Abstract
Hypoxia often occurs under various physiological and pathophysiological conditions, including solid tumors; it is linked to malignant transformation, metastatic progression, and treatment failure or resistance. Tip110 protein plays important roles in several known physiological and pathophysiological processes, including cancers. Thus, in the present study we investigated the regulation of Tip110 expression under hypoxia. Hypoxia led to Tip110 protein degradation through the ubiquitin-proteasome system. Under hypoxia, Tip110 stabilized p53, which in return destabilized Tip110. In addition, Tip110 regulated hypoxia-inducible factor 1α (HIF-1α), likely through enhancement of its protein stability. Furthermore, Tip110 upregulated p300, a known coactivator for both p53 and HIF-1α. Expression of a p53(22/23) mutant deficient in p300 binding accelerated Tip110 degradation under hypoxia. Tip110 knockdown resulted in the inhibition of cell proliferation and cell death in the presence of p53. Finally, significantly less Tip110, p53, and HIF-1α was detected in the hypoxic region of bone metastasis tumors in a mouse model of human melanoma cells. Taken together, these results suggest Tip110 is an important mediator in the cross talk between p53 and HIF-1α in response to hypoxic stress.
INTRODUCTION
Hypoxia is the common characteristic of many solid tumors. The adaptation of cells to hypoxia is mediated by hypoxia-inducible factor (HIF), a transcription factor, at the molecular level (1). Under normal oxygen conditions (normoxia), HIF-1α is hydroxylated, which promotes its binding to the ubiquitin ligase von Hippel-Lindau protein (pVHL), thereby targeting it for ubiquitin-proteasome system (UPS)-mediated degradation. However, under hypoxic conditions, HIF-1α becomes less hydroxylated, leading to its rapid accumulation and subsequent activation of hundreds of genes involved in cell survival, as well as genes involved in apoptosis (2). This opposing function of HIF in determining different cell fates is dependent on the physiopathological context and differential binding to other key partners, such as tumor suppressor protein p53.
Similarly to HIF-1α, p53 stability is also regulated by the hypoxic condition. p53 plays a crucial role in response to DNA damage, aberrant cell control, apoptosis, and senescence (3, 4). p53 function is constitutively regulated in different types of tumors under hypoxia by different mechanisms, such as p53 mutation, expression of inhibitors, or unknown host regulatory elements leading to induction of resistance to p53-mediated apoptosis. In normal cells, p53 protein expression is maintained at a low, often undetectable level due to ubiquitin-mediated proteasome degradation (5). Upon exposure to stress, such as oncogenic activation and certain hypoxic situations, p53 becomes stabilized. Consequently, p53 activates genes involved in cell cycle regulation and genes involved in apoptotic events (4).
HIV-1 Tat-interacting protein 110 (Tip110), also known as “squamous cell carcinoma antigen recognized by T cells 3” (SART3), is a nuclear protein and contains two RNA recognition motifs (RRMs) (6, 7). Tip110 regulates transcription of viral and several host genes and plays an important role in pre-mRNA splicing and spliceosome assembly (7–12). Tip110 expression is essential for embryonic development (13). Recently we have reported that UPS-mediated degradation of human Tip110 (hTip110) is regulated by oncogenic USP15 protein (14). Tip110 protein expression is very low in the normal tissues and nonproliferating cells (15) but becomes highly elevated in a number of malignant tumor cell lines and cancerous tissues as well as stem cells (16–25). Furthermore, Tip110 serves as a tumor antigen and could be used as a cancer immunotherapy adjuvant (26–28). The Tip110 protein expression level is also important for hematopoietic stem cell differentiation, which resides in the hypoxic bone marrow environment through reciprocal regulation of transcription factor c-Myc expression (19) and alternative splicing of OCT4 (20). In addition, Tip110 interacts with oncogenic transcription factor YB-1 and promotes the inclusion of exon 5 in CD44 alternative splicing (11). Both c-Myc and YB-1 proteins are regulated under hypoxic conditions (29, 30).
In the present study, we investigated Tip110 regulation under hypoxia and its relationship to HIF-1α and p53, two important regulators of hypoxia. We took advantage of a pair of osteosarcoma cell lines that differ in p53 status as an experimental model. We showed that Tip110 was degraded under hypoxia in vitro and in a mouse model of bone metastasis. The degradation was mainly mediated by the ubiquitin-proteasome system. The regulation of Tip110 protein level under hypoxia was p53 dependent; Tip110 overexpression enhanced HIF-1α protein stability. These findings together suggest an important role of Tip110 in the p53-HIF1α cross talk in a tumor hypoxic environment.
MATERIALS AND METHODS
Cell cultures and transfection.
U2OS (p53+/+), Soas-2 (p53−/−), and SW480 cells were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS). The parental 1205Lu melanoma cell line (31) was grown in a composite medium (W489) consisting of 3 parts MCBD153 (Sigma-Aldrich, St. Louis, MO) and 1 part L15 (Sigma-Aldrich), supplemented with 4% FBS and 2 mM l-glutamine (Sigma-Aldrich). The cell lines were supplemented with 100 IU/ml penicillin and 100 μg/ml streptomycin and cultured at 37°C, 21% O2, and 5% CO2 in a standard and humidified tissue culture chamber (normoxia). For hypoxia studies, the cells were cultured at 37°C under hypoxia (1% O2, 5% CO2, and 99% N2) or severe hypoxia (0.1% O2, 5% CO2, and 99.9% N2) in a humidified chamber of an INVIVO2 200 hypoxia workstation (Ruskinn Technologies, Bridgend, United Kingdom). Plasmid DNA and small interfering RNA (siRNA) were transfected using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA).
DNA plasmids and siRNAs.
