Abstract
The translation and degradation of mRNAs are two key steps in gene expression that are highly regulated and targeted by many factors, including microRNAs (miRNAs). While it is well established that translation and mRNA degradation are tightly coupled, it is still not entirely clear where in the cell mRNA degradation takes place. In this study, we investigated the possibility of mRNA degradation on the ribosome in Drosophila cells. Using polysome profiles and ribosome affinity purification, we could demonstrate the copurification of various deadenylation and decapping factors with ribosome complexes. Also, AGO1 and GW182, two key factors in the miRNA-mediated mRNA degradation pathway, were associated with ribosome complexes. Their copurification was dependent on intact mRNAs, suggesting the association of these factors with the mRNA rather than the ribosome itself. Furthermore, we isolated decapped mRNA degradation intermediates from ribosome complexes and performed high-throughput sequencing analysis. Interestingly, 93% of the decapped mRNA fragments (approximately 12,000) could be detected at the same relative abundance on ribosome complexes and in cell lysates. In summary, our findings strongly indicate the association of the majority of bulk mRNAs as well as mRNAs targeted by miRNAs with the ribosome during their degradation.
INTRODUCTION
The processes of mRNA degradation and translation play important roles in the regulation of gene expression. The general degradation of bulk cytoplasmic mRNAs is initiated by removal of the mRNA's poly(A) tail, followed by two alternative pathways (1–3). Predominantly, deadenylation is followed by decapping and exonucleolytic degradation by the 5′-to-3′ exonuclease XRN1. Alternatively, deadenylation can be followed by 3′-to-5′ degradation through the exosome (1–3). In eukaryotes the deadenylation step is often rate limiting and involves the consecutive action of two different cytoplasmic deadenylase complexes (4–6). Initially, the poly(A) tail is trimmed by the PAN2-PAN3 complex, followed by a rapid degradation by the CCR4-NOT complex (4, 5). Besides their general activity in the mRNA degradation pathway, both deadenylase complexes can get specifically recruited to mRNAs through RNA binding proteins. An important example is their interaction with the GW182 protein, a key factor in the microRNA (miRNA)-mediated mRNA degradation pathway (7–9). mRNA decapping is a critical step in the regulation of mRNA turnover, which makes mRNAs accessible for exonucleolytic degradation and interferes with translation initiation (10, 11). The cytoplasmic decapping complex is composed of the catalytic subunit DCP2 and its coactivator, DCP1. Additionally, several enhancers of decapping, such as Drosophila HPat (Pat1p in Saccharomyces cerevisiae and PatL1 in humans), Me31B (Dhh1 in yeast and DDX6 in mammals), EDC3, EDC4, or the Lsm1-7 proteins, are thought to modulate its decapping activity in vivo (10, 11).
In addition to the ARE-mediated mRNA decay (12–18) and the mRNA surveillance pathways, such as nonsense-mediated decay (NMD) (19), nonstop decay (NSD), or no-go decay (NGD) (20), the general mRNA degradation pathway has also been linked to translation for many years. First, the inhibition of translation with antibiotics such as cycloheximide can stabilize mRNAs (6, 21, 22). Second, in yeast XRN1 and mRNA degradation intermediates can be detected in polysome fractions (23, 24). Third, mutations of initiation factors leading to a decrease in translation initiation have been demonstrated to accelerate mRNA deadenylation and decapping rates (25). Fourth, the 7-methylguanosine (m7G) cap structure in eukaryotic mRNAs is generally not freely accessible, but in the cytoplasm it is bound by eukaryotic initiation factor 4E (eIF4E). eIF4E is a component of the eIF4F cytoplasmic translation initiation complex and can reduce the rate of decapping in vitro (26, 27). Moreover, decapping activators, such as Dhh1p and Pat1p, have been shown to inhibit translation (28, 29). Finally, many factors regulating specific mRNAs (e.g., miRNAs, CUP, Nanos, or PUF proteins) both repress translation and accelerate deadenylation (30–36).
However, mRNA degradation factors and mRNA degradation intermediates were found to localize in cytoplasmic P bodies, which are devoid of ribosomes. Therefore, it was postulated that mRNA decapping would require the dissociation of the mRNA from the ribosome followed by their accumulation in P bodies (11, 37–42). In contrast, more recently Hu et al. (43, 44) have shown decapped degradation intermediates of several mRNAs, including CYH2, ADH1, and RPL41a mRNAs, and the mRNA reporter PGK1 on active ribosomes. These findings indicate that in yeast mRNAs can get degraded while they are associated with ribosomes, thus supporting the idea of cotranslational mRNA degradation. In this study, we investigated the possibility of mRNA degradation on the ribosome in the higher eukaryote Drosophila. Using ribosome affinity purification, we could clearly demonstrate the copurification of mRNA degradation factors and factors of the miRNA pathway with ribosome complexes. Furthermore, we analyzed decapped intermediates of mRNA degradation from ribosome complexes by high-throughput sequencing. Interestingly, we detected a large fraction of the decapped transcriptome on the ribosome. Together these observations strongly argue for the ribosomes as a site of mRNA degradation in eukaryotic cells.
MATERIALS AND METHODS
Plasmids, antibodies, cell culture, RNA interference, and stable cell lines.
For the production of polyclonal rabbit antibodies (Pineda Company), the antigens were purified as recombinant His-tagged full-length versions of RpL10Ab (CG7283), RpS3 (CG6779), PABPC (CG5119), or enhanced green fluorescent protein (GFP). Polyclonal rabbit antibodies against HPat, AGO1, and GW182 were described elsewhere (45). Mouse monoclonal anti-NOT1 and polyclonal rabbit anti-NOT2 antibodies were kind gifts of E. Wahle (Halle, Germany), polyclonal rabbit anti-eIF4E and anti-Me31B antibodies and rat polyclonal anti-EDC4 antibodies were kind gifts of E. Izaurralde (Tübingen, Germany), and rabbit polyclonal anti-XRN1 antibody was a kind gift of S. Newbury (Sussex, United Kingdom). Monoclonal antitubulin antibody (clone DM1A) was purchased from Sigma.
Preparation of double-stranded RNA (dsRNA) against MBP, XRN1, and AGO1 and RNA interference in Drosophila S2 cells were performed as described in reference 45. The oligonucleotides used for the PCR step of dsRNA synthesis are listed in Table S1 in the supplemental material. For the knockdown of XRN1, dsRNA was added on day 0 and on day 4 and the cells were harvested on day 9. For AGO1 knockdown, dsRNA was added on day 0 and cells were harvested on day 4. Control cells were treated with dsRNA against maltose binding protein (MBP) instead of XRN1 or AGO1.
Drosophila S2 cells (Life Technologies) were cultured at 25°C in Schneider's Drosophila medium (Lonza) supplemented with 10% heat-inactivated fetal bovine serum (Sigma), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM glutamine. For the maintenance of stable cell lines, 150 μg/ml hygromycin B was added to the medium. Protein expression was induced by supplementing the medium with 0.5 mM copper sulfate for 3 days. For the selection of stable cell lines, plasmids were transfected into Drosophila S2 cells using the Effectene transfection reagent (Qiagen), followed by selection with hygromycin B. For the cell line coexpressing the BLRP-GFP-RpL10Ab fusion protein and the 3× Flag-tagged Escherichia coli BirA ligase, the plasmids were cotransfected in a ratio of 19:1, as previously described (46). For the expression of the N-terminal fusion protein of the ribosomal protein, a BirA recognition sequence, and GFP, we first cloned the gateway destination vector BLRP-GFP-Gateway as follows: the 23-amino-acid E. coli BirA biotin ligase recognition peptide (BLRP; MASSLRQILDSQKMEWRSNAGGS [47]) was cloned downstream of the metallothionein promoter into the pMK33/pMTHy backbone (48), followed in frame by a GFP-coding sequence and a gateway cassette (Life Technologies). RpL10Ab (CG7283) was PCR amplified from the cDNA of Drosophila S2 cells and recombined into pDONR221 (Life Technologies). This entry clone was further recombined into the destination vector described above (BLRP-GFP-Gateway) to create BLRP-GFP-RpL10Ab (termed GFP-RpL10Ab throughout the article), which was used for the selection of stable cell lines.
For the expression of a 3× Flag-tagged E. coli BirA ligase, a destination vector with a 3× Flag sequence downstream of the metallothionein promoter in the pMK33/pMTHy backbone, followed in frame by a gateway cassette (Life Technologies), was constructed. The E. coli BirA ligase was PCR amplified and recombined into the pDONR221 plasmid. Recombination of the BirA entry vector with the 3× Flag destination vector resulted in the expression vector used for the selection of stable cell lines coexpressing the BirA ligase.
