Abstract
To further our understanding of the RNAi machinery within the human nucleus, we analyzed the chromatin and RNA binding of Argonaute 2 (AGO2) within human cancer cell lines. Our data indicated that AGO2 binds directly to nascent tRNA and 5S rRNA, and to the genomic loci from which these RNAs are transcribed, in a small RNA- and DICER-independent manner. AGO2 chromatin binding was not observed at non-TFIIIC-dependent RNA polymerase III (Pol III) genes or at extra-TFIIIC (ETC) sites, indicating that the interaction is specific for TFIIIC-dependent Pol III genes. A genome-wide analysis indicated that loss of AGO2 caused a global increase in mRNA expression level among genes that flank AGO2-bound tRNA genes. This effect was shown to be distinct from that of the disruption of DICER, DROSHA, or CTCF. We propose that AGO2 binding to tRNA genes has a novel and important regulatory role in human cells.
INTRODUCTION
The RNA interference (RNAi) machinery has several important functions that are conserved from fission yeast to humans. In the cytoplasm of eukaryotic cells, Argonaute proteins promote degradation and/or translational repression of specific mRNAs through tethering by a complementary small RNA (1). The nuclear functions of the RNAi machinery appear to be diverse across species and include roles in promoting heterochromatin formation, regulating alternative splicing, and promoting insulator function (2–5). The ability of both Argonaute 1 (AGO1) and AGO2 to alter splicing in human cells depends upon small RNAs derived from exonic sequence, which tether Argonaute proteins to nascent transcripts (3, 6). Tethering of each Argonaute protein was shown to be capable of recruiting a histone lysine methyltransferase, which altered the RNA polymerase II (Pol II) elongation rate and thus facilitated alternative splicing. This interaction between Argonaute proteins and the transcriptional apparatus is conserved in Drosophila melanogaster (7).
Despite the similarities between Argonaute proteins, AGO2 is the only human Argonaute protein believed to have the endonuclease activity (Slicer) due to a unique amino-terminal region (8–10), and Ago2 but not Ago1 gene deletions are embryonic lethal in mice (11). AGO2 was shown to promote transcriptional gene silencing in a microRNA (miRNA)-dependent manner (12) and to affect nucleosome occupancy at certain transcription start sites through interaction with the SWI/SNF complex (13). Furthermore, AGO1 has been shown to interact with actively transcribed genes and enhancer regions (14, 15). Recent work has identified expanded binding capabilities of AGO2, which include binding to longer RNAs such as pre-miRNAs (65 to 75 nucleotides [nt]) and full-length tRNAs (∼75 nt) (16–20). To wit, AGO2 was recently shown to functionally interact with DICER in human nuclei but was unable to load duplex small interfering RNA (siRNA), indicating that nuclear RNAi processes may proceed through a mechanism distinct from that observed in the cytoplasm (21). These findings indicate that AGO2 may have roles beyond classical RNAi and that AGO1 and AGO2 may have divergent roles in the mammalian nucleus.
tRNAs are highly structured and highly modified RNA polymerase III (Pol III) transcripts that comprise roughly 15% of all transcripts in the cell (22). tRNA genes use a type 2 Pol III promoter which contains gene internal binding sites for the Pol III transcription factor, TFIIIC, and upstream contacts for the Pol III transcription factor, TFIIIB. The 5S rRNA genes use a type 1 Pol III promoter, which also requires TFIIIC and TFIIIB binding and additionally requires TFIIIA. Other Pol III genes, such as U6 and RNase P, utilize a type 3 Pol III promoter which, unlike type 1 and 2 promoters, is external and utilizes TFIIIB but not TFIIIC or TFIIIA (23, 24). In humans, there are 631 tRNA gene sequences (522 tRNA genes and 109 tRNA pseudogenes) that are dispersed across all chromosomes and are often found in clusters. Interestingly, only about half of these are actively transcribed, as evidenced by binding of the essential Pol III transcription factors: TFIIIC, TFIIIB, and Pol III (24, 25). Furthermore, the expression levels of individual tRNA genes vary among cell types and cellular conditions (24, 26–28).
Over the past 25 years it has come to light that tRNAs, as well as their gene sequences, have roles beyond translation (22). In yeast, mice, and humans, TFIIIC-bound tRNA gene sequences have been shown to function as both enhancer-blocking and barrier function chromatin insulators (29–31). Interestingly, the RNAi machinery has also been linked to chromatin insulator function in D. melanogaster, where it has been shown that AGO2 binds to insulator proteins and is critical for Fab-8 insulator function (4). In addition to roles in chromatin insulation, studies in yeast have shown that actively transcribed tRNA genes can repress neighboring Pol II gene transcription in a mechanism termed tRNA gene-mediated (tgm) silencing (32–34). Despite extensive work in yeast, it remains unknown whether tgm silencing is conserved in metazoans.
In this study, we expanded on the role of AGO2 in the human nucleus. We show that AGO2 interacts with nascent tRNA and the genes from which they are transcribed. The combination of AGO2, TFIIIC, and Pol III at active tRNA genes promotes the suppression of flanking Pol II genes in a distance-dependent manner, which functions in addition to the role of TFIIIC as a genomic insulator. We conclude that the AGO2/TFIIIC/Pol III-mediated suppression of neighboring Pol II genes represents a novel regulatory mechanism that likely contributes to cell type specificity.
MATERIALS AND METHODS
Cell culture and transfections.
All experiments were carried out in K562 and HEK293T cells. Culturing and transfection conditions were carried out essentially as previously described (5). The lentiviral shRNA constructs were acquired from GE Dharmacon (GIPZ lentiviral RHS4531-EG271611).
Immunoprecipitation. (i) ChIP-seq.
Chromatin from human K562 cells was harvested and sonicated, and chromatin immunoprecipitation (ChIP) was performed as previously described (90), with the exception that the cross-linking was performed with 0.5% formaldehyde. Immunoprecipitations were performed with two separate AGO2 antibodies (Abcam, catalog no. 57113, and Millipore, catalog no. 04-642). An IgG ChIP was carried out as a negative control (Abcam, ab37355). A 1% aliquot of chromatin was also sequenced without immunoprecipitation and used as the input control.
(ii) RNA immunoprecipitation (RIP)–RT-QPCR.
Cells were lysed in LB50 buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1% Triton X-100, 1 mM EDTA) supplemented with 1× Halt protease inhibitor cocktail (Thermo Scientific) and RNaseOUT (Invitrogen). The nuclei were pelleted at 5,000 × g and then resuspended in 5 ml of 1× phosphate-buffered saline (PBS). Formaldehyde was added to a final concentration of 1.2%, and the cells were rotated at room temperature for 15 min. Cross-linking was quenched by adding glycine to a final concentration of 0.125 M. Nuclei were pelleted, washed once with 1× PBS, and resuspended in radioimmunoprecipitation assay buffer. Nuclei were then sonicated in a Bioruptor 300 (Diagenode) for 60 cycles of 30 s on and 30 s off. Sonicated product was spun at 17,000 × g for 15 min to remove the insoluble fraction, and the supernatant was saved. The immunoprecipitation was performed as previously described (90), with the following exceptions. After the immunoprecipitation and final wash, beads were resuspended in TE elution buffer (1 × Tris-EDTA, 1% sodium dodecyl sulfate, RNaseOUT [Invitrogen], 4 μg of proteinase K) and incubated at 45°C for 1 h. Supernatant was then harvested and resuspended in TRIzol. Reverse transcriptase quantitative PCR (RT-QPCR) was carried out as previously described (5).
RT-QPCR.
RNA was harvested from K562 cells at 48 h posttransfection using TRIzol (Life Technology), and the RT-QPCR was carried out as previously described (5).
RNA Pol III inhibition studies.
293T cells were treated with a 25 μM concentration (i.e., a 1:1,000 dilution of a 25 mM stock) of an RNA Pol III inhibitor (Millipore, catalog no. 557403 EMD) under normal growth conditions for 4 h. Control cells were simultaneously incubated with an equal volume (5 μl) of dimethyl sulfoxide (DMSO). After the incubation, ChIP, RT-QPCR, and Western blotting were performed as described previously.
High-throughput sequencing.
ChIP-seq was performed on an Illumina Hi-seq 2000 at the NIDDK sequencing center and the Stem Cell Institute at UAB.
Data analysis. (i) Bioinformatics.
High-throughput sequencing FASTQ data were separately aligned to both hg18 and hg19 human genomes using Bowtie2 with default parameters (36). Both hg18 and hg19 aligned ChIP-seq and PAR-CLIP-seq data were analyzed to ensure similarity across the two genome annotations. Only hg18 aligned data are shown. Bowtie2 default parameters map each individual read once to the best possible mapping location. In the instance of a tie between two or more mapping locations, the read will be randomly assigned to one of the tied locations. The Bowtie2 default parameters in regard to mapping to repetitive loci are restricted by options ‘-R’ and ‘-D,’ which are set at 15 and 2, respectively. The -R option is an upper limit on the number of seed extensions that can fail in a row before Bowtie2 stops searching for better alignments and increasing this number increases the likelihood that Bowtie2 will report a correct alignment for a read that aligns many places. The -D option sets the maximum number of times Bowtie2 will attempt to align a read with repetitive seeds, and increasing this number increases the likelihood that Bowtie2 will report a correct alignment for a read that aligns many places. To ensure proper reporting of alignments, all AGO2 ChIP-seq experiments were aligned with the default settings as well as with 50 and 50 for -R and -D, respectively. These parameters did not affect the overall results and reads mapped with the default settings are shown. The Bowtie2 output SAM file was converted into the BAM format using the SAMtools view option (37). For all ChIP-seq data, PCR duplicates were removed, meaning that all identical reads were collapsed to one single read. This is a common and established processing step for ChIP-seq data (38–40). For all PAR-CLIP-seq data, PIPE-CLIP analysis was performed to identify reliable T→C mutations in BAM files as previously described (41). RNA-seq was aligned using Tophat2 under default parameters, with an ∼99% read alignment (42). We align to a custom hg18 build that contains a single copy of the 43-kb human rRNA gene, as previously described (43). For all RNA-seq, reads were not collapsed unless specifically stated. For single end reads, the total FASTQ data set was first filtered using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to return reads with a Qual score of ≥20 at 80% of the positions. RNA-seq data were processed by the Cufflinks pipeline (42, 44). BAM data were converted to BED format using the BEDtools bamToBed option (45). Randomly chosen loci were generated by BEDtools shuffleBed. Gene ontology analyses were performed using the online tool DAVID (46).
(ii) Visualization.
