Abstract
The interferon (IFN)-stimulated gene factor 3 (ISGF3) transcription factor with its Stat1, Stat2, and interferon regulatory factor 9 (IRF9) subunits is employed for transcriptional responses downstream of receptors for type I interferons (IFN-I) that include IFN-α and IFN-β and type III interferons (IFN-III), also called IFN-λ. Here, we show in a murine model of dextran sodium sulfate (DSS)-induced colitis that IRF9 deficiency protects animals, whereas the combined loss of IFN-I and IFN-III receptors worsens their condition. We explain the different phenotypes by demonstrating a function of IRF9 in a noncanonical transcriptional complex with Stat1, apart from IFN-I and IFN-III signaling. Together, Stat1 and IRF9 produce a proinflammatory activity that overrides the benefits of the IFN-III response on intestinal epithelial cells. Our results further suggest that the CXCL10 chemokine gene is an important mediator of this proinflammatory activity. We thus establish IFN-λ as a potentially anticolitogenic cytokine and propose an important role for IRF9 as a component of noncanonical Stat complexes in the development of colitis.
INTRODUCTION
Interferons (IFN) are subdivided into three distinct types, type I IFN (IFN-I; mainly IFN-α/β), type II IFN (IFN-II; IFN-γ), and type III IFN (IFN-III; IFN-λ/interleukin-28 [IL-28]/IL-29). Collectively, IFN are potent regulators of immune responses to pathogens (1, 2). In addition, they contribute to autoimmunity-related or other types of sterile inflammation (3–5). Biological responses to IFN require transcription of a large number of IFN-stimulated genes (ISGs), regulated by signal transducers and activators of transcription (Stat) and interferon regulatory factors (IRF). In their canonical signaling pathways, the type I and type III IFN receptors stimulate the assembly of the IFN-stimulated gene factor 3 (ISGF3) complex that contains tyrosine-phosphorylated Stat1 and Stat2 in association with IRF9, whereas the IFN-γ receptor employs Janus tyrosine kinases (Jaks) to produce the gamma interferon-activated factor (GAF), a homodimer of tyrosine-phosphorylated Stat1 (6). In addition, noncanonical complexes of Stat1/2 and IRF9 can be formed in response to IFN-I or IFN-γ signaling and contribute to gene selectivity of the transcriptional response (7–12). Stat1 homodimers bind to gamma interferon-activated sequences (GAS) in target promoters, whereas ISGF3 binds to IFN-stimulated response elements (ISRE), which can be found in a large number of antimicrobial and antiviral genes. Noncanonical complexes containing IRF9 would similarly be expected to associate with the ISRE, consistent with the DNA-binding specificity of this subunit.
In keeping with the common deployment of ISGF3, immunological activities of IFN-I and IFN-III appear to be very similar. However, the IFN-I receptor (IFNAR) is expressed on virtually all nucleated cells, whereas IFN-III receptor expression in mice is restricted to epithelial tissues (13). It is not entirely clear yet whether IFN-I and -III lead to completely identical signaling outputs, as their receptors may differ in their ability to activate Stats other than 1 and 2 or the mitogen-activated protein kinase (MAPK) pathway (14, 15).
Inflammatory bowel disease (IBD) is a health problem affecting a rising number of individuals, especially in the western world. A multistep process initiates this chronic disease, with a disturbance of the epithelial layer as an early event (16). Host factors as well as gut microbiota have been implicated in the development and maintenance of IBD, with specific innate and adaptive immune signaling pathways as well as specific bacterial species, such as members of the Enterobacteriaceae, shown to be associated with the disease (17, 18). IFN-I both reduce and enhance colitis in animal models, depending on the intensity of inflammation and whether the acute or resolution phases are examined (5, 19). Clinical studies testing IFN-I in the treatment of IBD support this view, as they led to conflicting results concerning their therapeutic potential (5, 20). Loss of IFN-γ signaling exacerbated disease in some reported animal studies (21, 22), while it was protective in others (23, 24). Blockade of IFN-γ caused a fairly subtle improvement of symptoms in human IBD patients (25, 26). IFN-III have not been studied in the context of colitis. However, such studies appear of high priority in light of recent reports showing they control rotavirus infections by targeting the intestinal epithelium (27). This study further demonstrated that polarized intestinal epithelial cells (IEC) respond to IFN-III from the basolateral and apical sides but to IFN-I only from the apical side. The authors suggested IFN-III might be more relevant for immune signaling in IEC than IFN-I. Earlier in vitro studies showed that IFN-III decrease proliferation and induce antiviral proteins in human IEC lines (28). Also, infection of IEC lines with various Gram-positive bacteria induced production of IFN-III (29).
The aim of our study was to determine the effect of simultaneous elimination of IFN-I and IFN-III responses by inducing colitis with the chemical dextran sodium sulfate (DSS) in mice lacking the ISGF3 subunit IRF9. We report that mice were strongly protected from colitis when deficient for IRF9. Surprisingly, the opposite effect was observed after combined deletion of IFN-I and IFN-III receptors. Our results suggest that the procolitogenic activity of IRF9 results from its participation in a noncanonical complex with Stat1 independently of IFN-I and IFN-III receptor signaling. In addition, they support the notion that IFN-III prevent damage of the gut mucosa after DSS treatment and might provide a novel therapeutic option.
MATERIALS AND METHODS
Mice, animal experiments.
Animal experiments were approved by the University of Veterinary Medicine Vienna institutional ethics committee and carried out in accordance with protocols approved by Austrian law (BMWF-66.006/002-II/10b/2010).
Mice lacking functional type III IFN receptors (IL28rα−/−) were provided by Bristol-Myers-Squibb Company (NJ). B6.A2G-Mx1 wild-type (WT) mice carrying intact Mx1 alleles (Mx1), B6.A2G-Mx1-IL28rα−/− mice lacking functional type III IFN receptors (Mx1-IL28rα−/−), and B6.A2G-Mx1-IL28rα−/−Ifnar1−/− double-knockout mice (Mx1-IL28rα−/− Ifnar1−/−) (30) lacking functional receptors for both type I and type III IFN were provided by Peter Stäheli (Freiburg, Germany). The mice were backcrossed to C57BL/6 mice as previously described for BALB/c mice (31). C57BL/6N, Ifnar1−/−, Irf9−/− (32), Stat1−/− (33), and Stat2−/− mice (34) backcrossed for more than 10 generations on a C57BL/6N background were housed in the same specific-pathogen-free (SPF) facility under identical conditions according to recommendations of the Federation of European Laboratory Animal Science Association and additionally monitored for being norovirus negative. Colitis experiments were performed in individually ventilated cage isolators.
To induce colitis, mice were provided with 2% dextran sodium sulfate (DSS; molecular mass of 36 to 50 kDa; MP Biomedicals) in autoclaved drinking water ad libitum for 7 days, after which they received DSS-free autoclaved drinking water for 2 days. Mice were weighed daily and sacrificed on day 0 (untreated), 5, or 10 of the colitis protocol. Upon sacrifice, the intestine was removed and flushed with phosphate-buffered saline (PBS) (pH 7.2). The colon was halved lengthwise, and one half was snap-frozen, while the other half was fixed in 2% paraformaldehyde, prepared as a Swiss role, and embedded in paraffin. Sections of whole intestine were stained with hematoxylin and eosin (H&E) using a standard protocol and blind-scored by a pathologist using an established method that evaluates inflammation severity, crypt damage, inflammation extent, and percent tissue involvement (35).
IHC.