The plasmids for expression of hTip110-His, Tip110–promoter-driven luciferase reporter gene (Tip110-Prom-Luc), and p300 promoter-driven luciferase reporter gene (p300-Prom-Luc) were described elsewhere (9, 10). The plasmid for expression of hemagglutinin-tagged ubiquitin (Ub-HA) was kindly provided by Mark Hannink of University of Missouri, Columbia, MO (32). p53 and p53(22/23) plasmids were kindly provided by Jiayuh Lin of The Research Institute at Nationwide Children's Hospital, Columbus, OH (33). Tip110 siRNA and control siRNA were purchased from Thermo Scientific (Lafayette, CO). p53 siRNA was purchased from Sigma-Aldrich.
Western blotting.
Cells were washed in phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 2 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail). Lysates were cleared of cell debris by centrifugation and fractionated by SDS-PAGE, followed by Western blotting. The antibodies were monoclonal anti-Tip110 (19), polyclonal anti-HIF-1α (H-206, sc-10790; Santa Cruz Biotechnologies, Santa Cruz, CA), polyclonal anti-p53 (FL-393, sc-6243; Santa Cruz Biotechnologies), monoclonal anti-p53 (DO-1, sc-126; Santa Cruz Biotechnologies), monoclonal anti-p300 (Novus Biological, CO), monoclonal anti-His tag (G020; ABM, Richmond, British Columbia, Canada), and monoclonal anti-β-actin (clone AC-15; Sigma, St. Louis, MO). ImageJ was used to quantitate protein expression levels.
qRT-PCR.
RNA was extracted from cells using TRizol (Invitrogen) according to the manufacturer's instructions. RNA (1 μg) was converted into cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and used as the template for PCR using Sso Advanced SYBR green Supermix and the CFX96 real-time PCR detection system (Bio-Rad). The quantitative reverse transcription-PCR (qRT-PCR) primers used and their sequences are as follows: for β-actin, 5′-AAA CTG GAA CGG TGA AGG TG-3′ and 5′-AGA GAA GTG GGG TGG CTT TT-3′; for Tip110, 5′-GGC TAG GAT TGA GGC TCG ACT G-3′ and 5′-GGG TGT CAC CAT GAG CTC TTT CC-3′; and for p53, 5′-CTC AGA TAG CGA TGG TCT GG-3′ and 5′-GGT GGT ACA GTC AGA GCC AA-3′. Threshold cycle (CT) values were calculated using Bio-Rad CFX manager software. The 2−ΔΔCT value was calculated to represent the fold change of the target gene mRNA under hypoxia compared to that under normoxia and normalized using β-actin as the reference.
Luciferase reporter gene assay.
The firefly luciferase activity was measured using the luciferase assay substrate (Promega, Madison, WI) according to the manufacturer's instructions. Briefly, cells were washed with ice-cold PBS and lysed with 120 μl 1× firefly luciferase lysis buffer (Promega) at room temperature for 15 min. The lysates were centrifuged at 12,000 × g for 2 min to remove cell debris. The cleared lysates (5 μl) were then mixed with 20 μl firefly luciferase substrate (Promega), and the luciferase activity was measured using an Opticomp luminometer (MGM Instruments, Hamden, CT).
Flow cytometry.
For cell cycle analysis, ∼70% confluent cells were grown on a 12-well tissue culture plate. On the following day, cells were transfected with an universal control siRNA (Si-Ctrl) or Tip110 siRNA (si-Tip110) and cultured for 48 h. Cells were washed with ice-cold PBS, trypsinized, harvested, and fixed in 1 ml of 70% ethanol and then stored at −20°C until the analysis. Cells were centrifuged at 700 × g for 5 min. The cell pellets were washed two times with PBS and suspended in 500 μl propidium iodide (PI) (Sigma) staining solution (40 μg/ml PI in 3.8 mM sodium citrate) and 50 μl RNase A (10 μg/ml) (Sigma) and incubated at 37°C for 30 min. Cell cycle was analyzed using FACScaliber (Becton Dickinson, CA) and ModFit LT software (Becton Dickinson, CA). A total of 50,000 cells were analyzed.
MTT assay.
Cells were seeded in a 12-well tissue culture plate and cultured overnight. The cells were transfected with the universal control siRNA (Si-Ctrl) or Tip110 siRNA (si-Tip110) for 48 h and cultured under normoxia (21% O2) or hypoxia (1% O2) for 24 h. MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] (120 μl of 50 mg/ml) was added to the culture and incubated for 4 to 5 h. The culture media were then removed, and the violet crystals in the well were dissolved by addition of 200 μl acid-isopropanol (3:22 ratio mixture of 0.2 N HCl and isopropanol), followed by gentle shaking at room temperature for 20 min. Optical densities at 655 and 595 nm (OD655 and OD595, respectively) were taken using an iMark microplate absorbance reader (Bio-Rad) and used to calculate the differences.
Data analysis.
Where appropriate, values are expressed as means ± standard deviations (SD) from triplicate samples. All comparisons were made based on the control using a two-tailed Student's t test. P values of <0.05 were considered statistically significant, <0.01 highly significant, and <0.001 strongly significant. All data are representative of multiple independent experiments.
RESULTS
Hypoxia promoted Tip110 protein degradation.
