, Ujihara T, Morisawa K, Ono M, Sugiyama Y, Ishida W, Fukushima A, Tsuda M, Agata Y, Taniguchi T. 2012. High expression of nuclear factor 90 (NF90) leads to mitochondrial degradation in skeletal and cardiac muscles. PLoS One 7:e43340. doi: 10.1371/journal.pone.0043340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 37.Sakamoto S, Taniguchi T. 2001. Identification of a phorbol ester-responsive element in the interferon-gamma receptor 1 chain gene. J Biol Chem 276:37237–37241. doi: 10.1074/jbc.M105543200. [DOI] [PubMed] [Google Scholar]
- 38.Song D, Sakamoto S, Taniguchi T. 2002. Inhibition of poly(ADP-ribose) polymerase activity by Bcl-2 in association with the ribosomal protein S3a. Biochemistry 41:929–934. doi: 10.1021/bi015669c. [DOI] [PubMed] [Google Scholar]
- 39.Goebel HH. 2003. Congenital myopathies at their molecular dawning. Muscle Nerve 27:527–548. doi: 10.1002/mus.10322. [DOI] [PubMed] [Google Scholar]
- 40.Zammit PS, Partridge TA, Yablonka-Reuveni Z. 2006. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem 54:1177–1191. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. 1999. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev 13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 43.Bentzinger CF, Wang YX, Rudnicki MA. 2012. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol 4:a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams GR, Haddad F, Baldwin KM. 1999. Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. J Appl Physiol (1985) 87:1705–1712. [DOI] [PubMed] [Google Scholar]
- 45.Gundersen K, Rabben I, Klocke BJ, Merlie JP. 1995. Overexpression of myogenin in muscles of transgenic mice: interaction with Id-1, negative crossregulation of myogenic factors, and induction of extrasynaptic acetylcholine receptor expression. Mol Cell Biol 15:7127–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. 2011. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem 286:19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. 2012. MicroRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122:2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu N, Bezprozvannaya S, Shelton JM, Frisard MI, Hulver MW, McMillan RP, Wu Y, Voelker KA, Grange RW, Richardson JA, Bassel-Duby R, Olson EN. 2011. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J Clin Invest 121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowling BS, Chevremont T, Prokic I, Kretz C, Ferry A, Coirault C, Koutsopoulos O, Laugel V, Romero NB, Laporte J. 2014. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J Clin Invest 124:1350–1363. doi: 10.1172/JCI71206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. 2007. Essential role for Dicer during skeletal muscle development. Dev Biol 311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. 2010. MicroRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol 190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, Tassone F, Willemsen R, Disney MD, Hagerman PJ, Todd PK, Charlet-Berguerand N. 2013. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep 3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, Wahbi K, Day JW, Fujimura H, Takahashi MP, Auboeuf D, Dreumont N, Furling D, Charlet-Berguerand N. 2011. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol 18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 55.Guo NL, Wan YW, Tosun K, Lin H, Msiska Z, Flynn DC, Remick SC, Vallyathan V, Dowlati A, Shi X, Castranova V, Beer DG, Qian Y. 2008. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res 14:8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y, Fu P, Zhu H, Reed E, Remick SC, Petros W, Mueller MD, Yu JJ. 2012. Correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol Rep 27:286–292. doi: 10.3892/or.2011.1483. [DOI] [PubMed] [Google Scholar]
- 57.Hu Q, Lu YY, Noh H, Hong S, Dong Z, Ding HF, Su SB, Huang S. 2013. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene 32:3933–3943. doi: 10.1038/onc.2012.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. 2011. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, Gupta M. 2012. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem 287:12913–12926. doi: 10.1074/jbc.M111.331751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. 2008. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. 2008. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boutz PL, Chawla G, Stoilov P, Black DL. 2007. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev 21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, Guo X, Wang G, Yang R, Zhu L, Zhang Y, Wang J, Xiang Y, Weng C, Zen K, Zhang J, Zhang CY. 2009. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem 284:5362–5369. doi: 10.1074/jbc.M807523200. [DOI] [PubMed] [Google Scholar]
- 64.Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van Ijcken WF, Grosveld F, Rudnicki MA. 2012. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell 47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ketley A, Warren A, Holmes E, Gering M, Aboobaker AA, Brook JD. 2013. The miR-30 microRNA family targets smoothened to regulate hedgehog signalling in zebrafish early muscle development. PLoS One 8:e65170. doi: 10.1371/journal.pone.0065170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reddy AM, Zheng Y, Jagadeeswaran G, Macmil SL, Graham WB, Roe BA, Desilva U, Zhang W, Sunkar R. 2009. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genomics 10:65. doi: 10.1186/1471-2164-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Wang R, Du W, Wang S, Yang L, Pan Z, Li X, Xiong X, He H, Shi Y, Liu X, Yu S, Bi Z, Lu Y, Shan H. 2013. Downregulation of miR-151-5p contributes to increased susceptibility to arrhythmogenesis during myocardial infarction with estrogen deprivation. PLoS One 8:e72985. doi: 10.1371/journal.pone.0072985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson C, Catoe H, Werner R. 2006. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan D, Dong XD, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. 2009. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem 284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M, Nicoletti C, Santini T, Sthandier O, Barberi L, Auricchio A, Musarò A, Bozzoni I. 2010. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab 12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.