For meta-gene analyses, the coverage of each nucleotide across the tRNA genes was determined using coverageBed against a BED file of 631 hg18 tRNAscan-SE tRNA gene annotations (47–51) and the “–d” option of the BEDTools Suite (45). In each case, the coverage was aligned at the +1 position and scaled according to reads per million mapped reads (RPM). Average coverage across all tRNA genes was generated by the BEDTools groupBy tool. The groupBy output was visualized using a line graph in Microsoft Excel. Binding profile heat maps were generated essentially as previously described, except without binning the genes (52). Briefly, the coverage of each nucleotide across the tRNA genes was determined using coverageBed against a BED file of the tRNAscan-SE tRNA gene annotations and the “–d” option of the BEDTools. In each case, the coverage was aligned at the +1 position and scaled according to the RPM. Outputs were converted to data matrices using a custom script and then visualized using conditional formatting options in Microsoft Excel (2010 version). All heat maps are internally scaled from 0 to the 95th percentile, unless otherwise noted. Line graphs, scatter plots, and bar plots in Fig. 1 to 6, with the exception of Fig. 6G and H were made using Microsoft Excel. The box-and-whisker and bar plots in Fig. 6 and 7 were generated using the ggplot2 package (version 1.0.0) with color palettes from the RColorBrewer package (version 1.0-5) in the R statistical environment (53–55). In all box plots, the black line in the middle of the box represents the median, the top and bottom of the box represent the 75th and 25th percentiles, respectively, and the upper and lower whiskers represent the 95th and 5th percentiles, respectively.
FIG 1.
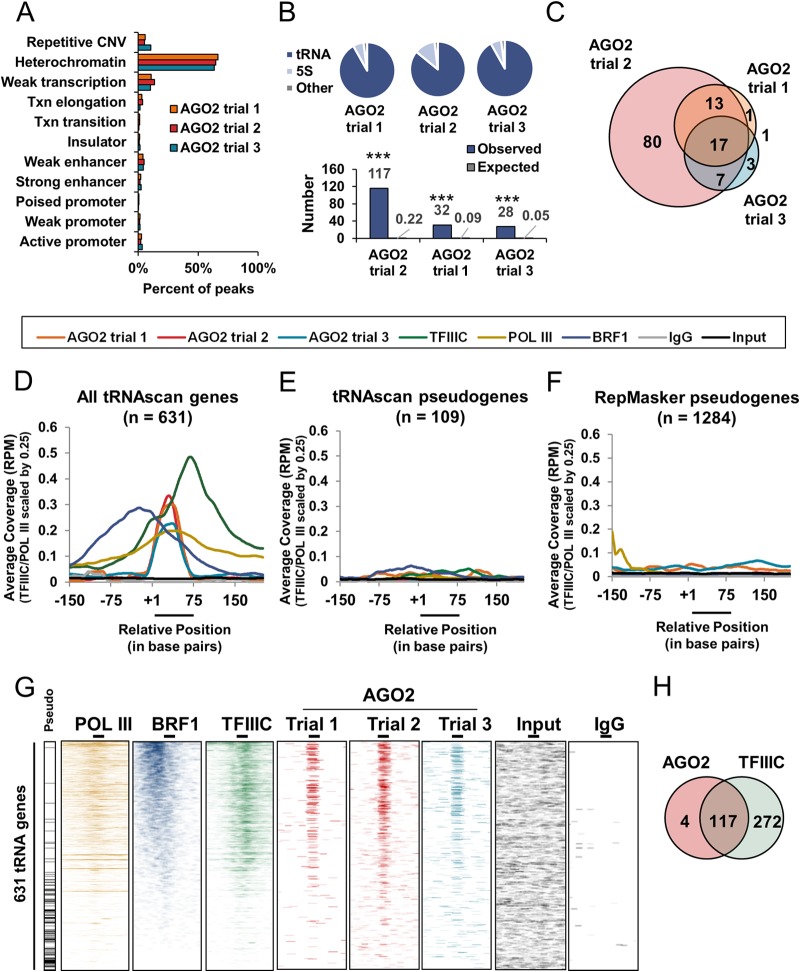
Human Argonaute 2 binds to actively transcribed tRNA genes. (A) Bar graph representing the percentage of AGO2 peaks intersecting with K562 annotated Broad ChromHMM regions for three independent trials of AGO2 ChIP-seq (trials 1 and 2 [Millipore, catalog no. 04-642]; trial 3 [Abcam, catalog no. ab57113) (see Materials and Methods). (B, top) Pie charts of AGO2 peak binding normalized to total base pair coverage for each annotated region. All annotated Broad ChromHMM regions are combined into “Other” category. All “tRNA” and “5S” regions that intersected with Broad ChromHMM regions were removed from the “Other” category. (B, bottom) Bar graph showing observed number of AGO2 peaks intersecting with tRNA genes from three independent trials of AGO2 ChIP-seq compared to the average number of randomly chosen peaks intersecting with tRNA genes across 1,000 randomized trials. The observed number of peaks and average number of peaks across 1,000 randomized trials is indicated by gray number next to each bar. (C) Venn diagram depicting the overlapping tRNA peaks across the three independent trials of AGO2 ChIP-seq. (D to F) Meta-analyses showing the binding of Pol III (RPC32; scaled by 0.25), BRF1, TFIIIC (110-kDa subunit; scaled by 0.25), AGO2 (trials 1 and 2 [Millipore, catalog no. 04-642]; trial 3 [Abcam, catalog no. ab57113]), IgG, and Input across 631 tRNAscan-SE tRNA gene sequences (D), 109 tRNAscan-SE annotated pseudogenes (E), or 1,284 Repeat Masker annotated pseudogenes (F) (see also Materials and Methods). The y axis indicates the coverage scaled according to tRNA reads per total million mapped reads (RPM). The x axis indicates the position in base pairs, with position +1 indicating the first nucleotide of the tRNA. (G) Binding profile heat maps of indicated protein binding across 631 tRNAscan-SE annotated tRNA gene sequences (y axis). The location of tRNAscan-SE annotated pseudogenes are indicated by horizontal black lines on the panel labeled “Pseudo”. Thick lines above heat maps indicate the average tRNA gene size, with the left end representing the +1 position and the right end representing the average 3′ end site (3′). Increasing intensity of color represents increasing read coverage. The coverage density for each heat map was internally scaled according to the 1st to the 99th percentiles. (H) Venn diagram depicting the overlap between AGO2 tRNA peaks (combination of all three trials) and TFIIIC tRNA peaks.
FIG 6.
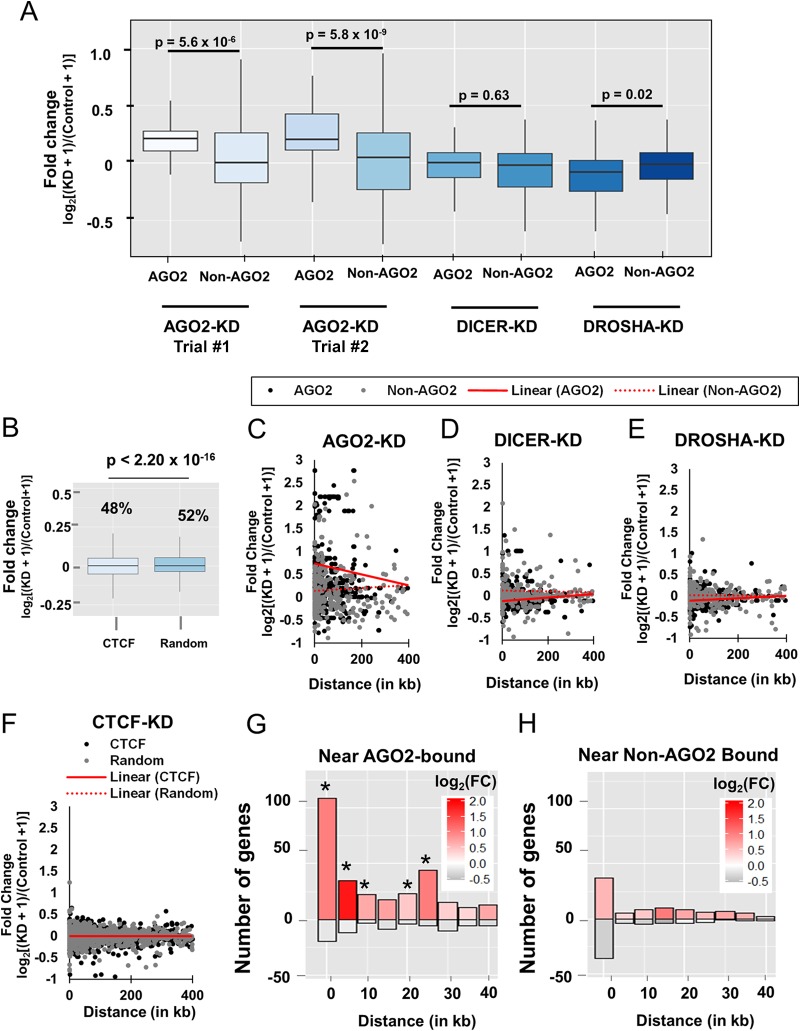
AGO2-KD, but not DICER-KD or DROSHA-KD, caused increased expression of Pol II genes near AGO2-bound tRNA genes. (A) Box-and-whisker plot of the indicated fold change (knockdown/control) in FPKM of the closest gene to AGO2-bound tRNA genes (AGO2) compared to non-AGO2-bound tRNA genes (Non-AGO2). (B) Box-and-whisker plot of the fold change (CTCF knockdown/control) in FPKM of the nearest upstream and downstream gene from CTCF-bound sites (CTCF) and randomly chosen loci (Random). (C to E) Scatter plot of the fold change of the nearest upstream and downstream gene from an AGO2-bound tRNA gene or a non-AGO2-bound tRNA gene for AGO2-KD RNA-seq (C), DICER-KD RNA-seq (D), and DROSHA-KD RNA-seq (E). Genes that were the nearest neighbor to both an AGO2-bound tRNA gene and a non-AGO2 tRNA gene were treated as the nearest gene to an AGO2-bound tRNA gene (AGO2, n = 684) and excluded from the non-AGO2-bound tRNA gene set (non-AGO2, n = 349). The distance is indicated in kilobases, and the fold change is calculated as the log2[(KD + 1)/(control + 1)]. Linear regressions for both AGO2 and non-AGO2 were calculated using the linear regression function in Microsoft Excel. The slope of the linear regression of AGO2 is −0.0012 for AGO2-KD (C), 0.0004 for DICER-KD (D), and 0.0003 for DROSHA-KD (E). The y intercepts of the linear regression of AGO2 are 0.738 for AGO2-KD (C), −0.112 for DICER-KD (D), and −0.114 for DROSHA-KD (E). The slope of the linear regression of non-AGO2 is 0.0003 for AGO2-KD (C), −0.0001 for DICER-KD (D) and −5 × 10−5 for DROSHA-KD (E). The y intercepts of the linear regression of non-AGO2 are 0.141 for AGO2-KD (C), 0.120 for DICER-KD (D), and 0.0154 for DROSHA-KD (E). Linear regressions were calculated based on the entire data set. Regression lines were tested for significant interaction by means of ANCOVA testing run in R statistical environment (see Materials and Methods). The P values for differences in the slopes of AGO2-KD, DICER-KD, and DROSHA-KD are 2.35 × 10−5, 0.046, and 0.108, respectively. The P values for differences in y intercepts of AGO2-KD, DICER-KD, and DROSHA-KD are 1.73 × 10−12, 6.57 × 10−5, and 8.3 × 10−3, respectively. (F) Scatter plot of the fold change of the nearest upstream and downstream gene from a CTCF site or a random site (CTCF, n = 191,129 genes; Random, n = 191,129 genes) versus the distance from the tRNA gene. A representative set of 2,000 randomly chosen genes from each set (CTCF or Random) is plotted. Randomly chosen sites that overlapped CTCF sites were excluded from both data sets and genes that were the nearest neighbor to both a CTCF site and a random site were treated as the nearest gene to a CTCF site (CTCF, n = 1,999) and excluded from the random site set (Random, n = 1,965). The distance is indicated in kilobases, and the fold change was calculated as the log2[(KD + 1)/(Control + 1)]. The slope of the linear regression of CTCF is −3 × 10−6, and the y intercept is −3.5 × 10−3. The slope of the linear regression of random sites is −1.0 × 10−7, and the y intercept is 9 × 10−4. Linear regressions were calculated based on the entire data set. Regression lines were tested for significant interaction as described in panels C to E. The P value for the difference in slopes was 0.453. The P value for the difference in y intercepts was 0.548. (G and H) Bar plot indicating the number of genes that are upregulated (y axis positive numbers) and downregulated (y axis negative numbers) in 5-kb bins ranging from 0 to 40 kb distal from AGO2-bound tRNA genes (G) and non-AGO2-bound tRNA genes (H). Significance for the number of genes showing upregulation in each bin was tested by chi-square test (*, P < 0.05). The intensity of the log2 of the average fold change in expression (FC) is color-coded according to the scale in the upper right of each plot.