Tissue sections were rehydrated and boiled 20 min in a citric buffer (pH 6) to unmask antigens. For proliferation assessment, slides were incubated with rabbit polyclonal anti-Ki67 antibody (NovoCastra) diluted 1:1,000 and, to evaluate apoptosis, with rabbit polyclonal anti-cleaved caspase-3 antibody (Cell Signaling) diluted 1:200. Epitope binding was revealed with peroxidase-conjugated secondary antibodies. Sections were counterstained with hematoxylin and imaged on an epifluorescence microscope (Zeiss AxioImager). Immunohistochemistry (IHC) images were photographed and quantified with the HistoQUEST software (TissueGnostics GmbH). At least 3 pictures per colon were taken, and the percentage of positive-stained cells per number of nucleated cells was calculated.
RNA isolation, cDNA synthesis, and quantitative PCR (qPCR).
For RNA preparation, colon tissue was homogenized and bone marrow-derived macrophages (BMDMs) were lysed in 700 μl RA1 buffer of the NucleoSpin II RNA isolation kit (Macherey and Nagel) and processed according to the manufacturer's protocol. cDNA was prepared as described previously (36). PCRs were performed with 60°C annealing temperature on an Eppendorf cycler. Expression of every sample was calculated relative to the OAZ1 housekeeping gene. Primer sequences are summarized in Table S1 in the supplemental material.
ChIP and ChIP sequencing (ChIP-Seq).
Chromatin immunoprecipitation (ChIP) was performed as described previously (37). A total of 2 μl (anti-Stat1 and anti-Stat2; Santa Cruz) or 4 μl (anti-IRF9; Santa Cruz) antiserum was used per 50 μg chromatin. ChIP data were normalized to and expressed as a percentage of input. Primers used for qPCR of the CXCL10 gene enhancer regions (E1 and E2) and IFN response region of Mx1 are listed in Table S1 in the supplemental material. Next-generation sequencing was carried out by the Vienna Biocenter Campus Support Facilities. A total of 5 to 10 ng of DNA precipitate was used for the generation of sequencing libraries using the KAPA library preparation kit for Illumina systems. Libraries were quantified with a Bioanalyzer double-stranded DNA (dsDNA) 1000 assay kit (Agilent) and a qPCR NGS library quantification kit (KAPA). Cluster generation and sequencing were performed with a HiSeq 2000 system with a read length of 100 nucleotides according to the manufacturer's guidelines (Illumina).
In the re-ChIP experiments (12), the immune complexes were eluted by adding 10 mM dithiothreitol (DTT) and incubating for 40 min at room temperature. The samples were diluted 40-fold and reimmunoprecipitated.
Cell isolation and treatment.
For IEC isolation, colons were washed in cold PBS containing 50 μg/ml gentamicin, cut into pieces, and incubated for 10 min in 15 mM EDTA-PBS at 37°C with shaking. After 1 g sedimentation to remove cell sheets, supernatants were collected, spun down, washed in PBS, and resuspended in buffer RA1 for RNA isolation. For Western blotting, cells were resuspended in RPMI 1640 with or without 5 ng/ml IFN-γ (eBioscience) for 15 min and processed as described previously (36).
For lamina propria mononuclear cell (LPMC) isolation, colons where incubated in RPMI 1640 containing gentamicin and 5 mM EDTA 3 times for 15 min at 37°C with shaking to fully remove IEC. Remaining tissue pieces were washed in RPMI and incubated for 90 min in RPMI containing 15 mM HEPES and 0.2 mg/ml type VIII collagenase (Sigma). The digest was filtered through a 70-μm-pore-size cell strainer, spun down, washed in PBS, and resuspended in buffer RA1. These fractions contained >50% LPMC.
Bone marrow-derived macrophages (BMDMs) were differentiated from bone marrow isolated from femurs and tibias of 6- to 8-week-old mice. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS) (Sigma-Aldrich), 10% L929 cell-conditioned medium as a source of colony-stimulating factor 1 (CSF-1), 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich) as previously described (36). The culture contained >99% F4/80+ cells. BMDMs were treated with 5 ng/ml of IFN-γ (Affymetrix; eBioscience).
CXCL10 determination.
Frozen tissue was homogenized in PBS containing proteinase inhibitors (phenylmethylsulfonyl fluoride; Roche cOmplete protease inhibitor cocktail), 1 mM vanadate, and 1 mM DTT. The homogenate was freeze-thawed twice. Total protein in the supernatants was determined by bicinchoninic acid (BCA) assay (Pierce), and CXCL10 levels were determined using a FlowCytomix kit (eBioscience) on 25 μl supernatant according to the manufacturer's instructions. The CXCL10 amount was normalized to total protein.
Western blotting.
Organs or isolated IEC where homogenized in Frackelton buffer containing protease and phosphatase inhibitors and centrifuged at 12,000 × g. The supernatant was used for Western blotting as described previously (36). Total Stat1 was detected using monoclonal antibody (MAb) to the Stat1 N terminus (BD Biosciences) at a dilution of 1:1,000. Tyrosine 701 phosphorylated Stat1 was detected using a 1:1,000 diluted antibody (Cell Signaling). As a loading control, the blot was probed with Abs to the Erk1 and Erk2 kinases (panErk) (BD Transduction Laboratories) at a dilution of 1:2,000.
Statistics.
Statistical analysis was done using the Mann-Whitney U test, considering P values of <0.05 as significant.
RESULTS
DSS-induced colitis in mice lacking IRF9, comparison to IFN receptor deficiency.
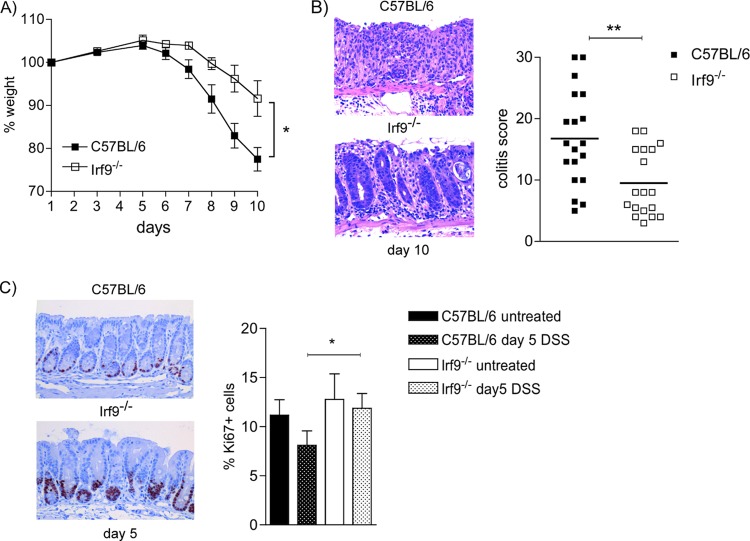
To examine the importance of ISGF3 target genes in intestinal inflammation without affecting transcriptional activity of Stat1 homodimers, we examined DSS colitis in mice deficient for IRF9. We expected the dominant effect of IRF9 deficiency to reflect the loss of canonical ISGF3 signaling. Following treatment with 2% DSS for 7 days, Irf9−/− mice started losing weight with a delay of up to 2 days compared to C57BL/6 (WT) controls and lost significantly less weight than WT mice (Fig. 1A). Pathology scoring of H&E-stained tissue confirmed a significant protection of Irf9−/− mice from DSS-induced colitis (Fig. 1B). Proliferation, as determined by the amount of Ki67-positive cells, decreased in WT colons during the early phase of colitis, consistent with previous reports (23). This decrease was not observed in IRF9-deficient colons. The difference between the Irf9−/− and WT genotypes was readily apparent in immunohistochemistry and confirmed by quantitative analysis (graph in Fig. 1C). Thus, the loss of IRF9 leads to profound protection from intestinal inflammation.