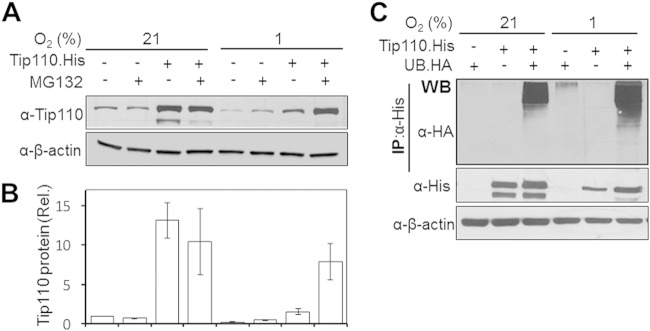
Our first attempt was to investigate the status of the Tip110 protein level under hypoxia. Cells of the colon adenocarcinoma line SW480 were transfected with Tip110-His or its backbone cDNA3. One set of transfected cells was cultured under hypoxia (1% O2), and the other set of cells was cultured under normoxia (21% O2). After being cultured for 24 h, the cells were harvested for Tip110 expression by Western blotting. Compared to normoxia, hypoxia showed considerable decreases in both endogenous and exogenous Tip110 proteins (Fig. 1A and B). To determine whether UPS was involved in hypoxia-induced Tip110 degradation, the transfected cells were treated with MG132, a well-characterized proteasomal inhibitor of the 26S proteasome within the UPS that detects and degrades ubiquitinated proteins in the cells, followed by Tip110 analysis by Western blotting. MG132 treatment led to stabilization of endogenous and exogenous Tip110 protein under hypoxic conditions (Fig. 1A and B). Previously, we reported that Tip110 is ubiquitinated (14). Thus, to determine whether hypoxia would affect Tip110 ubiquitination, cells were transfected with Tip110-His, Ub-HA, or both. Immunoprecipitation by an anti-His antibody, followed by Western blotting by an anti-HA antibody, detected more ubiquitinated Tip110 under hypoxia than that under normoxia (Fig. 1C). These results suggest that hypoxia induced Tip110 protein degradation through the UPS. Similar results were obtained using human embryonic kidney fibroblasts of the 293T line (data not shown).
FIG 1.
Tip110 degradation under hypoxia. (A and B) SW480 cells were transfected with Tip110-His, treated with 20 μM MG132, cultured at 21 or 1% O2 for 24 h, and harvested for Tip110 expression by Western blotting (A). The Tip110 protein level was quantitated using the loading control β-actin as a reference (B). Rel., relative. (C) SW480 cells were transfected with Ub-HA, Tip110-His, or both, treated with 20 μM MG132, and cultured at 21 or 1% O2 for 24 h. Cell lysates were immunoprecipitated (IP) using an anti-His antibody, followed by Western blotting using an anti-HA antibody. β-Actin was included as a loading control for Western blotting. The data are representative of three independent experiments.
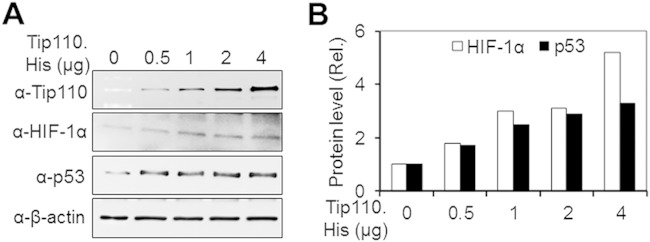
p53 and HIF-1α are two transcription factors that have major roles in numerous cellular pathways in response to hypoxia (34). Thus, we next determined whether Tip110 expression would alter p53 and HIF1-α protein levels. SW480 cells were transfected with increasing amounts of Tip110-His and monitored for HIF-1α and p53 expression by Western blotting. Increasing Tip110 expression resulted in upregulation of both p53 and HIF-1α protein in a dose-dependent manner (Fig. 2), suggesting possible roles of Tip110 in the regulation of both p53 and HIF-1α under limited-oxygen conditions.
FIG 2.
Relationship between Tip110, p53, and HIF-1α. (A and B) SW480 cells were transfected with increasing amounts of Tip110-His, cultured at 21% O2 for 24 h, and harvested for Tip110, p53, and HIF-1α expression by Western blotting. cDNA3 was used to equalize the total amount of DNA among all transfections (A). HIF-1α and p53 protein levels were quantitated using β-actin as a reference (B). β-Actin was included as a loading control for Western blotting.
Increased Tip110 degradation by p53 expression under hypoxia.
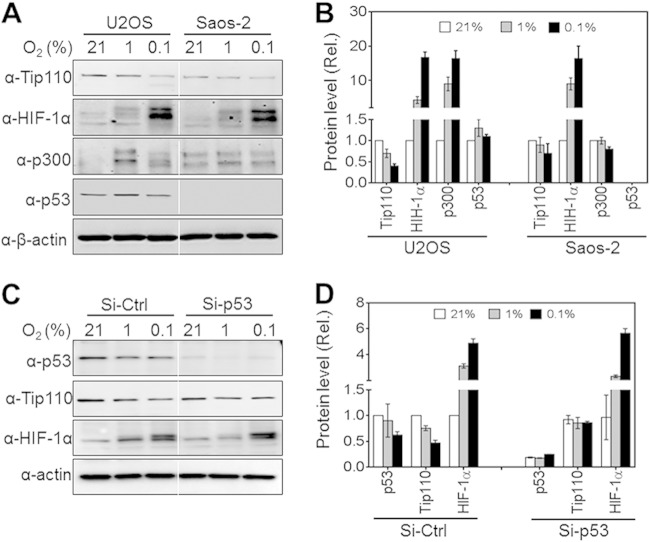
SW480 cells express a mutated p53 (35–37). Because of the possible roles of Tip110 in the regulation of p53, we decided to use cell line U2OS, a human bone osteosarcoma line that expresses wild-type p53, and its cognate p53-null cell line, Saos-2, for the rest of this study. To determine the effects of different hypoxic conditions on the endogenous Tip110 protein stability in the context of p53, U2OS and Saos-2 cells were cultured under 21, 1, and 0.1% (severe hypoxia) O2 for 24 h, and the cells were harvested for Tip110 expression by Western blotting. In U2OS cells, hypoxia led to lower endogenous Tip110 protein than normoxia, and severe hypoxia showed even more lower endogenous Tip110 protein than normoxia (Fig. 3). In Saos-2 cells, there was little change of endogenous Tip110 protein from normoxia to hypoxia, and there was only a slight decrease of endogenous Tip110 protein even under severe hypoxia. Quantitation showed that U2OS exhibited more rapid endogenous Tip110 degradation than Saos-2 (Fig. 3B). Meanwhile, the levels of HIF-1α, p300, and p53 in those cells were also determined by Western blotting. As expected, HIF-1α protein was stabilized in both U2OS and Saos-2 under hypoxic and severely hypoxic conditions but with different kinetics (38). Higher levels of p300 and p53 proteins were also detected in U2OS under conditions of hypoxia and severe hypoxia. To exclude the possibility that different cellular backgrounds other than p53 expression affect the Tip110 expression level, U2OS cells were transfected with control or p53 siRNA, cultured under 21, 1, and 0.1% O2 for 24 h, and analyzed for p53, Tip110, and HIF-1α expression by Western blotting. Similarly, hypoxia led to a significantly lower level of endogenous Tip110 in U2OS cells but not in USOS cells with p53 knocked down (Fig. 3C and D). These results together suggest that p53 regulates Tip110 degradation under hypoxic conditions.