FIG 7.
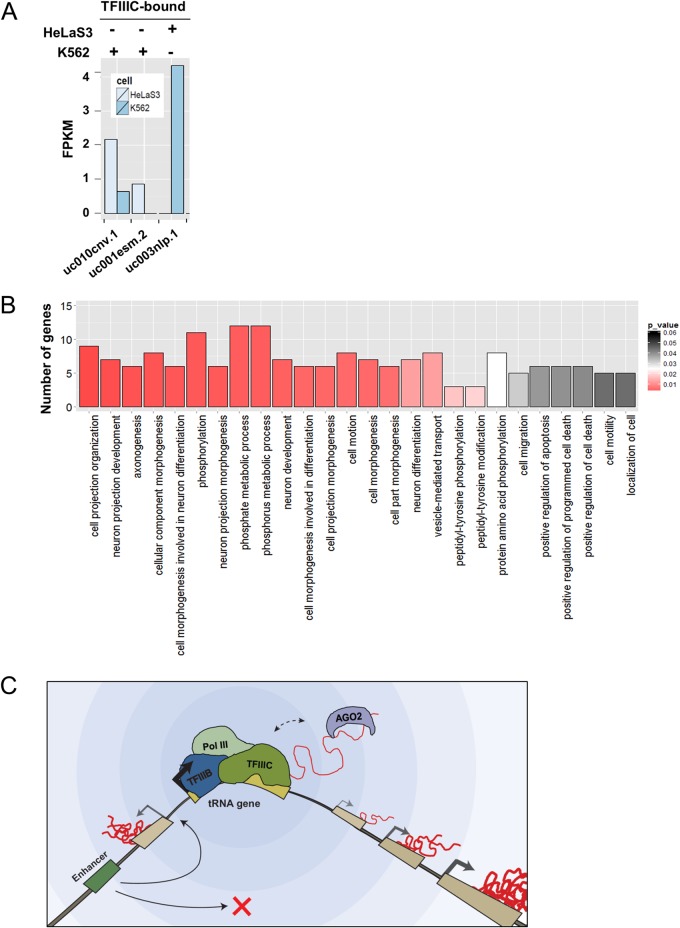
Cell-type-specific tRNA gene expression correlates with Pol II gene suppression. (A) Bar plot showing the FPKM of genes in K562 and HeLa S3 cells that are within 500 bp of a tRNA gene that is differentially bound by TFIIIC in K562 and HeLa S3. The two genes bound by TFIIIC in K562, but not in HeLa S3, are shown on the left, and the one gene bound by TFIIIC in HeLa S3, but not in K562, is shown on the right, as labeled at the top of the plot. (B) Bar graph summarizing results of a gene ontology (GO) analysis generated by DAVID (60, 61) for genes within 500 bp of any tRNA gene. The x axis shows all functional classifications with a P value of <0.05, ordered from left to right by increasing P value. The y axis shows number of genes represented within each functional classification. (C) Active tRNA genes can regulate Pol II gene expression in two distinct ways. (i) The binding of TFIIIC to a tRNA gene can create both an enhancer block and a barrier function insulator (49), and (ii) AGO2 binding to nascent tRNA facilitates the suppression of Pol II genes in a distance-dependent manner. This is similar to the tgm-silencing phenomena described in S. cerevisiae (29, 63). In the illustration, the enhancer-blocking function is shown upstream of the tRNA gene, and tgm silencing is shown downstream for clarity. In actuality, these processes occur independently of the orientation of the tRNA gene. Both processes can occur upstream, downstream, in both directions, or not at all for any given tRNA gene.
(iii) Linear regressions.
Linear regressions for AGO2, Non-AGO2, CTCF, and Random in Fig. 6C to F were calculated using the built-in linear regression function in Microsoft Excel (2013 version). The regression lines shown in Fig. 6C to F were tested for significant interaction by means of analysis of covariance (ANCOVA) testing run in R statistical environment. An interaction model was generated by assigning type (AGO2 or Non-AGO2) to the categorical variable, distance to the dependent variable, and fold change to the y response variable. This model was tested for differences in slopes and y intercepts by using the built-in lm() function in R.
(iv) Data acquisition.
All acquired data were taken from the Gene Expression Omnibus (56). ChIP-seq data for TFIIIC, Pol III, and BRF1 were sample sets GSM935343, GSM509047, and GSM935595, respectively. Small RNA immunoprecipitation-sequencing (sRIP-seq) data for endogenous AGO2 and IgG were data sets SRR529100 and SRR529099, respectively, from a previously published manuscript (3). Human PAR-CLIP-seq data were taken from previously published manuscripts (57–59). Specifically, wild-type (WT) AGO2 from Fig. 3 was a concatenation of data sets SRR189784 and SRR189785. The HuR data were a concatenation of data sets SRR189777 and SRR189778. The noDice PAR-CLIP-seq was data set SRR1241607. The small RNA-seq data sets were acquired from the GSE24565 series on GEO. ChIP-seq peaks for CTCF were obtained from a previously published manuscript and were sample set GSM749668 (60). RNA-seq data for CTCF knockdown and control were from a previous publication and were sample sets GSM1081540 and GSM1081538, respectively (61). RNA-seq data for DICER knockdown, DROSHA knockdown, and control were from a previous publication and were sample sets GSM1550537, GSM1550539, and GSM1550536, respectively (62). TFIIIC ChIP-seq data for K562 and HeLa S3 analyzed for Fig. 7A were sample sets GSM935343 and GSM935342, respectively. RNA-seq for K562 and HeLa S3 cells analyzed in Fig. 7A were sample sets GSM958731 and GSM958739, respectively. tRNAscan-SE tRNA gene annotations, UCSC gene annotations, RepMasker annotations, Broad ChromHMM annotations, and ENSEMBL gene annotations were all downloaded through Galaxy from the UCSC table browser (63, 64). The 631 hg18 tRNAscan-SE tRNA gene annotations include both functional tRNA genes and tRNA pseudogenes (47–51) The tRNAscan-SE pseudogene data set (n = 109) is a set of tRNA sequences included within the tRNAscan-SE tRNA gene set that are annotated as pseudogenes. The RepMasker pseudogene data set (n = 1,284) are tRNA repeats that are annotated by RepeatMasker (http://repeatmasker.org) but are not annotated as putative tRNA genes by tRNAscan-SE (50, 65).
FIG 3.