Polysome analysis.
Cycloheximide (0.1 mg/ml) was added to the growth medium, and the cells were incubated for 10 min at 25°C, washed once with ice-cold phosphate-buffered saline (PBS) supplemented with 0.1 mg/ml cycloheximide, and lysed for 10 min on ice in buffer A (20 mM HEPES-KOH [pH 8.0], 150 mM NH4Cl, 5 mM MgCl2, 2 mM dithiothreitol [DTT], 1% Igepal, 0.1 mg/ml heparin [Sigma], 0.5 U/μl RiboLock RNase inhibitor [Fermentas], cOmplete Ultra protease inhibitor [EDTA free; Roche], 1 mg/ml cycloheximide). The clarified lysate of about 100 Mio cells was layered on top of a linear 15 to 45% sucrose gradient in buffer B (20 mM HEPES-KOH [pH 8.0], 150 mM NH4Cl, 10 mM MgCl2, 1 mM DTT, 1 mg/ml heparin, 0.5 U/μl RiboLock [Fermentas], 0.1 mg/ml cycloheximide). The gradients were centrifuged in an SW40Ti rotor (Beckman) for 2 h at 36,000 rpm and 4°C, and the absorbance at 254 nm was recorded during fractionation.
Ribosome affinity purification.
Cell lysates were prepared as described above for the polysome analysis using 10 Mio cells in 250 μl buffer C (20 mM HEPES-KOH [pH 8.0], 50 mM NH4Cl, 5 mM MgCl2, 2 mM DTT, 1% Igepal, 0.1 mg/ml heparin, 0.5 U/μl RiboLock [Fermentas], cOmplete Ultra protease inhibitor [EDTA free Roche], 1 mg/ml cycloheximide). The ribosomes were pelleted from clarified lysate by ultracentrifugation for 1 h at 52,000 rpm in a TLA100 rotor (Beckman). The pellet was resuspended in buffer C and incubated with MyOne streptavidin T1 Dynabeads (Life Technologies). The Dynabeads had been prewashed three times with PBS and resuspended in buffer C, and 100 μl slurry was used per 250 μl lysate. After overhead rotation for 30 min at 4°C, the beads were washed 3 times with buffer D (50 mM HEPES-KOH [pH 8.0], 300 mM NH4Cl, 5 mM MgCl2, 2 mM DTT, 0.5% Igepal, 0.1 mg/ml cycloheximide) and once with PBS supplemented with 0.1 mg/ml cycloheximide and 2 mM MgCl2. The washed beads were resuspended in 25 μl SDS protein loading buffer, heated at 95°C, separated on an SDS-polyacrylamide gel, and analyzed by Western blot analysis. For quantitative Western blot analysis, secondary antibody Alexa Fluor 680-labeled goat antirabbit antibody (Life Technologies), IRDye700CW–goat anti-rabbit antibody (LI-COR), or IRDye800CW–goat anti-rabbit antibody (LI-COR) was used, and the membrane was scanned using an Odyssey CLx instrument (LI-COR). The Western blots were quantitated using ImageStudio software (LI-COR). Experiments were performed at least in biological triplicate.
For the optional micrococcal nuclease treatment, CaCl2 to 5 mM final concentration (f.c.) and 2 Kunitz units micrococcal nuclease (NEB) per one Mio cell were added to clarified lysate (buffer C with 20 U/ml RiboLock but without heparin) and incubated for 15 min on ice. The nuclease digestion was quenched by adding EGTA to 6.25 mM f.c. Lysate (200 μl) was layered on top of a sucrose cushion (550 μl 1.1 M sucrose in buffer B with 20 U/ml RiboLock but without heparin). The ribosomes were pelleted by centrifugation in a TLA100.2 rotor (Beckman) at 53,000 rpm for 60 min at 4°C. The pellet was resuspended in 200 μl lysis buffer.
Library preparation and sequencing.
For library preparation, RNA was isolated from input (IN) samples (10% of the lysate; for the expression and decapped libraries) or beads after ribosome pulldown (PD; for the decapped libraries) using an RNeasy kit (Qiagen). The RNA quantity was determined by measuring the absorbance at 260 nm (NanoDrop 1000 spectrophotometer; Peqlab), and the RNA integrity was determined using a Bioanalyzer 2100 system (RNA 6000 Pico kit; Agilent Technologies). The yield of total RNA isolated from the beads was calculated as a percentage of the total RNA used for the pulldown experiment: [(number of micrograms of RNA from the beads)/(9 × number of micrograms of input RNA)] × 100. The RNA was further treated twice with Turbo DNase I (Life Technologies), and three biological replicates were pooled for the preparation of one library. Two independent libraries were prepared and analyzed by Illumina sequencing. Total RNA was further subjected to two rounds of rRNA depletion using a Ribo-Zero kit (human/mouse/rat; Epicentre) together with three additional biotinylated oligonucleotides against 5.8S rRNA and 28S rRNA (the sequences are listed in Table S1 in the supplemental material).
For the expression libraries, 10 μg total RNA was used for rRNA depletion, fragmented to about 200 nucleotides (nt) in length, and reverse transcribed with random hexamers and SuperScript III reverse transcriptase (RT; Life Technologies). cDNA was purified using an Agencourt RNAClean XP system (Beckman Coulter). Second-strand DNA synthesis, end repair, dA tailing, and adapter ligation were performed using a NEBNext module (NEB). After triple solid-phase reversible immobilization (SPRI)-bead purification/size selection (Agencourt AMPure XP; Beckman Coulter) and 10 rounds of PCR amplification, the library was size selected over an agarose gel and eluted by use of the MinElute kit (Qiagen).
For decapped libraries, 20 μg total RNA was used for rRNA depletion and 50 pmol of a modified Solexa 5′ RNA linker (see Table S1 in the supplemental material) was ligated with T4 RNA ligase 1 (NEB). RNA was purified twice with the Agencourt RNAClean XP system, fragmented to about 200 nucleotides in length, and dephosphorylated with Antarctic phosphatase (NEB). A preadenylated and 3′ end-blocked DNA linker (see Table S1 in the supplemental material) was ligated with truncated T4 RNA ligase 2 KQ (NEB) and purified twice with the Agencourt RNAClean XP system. Reverse transcription was performed using the TruSeq RT primer (Illumina) and SuperScript III RT. After purification with the Agencourt RNAClean XP system, second-strand synthesis was performed with the TruSeq universal primer (Illumina), followed by triple SPRI-bead purification/size selection. The library was amplified in 12 PCR cycles using the TruSeq universal primer and the index primer (Illumina), size selected on an agarose gel, and eluted with the MinElute kit.
The libraries were multiplexed in three lanes (see Table S3 in the supplemental material) and subjected to 100-nt paired-end Illumina sequencing, resulting in 120 million to 180 million reads per lane. Sequencing was performed at the Campus Science Support Facility (CSF; NGS Unit). Demultiplexing yielded 25 million to 52 million reads per sample (see Table S3 in the supplemental material for the exact numbers of each sample).
For each control library in Fig. 5, 20 μg total RNA was depleted of rRNA as described above. For tobacco acid pyrophosphatase (TAP)-treated libraries, rRNA-depleted RNA was treated with 10 U TAP (Epicentre) for 3 h at 37°C. For Terminator-treated libraries, rRNA-depleted RNA was treated with 3 U Terminator 5′-phosphate-dependent exonuclease (Epicentre) for 3 h at 37°C. RNA was purified through MicroSpin S-300 HR columns (GE Healthcare), and 50 pmol of 5′ RNA linker (see Table S1 in the supplemental material) was ligated with T4 RNA ligase 1 (NEB). After purification through MicroSpin S-300 HR columns, the RNA was reverse transcribed using random hexamers and Moloney murine leukemia virus reverse transcriptase (Promega). PCR was performed using GoTaq polymerase (Promega) and quantitative PCR (qPCR) using a GoTaq qPCR master mix (SYBR green I; Promega) on a StepOnePlus qPCR cycler (Applied Biosystems). Primers are listed in Table S1 in the supplemental material. Biological triplicates were performed for each control library. Controls without RT were negative for all reactions. qPCR analysis was performed using the ΔΔCT threshold cycle (CT) method (49), and the error was carried through to the final relative value using standard error propagation methods.