FIG 3.
Effects of p53 on Tip110 degradation under hypoxia. (A and B) U2OS cells expressing wild-type p53 (p53+/+) or Saos-2 cells expressing no p53 (p53−/−) were cultured under 21, 1, or 0.1% O2 for 24 h and harvested for Tip110, HIF-1α, p300, and p53 expression by Western blotting (A). Levels of Tip110, HIF-1α, p300, and p53 protein expression were quantitated using β-actin as a reference (B). (C and D) U2OS cells were transfected with a universal siRNA control (Si-Ctrl) or p53 siRNA (Si-Tip110), cultured under 21, 1, or 0.1% O2 for 24 h, and harvested for Western blotting (C). Levels of Tip110, HIF-1α, p300, and p53 protein expression were quantitated using β-actin as a reference (D). The data are representative of three independent experiments.
Transactivation of Tip110 expression by p53.
To further verify the relationship between p53 and Tip110, Saos-2 cells were transfected with increasing amounts of p53 expression plasmid, cultured under normoxia and hypoxia, and harvested for Tip110 expression by Western blotting. p53 expression increased the endogenous Tip110 level under normoxia in a dose-dependent manner (Fig. 4A). In agreement with the previous findings (Fig. 3), p53 expression promoted Tip110 degradation under hypoxia. To determine whether p53 would transactivate Tip110 transcription, Saos-2 cells were transfected with a Tip110 promoter-driven luciferase reporter plasmid, pGL3.Tip110-Prom-Luc, and increasing amounts of p53 expression plasmids, cultured under normoxia and hypoxia, and harvested for the luciferase reporter gene activity assay. p53 expression transactivated the Tip110 promoter-driven luciferase reporter gene in a dose-dependent manner under both normoxia and hypoxia, although the transactivation was greater under normoxia than that under hypoxia (Fig. 4B). These results showed that p53 transactivated Tip110 transcription and increased Tip110 protein levels under normoxia and confirmed that p53 promoted Tip110 degradation under hypoxia.
FIG 4.
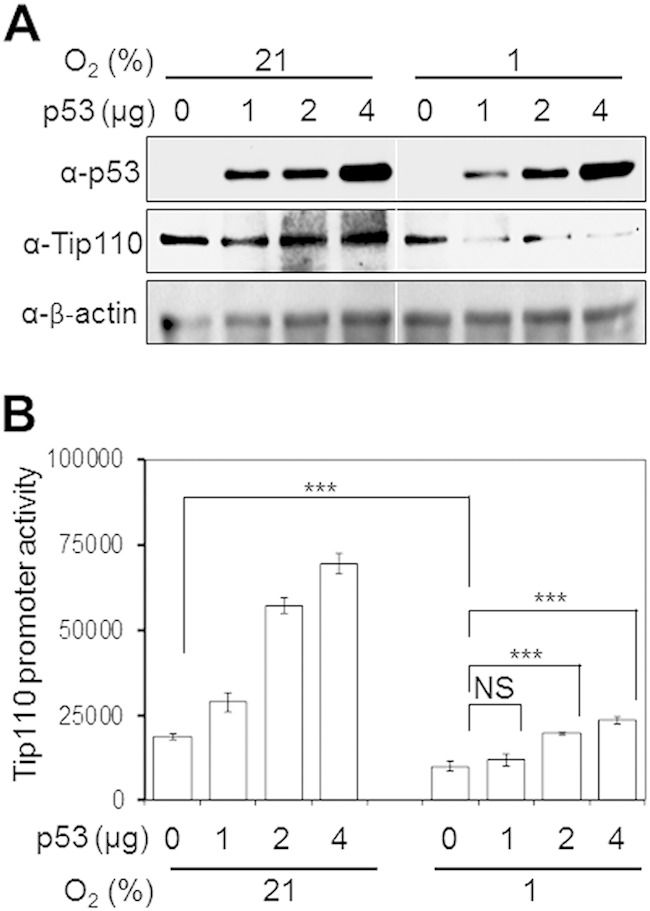
Effects of p53 expression on Tip110 protein and transcription under hypoxia. (A) Saos-2 cells were transfected with the indicated amounts of p53, cultured at 21 or 1% O2 for 24 h, and harvested for Tip110 and p53 protein expression by Western blotting. cDNA3 was used to equalize the total amount of DNA among all transfections. β-Actin was included as a loading control. (B) Saos-2 cells were transfected with 250 ng pGL3-Tip110 Prom-Luc alone or in combination with 0.5, 1, and 2 μg p53 for 24 h and cultured under 21 or 1% O2 for 24 h. Cells were harvested for the luciferase reporter gene assay. cDNA3 was used to equalize the total amount of DNA among all transfections. pTK-βgal was included to normalize the transfection efficiency variations among all transfections. The data are means ± standard errors (SE) of triplicate samples and representative of three independent experiments. ***, strongly significant (P < 0.001); NS, not significant.