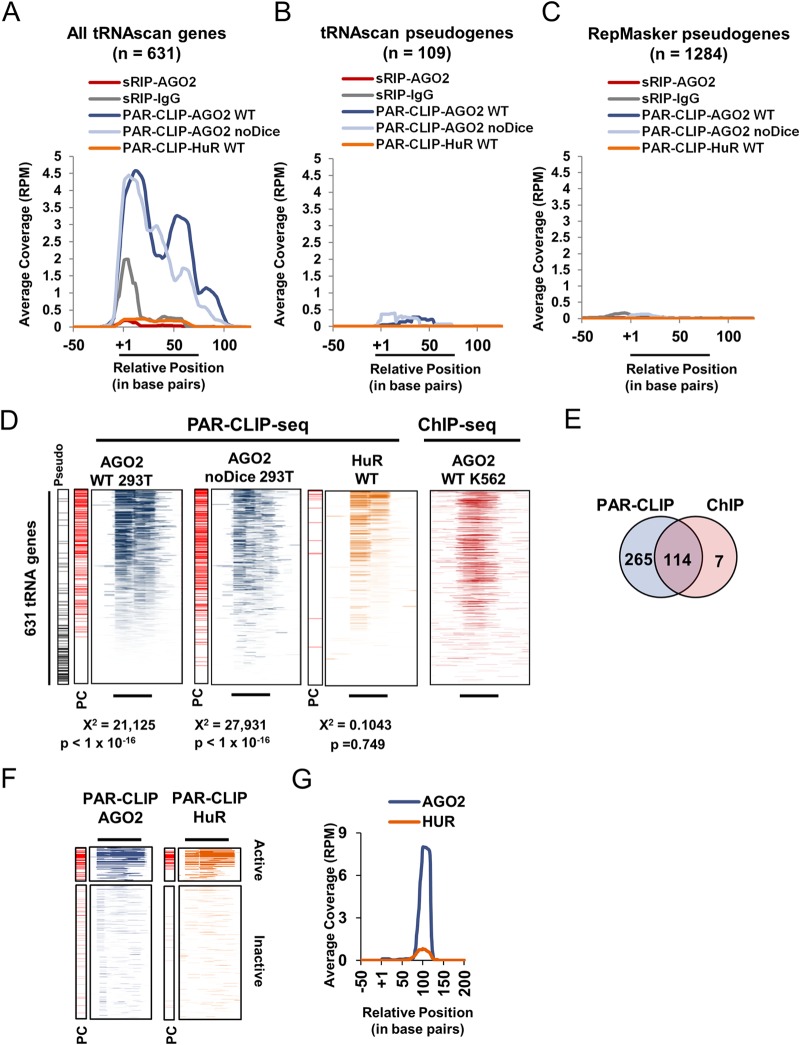
AGO2 binding is small RNA independent and is mediated through longer tRNA transcripts. (A to C) Meta-analyses showing the binding of AGO2 sRIP-seq, IgG sRIP-seq, and AGO2 PAR-CLIP-seq in WT 293T cells, AGO2 PAR-CLIP-seq in noDice 293T cells, and HuR PAR-CLIP-seq in WT 293T cells across 631 tRNAscan-SE tRNA gene sequences (A), 109 tRNAscan-SE annotated pseudogenes (B), or 1,284 Repeat Masker annotated pseudogenes (C) (see Materials and Methods). (D) Binding profile heat maps of indicated protein binding across 631 individual tRNA genes. Conditional formatting was scaled according to a 0-RPM minimum to 75-RPM maximum for PAR-CLIP-seq and according to the 1st to the 99th percentiles for ChIP-seq. The PC panel on the left hand side of each heat map indicates tRNA genes exhibiting a PIPE-CLIP-identified T-to-C mutation (in red). The location of tRNAscan-SE annotated pseudogenes are indicated by horizontal black lines on the panel labeled “Pseudo”. Significance was tested using the chi-square test comparing the observed number of tRNAs exhibiting a PIPE-CLIP verified mutation with the expected number of PIPE-CLIP verified mutations based on 1,000 randomized trials. (E) Venn diagram showing intersection between tRNAs exhibiting AGO2 PAR-CLIP signal (average coverage per base pair of tRNA gene greater than or equal to 1 RPM). (F) Binding profile heat maps of indicated protein binding across active and inactive 5S rRNA gene sequences (y axis). Thick lines above heat maps indicate average 5S rRNA gene size with the left end representing the +1 position and the right end representing the average 3′ end site (3′). Increasing intensity of color represents increasing read coverage. The coverage density for each heat map was internally scaled according to the 1st to the 99th percentiles. PC panel on the left hand side of each heat map indicates tRNA genes exhibiting a PIPE-CLIP-identified T-to-C mutation (in red). (G) Meta-analysis showing the binding of AGO2 and HuR by PAR-CLIP-seq across 60 active 5S rRNA genes (see Materials and Methods). The y axis indicates the coverage scaled according to 5S rRNA reads per total million mapped reads (RPM). The x axis indicates the position in base pairs, with position +1 indicating the first nucleotide of the 5S rRNA.
Accession number.
All ChIP-seq and control data sets (see Fig. 1), as well as RNA-seq and additional metadata (see Fig. 5 and 6), are available at the Gene Expression Omnibus under accession number GSE68813.
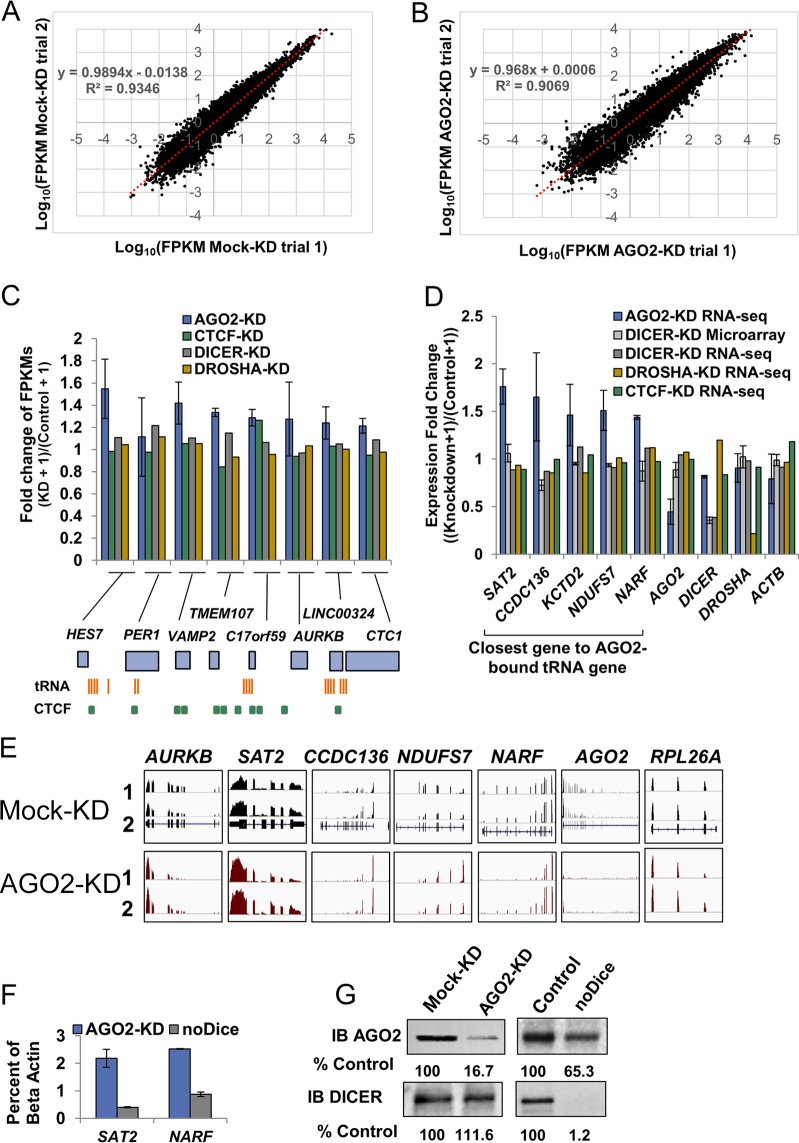
FIG 5.
AGO2 depletion leads to increased expression of genes in a tRNA gene insulator region on chromosome 17. (A and B) Scatter plot comparing the log10 FPKM values for trial 1 (x axis) versus trial 2 (y axis) for Mock-KD RNA-seq (A) or AGO2-KD RNA-seq (B). Linear regressions were calculated for each plot and are represented by a red dotted line. The equation and R2 value for each linear regression is shown on the plot. (C) Average fold change (knockdown/control) of FPKMs of AGO2-KD (averages and standard deviations of two biological replicates), DICER-KD, DROSHA-KD, and CTCF-KD RNA-seq for eight genes across select region of chromosome 17. A diagram of the locations of tRNA genes, CTCF binding sites, and RefSeq genes within this region is depicted below the graph. (D) Bar plot of the five genes with the highest fold change (AGO2 knockdown/control), while having a standard deviation of <0.25 for the fold change across two trials of AGO2 knockdown and an annotation across all experiments shown (left, “Closest gene to AGO2-bound tRNA gene”). Averages and standard deviations of the fold change across two trials of AGO2-KD RNA-seq and DICER knockdown microarray, as well as the fold change of one trial of DICER knockdown RNA-seq, DROSHA knockdown RNA-seq, and CTCF-KD RNA-seq, are shown. The fold change in FPKM of AGO2, DICER, DROSHA, and ACTB (control gene) across all experiments are shown (right). (E) Screenshots from IGV of trials 1 and 2 of the Mock and AGO2-KD RNA-seq experiments. The gene name is indicated above each box, and each gene is scaled identically for all four tracks. (F) Fold change (AGO2-KD/Mock-KD or noDice/Control) in mRNA levels measured by RT-QPCR for SAT2 and NARF presented as a percentage of ACTB. (G) Western blot for AGO2 and DICER in Mock-KD, AGO2-KD, noDice, and control cells. The percentage of the control is shown below blot images and was calculated by using ImageJ and normalizing to the total protein levels, as measured by Ponceau S staining.
RESULTS
Human Argonaute 2 binds to actively transcribed tRNA genes.
To investigate the chromatin binding preferences of AGO2, we performed AGO2 ChIP-seq in K562 cells using two different, well-established monoclonal antibodies (see Materials and Methods) (66–70). Peaks were called using MACS for each trial (see Materials and Methods) (71). Replicates 1 and 2 for monoclonal antibody 1 (trials 1 and 2) had 1,236 and 7,364 peaks, respectively. Replicate 1 for monoclonal antibody 2 (trial 3) had 991 peaks. The difference in peak number between trials 1, 2, and 3 reflects the difference in total numbers of mapped reads per trial, which were 2,541,318, 12,066,168, and 4,398,595, respectively. To understand where AGO2 binds in the genome, the peaks from each trial were intersected with Broad ChromHMM annotations for K562 cells. All three trials showed similar binding across each annotation and were highly enriched for heterochromatin (Fig. 1A). Upon closer inspection of AGO2 peaks, we noticed that many fell within annotated tRNA genes and 5S rRNA genes. These genomic regions were not separately classified within the Broad ChromHMM annotation but instead were dispersed throughout various regions. To quantify this observation separately, AGO2 peak enrichment for tRNA genes, 5S rRNA genes, and all other Broad ChromHMM annotations was calculated by comparing the base pair coverage for each AGO2 enriched region to the total base pair coverage of that annotation. AGO2 peaks were found in a high proportion of tRNA genes and 5S rRNA genes compared to any other Broad ChromHMM annotation (Other) (Fig. 1B, top). Furthermore, when the observed number of AGO2 peaks found within tRNA genes was compared to the number expected at random (average of 1,000 random trials), the observed peaks were highly enriched in all three trials (P < 0.0001, Fig. 1B, bottom). Considering that trial 1 and trial 3 had lower numbers of mapped reads than trial 2, we expected that trial 2 would have peaks at tRNA genes that were not called in trial 1 and trial 3 due to the overall difference in coverage. Accordingly, intersection of peaks across all trials showed trial 2 had 80 peaks that did not overlap with peaks from trial 1 and trial 3 (Fig. 1C). However, for the peaks called in trial 1 and trial 3 (n = 32 and n = 28, respectively) there was high overlap with the peaks called for trial 2 (>85%) (Fig. 1C). Meta-analysis of AGO2 binding across all tRNAscan-SE annotated tRNA gene sequences (see Materials and Methods) revealed enrichment of AGO2 compared to negative controls in the three independent trials (Fig. 1D). We applied the same meta-analysis to ChIP-seq data sets for known Pol III factors (GEO accession numbers: BRF1, GSM935595; TFIIIC, GSM935343; and Pol III, GSM509047) and found the expected binding profiles for these factors at tRNA genes, validating our analytical approach (Fig. 1D). TFIIIC (110-kDa subunit) was centered just downstream of the B-box, its well-established binding site. BRF1, a subunit of the TFIIIB complex, was bound just upstream of the +1 position (+1). Pol III binding was seen ∼100 bp upstream of the +1 position to ∼100 bp downstream of the average mature 3′ end. AGO2 binding peaked at the center of the tRNA genes. To understand whether this binding occurs exclusively at functional tRNA genes, we analyzed all ChIP experiments across tRNAscan-SE annotated pseudogenes and Repeat Masker annotated pseudogenes (see Materials and Methods) and found no enrichment for AGO2 or any of the Pol III factors (Fig. 1E and F) (72, 73).