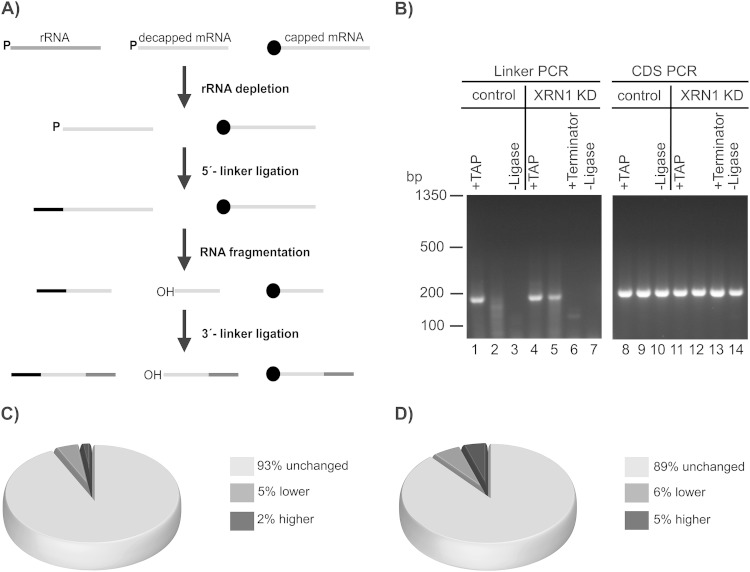
FIG 5.
Decapped mRNA degradation intermediates on the ribosome. (A) Cartoon outlining the initial steps of preparation of an RNA-seq library of decapped mRNA degradation intermediates. Only RNA fragments with both linker sequences get amplified in the RNA-seq library. (B) Ethidium bromide-stained agarose gel with RT-PCR fragments detecting linker-ligated firefly luciferase mRNA (lanes 1 to 7, Linker PCR) or total firefly luciferase mRNA (lane 8 to 14, CDS PCR) from control (lane 1 to 3 and 8 to 10) or XRN1 knockdown (KD) cells (lanes 4 to 7 and 11 to 14). In control reactions, RNA depleted of rRNA was treated with TAP (lanes 1, 4, 8, and 11) or Terminator exonuclease (lanes 6 and 13). In lanes 3, 7, 10, and 14, RNA ligase was omitted in the RNA linker ligation. (C, D) Pie charts depicting the percentages of all detected transcript species (C) or potential mRNA targets of miRNAs (D) with an abundance of decapped mRNA degradation fragments on ribosome complexes that was unchanged, lower, or higher compared to that in cell lysates in XRN1 knockdown cells.
Computational analysis.
Sequencing and index primers were removed from the demultiplexed samples with the cutadapt (v1.4) tool (50), and mapping against the Berkeley Drosophila Genome Project BDGPv5 reference genome (www.fruitfly.org) was performed with the segemehl (v0.1.7) program (51, 52). In a postprocessing step, the ViennaNGS toolbox (53) was applied to extract all uniquely mappable reads for the follow-up differential gene expression (DGE) analysis. The htseq-count utility from the HTSeq Python library (54) was used to determine read count numbers for each sample. DGE analysis under different conditions was finally performed with the DESeq package (55).
RESULTS
Sedimentation of NOT2, HPat, and AGO1 in polysome fractions.
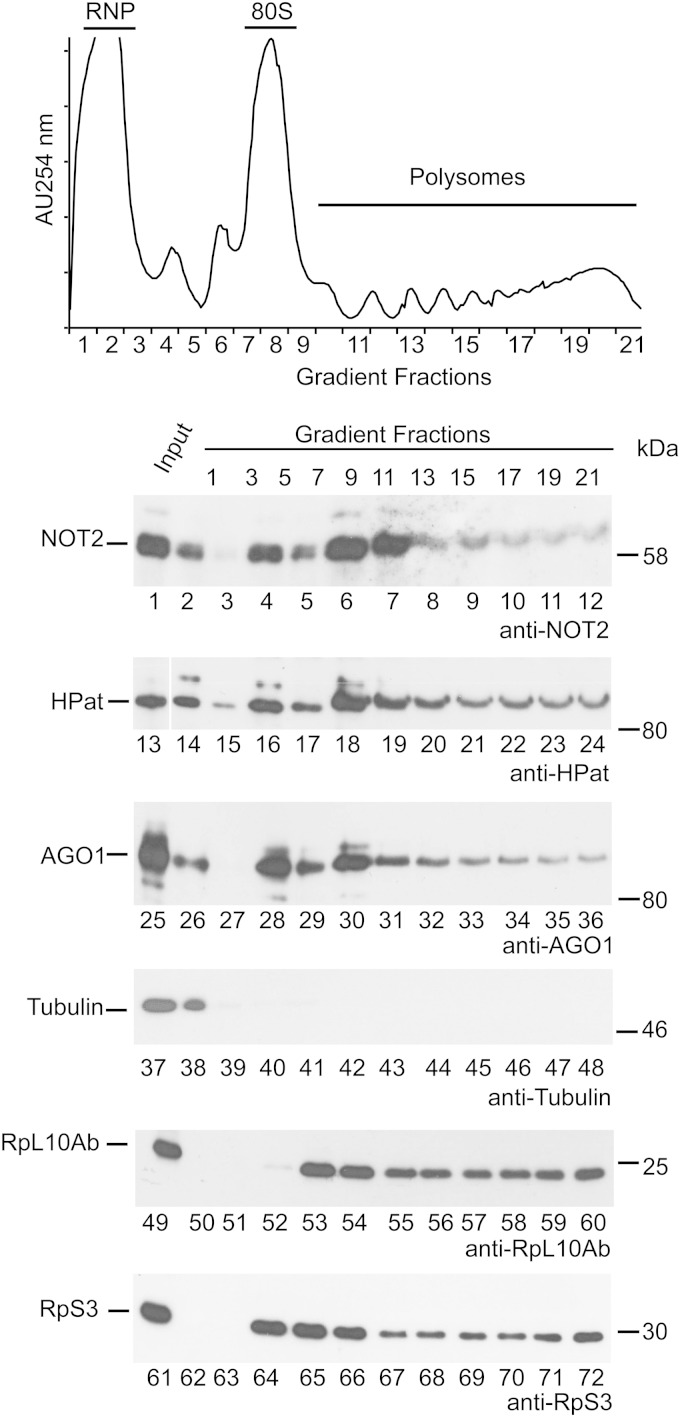
As an initial step to test for mRNA degradation on the ribosome in Drosophila cells, we investigated the sedimentation of NOT2 (a component of the deadenylation complex CCR4-NOT), HPat (a general decapping activator), and the miRNA effector component AGO1 in polysome gradients (Fig. 1). Cell lysates were separated on 15 to 45% sucrose density gradients, and fractions were collected and analyzed by Western blot analysis using anti-NOT2 (Fig. 1, lanes 1 to 12), anti-HPat (lanes 13 to 24), anti-AGO1 (lanes 25 to 36), or antitubulin (lanes 37 to 48) antibody. NOT2, HPat, and AGO1 comigrated with polysome fractions. As expected, tubulin, a cytoplasmic protein that is not implicated in an association with ribosomes, was detected in the light fractions of sucrose density gradients (mRNP-containing fractions; Fig. 1, lane 38). In addition, we raised antibodies against the large ribosomal subunit protein RpL10Ab and the small ribosomal subunit protein RpS3. Both antibodies were used to determine the sedimentation of ribosomes in the sucrose density gradient (Fig. 1, lanes 49 to 72). In summary, AGO1, NOT2, and HPat cosediment with polysome fractions, which is a prerequisite for mRNA degradation on the ribosome in Drosophila cells.
FIG 1.
Sedimentation of NOT2, HPat, and AGO1 in polysome profiles. Cell lysates were separated on 15 to 45% linear sucrose gradients, and the absorption at 254 nm (AU254 nm) was recorded during fractionation. Input and fractions were analyzed by Western blot analysis using antibodies against endogenous NOT2, HPat, AGO1, tubulin, RpL10Ab, or RpS3. The following amounts were separated on SDS-polyacrylamide gels: for NOT2, HPat, and AGO1, 0.1% input, 1 μl of fractions 1 and 3, 20 μl of fractions 5 and 7, and 60 μl of fractions 9 to 21; for tubulin, 0.5% input and the same amounts of the different fractions described for NOT2, HPat, and AGO1; and for RpL10Ab and RpS3, 0.03% input, 1 μl of fractions 1 and 3, and 10 μl of fractions 5 to 21. At least three biological replicates were analyzed throughout the study.
Ribosome affinity purification from Drosophila cell lysates.