FIG 1.
Decreased weight loss and pathology in IRF9-deficient animals upon 2% DSS treatment. (A) Representative weight loss curve (n = 4 mice/group); (B) representative H&E stainings from colons on day 10 of 2% DSS colitis (200-fold magnification) and pathology score data from 3 independent experiments (WT n = 19, Irf9−/− n = 18). (C) Proliferation rates are increased in IRF9-deficient colons. Representative pictures (×200 magnification) and quantification of colon tissue of WT and Irf9−/− mice after 5 days of 2% DSS treatment, stained for Ki67 (n = 10/group). Data are presented as means ± standard errors of the mean (SEM) and are representative of at least 3 independent experiments. *, P < 0.05; **, P < 0.01, Mann-Whitney U test.
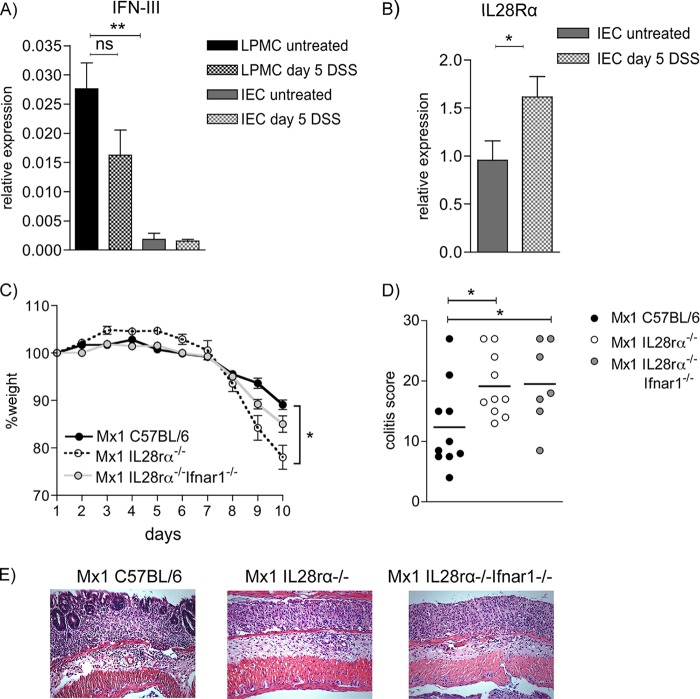
According to the Jak-Stat paradigm, ISGF3 formation results from signaling by the IFN-I and IFN-III receptors. We recently observed that the loss of IFN-I signaling has minor consequences for acute colitis after the moderate DSS dose used here (19). To seek an explanation for the impact of IRF9, we therefore examined the role of IFN-III in DSS colitis. Intestinal epithelial cells (IEC) from colon expressed low levels of IFN-λ mRNA if isolated from untreated or 5-day-DSS-treated mice (Fig. 2A). Fifteenfold-higher levels of IFN-III were detected in cell fractions strongly enriched for LPMC than in IEC. There was slight but not statistically significant reduction in expression upon DSS treatment. IFN-III receptors consist of the IL-10R2 chain shared with the IL-10 receptor and an IFN-III-specific IL-28Rα chain. We performed qPCR analysis of the IL-28Rα chain on IEC (Fig. 2B). In cells isolated from colons after 5 days of DSS treatment, we observed a small but significant increase of receptor expression over cells from healthy animals, indicating upregulation due to inflammatory signals. These results suggest that IFN-III are expressed in the colon and that IFN-III signaling might increase during colitis through enhanced receptor expression.
FIG 2.
Type III IFNs are expressed in colon cells, IL-28Rα is upregulated during inflammation, and loss of IL-28R signaling leads to increased pathology upon 2% DSS treatment. (A) qPCR analysis of IFN-III (IFN-λ2 and IFN-λ3) expression in isolated colon epithelial cells and crude purified lamina propria mononuclear cells from untreated and 5-day-DSS-treated WT mice (n = 4 to 6). (B) Colon epithelial cells from untreated and 5-day-DSS-treated (n = 6 or 7) WT mice were analyzed for IL-28Rα expression by qPCR. *, P < 0.05; **, P < 0.01, Student's t test. (C and D) Representative weight loss curve (n = 4 mice/group) (C) and pathology scoring (n = 10 for Mx1 C57BL/6 and Mx1 IL28rα−/− and n = 7 for Mx1 IL28rα−/− Ifnar1−/−) (D). (E) Representative H&E staining from colons after 10 days of 2% DSS treatment (200-fold magnification). Data shown are from male animals. Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01, Mann-Whitney U test.
To evaluate whether the protection from disease upon IRF9 deficiency is due to a loss of ISGF3 activity downstream of the IFN-III or combined IFN-I/IFN-III receptors, we analyzed mice lacking IFN-III receptors only or lacking IFN-I as well as IFN-III receptors. For studies with influenza virus (30), these animals were bred to contain an intact Mx1 gene, unlike normal C57BL/6 mice. They are designated –Mx1 and compared to their own Mx1+ C57BL/6 WT controls. When Mx1-WT, IL28rα−/−, and IL28rα−/− Ifnar1−/− mice were treated with 2% DSS, we recorded a pronounced and significant increase in weight loss for the IL28rα−/− genotype compared to that of the WT. Mice deficient for both receptors lost more weight than WT controls as well, but without differing significantly from the IL28rα−/− mice (Fig. 2C). Pathology scoring of colon H&E sections revealed a significant increase of damage in animals of both genotypes compared to their WT controls (Fig. 2D and E). Additional loss of IFN-I signaling did not change pathology scores with regard to the loss of IFN-III signaling alone. This confirms our earlier observations that IFN-I deficiency does not significantly affect acute colitis after treatment with moderate DSS concentrations (19).
Despite identical food and housing conditions for our mice, differences in intestinal microbiota composition cannot be ruled out (38). To minimize the possibility that microbiota variability accounts for the differences between Irf9−/− mice and those lacking IFN receptors, we performed cohousing experiments with Mx1-WT, Mx1-IL28rα−/−, Mx1-IL28rα−/− Ifnar1−/−, C57BL/6, and Irf9−/− mice. Three weeks of housing all the mentioned genotypes together did not change the results shown in Fig. 1 and 2 (not shown). This experiment corroborates our notion that the adverse impact of IRF9 on DSS-induced colitis does not reflect individual or combined signaling by IFN-I and IFN-III receptors during acute intestinal inflammation.
IFN-induced gene expression in mice lacking IFN receptors, Stats, or IRF9.
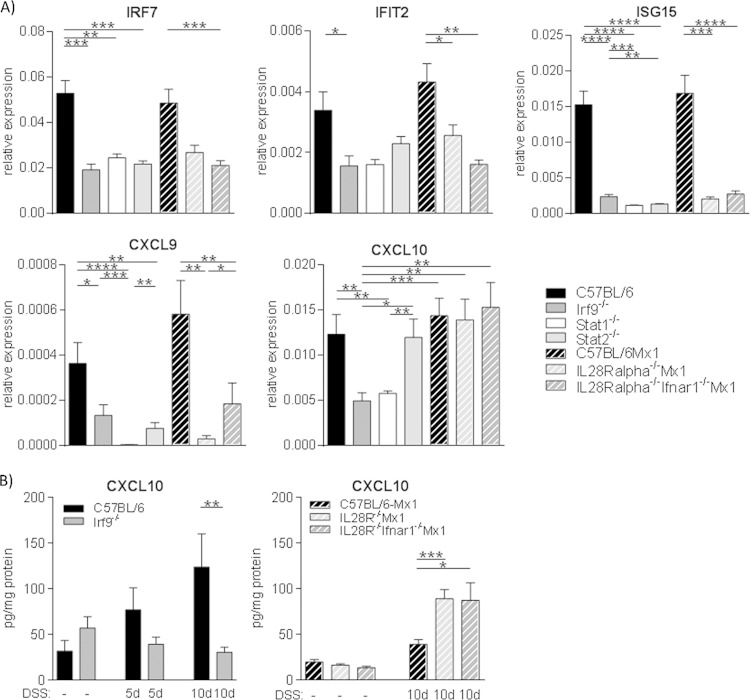
To seek an explanation for the phenotypic difference between IRF9 deficiency and the lack of IFN-I and IFN-III receptor signaling, we analyzed the expression of established IFN target genes in colon tissue. To obtain additional information about the Stat complexes involved, Stat1−/− and Stat2−/− mice were included in our analysis. According to the data of Fig. 1 and 2, the influence of IFN and IRF9 on colitis must be established before weight loss and inflammatory pathology occur; therefore, day 5 of the DSS protocol was chosen for the sampling of intestinal tissue.