In order to understand AGO2 binding on an individual tRNA gene basis, we generated binding profile heat maps for AGO2 in all three trials and found that AGO2 consistently bound to the same subset of tRNA genes (Fig. 1G). The binding profile heat maps indicated AGO2 binding occurring mostly at active tRNA genes, which are marked by Pol III, BRF1, and TFIIIC binding (Fig. 1G). Consistent with this model, >95% of AGO2 tRNA gene peaks overlapped with TFIIIC peaks (Fig. 1H). We also investigated whether AGO2 binding shows preference for certain tRNA isotypes, isoacceptors, or flanking sequences and found no correlation (data not shown).
AGO2 binds to tRNA genes and 5S rRNA genes but not to other Pol III and TFIIIC binding sites.
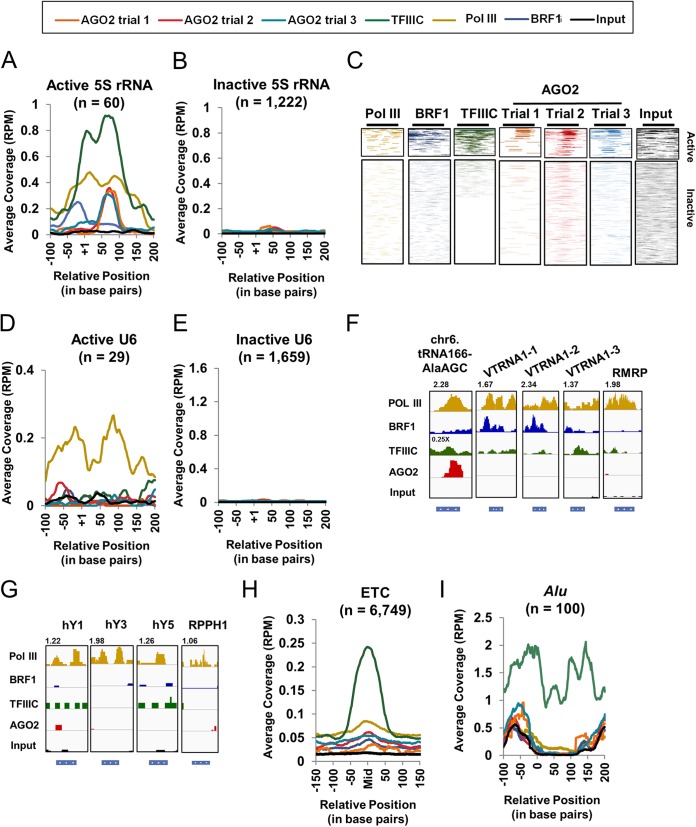
As observed in Fig. 1B, 5S rRNA genes had high proportion of AGO2 binding compared to other annotated regions. Similar to tRNA genes, meta-analysis and binding profile heat maps of AGO2 binding across all 5S rRNA gene sequences revealed enrichment of AGO2 at active, but not inactive, 5S rRNA genes for all three trials (Fig. 2A to C). Expected binding profiles for TFIIIC, BRF1, and Pol III were seen for both active and inactive 5S rRNA genes (Fig. 2A to C). We next sought to determine whether AGO2 exhibited binding at other Pol III genes. Analysis of ChIP-seq data for U6 RNA, vault RNAs (VTRNA), RNase MRP RNA (RMRP), hY RNAs, and RNase P RNA (RPPH1) exhibited no enrichment for AGO2 binding (Fig. 1D to G). Sites enriched for TFIIIC but not Pol III, called extra-TFIIIC (ETC) sites, also showed no enrichment for AGO2 binding (Fig. 1H). Furthermore, in light of reports indicating the expression and function of Alu- and Alu-related RNAs (74–76), we sought to investigate whether AGO2 binds to the genes from which these RNAs arise. Meta-analysis across potentially active Alu elements, which contain an internal promoter similar to that of tRNA genes, showed no enrichment for AGO2 binding (Fig. 2I).
FIG 2.
AGO2 binds to tRNA genes and 5S rRNA genes but not to other Pol III and TFIIIC binding sites. (A and B) Meta-analyses showing the binding of Pol III, BRF1, TFIIIC, AGO2, and Input across 60 active 5S rRNA genes (A) or 1,222 inactive 5S rRNA genes (B) (see also Materials and Methods). The y axis indicates the coverage scaled according to 5S rRNA reads per total million mapped reads (RPM). The x axis indicates the position in base pairs, with position +1 indicating the first nucleotide of the 5S rRNA. (C) Binding profile heat maps of indicated protein binding across active and inactive 5S rRNA gene sequences. Thick lines above heat maps indicate the average tRNA gene size, with the left end representing the +1 position and the right end representing the average 3′ end site (3′). Increasing intensity of color represents increasing read coverage. The coverage density for each heat map was internally scaled according to the 1st to the 99th percentiles. (D and E) Meta-analyses showing the binding of Pol III, BRF1, TFIIIC, AGO2, and Input across 29 active U6 genes (D) or 1,659 inactive U6 genes (E) (see also Materials and Methods). The y axis indicates the coverage scaled according to U6 RNA reads per total million mapped reads (RPM). The x axis indicates the position in base pairs, with position +1 indicating the first nucleotide of the U6 RNA. (F) Screenshots from the Integrative Genome Viewer (IGV) for Pol III, BRF1, TFIIIC, AGO2 trial 2, and Input across a tRNA gene, vault RNA genes (VTRNA), and an RNase MRP RNA gene (RMRP) (86). The scale is indicated above each screenshot. The gene size and directionality is indicated by the blue box below each screenshot. (G) Screenshots from the IGV for Pol III, BRF1, TFIIIC, AGO2 trial 2, and Input across hY RNA genes and RNase P RNA (RPPH1). The scale is indicated above each screenshot. The gene size and directionality is indicated by the blue box below each screenshot. (H) Meta-analysis showing the binding of Pol III, BRF1, TFIIIC, AGO2, and Input across 6,749 extra-TFIIIC (ETC) sites (see also Materials and Methods). The y axis indicates the coverage scaled according to reads per total million mapped reads (RPM). The x axis indicates the position in base pairs with position “Mid” being the middle of the ETC peaks. (I) Meta-analysis showing the binding of Pol III, BRF1, TFIIIC, AGO2, and Input across 100 potentially active Alu elements (see Materials and Methods). The y axis indicates the coverage scaled according to reads per total million mapped reads (RPM). The x axis indicates the position in base pairs, with position 0 indicating the start of the Alu element annotation.
AGO2 chromatin localization is small RNA independent and is mediated through binding to tRNA.
The well-characterized mechanism of AGO2 binding involves tethering to target RNAs through a small antisense RNA (2). In addition, there is evidence for a variety of small RNA species that are derived from tRNAs (reviewed in reference 77). To examine whether AGO2 binding to tRNA genes occurs through tethering via a small tRNA-derived fragment, we analyzed previously published sRIP-seq data to determine whether AGO2 was enriched for small RNAs-derived from tRNA sequence (3). Binding profile heat maps and meta-analysis showed no enrichment for tRNA sequence compared to negative control (Fig. 3A, gray and red lines). In addition, no antisense tRNA sequences were found in the AGO2 sRIP-seq (data not shown). These results suggest that AGO2 is bound to tRNA genes through an uncharacterized mechanism in vivo.
Previous work has shown that AGO2 is capable of binding directly to long RNAs (18). To investigate this as a possibility, we analyzed endogenous AGO2 photoactivatable ribonucleoside enhanced cross-linking immunoprecipitation, followed by deep sequencing (PAR-CLIP-seq) (58). Unlike sRIP-seq, which only allows analysis of small RNAs (<35 nt), PAR-CLIP-seq can demonstrate binding to any RNA, regardless of length. The binding of AGO2 was compared to HuR, which has an affinity for AU-rich elements found within the 3′ untranslated region of many mRNAs and tends to stabilize mRNAs, whereas AGO2 is a translational repressor (78). Meta-analysis of PAR-CLIP-seq signal across all tRNA genes showed strong enrichment for sense tRNA sequence compared to HuR (Fig. 3A, dark blue and orange lines). The analysis of the sRIP-seq suggested that AGO2 was not enriched for small RNAs derived from tRNA sequences (Fig. 3A, red line). Consistent with this, AGO2 was capable of binding to tRNA in Dicer-null (noDice) 293T cells (59) (Fig. 3A, light blue line). Furthermore, analysis of the sRIP-seq and PAR-CLIP-seq data sets across tRNAscan-SE annotated pseudogenes, and Repeat Masker annotated pseudogenes indicated that AGO2 binding was exclusive to authentic tRNAs (Fig. 3B and C). A heat map of the AGO2-bound tRNA genes indicated that AGO2 binding spanned the entire tRNA, was depleted among the pseudogenes, and had a similar distribution among active tRNA genes, as was observed with AGO2 ChIP (Fig. 3D). A Venn diagram illustrated that 114 of 121 AGO2 ChIP-seq peaks were bound to active tRNAs that were also bound by AGO2 via PAR-CLIP-seq (Fig. 3E; see also Materials and Methods). Further analysis of the PAR-CLIP-seq data indicated an authentic interaction with the 5S rRNA (Fig. 3F and G). The binding of AGO2 to 5S rRNA molecules was expected given the prevalence of ChIP-seq peaks within 5S rRNA genes, similar to what we observed for tRNA genes and tRNA.