Regulation of p53 and HIF-1α stability by Tip110.
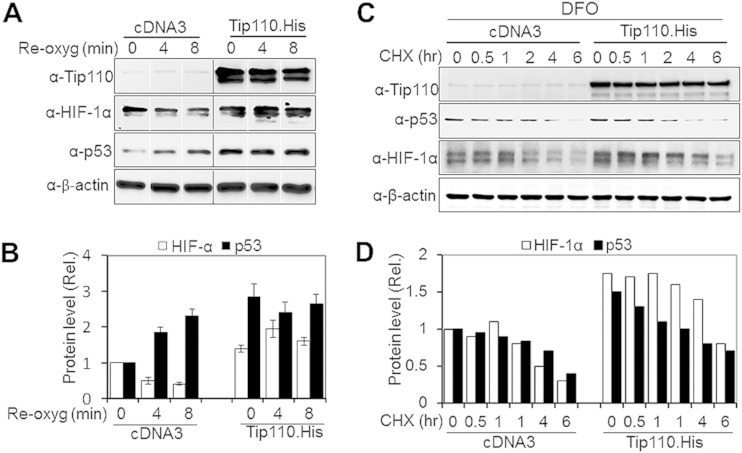
Next, we determined whether Tip110 expression would alter p53 and HIF-1α protein levels under both normoxic and hypoxic conditions. U2OS and Saos-2 were transfected with Tip110-His, cultured under normoxia and hypoxia, and then analyzed for p53 and HIF-1α protein levels. In U2OS, Tip110 expression showed more increases in p53, p300, and HIF-1α protein levels under hypoxia (Fig. 5A and B). In Saos-2 cells, Tip110 expression also led to increased HIF-1α and p300 protein levels under both normoxia and hypoxia (Fig. 5C and D). In addition, U2OS and Saos-2 were transfected with Tip110 siRNA or control siRNA, cultured under normoxia and hypoxia, and then analyzed for p53 and HIF-1α protein levels. Tip110 knockdown led to more decreases of the p53 and HIF-1α protein levels in U2OS cells under hypoxia (Fig. 6A and B) and lower levels of HIF-1α in Saos-2 cells under normoxia and hypoxia (Fig. 6C and D). To determine whether the increased p53 protein occurred at the transcriptional level, U2OS cells were transfected with Tip110-His, cultured under normoxia and hypoxia, and harvested for determination of Tip110 and p53 mRNA levels by qRT-PCR. Consistent with previous findings (Fig. 1 to 3), hypoxia showed little effects on the Tip110 mRNA level (Fig. 7A). As expected, hypoxia showed little increases in p53 mRNA (39, 40). Tip110 expression showed no effects on the p53 mRNA level under both normoxia and hypoxia (Fig. 7B). To determine whether Tip110 expression also led to increased stability of HIF-1α, we performed the reoxygenation experiment. U2OS cells were transfected with Tip110-His, cultured under hypoxia for 24 h, and then transferred to normoxia for different lengths of time and harvested for Tip110 and HIF-1α expression by Western blotting. In the absence of Tip110 overexpression, the hypoxia-induced HIF-1α protein became gradually degraded (41, 42) (Fig. 8A and B). In contrast, in the presence of Tip110 overexpression, there was little change in hypoxia-induced HIF-1α protein. In addition, the p53 protein level was also determined. Consistent with a previous report (43), reoxygenation led to p53 stabilization. As shown before (Fig. 1, 3, and 6), Tip110 overexpression also led to increased p53 stability after reoxygenation. HIF-1α and p53 protein turnover was further determined by treatment of Tip110-transfected U2OS or its control with deferoxamine mesylate (DFO), a chemical that has been widely used to mimic hypoxia by blocking HIF-1α protein degradation (44), for 24 h with addition of cycloheximide, an inhibitor of protein synthesis, for various lengths of time, and then harvested to determine HIF-1α and p53 protein expression by Western blotting. Consistent with our previous observations, overexpression of Tip110 increased the half-life of p53 and HIF-1α (Fig. 8C and D). These results together suggest that Tip110 expression increased p53 and HIF-1α protein stability.
FIG 5.
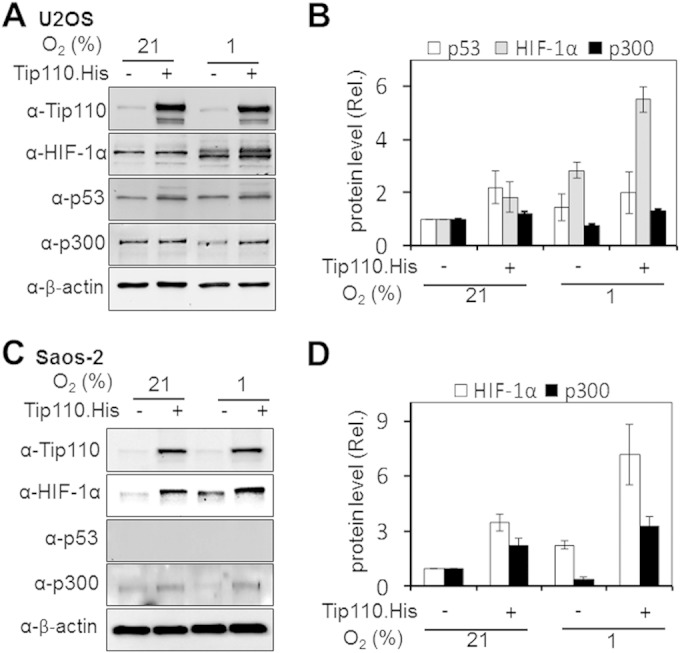
Effects of Tip110 expression on p53 and HIF-1α under hypoxia. (A to D) U2OS (A) and Saos-2 (C) cells were transfected with a Tip110-His or cDNA3 plasmid for 24 h and cultured under 21% or 1% O2 for 24 h. Cells were harvested and prepared for Tip110, p53, HIF-1α, and p300 protein expression by Western blotting. Levels of Tip110, HIF-1α, p300, and p53 protein expression were quantitated using β-actin as a reference (B and D). The data are representative of three independent experiments.