Sucrose density gradient fractionation is a size-dependent fractionation method. Thus, higher-molecular-weight structures, such as P bodies or pseudopolysomes, can cosediment in polysome fractions (31, 56). In order to distinguish between the association of NOT2, HPat, and AGO1 with ribosome complexes and other higher-molecular-weight structures, we set up ribosome affinity purification in Drosophila S2 cells. Similar to the methods used in mammalian cells and flies, we expressed a tagged version of the large ribosomal subunit protein RpL10Ab (the mammalian homolog of RpL10a [57, 58]) in Drosophila S2 cells, which allows the rapid purification of ribosomes from cell lysates. More specifically, we generated stable cell lines coexpressing the N-terminal fusion protein BLRP-GFP-RpL10Ab (termed GFP-RpL10Ab throughout the article) and the E. coli BirA ligase. The BirA ligase recognizes the BLRP (biotin ligase recognition protein) protein sequence and biotinylates the fusion protein (46, 47), which can be used for isolation with streptavidin beads. We first tested the expression of GFP-RpL10Ab and its integration into ribosomal complexes by polysome analysis. As an initial purification step, we prepared S100 extracts and fractionated the resuspended pellet in sucrose density gradients as described above (see Fig. S1A in the supplemental material). We detected GFP-RpL10Ab in the mRNP fractions as well as in the fractions of the 80S ribosome and polysomes of the sucrose density gradient. In addition, the signal for GFP-RpL10Ab shifts similarly to the signal for RpS3 and RpL10Ab after treatment with limited amounts of micrococcal nuclease (see Fig. S1B in the supplemental material). This mild nuclease treatment nicks the mRNA between ribosomes but keeps the ribosome itself intact (59). Thus, not all but a significant amount of GFP-RpL10Ab protein was integrated into functional ribosome complexes. Next, we tested the efficiency of biotinylation in cell lysates by addition of streptavidin. The strong binding affinity of streptavidin to biotin results in a shift of the GFP-RpL10Ab in the SDS-polyacrylamide gel (see Fig. S1C, lane 4, in the supplemental material). In contrast, when using a control cell line expressing BLRP-GFP-RpL10Ab but not the E. coli BirA ligase (minus-BirA cells), the protein is not biotinylated and therefore does not shift upon addition of streptavidin (see Fig. S1C, lane 8, in the supplemental material).
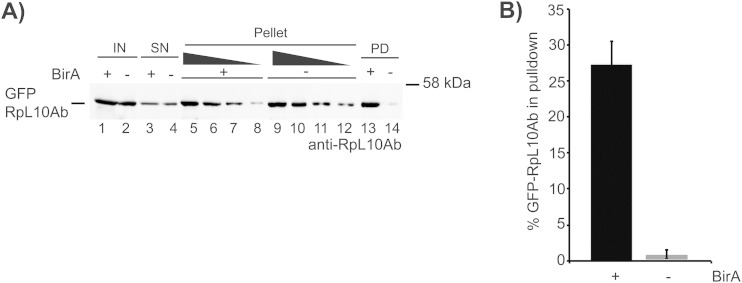
For pulldown experiments we resuspended the pellet of S100 extract preparations in lysis buffer and isolated the tagged ribosomes using streptavidin beads. The bead eluate was analyzed by Western blot analysis (Fig. 2A, lane 13). As a control for specificity we used cell lysate from minus-BirA cells (described above; Fig. 2A, lane 14). For the calculation of the pulldown efficiency, the signal intensities were quantitated and normalized to the signal intensity of the resuspended ribosome pellet (see Fig. S1D in the supplemental material). A total of 27.3% ± 3.5% of the GFP-RpL10Ab protein in the S100 pellet could be isolated on streptavidin beads in BirA-expressing cells, but less than 1% could be isolated in minus-BirA cells. Thus, the background binding of the protein to streptavidin beads is less than 1% of the input (Fig. 2B).
FIG 2.
Ribosome affinity purification. (A) Ribosome complexes were pelleted from cell extracts expressing GFP-RpL10Ab and purified using streptavidin-coated beads. Cells coexpressing BirA ligase (lanes +) were compared to control cells lacking BirA ligase (lanes −). Lysate (input [IN]; 4.9% lysate in lanes 1 and 2), supernatant (SN; 4.9% in lanes 3 and 4), decreasing amounts of resuspended ribosome pellet (4.6% in lanes 5 and 9, 2.3% in lanes 6 and 10, 1.1% in lanes 7 and 11, 0.6% in lanes 8 and 12), and pulldowns (PDs; 20% in lanes 13 and 14) were analyzed by Western blot analysis using anti-RpL10Ab antibody. (B) The amount of GFP-RpL10Ab in the Western blot in panel A was quantitated. The signal intensities were obtained using the Odyssey instrument (v2.1), plotted against the amount of resuspended pellet, and analyzed by linear regression analysis (see Fig. S1D in the supplemental material). The amount of GFP-RpL10Ab in the pulldown was calculated as a percentage of the total amount of GFP-RpL10Ab in the resuspended pellet and is plotted in the graph. The bars represent the mean values, and the error bars represent the standard deviations from at least three independent experiments.
Copurification of mRNA degradation factors and miRNA effector components with ribosome complexes.
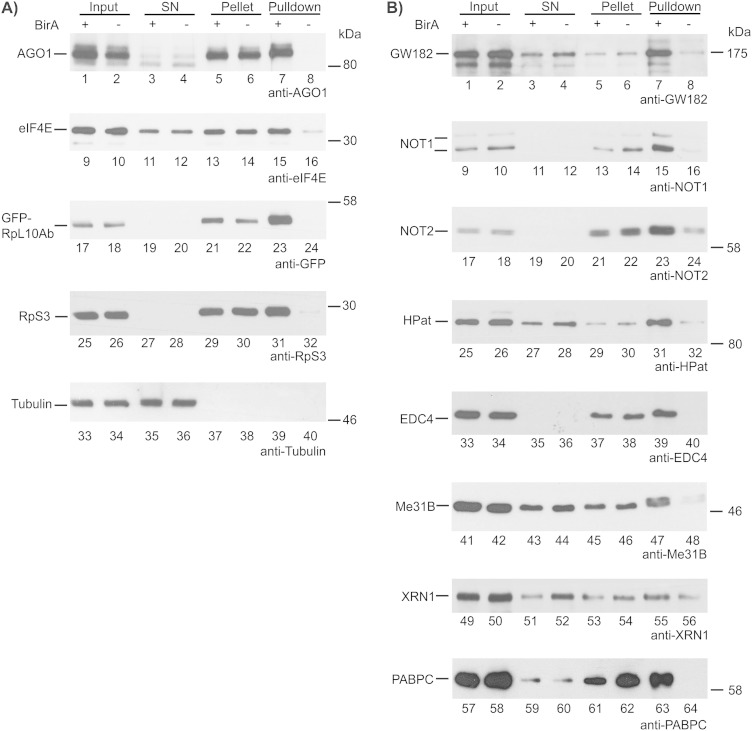
Using affinity-purified ribosome complexes, we further investigated the copurification of several mRNA degradation factors and factors of the miRNA effector complex. Since the initial step of purification of the cell lysate (S100 extract) was important for the specificity of the pulldown experiment, we analyzed the cell lysate (input [IN]), the supernatant (SN), the pellet of the S100 preparation, and the eluate of the streptavidin pulldown (PD) (Fig. 3). As a control for the efficient pulldown of GFP-RpL10Ab, we performed Western blot analysis using anti-GFP antibody (Fig. 3A, lanes 17 to 24). Western blot analysis with anti-RpS3 antibody was used to test for the copurification of the small ribosomal subunit protein and, hence, the isolation of 80S ribosome complexes (Fig. 3A, lanes 25 to 32). Furthermore, as a positive control we detected eIF4E, a eukaryotic translation initiation factor, which binds to the cap structure of mRNAs and is therefore expected to copurify with intact ribosome complexes (Fig. 3A, lanes 9 to 16). Tubulin was used as a negative control (Fig. 3A, lanes 33 to 40). In all pulldown experiments, the cell lysate of minus-BirA cells, expressing GFP-RpL10Ab but not the BirA ligase, was included as a control for specificity.
FIG 3.