Expression of the IRF7, IFIT2, ISG15, and CXCL9 ISGs in colon tissue was similarly decreased upon either the combined loss of IFN-I and -III receptors, Stat1, Stat2, or IRF9, suggesting their expression requires the canonical ISGF3 complex (Fig. 3A). IFIT1 and Stat1 genes were regulated in a similar fashion, while there was no decrease in expression of IRF1, IRF2, or CXCL2 in IRF9−/− or Mx1-IL28rα−/− Ifnar1−/− mice compared to that of their respective WT controls (not shown).
FIG 3.
CXCL10 expression in the colon is not dependent on IFN-I and -III signaling but requires IRF9 and Stat1. (A) qPCR analysis of IRF7, IFIT2, ISG15, CXCL9, and CXCL10 expression in colon tissue from 5-day-DSS-treated mice, normalized to the OAZ1 housekeeping gene (n = 6 to 14/genotype). (B) CXCL10 protein levels in healthy and inflamed colon tissue were determined by bead assay (n = 5 to 13/genotype). Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0,0001, Mann-Whitney U test.
The genes shown in Fig. 3 are a representative fraction of 27 ISGs examined in total. Compared to expression in WT mice the only ISG mRNA with significantly reduced expression in mice with IRF9−/− and Stat1−/− genotypes but with normal expression in IFN-I/IFN-III receptor double-deficient or Stat2−/− mice was that encoding the chemokine CXCL10, also known as IP10. (Fig. 3A). To examine whether the amount of CXCL10 chemokine corresponded to its mRNA expression, colon tissue was analyzed for protein. As in the case of mRNA, there were no significant differences in CXCL10 protein between healthy tissues of the investigated genotypes (Fig. 3B and not shown). On day 5, levels of CXCL10 were increased in WT colon tissue compared to those in untreated mice, while unchanged in Irf9−/− mice. This difference between WT and Irf9−/− mice was not statistically significant. On day 10, during full-blown colitis, CXCL10 amounts in WT mice were significantly higher than in Irf9−/− mice, where levels were still at baseline. In both Mx1-WT and Mx1-IL28rα−/− Ifnar1−/− mice, on the other hand, CXCL10 was increased on day 10 of colitis compared to that in healthy tissue. In keeping with the pathology scores of Fig. 2, CXCL10 levels were higher in mice lacking type III or both type I and type III IFN receptors (Fig. 3B).
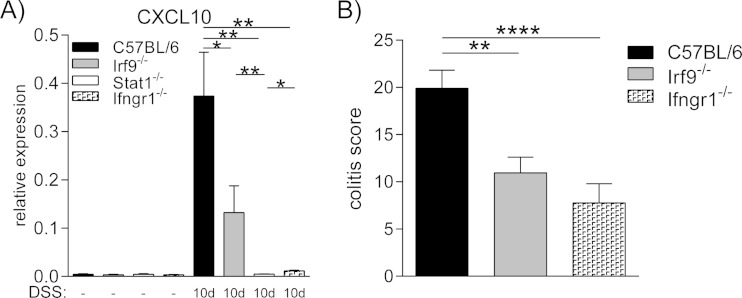
The CXCL10 gene has previously been identified as a target of IFN-γ signaling in colitic tissue (24). To directly compare the dependency of CXCL10 mRNA expression on IRF9 and IFN-γ signaling, we analyzed day 10 colitic tissues of Irf9-, Stat1-, and IFN-γ receptor-deficient animals (Fig. 4). While CXCL10 expression was strongly upregulated in WT tissue at the peak of colitis, its expression was significantly lower in the absence of IRF9 but still upregulated compared to healthy tissue. In the absence of Stat1 or IFN-γ signaling, however, CXCL10 expression was barely detectable. These findings suggest that IRF9, together with Stat1, plays a role in the regulation of the CXCL10 gene by IFN-γ, independently of Stat2, IFN-I, and IFN-III signaling. To examine whether IFN-γ is a driver of DSS-induced colitic pathology, scores of Ifngr1−/−, Irf9−/−, and WT mice at day 10 after DSS treatment were compared (Fig. 4B). In agreement with published results (23, 24), loss of IFN-γ responsiveness, like IRF9 deficiency, protected from DSS-induced colon inflammation.
FIG 4.
IFN-γ stimulates CXCL10 expression and increases the severity of DSS-induced colitis. (A) qPCR analysis of CXCL10 expression in healthy tissue (n = 4 to 11/genotype) compared to day 10 of colitis (n = 4 to 11/genotype), normalized to the OAZ1 housekeeping gene. (B) Pathology scoring of Ifngr1−/− mice and Irf9-deficient animals on day 10 after 2% DSS treatment. Data are presented as means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Mann-Whitney U test.
In some cell types, the loss of IFN-I responsiveness decreases Stat1 expression, due to the absence of autocrine or tonic signals from the IFN-I receptor (39). Reduced Stat1 expression may, in turn, decrease responsiveness to IFN-γ. While potential consequences of tonic signaling to the Stat1 gene should affect Ifnar1−/− cells as well, they might be particularly strong in Irf9−/− cells in case constitutive signals from the IFN-λ receptor maintain Stat1 expression. To investigate whether lack of IRF9 decreases Stat1, we first analyzed its mRNA in the colon of healthy as well as 5-day-DSS-treated mice (see Fig. S1A in the supplemental material). While the loss of IRF9 was without effect on Stat1 mRNA under tissue homeostasis, DSS treatment revealed an approximately 2-fold reduction in Irf9−/− mice. Western blot analysis of mesenteric lymph nodes, spleens, and colons confirmed that steady-state amounts of total Stat1 protein are not reduced in Irf9−/− organs, suggesting intact IFN-γ responsiveness (see Fig. S1B). In spite of its impact on mRNA induction during colitis, the loss of IRF9 did not cause a reduction of Stat1 protein in colonic tissue 5 days after DSS treatment. In keeping with these results, phospho-Stat1 levels in isolated intestinal epithelial cells treated with IFN-γ were highly similar (see Fig. S1C). In further agreement with intact IFN-γ responsiveness, immunity of Irf9−/− mice to Listeria monocytogenes was not decreased (not shown).
Reduced numbers of CXCR3+ cells in DSS-treated, IRF9-deficient mice.