FIG 6.
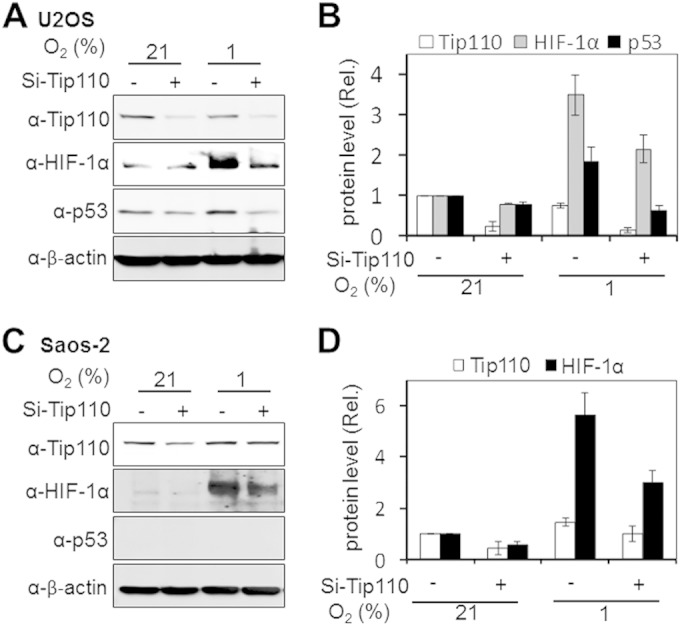
Effects of Tip110 knockdown on p53 and HIF-1α under hypoxia. (A to D) U2OS (A) and Saos-2 (C) cells were transfected with a universal siRNA control (Si-Ctrl) or Tip110 siRNA (Si-Tip110) for 48 h and cultured under 21 or 1% O2 for 24 h. Levels of Tip110, HIF-1α, and p53 protein expression were quantitated using β-actin as a reference (B and D). The data are representative of three independent experiments.
FIG 7.
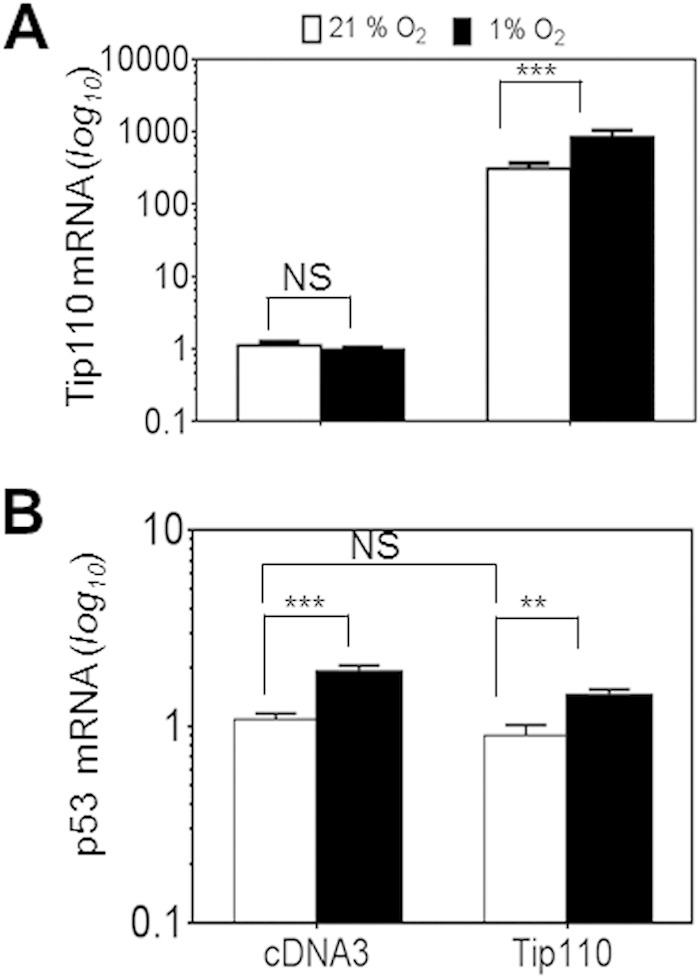
Effects of hypoxia on Tip110 and p53 mRNA levels. U2OS cells were transfected with a Tip110-His or cDNA3 plasmid for 24 h and cultured under 21 or 1% O2 for 24 h. The cells were harvested for total RNA isolation, and qRT-PCR was performed to determine the Tip110 mRNA level (A) and p53 mRNA level (B). β-Actin was included in the qRT-PCR to estimate relative Tip110 and p53 mRNA levels. Tip110 and p53 mRNA levels in cells that were transfected with cDNA3 and cultured under normoxia were set at 1. The data are means ± SE from triplicate samples and representative of three independent experiments. **, highly significant (P < 0.01); ***, strongly significant (P < 0.001); NS, not significant.
FIG 8.
Tip110's effects on HIF-1α protein stability. (A) U2OS cells were transfected with Tip110-His or cDNA3 and cultured under 1% O2 for 24 h. The cells were then cultured under 21% O2 (also known as reoxygenation) for the indicated lengths of time before being harvested for Tip110, HIF-1α, and p53 expression by Western blotting. (B) Levels of HIF-1α and p53 protein expression were quantitated by using a loading control for Western blotting, β-actin, as a reference. (C) U2OS cells were transfected with a Tip110-His or cDNA3 plasmid and then treated with DFO (250 μM) for 24 h. Cycloheximide (CHX) (40 μg/ml) was added at the indicated times before cells were harvested. (D) Levels of HIF-1α and p53 protein expression were quantitated using β-actin as a reference.