Copurification of components of mRNA degradation and the miRNA pathway with ribosome complexes. (A) AGO1 and controls; (B) GW182, NOT1, NOT2, HPat, EDC4, Me31B, XRN1, and PABPC. Ribosomes were pelleted from cell lysates expressing GFP-RpL10Ab. Cells coexpressing BirA ligase (lanes +) were compared to cells lacking BirA ligase (lanes −). The pellet with ribosomal complexes was resuspended and used for the pulldown experiment with streptavidin beads. For Western blot analysis, the cell lysate (input [IN]), the supernatant (SN) after pelleting of ribosomes, the resuspended ribosome pellet, and the eluate of the beads (pulldown [PD]) were analyzed using an anti-AGO1, anti-eIF4E, anti-GFP, anti-RpS3, or antitubulin antibody (A) or an anti-GW182, anti-NOT1, anti-NOT2, anti-HPat, anti-EDC4, anti-Me31B, anti-XRN1, or anti-PABPC antibody (B). The amount of IN, SN, or pellet loaded was calculated as a percentage of the total amount of lysate used for the PD, and for the PD the indicated percentage of the total eluate of the PD was loaded. Sample loading in panel A was as follows: for the anti-AGO1 Western blot, 1.0% IN, SN, and pellet and 32% PD; for anti-eIF4E probing, 0.6% IN, SN, and pellet and 14% PD; and for antitubulin, anti-GFP-RpL10Ab, and anti-RpS3, 0.4% IN, SN, and pellet and 4% PD. Sample loading in panel B was as follows: for anti-GW182 and anti-HPat, 0.4% IN, SN, and pellet and 36% PD; for anti-NOT1, 1.2% IN, SN, and pellet and 50% PD; for anti-NOT2, 0.5% IN, SN, and pellet and 50% PD; for anti-EDC4, 0.6% IN, SN, and pellet and 84% PD; for Me31B, 1.3% IN, SN, and pellet and 40% PD; for XRN1 1.2% IN, SN, and pellet and 50% PD; and for PABPC, 0.2% IN, SN, and pellet and 30% PD.
First, we tested for the copurification of miRNA effector components with ribosome complexes using anti-AGO1 and anti-GW182 antibodies. Both proteins copurified with ribosomes (Fig. 3A and B, lanes 7 and 8). Second, we tested for NOT1 and NOT2, two components of the deadenylation complex CCR4-NOT, both of which not only pelleted very efficiently (Fig. 3B, lanes 11 to 14 and 19 to 22) but also copurified with ribosome complexes (Fig. 3B, lanes 15 and 16 and lanes 23 and 24). Third, we selected three general decapping activators, HPat, EDC4, and Me31B. All three factors copurified very specifically with ribosome complexes in pulldown experiments (Fig. 3B, lanes 31 and 32, 39 and 40, and 47 and 48). Fourth, we tested for XRN1, which not only is the exonuclease responsible for mRNA degradation after decapping but also promotes decapping of mRNAs in Drosophila cells (60). XRN1 partly pelleted with ribosome complexes (Fig. 3B, lanes 51 to 54) and was purified in ribosome pulldown experiments (Fig. 3B, lanes 55 and 56). Lastly, we tested for the cytoplasmic poly(A)-binding protein PABPC, which pelleted very efficiently from cell lysates (Fig. 3B, lanes 59 to 62) and also copurified in the ribosome pulldown (Fig. 3B, lanes 63 and 64). In order to stabilize polysomes and thereby increase the amount of purified ribosome complexes, all pulldown experiments were performed with cells after treatment with cycloheximide. We therefore also tested the copurification without antibiotic treatment. As expected, due to the lower stability of ribosome complexes, the amount of pelleted protein was generally smaller, although the amounts varied. However, all of the tested factors copurified with ribosome complexes in pulldown experiments in which cycloheximide was omitted (see Fig. S2 in the supplemental material).
In summary, AGO1 and GW182, factors of the mRNA deadenylation machinery (NOT1 and NOT2), and the decapping machinery (HPat, EDC4, Me31B, and XRN1) copurify with ribosome complexes. The availability of mRNA degradation factors on ribosome complexes indicates the possibility of mRNA degradation on the ribosome.
RNA dependence of association of mRNA degradation factors and components of the miRNA effector complex with ribosome complexes.
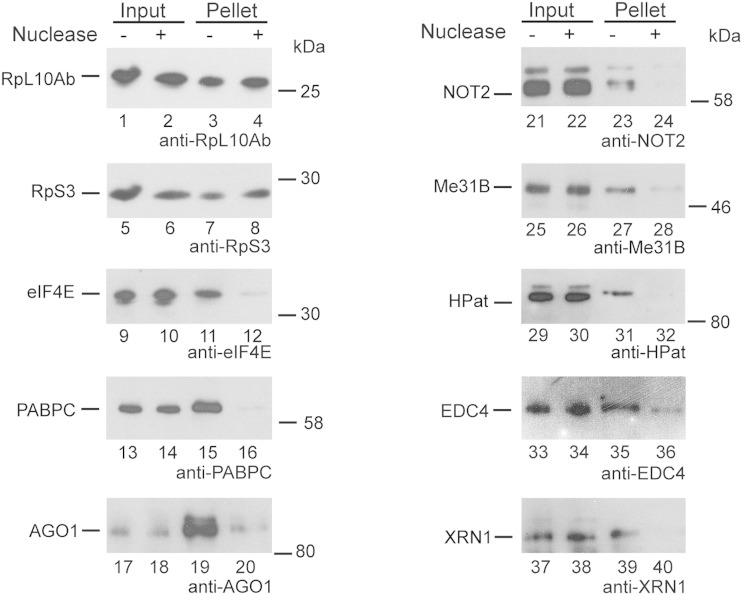
We further investigated whether factors copurifying with ribosome complexes are, rather, associated with the ribosome or the mRNA. Treatment of lysates with limiting amounts of micrococcal nuclease results in nicking of the mRNA but leaves ribosomes intact (59). Consequently, the ribosome complexes shifted from polysomes to the 80S peak in sucrose density gradients (see Fig. S1B in the supplemental material) under conditions which had been optimized for ribosome profiling in Drosophila S2 cells (59). In order to test for the cosedimentation of mRNA degradation factors and miRNA effector components, cell lysates were treated with micrococcal nuclease, and the ribosome complexes were pelleted and analyzed by Western blot analysis (Fig. 4). Since the ribosomes were still intact, we detected both RpL10Ab and RpS3 in the ribosome pellet independently of the nuclease treatment (Fig. 4, lanes 3 and 4 and lanes 7 and 8). In contrast, eIF4E and PABPC, which bind to the cap structure or the poly(A) tail of the mRNA, are dependent on an intact mRNA and were not sedimented after nuclease treatment (Fig. 4; compare lane 11 with lane 12 and compare lane 15 with lane 16). Similarly, AGO1 (lanes 19 and 20), NOT2 (lanes 23 and 24), Me31B (lanes 27 and 28), HPat (lanes 31 and 32), EDC4 (lanes 35 and 36), and XRN1 (lanes 39 and 40) were not detected in the ribosome pellet upon micrococcal nuclease treatment. This result suggests that the copurification of these factors with ribosome complexes is mediated through binding to the mRNA or a region on the surface of the ribosome which is accessible for the nuclease treatment.
FIG 4.
RNA dependence of the association of eIF4E, PABPC, AGO1, NOT2, Me31B, HPat, EDC4, and XRN1 with ribosome complexes. Cell lysates expressing GFP-RpL10Ab and BirA ligase were incubated with (lanes +) or without (lanes −) micrococcal nuclease, and ribosome complexes were sedimented by ultracentrifugation. Cell lysates (input [IN]) and ribosome pellets were analyzed by Western blot analysis using the indicated antibody. Loading was as follows: for anti-RpL10Ab and anti-RpS3, 0.8% IN and pellet; for anti-eIF4E: 0.3% IN and 2.5% pellet; for anti-PABPC, 0.5% IN and 3.8% pellet; for anti-AGO1, 0.1% IN and 7.5% pellet; for anti-NOT2, 0.5% IN and 4% pellet; for anti-Me31B, 0.1% IN and 1.5% pellet; for anti-HPat, 0.5% IN and 10% pellet; for anti-EDC4, 0.3% IN and 15% pellet; and for anti-XRN1 probing, 0.5% IN and 5% pellet.
Decapped mRNA degradation intermediates on the ribosome.