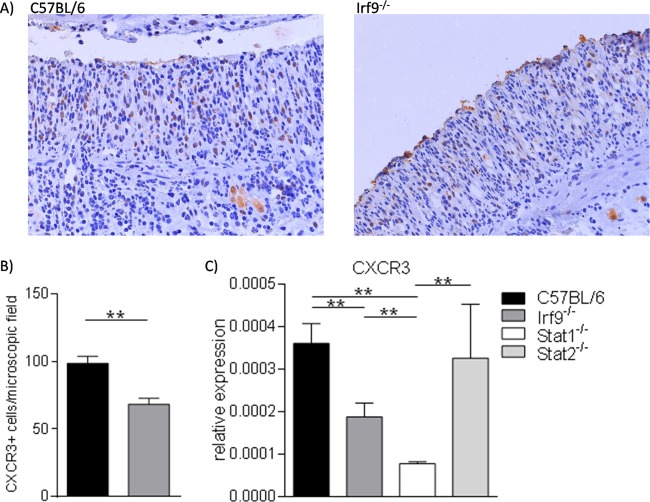
The CXCL10 chemokine attracts various inflammatory cell types expressing the CXCR3 receptor to sites of inflammation (40). We therefore tested whether the reduced CXCL10 expression in inflammatory intestinal tracts of IRF9−/− mice corresponded to a decrease of CXCR3+ cells. Visual inspection of the immunohistochemical analysis shown in Fig. 5A suggested this was the case, and this notion was confirmed by quantitative analysis of CXCR3+ cells (Fig. 5B). In keeping with reduced numbers of CXCL10-responsive cells, the total amount of Cxcr3 mRNA was reduced in colon tissue of IRF9−/− mice (Fig. 5C). Our data strengthen the idea that IRF9-dependent CXCL10 production plays an important role in shaping the inflammatory infiltrate during DSS-induced colitis.
FIG 5.
Colon tissue of IRF9-deficient mice contains reduced numbers of CXCR3-positive cells. (A) Representative images (×20 magnification) of immunohistochemistry with anti-CXCR3 antibodies. Counterstaining was with hematoxylin. Colon sections of WT and Irf9−/− mice at day 10 after treatment with 2% DSS are shown as indicated. (B) Means of CXCR3-positive cells in colon tissue from WT and Irf9−/− mice per microscopic field on day 10 after treatment with 2% DSS (8 to 12 mice per genotype and 5 slides per animal were evaluated). (C) Mean of the relative Cxcr3 mRNA expression in colon tissue on day 10 after treatment with 2% DSS colitis (mean expression of 5 to 8 mice per genotype).
IRF9-dependent gene regulation in IFN-γ-treated macrophages.
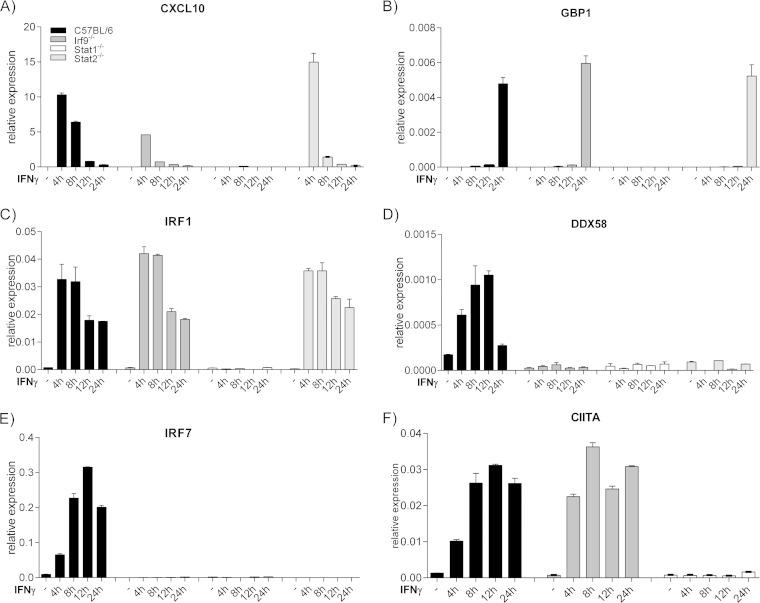
Macrophages are a prime source of CXCL10, particularly when activated by IFN-γ (3). Because it is virtually impossible to purify colon macrophages in sufficient quantities for biochemical experiments, we considered bone marrow macrophages (BMM) as suitable surrogates. qPCR analysis of IFN-γ-inducible genes confirmed that CXCL10 expression required Stat1 and was strongly dependent on IRF9 in this cell type (Fig. 6A). Stat2 independence was observed in the early phase of the CXCL10 gene response to IFN-γ, consistent with Stat2 independence of colonic CXCL10. At later time points, CXCL10 expression was reduced in Stat2−/− BMM. This finding conforms with the regulation of selected ISRE-containing genes by IFN-γ through a noncanonical, Stat2-containing complex designated ISGF3II, acting at delayed stages of the transcriptional response to IFN-γ (10). In keeping with our previous studies, IRF9 was needed for the induction of the IRF7 gene by IFN-γ (12). Contrasting the CXCL10 gene, the response of the Irf7 gene required Stat2 at all time points, suggesting it makes use of ISGF3 complexes (Fig. 6E). The same behavior was noted for the DDX58 gene encoding the Rig-I helicase (Fig. 6D). The other examined genes (Gbp1, Irf1, and CIIta genes) (Fig. 6B, C, and F) behaved like canonical responders, requiring Stat1 but not IRF9 or Stat2. The data suggest that IFN-γ-treated BMM reproduce the IRF9-dependent CXCL10 regulation observed in the gut. They suggest that ISRE-containing promoters select IRF9/Stat1 complexes either with or without the Stat2 subunit for the cellular response to IFN-γ.
FIG 6.
Stat-IRF9 dependence of IFN-γ-induced gene expression in bone marrow macrophages (BMM). (A to F) qPCR analysis of the indicated genes at various times after IFN-γ stimulation of cells with the indicated genotypes. The graph shows three technical replicates from one of three independent experiments.
Association of Stats and IRF9 with the CXCL10 promoter.
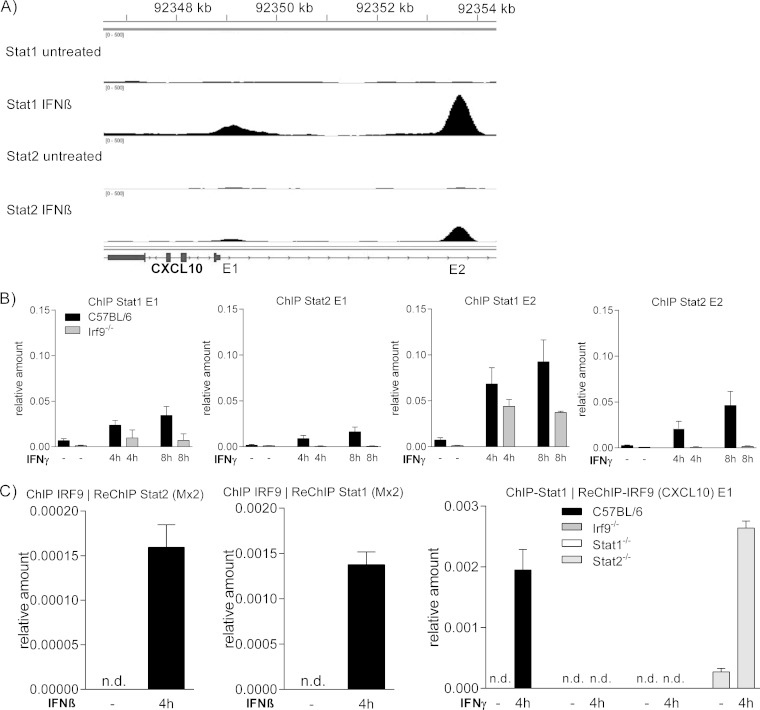
To strengthen the interpretation of our gene expression analysis, ChIP was performed to test protein binding to the IFN-responsive regions of the CXCL10 promoter. Two such regions have been described. One of these (henceforth called enhancer 1 [E1]) is located proximal to the transcription start and contains an ISRE sequence (8, 41). A second IFN response region around −4.6 kB containing both ISRE and GAS sequences was identified by a computer-assisted search (E2) (42). To confirm the presence of functional ISREs in both regions, we interrogated our ChIP-Seq data set from IFN-β-stimulated macrophages. These confirmed the association of Stat1 and Stat2 with both enhancers (Fig. 7A). Beyond that, they showed that the proximal element contains a weaker binding site, particularly for complexes with Stat2.