Involvement of p300 in Tip110-p53 mutual regulation.
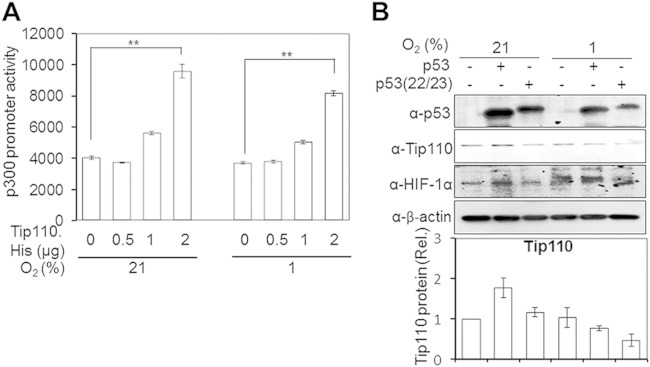
p300 is a multifunctional transcription coactivator for p53 and HIF-1α; its interaction with p53 and HIF-1α regulates a number of cellular genes (45, 46). The finding that Tip110 expression led to an increased p300 protein level (Fig. 5A to D) prompted us to investigate the roles of p300 in Tip110-p53 reciprocal regulation. We first determined whether Tip110 would activate p300 expression at the transcriptional level in the absence of p53 expression. To address this possibility, Saos-2 cells were transfected with the p300 promoter-driven luciferase reporter gene (47) and increasing amounts of Tip110-His, cultured under normoxia and hypoxia, and harvested for the luciferase reporter gene assay. Tip110 expression transactivated the p300 promoter-driven luciferase reporter gene expression in a dose-dependent manner under both normoxic and hypoxic conditions (Fig. 9A). The residues Gln22 and Ser23 in the N-terminal transactivation domain of p53 are required for p53 binding to p300/CBP transcriptional coactivators and critical for its interaction with Mdm2 (33, 45, 48). Thus, we then took advantage of the p53(22/23) double mutant and determined its effects on the Tip110 protein level. Saos-2 cells were transfected with wild-type p53 or the p53(22/23) mutant, cultured under normoxia and hypoxia, and harvested for Tip110 expression by Western blotting. Consistent with our previous results (Fig. 4A), overexpression of wild-type p53 led to increased Tip110 protein under normoxia but decreased Tip110 protein under hypoxia (Fig. 9B). In comparison, expression of p53(22/23) led to less increased Tip110 protein under normoxia but a significantly greater reduction of Tip110 protein under hypoxia. On the other hand, p53(22/23) had little effect on HIF-1α protein levels (38). These results provided evidence to support that p300 is involved in Tip110-p53 mutual regulation.
FIG 9.
p300 transactivation by Tip110. (A) Saos-2 cells were transfected with 250 ng pGL3.p300-Prom-Luc reporter alone or in combination with 0.5, 1, and 2 μg pTip110 for 24 h and cultured under 21% or 1% O2 for 24 h. Cells were harvested for the luciferase reporter gene assay. cDNA3 was used to equalize the total amount of DNA among all transfection. The pTK-βgal plasmid was included to normalize the transfection efficiency variation among all transfections. The data represent means ± SE from triplicate samples and are representative of three independent experiments. (B) Saos-2 cells were transfected with either p53 or the p53(22/23) mutant for 24 h. Cells were cultured under 21% or 1% O2 for 24 h and harvested for Tip110, HIF-1α, and p53 expression by Western blotting. Tip110 protein expression was quantitated by using β-actin as a reference. The data are representative of three independent experiments. **, highly significant (P < 0.01).
Tip110 knockdown inhibited cell proliferation.
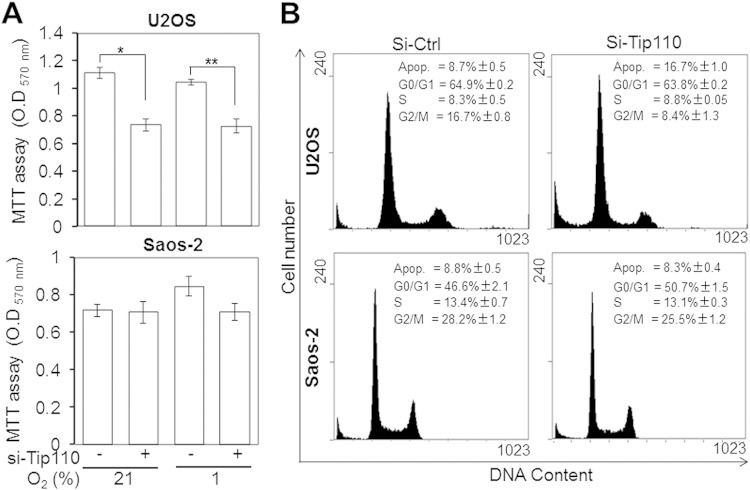
Altered regulation of p53 and HIF-1α protein levels under normoxia and hypoxia likely leads to changes in cell proliferation (34). To assess the outcomes of Tip110–p53–HIF-1α interaction, U2OS and Saos-2 were transfected with Tip110 siRNA, cultured under normoxia and hypoxia, and harvested for cell proliferation by MTT assay and for cell cycle analysis by flow cytometry. Compared to the control siRNA, Tip110 knockdown inhibited U2OS proliferation but showed no effects on Saos-2 under both normoxia and hypoxia (Fig. 10A). Meanwhile, compared with the control siRNA, Tip110 knockdown led to 2-fold increases in the sub-G0 (apoptotic) fraction and 2-fold decreases in the G2/M fraction in U2OS cells, while it showed no effects in Saos-2 cells (Fig. 10B). Nevertheless, Tip110 overexpression showed no effects on cell proliferation or apoptosis in both U2OS and Saos-2 cells (data not shown), likely due to a relatively higher level of constitutive Tip110 expression in those cells. These results indicate that Tip110 plays an important role in regulation of cell proliferation and survival in the context of p53 protein.