In addition to the copurification of mRNA degradation factors with ribosomes, we isolated intermediates of mRNA degradation from ribosome complexes. Not only general mRNA degradation but also miRNA- and ARE-mediated mRNA decay in Drosophila S2 cells are initiated mainly by deadenylation followed by decapping (18, 61–63). However, decapped mRNAs are very quickly degraded unless XRN1 activity is compromised (24, 64). Also, in the Drosophila S2 cell line, XRN1 knockdowns can be used to stabilize and detect deadenylated mRNA degradation fragments (60, 65). While it is known that XRN1 in Drosophila cells promotes decapping, only the knockdown of XRN1 together with another decapping factor causes the efficient inhibition of decapping (60, 66). Thus, we prepared mRNA libraries of decapped mRNA degradation products from XRN1 knockdown cells. The knockdown was verified by Western blot analysis using anti-XRN1 antibody and antitubulin antibody as a loading control (see Fig. S3B in the supplemental material). In addition, we verified the copurification of several proteins with ribosome complexes in XRN1 knockdown cells, as in control cells (see Fig. S3A in the supplemental material).
In order to specifically amplify RNAs lacking a cap structure, we ligated a 5′ RNA linker to the RNA, similar to methods used for mapping the 5′ termini of mRNAs or the preparation of small RNA libraries (67–70). The initial steps of library preparation are outlined in Fig. 5A. Hydrolysis of the m7G cap structure by the decapping complex releases 7-methyl-GDP (37, 71, 72). This leaves an mRNA with a 5′ monophosphate, which is a substrate for the template-independent T4 RNA ligase (73). In contrast, RNAs with an intact cap structure are not ligated to the 5′ RNA linker and, therefore, not amplified in the transcriptome sequencing (RNA-seq) library. In order to confirm the importance of the XRN1 knockdown for stabilizing mRNA degradation fragments, we compared control libraries of XRN1 knockdown cells and control cells. We used cell lines expressing a firefly luciferase reporter, in addition to GFP-RpL10Ab and BirA ligase. For control libraries, total RNA of lysates or pulldown samples was depleted of rRNA and an RNA linker was ligated to the 5′ end (first two steps of Fig. 5A). The RNA was further reverse transcribed using random hexamers and used for PCR or qPCR (Fig. 5B). First, we amplified the cDNA using a primer pair binding to the RNA linker sequence and the 5′ end of the firefly luciferase transcript (Fig. 5B, lanes 1 to 7), thus detecting only cDNAs with the RNA linker sequence at the 5′ end of firefly luciferase mRNA (linker PCR). The PCR fragments were cloned and their sequences were verified. Second, as a loading control we used a primer pair amplifying a fragment within the firefly luciferase-coding region (coding sequence [CDS] PCR) (Fig. 5B, lanes 8 to 14). As shown in Fig. 5B, we detected a linker PCR product using only libraries of XRN1 knockdown cells and not libraries of control cells (compare lane 5 to lane 2). When RNA ligase was omitted during the ligation step, we did not detect a linker PCR product (Fig. 5B, lanes 3 and 7), but we still detected a product for the coding region of firefly luciferase (Fig. 5B, lanes 10 and 14). In positive-control reactions, prior to RNA ligation the RNA was treated with tobacco acid pyrophosphatase (TAP), which cleaves the pyrophosphate bond at the cap structure, resulting in a 5′ monophosphate terminus (74), and thereby converts all mRNAs into substrates for T4 RNA ligase. As expected, a linker PCR product could be detected in libraries of both control and XRN1 knockdown cells (Fig. 5B, lanes 1 and 4). Additionally, we treated the RNA of XRN1 knockdown cells with Terminator 5′-phosphate-dependent exonuclease, which digests uncapped, 5′ monophosphorylated RNAs (24), prior to RNA ligation. After this treatment, the linker PCR fragment was no longer detected in libraries of XRN1 knockdown cells (Fig. 5B, lane 6), demonstrating the specificity for decapped, monophosphorylated RNAs. Furthermore, we performed qPCR analysis of the libraries described above and libraries of pulldown samples of XRN1 knockdown cells (see Table S2 in the supplemental material). This confirmed that in all samples comparable amounts of firefly luciferase cDNA were present, and therefore, the lack of a linker PCR fragment in the library of control cells (Fig. 5, lane 2) was not due to a lower abundance of firefly luciferase mRNA. Additionally, the CT values in Table S2A in the supplemental material demonstrate that similar amounts of linker PCR product and CDS PCR product can be detected in pulldown and input libraries, which had been prepared from the same amount of total RNA. In summary, we could confirm the importance of the XRN1 knockdown for the stability of decapped mRNA degradation intermediates and the specific ligation of monophosphorylated RNAs to an RNA linker.
Next, we used the linker ligation method to prepare libraries for high-throughput sequencing to analyze the copurification of decapped mRNA degradation intermediates on ribosome complexes for the whole transcriptome. For these RNA libraries of decapped mRNA degradation fragments (decapped RNA libraries), we isolated the total RNA from XRN1 knockdown cells expressing GFP-RpL10Ab and BirA ligase from ribosome pulldown experiments. Of the total amount of RNA used for the pulldown experiment, 14.6% ± 1.3% (mean ± standard deviation from at least three biological replicates) could be isolated from the beads. As a control for specificity, we isolated RNA from cells expressing GFP-RpL10Ab but lacking BirA ligase from pulldown experiments. In this case, only 0.9% ± 0.2% of the total amount of RNA in the pulldown experiment was isolated from the beads. Thus, the RNA background also is very low in the pulldown experiments. For control cells treated with dsRNA against maltose binding protein (MBP), the values were similar: after the pulldown, 14.0% ± 1.9% of total RNA from the beads from cell lysates of cells containing BirA ligase and 0.9% ± 0.2% from cells lacking BirA ligase. RNA libraries were prepared from cell lysates (input) and ribosome PDs and analyzed by high-throughput sequencing (decapped RNA libraries D1 and D2, respectively, in Table S3 in the supplemental material). Strikingly, for about 12,000 (93%) transcript species detected, their relative abundance did not change between the input (D1) and PD (D2) RNA libraries, only about 680 transcripts (5%) were less abundant, and about 210 transcripts (2%) were slightly enriched on the ribosome (Fig. 5C; see also Table S4 in the supplemental material). All transcripts with a slightly higher abundance on the ribosome were protein-coding genes. In contrast, among the transcripts with a lower abundance on the ribosome, only about 40% coded for proteins and the others annotated as noncoding transcripts, including, e.g., the RNase MRP RNA (−5.09 log2-fold change). The RNA component of RNase MRP is a polymerase III transcript with a nucleolar localization in Drosophila (75). Thus, MRP RNA is a good substrate for RNA ligation but is expected to be present at a much lower abundance in the ribosome pulldown, which also demonstrates the specificity of the method. In order to confirm the results for a few transcripts, we randomly picked three transcripts with unchanged abundance in the RNA-seq library and RNase MRP RNA and performed qPCR analysis on the control libraries of the XRN1 knockdown cells also used for results described in Fig. 5B using primers specific for the ligated linker and the 5′ end of each transcript (see Tables S2B and C in the supplemental material). For the transcripts with an unchanged abundance in the RNA-seq library, the CT values of the fragments detected in the control libraries were also very similar (see Table S2B in the supplemental material). Thus, we amplified similar amounts of linker-ligated fragments from input and pulldown libraries. Next we performed a very simplified analysis of the relative abundance of the linker-ligated fragments and randomly picked one of the genes, FBgn0039857, and used it for normalization. Using the ΔΔCT method (49), we compared the relative abundance between input and pulldown samples (see Table S2C in the supplemental material). As for the RNA-seq results, for FBgn0003279 and FBgn0000556, the relative abundance of the linker fragment in input libraries compared to that in the pulldown library was not significantly changed when applying our cutoff ±0.568 log2-fold. However, for RNase MRP RNA (FBgn0046696), we detected a reduction (−6.81 ± 0.21 log2-fold; see Table S2C in the supplemental material) in the pulldown of the control library, similar to that in the RNA-seq library. Thus, for this limited set of transcripts, we could recapitulate the results of the RNA-seq analysis using qPCR. Furthermore, since all libraries were prepared with the same amount of input RNA, the CT values (see Table S2B in the supplemental material) indicate that a large fraction of the linker fragment of the lysate can be detected in the pulldown library.
In order to ensure that we detected a significant amount of transcript species in our decapped RNA libraries, we additionally prepared a regular expression library from total RNA of input samples (see library E6 in Table S3 in the supplemental material). Only 21 transcripts could not be detected in the decapped library but could be detected in the expression library (compare library D1 to library E6). Thus, we detected almost the same number of transcripts in the decapped library that we detected in a regular expression library. Furthermore, we tested the effect of the XRN1 knockdown, which we used to stabilize decapped mRNA degradation intermediates, comparing the expression libraries of input samples (library E6) and cells not treated with dsRNA against XRN1 (see library E4 in Table S3 in the supplemental material). Interestingly only 77 transcripts had different expression levels (see Table S5 in the supplemental material). Since we always included cycloheximide in our cell lysates, we also analyzed expression libraries of cell lysates not treated with cycloheximide (see library E3 in Table S3 in the supplemental material). We could not detect any significant difference in the transcript levels between the two expression libraries (compare library E4 to library E3).