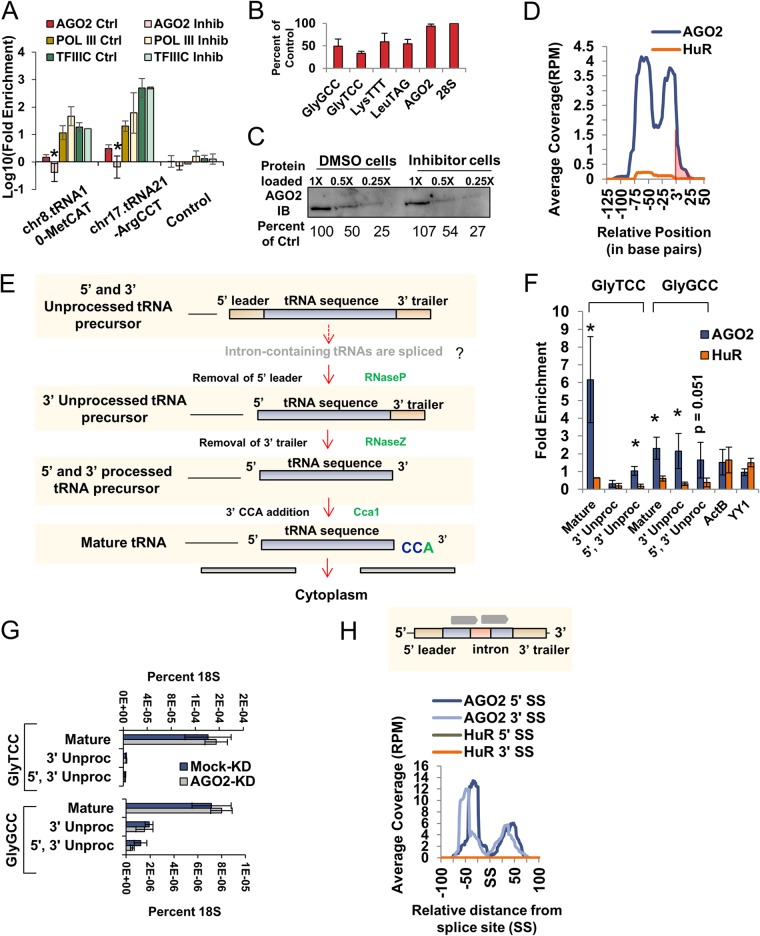
The combination of ChIP and PAR-CLIP of AGO2 to these type I and II Pol III genes (5S rRNA and tRNA genes) led us to hypothesize that AGO2 binding to nascent transcripts creates a chromatin tether, which allows detection by ChIP methods. To test this hypothesis, we performed ChIP-QPCR for AGO2, Pol III, and TFIIIC in cells treated with a Pol III inhibitor (see Materials and Methods) or DMSO alone (Fig. 4A, Ctrl). In control cells, AGO2 binding was enriched at the two tRNA genes tested (P = 0.000681 and P = 0.00371). Treatment with a Pol III inhibitor caused a specific and significant reduction in AGO2 ChIP enrichment at the two tRNA genes tested (Fig. 4A), whereas Pol III and TFIIIC ChIP enrichments were unchanged, indicating that the inhibitor does not affect PIC formation. RT-QPCR of control and Pol III inhibitor-treated cells confirmed that the concentration of inhibitor used specifically reduced tRNA levels by ∼50%, while not affecting AGO2 mRNA levels (Fig. 4B). Furthermore, AGO2 Western blot confirmed that protein levels of AGO2 were unchanged upon Pol III inhibitor treatment (Fig. 4C). A close examination of the PAR-CLIP-seq data indicated that that AGO2 appeared to be enriched from the +1 position to ∼25 nt downstream of the average mature 3′ end. To examine this enrichment more directly, we realigned the data from each mature 3′ end, rather than the +1 position, and found that AGO2 is indeed enriched ∼25 nt downstream compared to HuR (Fig. 4D, shaded region indicates 3′ unprocessed tRNA). tRNAs undergo extensive processing prior to nuclear export and maturation (Fig. 4E). To confirm directly that AGO2 was binding to both mature and unprocessed tRNA (as suggested by the PAR-CLIP meta-analysis and the ChIP dependence on active Pol III transcription), we performed AGO2 RNA immunoprecipitation, followed by quantitative RT-PCR (RIP–RT-QPCR), on cross-linked nuclei using primers that amplify 5′ and 3′ unprocessed tRNA, 3′ unprocessed tRNA, and mature tRNA. AGO2 RIP–RT-PCR was enriched for both unprocessed and mature tRNA forms, but not for two mRNAs tested (Fig. 4F). To assess whether AGO2 binding to tRNA had any effect on tRNA processing, we performed RT-QPCR in Mock-KD and AGO2-KD cells using primers that amplify 5′ and 3′ unprocessed tRNA, 3′ unprocessed tRNA, and mature tRNA. This revealed that AGO2 knockdown does not drastically alter processing of the two tRNAs tested (Fig. 4G). The human genome contains 31 intron-containing tRNAs, which are thought to be spliced in the nucleus prior to end processing of tRNAs in metazoans (Fig. 4E) (79, 80). Although AGO2 was verified to interact with spliced tRNA, we were unable to detect an interaction between AGO2 and the intronic region of any tRNA (Fig. 4H). We conclude that AGO2 exhibits binding to both unprocessed and mature, full-length, nuclear, tRNA but that this interaction does not alter processing. This is consistent with the interaction forming on nascent tRNA transcripts.
FIG 4.
AGO2 binds to nascent tRNAs. (A) Bar graph representing the log10 fold enrichment of AGO2, Pol III, and TFIIIC ChIP-QPCR in Pol III inhibitor-treated cells (Inhib) and DMSO-treated cells (Ctrl). Significance for the reduction of enrichment upon Pol III inhibitor-treated cells was calculated by using a Student t test (*, P < 0.05). (B) Bar graph of RT-QPCR from inhibitor-treated cells normalized to 28S rRNA and presented as a percentage of control for four tRNAs and AGO2. (C) Western blot for AGO2 in DMSO-treated cells and Pol III inhibitor-treated cells. The percentage of control is indicated below the blot images and was calculated using ImageJ and normalized to the total protein levels, as measured by Ponceau S staining. (D) Meta-analysis illustrating the average coverage for PAR-CLIP-seq experiments using anti-AGO2 (dark blue) and anti-HuR (orange) in WT 293T cells aligned at the mature 3′ end of the tRNA sequence (3′). The shaded region indicates portions of a tRNA downstream of the mature 3′ end. (E) Schematic depicting sequential steps in tRNA processing. The primary transcript of non-intron-containing tRNAs is initially processed by cleavage of the 5′ leader sequence by RNase P. This cleavage creates position +1 in the tRNA. For intron-containing tRNAs, splicing is thought to precede 3′ end processing. After the completion of transcription, the RNA is cleaved a second time at its 3′ end by RNase Z to liberate the 3′ trailer sequence. After the 5′ and 3′ cleavage events, a nonencoded –CCA is added to the 3′ end, and the transcript is exported from the nucleus. (F) RIP-QPCR for AGO2 and HuR presented as the fold enrichment compared to input (y axis) across mature, 3′ unprocessed, and 5′, 3′ unprocessed forms of two different tRNAs (x axis). Significance for enrichment compared to HuR was tested by using a Student t test (*, P < 0.05, n = 3). (G) RT-QPCR levels of mature, 3′ unprocessed, and 5′, 3′ unprocessed tRNA forms normalized to 18S rRNA for Mock-KD and AGO2-KD cells. (H) Meta-analysis of average coverage of AGO2 and HuR PAR-CLIP-seq across all intron-containing tRNA genes aligned at either the 5′ or 3′ splice site (SS). A diagram of intron-defining tags is depicted above the graph. No intron-defining tags were found in AGO2 or HuR PAR-CLIP-seq (data not shown).
AGO2 depletion leads to increased expression of genes in a tRNA gene insulator region on chromosome 17.
Some active tRNA genes have been shown to possess chromatin insulator activity (35, 81, 82). In addition, extensive work in yeast has demonstrated that active tRNA genes can repress neighboring Pol II genes through tgm silencing (34, 83, 84). To understand whether AGO2 plays a role in either of these processes, we focused on a region on the p arm of human chromosome 17 that has been previously characterized to have active tRNA genes that can act as insulators (35). The nine annotated genes in this region are separated by several clusters of tRNA genes that are capable of functioning as both enhancer block and barrier function insulators (35). To test whether AGO2 has any effect on the expression levels of the genes within this locus, we performed RNA-seq. The expression levels of all transcripts were analyzed using the Cufflinks pipeline, and expression levels are represented as fragments per kilobase of transcript per million mapped reads (FPKM) (44, 85). Two replicates of RNA-seq from Mock-KD and AGO2-KD 293T cells demonstrated similar results (Fig. 5A and B). Interestingly, seven of the nine genes in the selected region of chromosome 17 showed a reproducible increase in expression upon AGO2 knockdown (Fig. 5C, blue bars). One of the nine genes, ALOXE3, was expressed at such a low level that FPKM could not be reliably calculated (data not shown). Considering that AGO2 is integral in miRNA-mediated repression, we reasoned that these results could be explained by derepression of miRNA-mediated targets. To test this, we calculated the fold change in FPKM from previously published DICER-KD and DROSHA-KD RNA-seq data sets (GEO accession numbers: Mock-KD, GSM155053; DICER-KD, GSM1550537; and DROSHA-KD, GSM1550539) (62). Interestingly, we found that neither DICER nor DROSHA knockdown leads to the same effect on gene expression seen upon AGO2 knockdown (Fig. 5C, gray and gold bars). Since AGO2 binds tRNA genes that have known insulator properties, we reasoned that the effect on nearby gene expression upon AGO2 knockdown could be similar or related to that of insulator disruption. Considering that CTCF is a well-characterized insulator protein that has various binding sites throughout this region of chromosome 17 (Fig. 5C bottom, green boxes), we analyzed the fold change in FPKM from previous published CTCF-KD data (Fig. 5C, green bars). Interestingly, although CTCF-KD did cause some slight changes in gene expression, it did not mimic the effect of AGO2-KD (Fig. 5C).
AGO2 regulates the expression of Pol II genes that flank active tRNA genes.