FIG 7.
Stat-IRF9 association with IFN-responsive elements of the CXCL10 gene. (A) Representation of the CXCL10 gene binding sites for Stat1 and Stat2 as determined by ChIP-Seq. ChIP with antibodies to Stat1 and Stat2 was performed using BMM after stimulation with IFN-β for 2 h, followed by next-generation sequencing. (B) BMM with indicated genotypes, treated with IFN-γ for the indicated times, were subjected to ChIP with antibodies to Stat1 and Stat2. E1 and E2 IFN response regions were amplified by qPCR. (C) Left and middle panels, amplification of the Mx promoter containing the ISRE region after ChIP with antibodies against IRF9 and re-ChIP with Stat1 and Stat2 antibodies. Macrophages were treated with IFN-β for the indicated time. Right panel, amplification of the CXCL10 promoter region after ChIP with an antibody against Stat1 and reprecipitation with an antibody against IRF9 from chromatin of macrophages with indicated genotypes, treated for 4 h with IFN-γ. Three technical replicates from one of three independent experiments are shown. n.d., not detectable.
Consistent with the ChIP-Seq data, Stat1 and Stat2 binding to E2 was stronger than to E1 4 h after treatment with IFN-γ (Fig. 7B). Particularly, Stat2 association with E1 was weak. However, Stat2 binding to both enhancers after 4 h was surprising given the robust Stat2-independent induction of the CXCL10 gene by IFN-γ at this time point. Stat1 association was strongly reduced in the absence of IRF9, particularly at E1. In contrast, Stat2 binding was completely abrogated. The data agree with the idea that Stat2 binding occurs in the context of the ISGF3 or ISGF3II complexes, whereas a fraction of Stat1 binds as homodimers. This notion is in line with the GAS sequence of E2, whereas no overt GAS consensus in E1 has been found by us or others.
The ChIP data summarized in Fig. 7B explain the clear IRF9 dependence of IFN-γ-induced CXCL10 gene expression. However, they do not explain the Stat2 independence of expression 4 h after treatment. To demonstrate the formation of Stat2-independent Stat1-IRF9 complexes, ChIP-re-ChIP analysis with antibodies to Stat1 and IRF9 was performed. Since commercial IRF9 antibodies produce poor signal-to-noise ratios in simple ChIP, the feasibility of the approach was tested using the strong ISGF3 binding site of the Mx1 promoter and IFN-β-stimulated BMM. The left and middle panels of Fig. 7C demonstrate that the technique reveals the simultaneous presence of Stat2 and IRF9 as well as Stat1 and IRF9 at the Mx ISRE. Applying this approach to IFN-γ-stimulated BMM and the CXCL10 gene E1 region, strong ChIP-reChIP signals were obtained in wt and Stat2−/− cells but not in BMM lacking Stat1 or IRF9 (Fig. 7C, right). The data support the notion that Stat2-independent Stat1-IRF9 complexes form at the CXCL10 gene promoter and exert transcriptional activity in the context of the cellular response to IFN-γ.
DISCUSSION
The original aim of this study was to investigate DSS-induced colitis in Irf9−/− animals as a model for the combined loss of IFN-I and IFN-III responses. In the course of our experiments, we realized that the activity of IRF9 was not restricted to the classical ISGF3 complex downstream of the IFN-I and IFN-III receptors. Rather, the dominant function of the protein was to act in concert with Stat1 during IFN-γ signaling. We clearly demonstrate that IFN-III are beneficial by suppressing acute colitis, and this effect is maintained in the absence of the IFN-I receptor. In striking contrast to IFN-III signaling, IRF9 exacerbated colitis, suggesting its proinflammatory role overrides the benefits of its activity in the IFN-III pathway.
The search among candidate genes for the relevant target of IRF9's proinflammatory activity led us to the CXCL10 chemokine. Our results suggest that during colon inflammation, IRF9 is necessary for induction of CXCL10, as in the absence of IRF9, CXCL10 amounts remain at the level of healthy tissue. CXCL10 levels correlate very well with the pathology scores, leading us to the conclusion that the protective effect of IRF9 deficiency is to a large extent due to this decrease. Data presented in Fig. 5 are consistent with an important role of IRF9/CXCL10 signaling for the recruitment of CXCR3+ inflammatory cells. While we cannot rule out a participation of other Stat1-IRF9 target genes, a major role of CXCL10 is strengthened by several reports demonstrating the importance of CXCL10 in immune cell recruitment during inflammation of various organs (40). In different experimental models, blockade of CXCL10 protected from colitis (43–47). Increased proliferation of epithelial cells contributes to this effect (47). Consistently, loss of the CXCL10 receptor, CXCR3, strongly protected mice from DSS-induced colitis (48). Based on these results, anti-CXCL10 treatment appears as an attractive option for clinical therapy. In fact, clinical trials exploring anti-CXCL10 treatment of a patient cohort suffering from ulcerative colitis have been reported (49). Although the prespecified endpoints were not met, the study suggested that alterations of the dose regimen hold promise for future clinical benefit. Various treatment protocols are currently subject to further clinical trials (https://clinicaltrials.gov/ct2/show/results/NCT01294410). The anticolitogenic activity of IFN-λ noted in our study emphasizes the importance of assessing their clinical potential as well.
Originally described as the gene encoding IFN-γ-inducible protein 10 (IP-10) (50), the CXCL10 gene is regulated via ISRE and nuclear factor kappa B (NF-κB) elements (40). Strong transcriptional activation was described for IFN-γ as well as IFN-I. A weaker response is stimulated by tumor necrosis factor (TNF), although TNF can synergize with the IFNs for induction (40). Our results with IFN-γ-treated macrophages show that as in colitic mice, both Stat1 and IRF9 are necessary for full-blown CXCL10 expression. This is in line with earlier studies in human fibrosarcoma cells, leading the authors to propose a nonconventional Stat1-IRF9 complex as the main inducer of expression (8). Our studies in macrophages cannot answer the question of why IFN-γ but not IFN-I or IFN-III regulates CXCL10 expression. The potential to form Stat1-IRF9 complexes exists after activation of the IFN-γ, IFN-I, and IFN-III receptors. In spite of this, we demonstrate in animals with combined loss of IFN-I and -III signaling that neither IFN-I nor IFN-III regulate the CXCL10 gene during DSS-induced colon inflammation. Additionally, the data with the double-deficient mice defy the canonical ISGF3 complex as the dominant regulator of CXCL10 expression. In contrast, IFN-γ signaling creates the IRF9 complexes required for the upregulation of CXCL10. In support of our findings, a study inducing DSS colitis in IFN-γ-deficient mice reported not only a protective effect but also significantly reduced CXCL10/IP-10 levels in inflamed colon tissue (24).