Another interesting aspect is whether mRNAs, which are potential targets of miRNAs, are found in the decapped mRNA degradation libraries (libraries D1 and D2) and whether their abundance changes between ribosome pulldowns and cell lysates. In order to address this question, we first identified mRNAs, which are potential targets of miRNAs, in our cell line. Thus, we compared the expression libraries of AGO1 knockdown cells to those of control cells (see libraries E3 and E7 in Table S3 in the supplemental material). The knockdown of AGO1 was verified by Western blot analysis (see Fig. S3C in the supplemental material). We found 1,115 transcripts upregulated in AGO1 knockdown cells (see Table S6 in the supplemental material). In order to verify some of the targets, we compared the transcripts to the data sets available from Hong et al. (76), and 256 transcripts had been described to be AGO1 targets. Next, we were interested in whether we had also detected these 1,115 transcripts in our decapped RNA-seq libraries of XRN1 knockdown cells (Fig. 5C, libraries D1 and D2; see also Table S4 in the supplemental material) and, indeed, could find 1,110 out of 1,115 transcripts (see the rows highlighted in blue in Table S4 in the supplemental material). Moreover, the relative abundance for 89% of these 1,110 transcripts was unchanged, for 5% it was higher, and for 6% it was lower in the pulldown library than in the input library of decapped mRNA degradation fragments of XRN1 knockdown cells (Fig. 5D). Thus, mRNA degradation products of potential mRNA targets of miRNAs are also found at a high abundance on the ribosome.
In summary, we could isolate the majority of decapped mRNA degradation intermediates present in cell lysates from ribosome complexes. Together with the copurification of mRNA degradation factors and miRNA effector components, this strongly argues for the ribosome as a site for general and miRNA-mediated mRNA degradation.
DISCUSSION
mRNA degradation and translation are two processes crucial for posttranscriptional gene regulation. The strong interconnection of mRNA degradation and translation has been noted for many years and has led to the hypothesis that mRNAs could be degraded on the ribosome (6, 21–24). However, with the finding of cytoplasmic P bodies, which accumulate, e.g., factors of the general mRNA degradation pathway but no ribosomes, it was proposed that mRNA decapping could occur only in P bodies after dissociation of the mRNA from ribosomes (11, 37–42). More recently, Coller and coworkers provided additional experimental evidence to support the initial hypothesis of cotranslational degradation of some mRNAs in yeast (43, 44). In this study, we significantly extend the experimental support for the hypothesis that the ribosome is a very general site not only for general 5′-to-3′ mRNA degradation in Drosophila but also for the miRNA-mediated mRNA degradation pathway.
Several reports have already demonstrated the cosedimentation of the mRNA degradation factors DCP2 (71), Dhh1 (77), and XRN1 (23) with polysomes. In agreement with these observations, we could detect all tested mRNA degradation factors, including NOT2 and HPat, as well as the miRNA effector component AGO1, in polysome fractions (Fig. 1). We included the miRNA effector components, since it is well established that, upon targeting by miRNAs, many mRNAs are degraded by the 5′-to-3′ mRNA degradation pathway (62, 66, 78), and human AGO2 (79) was also reported to cosediment with polysomes. Thus, the ribosome could be a site not only for general mRNA degradation but also for miRNA-mediated mRNA degradation.
We adapted the affinity purification of ribosome complexes for Drosophila cells to ensure that the factors are not cosedimenting in high-molecular-weight structures, such as pseudopolysomes (31, 56) or P bodies (Fig. 2), but they are indeed associated with ribosome complexes. More specifically, we tested for the copurification of components of the deadenylase complex CCR4-NOT (NOT1 and NOT2), several decapping activators (HPat, EDC4, Me31B), the exonuclease XRN1, and the miRNA effector components AGO1 and GW182. All of the factors clearly copurified with the ribosomes, demonstrating an association with ribosome complexes (Fig. 3). The loss of the copurification of these factors upon limited micrococcal nuclease treatment indicates their association with mRNAs or with regions of the ribosome which are accessible to nuclease digestion (Fig. 4). The copurification of mRNA degradation and miRNA effector components is a good indication that mRNA degradation occurs on ribosomes and is a prerequisite for mRNA degradation to occur on ribosomes.
While experimental evidence for decapping on the ribosome in yeast has been provided for several reporter constructs and a few endogenous genes (43, 44), we aimed to test how general the phenomenon is in Drosophila cells on a transcriptome-wide level. Since the decapping step is the final step committing the mRNA for degradation, we prepared libraries of decapped mRNA degradation intermediates isolated from ribosome complexes and cell lysates for high-throughput sequencing. Interestingly, for about 93% of the detected transcripts, their abundance on the ribosome compared to that in lysates did not change. About 5% of the transcripts were less abundant and 2% were slightly more abundant on the ribosome than in lysates (Fig. 5C). The transcripts with a lower abundance included those which, due to their known function, are expected to copurify less efficiently with the ribosome (e.g., RNase MRP RNA). Based on additional qPCR analysis of transcripts with an unchanged abundance, we detected a large fraction of mRNA degradation fragments of the lysate on ribosome complexes. This clearly indicates that the aggregation into a ribosome free state is not a prerequisite for their degradation. This finding is also consistent with the observation that mRNA degradation can be uncoupled from P-body formation (80–83). Future experiments investigating the kinetics of mRNA degradation and translation on a transcriptome-wide level will allow a precise quantitative conclusion of which fraction of each transcript is degraded on the ribosome. Furthermore, it will be interesting to compare the extent of mRNA degradation on the ribosome under various stress conditions and at various developmental stages.
The close relation of mRNA degradation and translation is also reflected in the observation that each step of mRNA degradation limits and potentially regulates translational events. First, deadenylation beyond a minimal poly(A) tail length results in reduced translation efficiency, most likely due to the loss of PABPC, resulting in the inhibition of the formation of the closed-loop conformation between eIF4F, eIF4G, and PABPC, which is important for efficient translation (84–86). Second, the yeast orthologs of the decapping activators Me31B and HPat have been demonstrated to inhibit translation initiation and have been further proposed to render the cap accessible to the decapping machinery (28, 29). Moreover, recent studies attribute to HPat and Me31B important roles in the coupling of mRNA deadenylation and decapping. HPat binds to both the deadenylase complex and decapping factors, including DCP2, Me31B, and the Lsm1-7 ring (87). Recent crystal structures show the direct binding of DDX6 (a homolog of Me31B in mammals) to CNOT1 of the deadenylation complex CCR4-NOT (88, 89). Third, once the mRNA is decapped, it can no longer bind the initiation factor eIF4E, and therefore, the assembly of the eIF4F translation initiation complex and further translation are prevented (90). Finally, following decapping, bulk mRNAs are degraded by digestion with the exonuclease XRN1. In case the translation of the mRNA is not completed, the digestion in the 5′-to-3′ direction would not interfere and would ensure the production of a complete protein.
An important mechanism of posttranscriptional gene regulation in many eukaryotes, including mammals, flies, and worms but not yeast, is regulation by miRNAs. Although miRNAs can act as translational repressors, the common destabilization of mRNA targets is a consequence of miRNA-mediated mRNA deadenylation (62, 91, 92). Deadenylation is followed by decapping and 5′-to-3′ mRNA degradation (62, 78) utilizing the general mRNA degradation machinery. Recent studies indicate that mRNA degradation is the dominant effect of mammalian miRNAs (93–98). While biochemical analysis had already demonstrated earlier the direct recruitment of GW182 to the CCR4-NOT complex in miRNA-mediated gene silencing (7–9), the structures of CCR4-NOT provided important insight into the binding of GW182 to the CCR4-NOT complex (88, 89). Interestingly, we could detect the two crucial miRNA effector components AGO1 and GW182 and the decapped mRNA degradation products of the majority of potential targets of miRNAs on ribosome complexes. These findings suggest that mRNAs targeted by miRNAs can also get degraded while they are still associated with the ribosome, and consequently, their aggregation into P bodies is not a prerequisite for degradation.