We next sought to investigate whether the increase in FPKM values seen throughout the p arm of chromosome 17 upon AGO2 knockdown was a general feature of all AGO2-bound tRNA genes. To test this, we classified tRNA genes as either “AGO2 bound” or “non-AGO2 bound”, based on ChIP-seq coverage and then identified the FPKM (or microarray expression level) of the nearest upstream and downstream gene from each tRNA gene. We then compared the FPKM (or microarray expression level) of the genes near AGO2-bound or non-AGO2-bound tRNA genes before and after knockdown of AGO2, DICER, DROSHA, and CTCF. Similarly to the genes analyzed on the p arm of chromosome 17, the five genes with the highest fold changes upon AGO2 knockdown across two trials showed little to no change in gene expression upon knockdown of DICER, DROSHA, or CTCF (Fig. 5D). The alignment of a subset of genes from each RNA-seq trial is shown as an Integrative Genome Viewer (IGV) screenshot (Fig. 5E) (86).
To confirm these results, we performed RT-QPCR on AGO2 knockdown (AGO2-KD) and DICER-deficient (noDice) (59) cells using primers that amplify the two genes (SAT2 and NARF) that had the highest fold changes and a standard deviation of <0.2 across the two trials of AGO2-KD RNA-seq (Fig. 5F). To ensure specific reduction of targeted proteins, AGO2 and DICER were probed by Western blotting in AGO2-KD and noDice cells (Fig. 5G). Consistent with our genome-wide observations, the expression levels of SAT2 and NARF were upregulated upon AGO2-KD but not within the noDice cell line (Fig. 6F). AGO2 levels were reduced to 16.7% of the wild type in our AGO2-KD cells (Fig. 6G). When considering the closest gene to all AGO2-bound and non-AGO2-bound tRNA genes, we found that the mean, median, 5th, 25th, 50th, and 75th percentiles of the fold changes in expression level were greater for AGO2-bound tRNA genes than for non-AGO2-bound tRNA genes across two trials (Fig. 6A, Mann-Whitney U test, P = 5.60 × 10−6 and P = 5.80 × 10−9). The AGO2 knockdown treatment caused 80 and 88% of all Pol II genes that flank an AGO2-bound tRNA gene to increase in FPKM values across two trials, while only 49 and 56% of Pol II genes that flank non-AGO2-bound tRNA genes had an increase in FPKM across two trials (Fig. 6A). To confirm that the results were not solely a consequence of derepression of miRNA targets, the fold change in Pol II gene expression among genes flanking both AGO2-bound and non-AGO2-bound tRNA genes was calculated as described above for both DICER and DROSHA knockdown RNA-seq (Fig. 6A) (62). The pattern of gene expression was both quantitatively and qualitatively different when comparing AGO2 knockdown to that of either DICER or DROSHA. When considering the closest gene to AGO2-bound tRNA genes, the fold changes across all percentiles were higher for AGO2-KD than for DICER- or DROSHA-KD. Furthermore, the median fold change in expression level was not significantly higher for Pol II genes nearest to AGO2-bound tRNA genes compared to Pol II genes nearest to non-AGO2-bound tRNA genes (Fig. 6A, Mann-Whitney U test, P = 0.63 and P = 0.02, note directionality). In addition, only 53 and 36% of the closest Pol II genes to AGO2-bound tRNA genes were increased for DICER and DROSHA knockdowns, respectively. We were unable to do a similar analysis at the 5S rRNA genes due to the small number of Pol II genes that flank an AGO2-bound 5S rRNA gene (data not shown).
We next investigated the effects on gene expression of nearby genes when knocking down a well-characterized insulator protein, CTCF. An analysis similar to that in Fig. 6A was performed for all CTCF binding sites (n = 94,668) after CTCF knockdown (Fig. 6C; see also Materials and Methods for the GEO accession numbers) (60, 61). The knockdown of CTCF did cause a significant change in gene expression of Pol II genes that immediately flank the CTCF binding sites (relative to a sample of Pol II genes that flank 94,668 randomly chosen non-CTCF bound genomic loci), but these changes were equally likely to be increases or decreases, and the median fold change was 1.0 (Fig. 6B, Mann-Whitney U test, P < 2.20 × 10−16, 48% of genes increased for CTCF versus 52% of genes increased for random sequence). Thus, AGO2 knockdown has an effect on gene expression that is distinct from that of putative enhancer-blocking insulator sequences (the CTCF binding sites).
AGO2-bound tRNA genes suppress Pol II gene expression in cis.
The increase in gene expression (FPKM) among genes that flank AGO2-bound tRNA genes upon AGO2 knockdown suggested a cis effect exerted by the actively transcribed tRNA gene upon nearby Pol II genes, similar to tgm silencing in yeast (34, 83, 84). To address this possibility, we quantified the fold change in each upstream and downstream Pol II gene as a function of its distance from the nearest tRNA gene (Fig. 6C to F). This was done for both AGO2-bound and non-AGO2-bound tRNA genes (or CTCF and Random) for AGO2, DICER, DROSHA, and CTCF knockdowns. A linear regression and two-way ANCOVA statistical analysis of the fold changes upon AGO2 knockdown confirmed a negative slope for AGO2-bound tRNA genes upon AGO2-KD (slope = −0.0012), indicating that the magnitude of the effect of the AGO2-bound tRNA gene on Pol II gene expression dissipated with increasing distance from the nearest tRNA gene (Fig. 6C, P = 2.35 × 10−5, 2-way ANCOVA comparing AGO2-bound with non-AGO2-bound slope). The fold changes in FPKM upon either DICER, DROSHA, or CTCF knockdown appeared unrelated to the genes proximity to an AGO2-bound tRNA gene or CTCF binding site (Fig. 6D to F, DICER-KD slope: 0.0004, DROSHA-KD slope: 0.0003, CTCF-KD slope: −3 × 10−6). The absolute value of the slope of the inductions of FPKM as a function of distance from an AGO2-bound tRNA gene was 3-fold greater upon AGO2 knockdown compared to DICER knockdown, 4.3-fold greater than DROSHA knockdown, and 485-fold greater than CTCF knockdown. For genes near AGO2-bound tRNA genes, the y intercept was only positive upon AGO2-KD and the magnitude of difference from no fold change was ∼6.5-fold greater than for that of DICER- or DROSHA-KD. Furthermore, for genes near an AGO2-bound tRNA gene, we only observed a negative slope upon AGO2 knockdown. These data suggest that the effect of AGO2 on FPKM levels is dependent on the distance from an AGO2-bound tRNA gene, and predominantly suppressive to mRNA level. In contrast, the miRNA-related proteins DICER and DROSHA and the insulator protein CTCF affect FPKM levels independent of distance from an AGO2-bound tRNA gene or CTCF site and are equally likely to result in increased or decreased mRNA levels.
To further illustrate the distance dependence of the effect that AGO2-bound tRNA genes have on Pol II gene expression, each Pol II gene was binned according to its distance from AGO2-bound tRNA genes and the number of genes that showed an increase or decrease in expression was quantified (Fig. 6G). Genes within 5 kb of an AGO2-bound tRNA gene were significantly more likely to have increased than decreased expression (Fig. 6G, first bin, note up/red versus down/gray bar; P = 3 × 10−8, chi-square test). This tendency lessened with increasing distance from an AGO2-bound tRNA gene and lost significance at bins at a distance greater than 30 kb away (Fig. 6G). This result was in contrast to that seen for Pol II genes that flank non-AGO2-bound tRNA genes, where the changes in FPKM were equally likely to be upregulated or downregulated upon AGO2 knockdown, and were independent of distance from the tRNA gene (Fig. 6H).
The ability of tRNA genes to facilitate higher-order chromatin structure and/or regulate the activity of nearby Pol II genes has potential to be physiologically relevant in light of previous work demonstrating that some tRNA genes have cell-type-specific expression patterns (24). To test whether variability in which tRNA genes are active across cell types regulates nearby Pol II gene expression, we focused on tRNA genes that were differentially bound by TFIIIC in HeLa S3 and K562 cells (GEO accession numbers: HeLa S3, GSM935342; and K562, GSM935343). We then compared the FPKM of genes within 500 bp of these differentially bound tRNA genes across the two cell types (K562 RNA-seq [GSM958731] and HeLa S3 [GSM958739]). Only genes that showed expression in at least one of the two cell types (FPKM > 0.5) and did not overlap with any constitutively expressed tRNA genes were included. Two genes were within 500 bp of a tRNA gene that was bound by TFIIIC in K562 but not in HeLa S3, and both were repressed in K562 cells (Fig. 7A). Only one gene was within 500 bp of a tRNA gene that was bound by TFIIIC in HeLa S3 but not in K562 cells, and it was expressed in K562 but fully silent in HeLa S3 (Fig. 7A). For each of these three genes, juxtaposition next to an active tRNA gene correlated with repression or full silencing. To further probe the regulatory potential of tRNA genes, a gene ontology (GO) analysis was run using DAVID for all genes that are within 500 bp of a tRNA gene to identify enriched functional classifications (46, 87). The number of genes within each enriched annotation (P < 0.05) is illustrated in Fig. 7B. Enriched functional classifications include cell morphogenesis during differentiation, phosphate metabolic processes, neuron projection development, neuron differentiation, and protein amino acid phosphorylation. Enrichment of these functional classifications is consistent with genes near tRNA genes being involved in cell type specification.
DISCUSSION
Our results demonstrate that AGO2 localizes to human tRNA and 5S rRNA genes through direct binding (Fig. 7C). This is facilitated by direct protein-RNA contact with nascent RNA, which allows AGO2 to be cross-linked to these gene loci in ChIP assays. AGO2 chromatin binding was specific to TFIIIC-dependent Pol III genes, since it was not observed at non-TFIIIC-dependent Pol III genes or ETC sites, which are bound by TFIIIC but not transcribed. The loss of AGO2 at tRNA genes causes an increase in FPKM values for genes throughout a previously characterized tRNA gene insulator region (35). This result suggests that the function of AGO2 at these tRNA genes is distinct from simple disruption of enhancer-promoter interactions, which would be equally likely to cause increase or decrease in expression of nearby genes. Genome-wide analysis indicates that the loss of AGO2 causes a global increase in expression of Pol II genes flanking AGO2-bound tRNA genes and that this effect on gene expression is distinct from that of non-AGO2-bound tRNA genes and from the loss of CTCF in two ways: (i) loss of AGO2 specifically increases Pol II genes that flank AGO2-bound tRNA genes, and (ii) the magnitude of this increase lessens as a function of distance from the nearest AGO2-bound tRNA gene. The effect on FPKM values was also distinct from both DICER and DROSHA, indicating a novel mechanism not related to the traditional RNAi pathway.