Our in vitro experiments further support the idea that IRF9 is necessary for full CXCL10 induction by IFN-γ and that IRF9 and Stat1 are simultaneously present at the CXCL10 promoter. Noncanonical assemblies of Stats and their associates have been described in other situations as well. For example, Stat2-IRF9 complexes exert transcriptional activity independently of Stat1 in the antiviral response (7, 9, 11). In addition, unphosphorylated Stat complexes were described (U-STATs) (51), and the ISGF3II complex containing phosphorylated Stat1 in association with unphosphorylated Stat2 and IRF9 subunits was described as a mediator of delayed gene induction by IFN-γ (10). The ChIP-re-ChIP analysis of Fig. 7 shows that Stat1-IRF9 complexes form in the absence of Stat2. In addition, however, our experiments in Stat2−/− macrophages suggest that ISGF3 or, more likely, ISGF3II contributes to the delayed transcriptional response of CXCL10 to IFN-γ (Fig. 6 and 7, model in Fig. 8). Initially, Stat2 deficiency produces increased expression, most likely because more Stat1 is available for the formation of Stat1 dimers and Stat1-IRF9 complexes. At later time points, expression decreases with regard to the WT, which we interpret as a need for ISGF3II in maintaining high levels of transcription (Fig. 8) (10). A contribution of Stat1-IRF9 complexes to the delayed IFN-γ response cannot be deduced from our experiments, as both respective knockouts would affect ISGF3II as well. Cumulative CXCL10 mRNA expression over all time points is very similar between WT and Stat2−/− macrophages, and this may account for the lack of a Stat2 effect on steady-state CXCL10 expression in colon tissue of mice at day 5 after DSS treatment (Fig. 3A). As an alternative explanation, different cells and tissues might use different ratios of Stat1 dimers, Stat1-IRF9, ISGF3, or ISGF3II complexes for the IFN-γ response. While being beyond the scope of this study, examination of noncanonical complex formation in primary tissues and different cell types and assessment of its biological relevance are important scientific problems for future research.
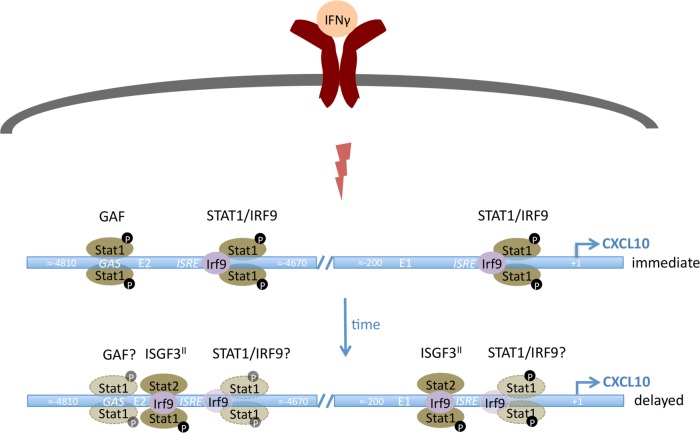
FIG 8.
Graphic representation of CXCL10 regulation by IFN-γ. In the early phase of the transcriptional response, Stat1-IRF9 complexes appear at both response regions. IRF9 deficiency causes a strong but incomplete reduction of transcription, indicating a contribution of GAF, the Stat1 homodimer. Stat2 deficiency increases transcription, demonstrating that in WT, Stat2-containing complexes may be present but of no relevance for promoter activation. At a delayed stage, transcription requires Stat1, Stat2, and IRF9. By inference from data shown by Morrow et al. (10), a complex designated ISGF3II, containing phosphorylated Stat1, unphosphorylated Stat2, and IRF9, may explain the Stat2 dependence. A contribution by Stat1-IRF9 complexes or GAF at this stage is likely but speculative.
Together with our earlier observations of phase and intensity-dependent IFN-I activity during colitis (19), our current study extends the complex impact of the IFN system on colitis to activities of IFN-III, IFN-γ, and noncanonical Stat1-IRF9 signaling, thus demanding further research of new strategies for colitis treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Staeheli, Institute of Virology, University Medical Center Freiburg, Freiburg, Germany, for provision of Mx1-WT, IL28rα−/−, and IL28rα−/− Ifnar1−/− mice.
This work was supported by the Austrian Science Fund (FWF) through SFB-F28 (to B.S., M.M., and T.D.), grant P25186-B22 to T.D., and grant P26011 to L.K. Further support was provided by the Austrian Federal Ministry of Science and Research through GEN-AU III (project InflammoBiota to M.M., L.K., and T.D.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01498-14.
REFERENCES
- 1.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donnelly RP, Kotenko SV. 2010. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res 30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rauch I, Müller M, Decker T. 2013. The regulation of inflammation by interferons and their STATs. JAKSTAT 2:e23820. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moschella F, Torelli GF, Valentini M, Urbani F, Buccione C, Petrucci MT, Natalino F, Belardelli F, Foà R, Proietti E. 2013. Cyclophosphamide induces a type I interferon-associated sterile inflammatory response signature in cancer patients' blood cells: implications for cancer chemoimmunotherapy. Clin Cancer Res 19:4249–4261. doi: 10.1158/1078-0432.CCR-12-3666. [DOI] [PubMed] [Google Scholar]
- 5.González-Navajas JM, Lee J, David M, Raz E. 2012. Immunomodulatory functions of type I interferons. Nat Rev Immunol 12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy DE, Darnell JEJ. 2002. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 7.Fink K, Grandvaux N. 2013. STAT2 and IRF9: beyond ISGF3. JAKSTAT 2:e27521. doi: 10.4161/jkst.27521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majumder S, Zhou LZ, Chaturvedi P, Babcock G, Aras S, Ransohoff RM. 1998. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J Immunol 161:4736–4744. [PubMed] [Google Scholar]
- 9.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. 2011. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog 7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrow AN, Schmeisser H, Tsuno T, Zoon KC. 2011. A novel role for IFN-stimulated gene factor 3II in IFN-γ signaling and induction of antiviral activity in human cells. J Immunol 186:1685–1693. doi: 10.4049/jimmunol.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fink K, Martin L, Mukawera E, Chartier S, De Deken X, Brochiero E, Miot F, Grandvaux N. 2013. IFN-β/TNFα synergism induces a noncanonical STAT2/IRF9-dependent pathway triggering a novel DUOX2 NADPH Oxidase-mediated airway antiviral response. Cell Res 23:673–690. doi: 10.1038/cr.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farlik M, Rapp B, Marie I, Levy DE, Jamieson AM, Decker T. 2012. Contribution of a TANK-binding kinase 1-interferon (IFN) regulatory factor 7 pathway to IFN-γ-induced gene expression. Mol Cell Biol 32:1032–1043. doi: 10.1128/MCB.06021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommereyns C, Paul S, Staeheli P, Michiels T. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog 4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. 2007. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol 81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy DE, Marié IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maloy KJ, Powrie F. 2011. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 17.Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, Kenner L, Muller M, Rauch I, Strobl B, Wagner M, Schleper C, Urich T, Loy A. 2012. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J 6:2091–2106. doi: 10.1038/ismej.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rauch I, Hainzl E, Rosebrock F, Heider S, Schwab C, Berry D, Stoiber D, Wagner M, Schleper C, Loy A, Urich T, Müller M, Strobl B, Kenner L, Decker T. 2014. Type I interferons have opposing effects during the emergence and recovery phases of colitis. Eur J Immunol 44:2749–2760. doi: 10.1002/eji.201344401. [DOI] [PubMed] [Google Scholar]
- 20.Salk A, Stobaugh DJ, Deepak P, Ehrenpreis ED. 2013. Ischemic colitis with type I interferons used in the treatment of hepatitis C and multiple sclerosis: an evaluation from the food and drug administration adverse event reporting system and review of the literature. Ann Pharmacother 47:537–542. doi: 10.1345/aph.1R526. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y, Lin Y, Lin L, Zheng C. 2012. IL-17/IFN-γ interactions regulate intestinal inflammation in TNBS-induced acute colitis. J Interferon Cytokine Res 32:548–556. doi: 10.1089/jir.2012.0030. [DOI] [PubMed] [Google Scholar]