Cytoplasmic P bodies and stress granules are both dependent on nontranslating mRNAs, and under stress conditions mRNAs accumulate in these bodies (99–101). mRNAs in these granules can either get degraded or return to the pool of actively translating mRNAs (102, 103). Thus, they facilitate the adaption of the cell to various environmental changes. In contrast, under regular growth conditions the degradation of mRNAs at the same site as translation would be of advantage. In particular, mRNAs could get degraded very efficiently at the end of their life cycle without the need to aggregate into granules.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Izaurralde, S. Newbury, and E. Wahle for antibodies and members of the labs of A. Barta and J. Brennecke for help during library preparation.
This work was funded by the Austrian Science Fund (FWF; grants P22124-B09 and P23884-B09 to S.D. and F43 to M.T.W.) and the Vienna Science and Technology Fund (WWTF; LS09-044 to S.D.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01346-14.
REFERENCES
- 1.Meyer S, Temme C, Wahle E. 2004. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 2.Parker R, Song H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 3.Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 5.Wahle E, Winkler GS. 2013. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta 1829:561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson A, Peltz SW. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem 65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 7.Braun JE, Huntzinger E, Fauser M, Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 9.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arribas-Layton M, Wu D, Lykke-Andersen J, Song H. 2013. Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim Biophys Acta 1829:580–589. doi: 10.1016/j.bbagrm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol Cell 32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aharon T, Schneider RJ. 1993. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol Cell Biol 13:1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird-Offringa IA, de Wit CL, Elfferich P, van der Eb AJ. 1990. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol 10:6132–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wisdom R, Lee W. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev 5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 15.Savant-Bhonsale S, Cleveland DW. 1992. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev 6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 16.Winstall E, Gamache M, Raymond V. 1995. Rapid mRNA degradation mediated by the c-fos 3′ AU-rich element and that mediated by the granulocyte-macrophage colony-stimulating factor 3′ AU-rich element occur through similar polysome-associated mechanisms. Mol Cell Biol 15:3796–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiavi SC, Wellington CL, Shyu AB, Chen CY, Greenberg ME, Belasco JG. 1994. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J Biol Chem 269:3441–3448. [PubMed] [Google Scholar]
- 18.Vindry C, Lauwers A, Hutin D, Soin R, Wauquier C, Kruys V, Gueydan C. 2012. dTIS11 protein-dependent polysomal deadenylation is the key step in AU-rich element-mediated mRNA decay in Drosophila cells. J Biol Chem 287:35527–35538. doi: 10.1074/jbc.M112.356188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popp MW-L, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graille M, Séraphin B. 2012. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol 13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 21.Herrick D, Parker R, Jacobson A. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 10:2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peltz SW, Donahue JL, Jacobson A. 1992. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 12:5778–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangus DA, Jacobson A. 1999. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods 17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 24.Hsu CL, Stevens A. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13:4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz DC, Parker R. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 19:5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz DC, Parker R. 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20:7933–7942. doi: 10.1128/MCB.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez CV, Vilela C, Berthelot K, McCarthy JEG. 2002. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J Mol Biol 318:951–962. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- 28.Coller J, Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nissan T, Rajyaguru P, She M, Song H, Parker R. 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huntzinger E, Kuzuoğlu-Öztürk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. 2013. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 41:978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukaya T, Tomari Y. 2012. MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol Cell 48:825–836. doi: 10.1016/j.molcel.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 32.Wickens M, Bernstein DS, Kimble J, Parker R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 18:150–157. doi: 10.1016/S0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 33.Igreja C, Izaurralde E. 2011. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev 25:1955–1967. doi: 10.1101/gad.17136311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. 2014. Structural basis for the Nanos-mediated recruitment of the CCR4-NOT complex and translational repression. Genes Dev 28:888–901. doi: 10.1101/gad.237289.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blewett NH, Goldstrohm AC. 2012. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol 32:4181–4194. doi: 10.1128/MCB.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 37.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489–1501. doi: 10.1017/S1355838202021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eystathioy T, Chan EKL, Mahler M, Luft LM, Fritzler ML, Fritzler MJ. 2003. A panel of monoclonal antibodies to cytoplasmic GW bodies and the mRNA binding protein GW182. Hybrid Hybridomics 22:79–86. doi: 10.1089/153685903321947996. [DOI] [PubMed] [Google Scholar]
- 40.Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cougot N, Babajko S, Séraphin B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eystathioy T, Jakymiw A, Chan EKL, Séraphin B, Cougot N, Fritzler MJ. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu W, Petzold C, Coller J, Baker KE. 2010. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol 17:244–247. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barišić-Jäger E, Krêcioch I, Hosiner S, Antic S, Dorner S. 2013. HPat a decapping activator interacting with the miRNA effector complex. PLoS One 8:e71860. doi: 10.1371/journal.pone.0071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mito Y, Henikoff JG, Henikoff S. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 47.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. 2003. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A 100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS. 1991. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67:59–77. doi: 10.1016/0092-8674(91)90572-G. [DOI] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 51.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermüller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt L, Teupser D, Hackermüller J, Stadler PF. 2014. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing, and fusion detection. Genome Biol 15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfinger MT, Fallmann J, Eggenhofer F, Amman F. 2015. ViennaNGS: a toolbox for building efficient next-generation sequencing analysis pipelines. F1000Res 4:50. doi: 10.12688/f1000research.6157.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thermann R, Hentze MW. 2007. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 57.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas A, Lee P-J, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA. 2012. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS One 7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun JE, Truffault V, Boland A, Huntzinger E, Chang C-T, Haas G, Weichenrieder O, Coles M, Izaurralde E. 2012. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat Struct Mol Biol 19:1324–1331. doi: 10.1038/nsmb.2413. [DOI] [PubMed] [Google Scholar]
- 61.Bonisch C, Temme C, Moritz B, Wahle E. 2007. Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J Biol Chem 282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 62.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lauwers A, Twyffels L, Soin R, Wauquier C, Kruys V, Gueydan C. 2009. Post-transcriptional regulation of genes encoding anti-microbial peptides in Drosophila. J Biol Chem 284:8973–8983. doi: 10.1074/jbc.M806778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muhlrad D, Decker CJ, Parker R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev 8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 65.Gatfield D, Izaurralde E. 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 66.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang S-F, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. 2007. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malone C, Brennecke J, Czech B, Aravin A, Hannon GJ. 2012. Preparation of small RNA libraries for high-throughput sequencing. Cold Spring Harb Protoc 2012:1067–1077. doi: 10.1101/pdb.prot071431. [DOI] [PubMed] [Google Scholar]
- 68.Fromont-Racine M, Bertrand E, Pictet R, Grange T. 1993. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res 21:1683–1684. doi: 10.1093/nar/21.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maruyama K, Sugano S. 1994. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 70.Volloch V, Schweitzer B, Rits S. 1994. Ligation-mediated amplification of RNA from murine erythroid cells reveals a novel class of beta globin mRNA with an extended 5′-untranslated region. Nucleic Acids Res 22:2507–2511. doi: 10.1093/nar/22.13.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A 99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silber R, Malathi VG, Hurwitz J. 1972. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci U S A 69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shinshi H, Miwa M, Sugimura T. 1976. Enzyme cleaving the 5′-terminal methylated blocked structure of messenger RNA. FEBS Lett 65:254–257. doi: 10.1016/0014-5793(76)80492-9. [DOI] [PubMed] [Google Scholar]
- 75.Schneider MD, Bains AK, Rajendra TK, Dominski Z, Matera AG, Simmonds AJ. 2010. Functional characterization of the Drosophila MRP (mitochondrial RNA processing) RNA gene. RNA 16:2120–2130. doi: 10.1261/rna.2227710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong X, Hammell M, Ambros V, Cohen SM. 2009. Immunopurification of Ago1 miRNPs selects for a distinct class of microRNA targets. Proc Natl Acad Sci U S A 106:15085–15090. doi: 10.1073/pnas.0908149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sweet T, Kovalak C, Coller J. 2012. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol 10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chu C-Y, Rana TM. 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Decker CJ, Teixeira D, Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. 2007. Microtubule disruption stimulates P-body formation. RNA 13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. 2009. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amrani N, Ghosh S, Mangus DA, Jacobson A. 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gallie DR. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 86.Tarun SZ, Sachs AB. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 87.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. 2010. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y, Boland A, Kuzuoğlu-Öztürk D, Bawankar P, Loh B, Chang C-T, Weichenrieder O, Izaurralde E. 2014. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 89.Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. 2014. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 90.Jackson RJ, Hellen CUT, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu L, Fan J, Belasco JG. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 93.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu S-H, Ghoshal K, Villén J, Bartel DP. 2014. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. 2009. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 98.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Decker CJ, Parker R. 2012. P bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.