We observed binding of AGO2 both to the tRNA genes (ChIP-seq and ChIP-QPCR) and to tRNAs themselves (PAR-CLIP-seq and RIP-QPCR). Our results imply that AGO2 does not bind to DNA directly but instead does so via interactions with nascent tRNA. We did not observe antisense small RNAs that could potentially target AGO2 to the tRNA species, and the direct binding to tRNA is maintained in DICER knockout cells. Furthermore, a careful analysis of the PAR-CLIP-seq data indicates that an excess of PAR-CLIP-induced mutations, specific for the AGO2 binding sites, exist throughout the tRNA. In addition, RIP-QPCR shows enrichment for AGO2 binding to at least full-length tRNA compared to HuR. Thus, we conclude that AGO2 is binding directly to tRNAs. The binding of AGO2 to long RNAs without a targeting small RNA is not without precedent. Work in vitro has shown that recombinant, purified AGO2 can specifically bind to pre-miRNAs and long unstructured RNAs (18, 20). Furthermore, it was shown in vivo that AGO2 can immunoprecipitate a pre-miRNA in the complete absence of DICER and miRNAs (18). The fact that AGO2 binds only to active tRNA genes led us to conclude that AGO2 must be interacting with nascent tRNA precursors or tRNA precursors shortly after the completion of transcription, which was confirmed by our studies using an inhibitor of RNA Pol III. According to this model, AGO2 would be in close physical proximity to the chromatin and allow for cross-linking to the DNA and subsequent detection by ChIP methods.
Many molecular details of this novel role for AGO2 remain to be elucidated. TFIIIC-bound tRNA genes have been shown to function as both an enhancer-blocking and barrier function insulators in heterologous reporter constructs in humans (35). However, the effect on FPKM values of nearby Pol II genes is distinct from that of a more well-established metazoan insulator protein, CTCF. Considering this, our results may be more consistent with the phenomena of tRNA gene-mediated (tgm) silencing. This effect, which has been characterized in Saccharomyces cerevisiae, causes the suppression of most Pol II genes that are within 500 bp of an active tRNA gene, requires factors distinct from those needed for TFIIIC boundary element function, and also requires nucleolar localization (32–34, 83, 84, 88, 89). It remains unclear whether the human homologs to CBF5 and Mod5, yeast factors required for tgm silencing, or nucleolar localization, are also required for our observations.
A great deal of additional work is required to fully understand both the molecular mechanisms by which AGO2 recognizes tRNAs and how these complexes promote genome-wide repression in cis. In addition, the cell-type-specific expression of tRNA genes and its suppressive effect on nearby Pol II genes underscores an unexpected layer of regulatory complexity that may be crucial for many biological processes. Future studies should be aimed at discerning the role of tRNA gene-mediated suppression during normal differentiation.
ACKNOWLEDGMENTS
We thank Michael Crowley at the UAB Heflin Center for Genomic Sciences for assistance with ChIP-seq and RNA-seq. We also thank Bryan R. Cullen at the Duke University Medical Center for kindly providing us with the noDice cell line.
REFERENCES
- 1.Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Turcu FE, Grewal SI. 2012. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev 22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsche E, Harel-Bellan A. 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 4.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. 2011. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev 25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giles KE, Ghirlando R, Felsenfeld G. 2010. Maintenance of a constitutive heterochromatin domain in vertebrates by a Dicer-dependent mechanism. Nat Cell Biol 12:91–99. doi: 10.1038/ncb2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 7.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, Siomi MC, Carninci P, Gilmour DS, Corona DF, Orlando V. 2011. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 9.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 11.Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. 2007. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. 2012. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carissimi C, Laudadio I, Cipolletta E, Gioiosa S, Mihailovich M, Bonaldi T, Macino G, Fulci V. 2015. Argonaute2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human transcription start sites. Nucleic Acids Res 43:1498–1512. doi: 10.1093/nar/gku1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, Blaustein M, Minana B, Dujardin G, Pozzi B, Pelisch F, Bechara E, Agafonov DE, Srebrow A, Luhrmann R, Valcarcel J, Eyras E, Kornblihtt AR. 2014. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc Natl Acad Sci U S A 111:15622–15629. doi: 10.1073/pnas.1416858111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC. 2013. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS genetics 9:e1003821. doi: 10.1371/journal.pgen.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loss-Morais G, Waterhouse PM, Margis R. 2013. Description of plant tRNA-derived RNA fragments (tRFs) associated with Argonaute and identification of their putative targets. Biol Direct 8:6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, McManus MT, Kiriakidou M. 2009. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res 37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maniataki E, Mourelatos Z. 2005. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. 2009. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16:1259–1266. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 21.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. 2014. RNAi factors are present and active in human cell nuclei. Cell Reports 6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebetsberger J, Polacek N. 2013. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schramm L, Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 24.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernens I, Goodfellow SJ, Innes F, Kenneth NS, Derblay LE, White RJ, Scott PH. 2006. Hypoxic stress suppresses RNA polymerase III recruitment and tRNA gene transcription in cardiomyocytes. Nucleic Acids Res 34:286–294. doi: 10.1093/nar/gkj402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen VC, Clelland BW, Hockman DJ, Kujat-Choy SL, Mewhort HE, Schultz MC. 2010. Replication stress checkpoint signaling controls tRNA gene transcription. Nat Struct Mol Biol 17:976–981. doi: 10.1038/nsmb.1857. [DOI] [PubMed] [Google Scholar]
- 28.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, Andersen LD, Andersen CL, Hulleman E, Wurdinger T, Ralfkiaer E, Helin K, Gronbaek K, Orntoft T, Waszak SM, Dahan O, Pedersen JS, Lund AH, Pilpel Y. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, Larionov V. 2011. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle 10:2779–2791. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donze D, Adams CR, Rine J, Kamakaka RT. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott KC, Merrett SL, Willard HF. 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 32.Hull MW, Erickson J, Johnston M, Engelke DR. 1994. tRNA genes as transcriptional repressor elements. Mol Cell Biol 14:1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. 2000. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci U S A 97:13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, McLeod IX, Yates JR 3rd, Hopper AK, Engelke DR. 2013. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci U S A 110:E3081–E3089. doi: 10.1073/pnas.1219946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. 2012. Human tRNA genes function as chromatin insulators. EMBO J 31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey T, Krajewski P, Ladunga I, Lefebvre C, Li Q, Liu T, Madrigal P, Taslim C, Zhang J. 2013. Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput Biol 9:e1003326. doi: 10.1371/journal.pcbi.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B, Yun J, Kim MS, Mendell JT, Xie Y. 2014. PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol 15:R18. doi: 10.1186/gb-2014-15-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC. 2011. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res 39:4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 47.Eddy SR, Durbin R. 1994. RNA sequence analysis using covariance models. Nucleic Acids Res 22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fichant GA, Burks C. 1991. Identifying potential tRNA genes in genomic DNA sequences. J Mol Biol 220:659–671. doi: 10.1016/0022-2836(91)90108-I. [DOI] [PubMed] [Google Scholar]
- 50.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavesi A, Conterio F, Bolchi A, Dieci G, Ottonello S. 1994. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res 22:1247–1256. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, Lee YH, Jones AE, Woolnough JL, Zhou D, Dai Q, Wu Q, Giles KE, Townes TM, Wang H. 2014. The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat Commun 5:3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R Core Team. 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 54.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 55.Neuwirth E. 2011. RColorBrewer: ColorBrewer palettes. R package version 1.0–5. [Google Scholar]
- 56.Edgar R, Domrachev M, Lash AE. 2002. Gene Expr. Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. 2011. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods 8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 59.Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. 2014. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, Grosveld FG, Ren B, Wendt KS. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, Rajewsky N. 2014. A variety of Dicer substrates in human and Caenorhabditis elegans. Cell 159:1153–1167. doi: 10.1016/j.cell.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 63.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC table browser data retrieval tool. Nucleic Acids Res 32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coughlin DJ, Babak T, Nihranz C, Hughes TR, Engelke DR. 2009. Prediction and verification of mouse tRNA gene families. RNA Biol 6:195–202. doi: 10.4161/rna.6.2.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. 2009. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell 20:428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flores O, Kennedy EM, Skalsky RL, Cullen BR. 2014. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res 42:4629–4639. doi: 10.1093/nar/gkt1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farazi TA, Ten Hoeve JJ, Brown M, Mihailovic A, Horlings HM, van de Vijver MJ, Tuschl T, Wessels LF. 2014. Identification of distinct miRNA target regulation between breast cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets. Genome Biol 15:R9. doi: 10.1186/gb-2014-15-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lipchina I, Elkabetz Y, Hafner M, Sheridan R, Mihailovic A, Tuschl T, Sander C, Studer L, Betel D. 2011. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev 25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR. 2012. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog 8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smit AF. 1996. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6:743–748. doi: 10.1016/S0959-437X(96)80030-X. [DOI] [PubMed] [Google Scholar]
- 73.Smit AF. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9:657–663. doi: 10.1016/S0959-437X(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 74.Oler AJ, Traina-Dorge S, Derbes RS, Canella D, Cairns BR, Roy-Engel AM. 2012. Alu expression in human cell lines and their retrotranspositional potential. Mobile DNA 3:11. doi: 10.1186/1759-8753-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baryakin DN, Semenov DV, Savelyeva AV, Koval OA, Rabinov IV, Kuligina EV, Richter VA. 2013. Alu- and 7SL RNA analogues suppress MCF-7 cell viability through modulating the transcription of endoplasmic reticulum stress response genes. Acta Naturae 5:83–93. [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Q, Tanasa B, Trabucchi M, Li W, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. 2012. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat Struct Mol Biol 19:1168–1175. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sobala A, Hutvagner G. 2011. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA 2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 78.Simone LE, Keene JD. 2013. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev 23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Melton DA, De Robertis EM, Cortese R. 1980. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature 284:143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- 80.Lund E, Dahlberg JE. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 81.Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, Edwards DJ, Donze D. 2008. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell 7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noma K, Cam HP, Maraia RJ, Grewal SI. 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 83.Good PD, Kendall A, Ignatz-Hoover J, Miller EL, Pai DA, Rivera SR, Carrick B, Engelke DR. 2013. Silencing near tRNA genes is nucleosome-mediated and distinct from boundary element function. Gene 526:7–15. doi: 10.1016/j.gene.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. 2005. Silencing near tRNA genes requires nucleolar localization. J Biol Chem 280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bolton EC, Boeke JD. 2003. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res 13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]