- 22.Sheikh SZ, Matsuoka K, Kobayashi T, Li F, Rubinas T, Plevy SE. 2010. Cutting edge: IFN-β is a negative regulator of IL-23 in murine macrophages and experimental colitis. J Immunol 184:4069–4073. doi: 10.4049/jimmunol.0903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nava P, Koch S, Laukoetter MG, Lee WY, Kolegraff K, Capaldo CT, Beeman N, Addis C, Gerner-Smidt K, Neumaier I, Skerra A, Li L, Parkos CA, Nusrat A. 2010. Interferon-gamma regulates intestinal epithelial homeostasis through converging beta-catenin signaling pathways. Immunity 32:392–402. doi: 10.1016/j.immuni.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito R, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, Imanishi J, Kita M, Ueda Y, Iwakura Y, Kataoka K, Okanoue T, Mazda O. 2006. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol 146:330–338. doi: 10.1111/j.1365-2249.2006.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obermeier F, Kojouharoff G, Hans W, Scholmerich J, Gross V, Falk W. 1999. Interferon-gamma (IFN-γ)-and tumour necrosis factor (TNF)-induced nitric oxide as toxic effector molecule in chronic dextran sulphate sodium (DSS)-induced colitis in mice. Clin Exp Immunol 116:238–245. doi: 10.1046/j.1365-2249.1999.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hans W, Scholmerich J, Gross V, Falk W. 2000. Interleukin-12 induced interferon-gamma increases inflammation in acute dextran sulfate sodium induced colitis in mice. Eur Cytokine Netw 11:67–74. [PubMed] [Google Scholar]
- 27.Pott J, Mahlakõiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. 2011. IFN-λ determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A 108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte J-M, Diebold J, Diepolder H, Adler B, Auernhammer CJ, Göke B, Dambacher J. 2005. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol 289:G960–G968. doi: 10.1152/ajpgi.00126.2005. [DOI] [PubMed] [Google Scholar]
- 29.Bierne H, Travier L, Mahlakoiv T, Tailleux L, Subtil A, Lebreton A, Paliwal A, Gicquel B, Staeheli P, Lecuit M, Cossart P. 2012. Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS One 7:e39080. doi: 10.1371/journal.pone.0039080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mordstein M, Kochs G, Dumoutier L, Renauld J-C, Paludan SR, Klucher K, Staeheli P. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog 4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horisberger MA, Staeheli P, Haller O. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc Natl Acad Sci U S A 80:1910–1914. doi: 10.1073/pnas.80.7.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan RS, Takasugi T, Matsuyama T, Mak TW, Noguchi S, Taniguchi T. 1996. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells 1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 33.Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450. doi: 10.1016/S0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 34.Park C, Li S, Cha E, Schindler C. 2000. Immune response in Stat2 knockout mice. Immunity 13:795–804. doi: 10.1016/S1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 35.Williams KL, Fuller CR, Dieleman LA, DaCosta CM, Haldeman KM, Sartor RB, Lund PK. 2001. Enhanced survival and mucosal repair after dextran sodium sulfate-induced colitis in transgenic mice that overexpress growth hormone. Gastroenterology 120:925–937. doi: 10.1053/gast.2001.22470. [DOI] [PubMed] [Google Scholar]
- 36.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T. 2004. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol 173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 37.Wienerroither S, Rauch I, Rosebrock F, Jamieson AM, Bradner J, Muhar M, Zuber J, Müller M, Decker T. 2014. Regulation of NO synthesis, local inflammation, and innate immunity to pathogens by BET family proteins. Mol Cell Biol 34:415–427. doi: 10.1128/MCB.01353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers GB, Kozlowska J, Keeble J, Metcalfe K, Fao M, Dowd SE, Mason AJ, McGuckin MA, Bruce KD. 2014. Functional divergence in gastrointestinal microbiota in physically separated genetically identical mice. Sci Rep 4:5437. doi: 10.1038/srep05437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gough DJ, Messina NL, Clarke CJP, Johnstone RW, Levy DE. 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groom JR, Luster AD. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohmori Y, Hamilton TA. 1993. Cooperative interaction between interferon (IFN) stimulus response element and kappa B sequence motifs controls IFN gamma- and lipopolysaccharide-stimulated transcription from the murine IP-10 promoter. J Biol Chem 268:6677–6688. [PubMed] [Google Scholar]
- 42.Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F, Owen MR, Naumann R, Decker T, Vinkemeier U. 2014. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat Immunol 15:168–176. doi: 10.1038/ni.2794. [DOI] [PubMed] [Google Scholar]
- 43.Hyun JG, Lee G, Brown JB, Grimm GR, Tang Y, Mittal N, Dirisina R, Zhang Z, Fryer JP, Weinstock JV, Luster AD, Barrett TA. 2005. Anti-interferon-inducible chemokine, CXCL10, reduces colitis by impairing T helper-1 induction and recruitment in mice. Inflamm Bowel Diseases 11:799–805. doi: 10.1097/01.MIB.0000178263.34099.89. [DOI] [PubMed] [Google Scholar]
- 44.Singh UP, Singh S, Singh R, Cong Y, Taub DD, Lillard JW. 2008. CXCL10-producing mucosal CD4+ T cells, NK cells, and NKT cells are associated with chronic colitis in IL-10(-/-) mice, which can be abrogated by anti-CXCL10 antibody inhibition. J Interferon Cytokine Res 28:31–43. doi: 10.1089/jir.2007.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh UP, Singh S, Taub DD, Lillard JW. 2003. Inhibition of IFN-gamma-inducible protein-10 abrogates colitis in IL-10-/- mice. J Immunol 171:1401–1406. doi: 10.4049/jimmunol.171.3.1401. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki K, Kawauchi Y, Palaniyandi SS, Veeraveedu PT, Fujii M, Yamagiwa S, Yoneyama H, Han GD, Kawachi H, Okada Y, Ajioka Y, Watanabe K, Hosono M, Asakura H, Aoyagi Y, Narumi S. 2007. Blockade of interferon-gamma-inducible protein-10 attenuates chronic experimental colitis by blocking cellular trafficking and protecting intestinal epithelial cells. Pathol Int 57:413–420. doi: 10.1111/j.1440-1827.2007.02117.x. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki S, Yoneyama H, Suzuki K, Suriki H, Aiba T, Watanabe S, Kawauchi Y, Kawachi H, Shimizu F, Matsushima K, Asakura H, Narumi S. 2002. Blockade of CXCL10 protects mice from acute colitis and enhances crypt cell survival. Eur J Immunol 32:3197–3205. doi:. [DOI] [PubMed] [Google Scholar]
- 48.Chami B, Yeung AWS, van Vreden C, King NJC, Bao S. 2014. The role of CXCR3 in DSS-induced colitis. PLoS One 9:e101622. doi: 10.1371/journal.pone.0101622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer L, Sandborn WJ, Stepanov Y, Geboes K, Hardi R, Yellin M, Tao X, Xu LA, Salter-Cid L, Gujrathi S, Aranda R, Luo AY. 2014. Anti-IP-10 antibody (BMS-936557) for ulcerative colitis: a phase II randomised study. Gut 63:442–450. doi: 10.1136/gutjnl-2012-303424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luster AD, Unkeless JC, Ravetch JV. 1985. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 51.Cheon H, Stark GR. 2009. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A 106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.