FIG 10.
Effects of Tip110 expression on cell proliferation and cell cycle. U2OS and Saos-2 cells were transfected with a universal control siRNA (Si-Ctrl) or Tip110 siRNA (si-Tip110) for 48 h and then cultured under 21% or 1% O2 for 24 h. The cells were harvested for cell proliferation by MTT assay (A) or for cell cycle analysis by flow cytometry (B). O.D, optical density; Apop., apoptosis. The data are means ± SE from triplicate samples (A) and are representative of three independent experiments (A and B). *, statistically significant (P < 0.05); **, highly significant (P < 0.01).
Decreased Tip110 expression in the core region of bone cancer tissues.
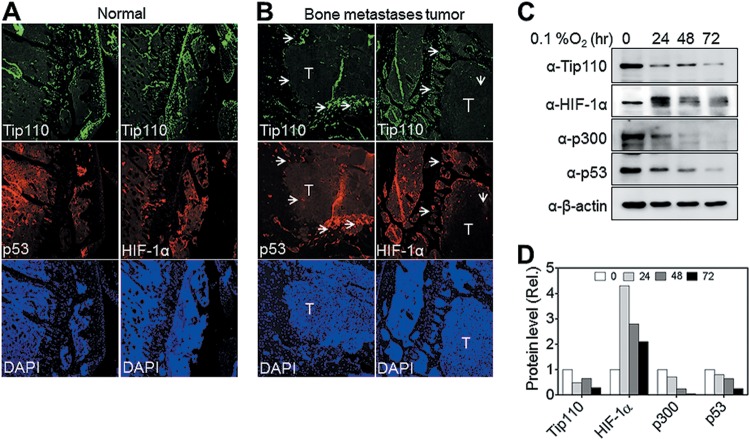
To further investigate the relationship among Tip110, p53, and HIF-1α expression, we used a bone metastasis mouse model derived from inoculation of the human bone metastatic cancer cell line 1205L, a melanoma line expressing wild-type p53, into athymic nude mice (31, 49). Immunofluorescence staining was performed for Tip110, HIF-1α, and p53 expression on the longitudinal section of the bone metastases from osteolytic bone tumor (humerus region). Tip110 was detected throughout the normal bone tissues (Fig. 11A), but there was little Tip110 expression detected in the central hypoxic region of the tumor (labeled “T” in Fig. 11B) compared to that in the peripheral region (Fig. 11B, Tip110 panels, arrowheads). In addition, significantly less HIF-1α and p53 were also detected in the same regions where less Tip110 was detected (Fig. 11B, p53 and HIF-1α panels). To validate these findings, we directly cultured 1205Lu cells under severe hypoxia (0.1% O2) for up to 72 h and then harvested them for protein expression analysis. Endogenous Tip110, p53, and HIF-1α protein levels were significantly downregulated under severe hypoxia in a similar fashion (Fig. 11C and D). Interestingly, p300 protein expression was also downregulated in those cells under the same conditions. These results support the notion that there is a complicate intertwined regulatory network among Tip110, p53, HIF-α, and p300 under hypoxia.
FIG 11.
Tip110, p53, and HIF-α expression in mouse bone metastases. (A and B) Mouse osteolytic bone metastasis tissues were obtained by inoculating cells of the human bone metastatic cancer melanoma cell line, 1205Lu (which contains wild-type p53), into the bones of athymic nude mice. Longitudinal sections of control (A) or metastasis (B) bone were prepared and immunostained for Tip110, p53, and HIF-1α expression. DAPI was used to visualize the nuclear DNA. T, tumor core region; arrowheads, peripheral tissues. (C and D) 1205Lu cells were cultured under severe hypoxia (0.1% O2) for the indicated lengths of time and then harvested for expression of the Tip110, p53, HIF-1α, and p300 proteins by Western blotting (C). Levels of protein expression were quantitated by using β-actin as a reference (D).
DISCUSSION
The Tip110 protein level is regulated during hematopoietic stem cell proliferation and differentiation (19), which are known to reside in bone marrow hypoxic microenvironments (partial O2 pressure [pO2] of between 1 and 7%) (50). Furthermore, Tip110 interacted with oncogenic proteins that play an important role during hypoxia (11, 19). Thus, these observations prompted us to investigate the regulatory mechanism of Tip110 under limited-oxygen conditions. In this study, we showed that Tip110 protein levels were downregulated in human cancer cell lines cultured under hypoxic conditions (Fig. 1 to 4) (other data not shown). Downregulation of Tip110 protein was mainly achieved by the ubiquitin proteasome system (UPS)-mediated protein degradation pathway (Fig. 1 and 7). In addition, Tip110 regulated the p53 and HIF-1α proteins' stability, whereas p53 expression was responsible for Tip110 downregulation under hypoxia (Fig. 1 to 7). Furthermore, we showed that p300 was involved in Tip110-p53 reciprocal regulation (Fig. 9 and 11). Finally, we showed that Tip110 expression altered cell proliferation and survival in the context of p53 protein (Fig. 10).
We have previously reported that Tip110 and c-Myc reciprocally regulate each other's gene expression, suggesting a positive-feedback-loop regulation between Tip110 and c-Myc (19, 20). Similar to Tip110, the expression of c-Myc protein levels in cancer cells is significantly downregulated under hypoxia and glucose-deprived conditions through UPS-mediated protein degradation (51). Suppression of c-Myc decreases necrotic cell death induced by oxygen and glucose deprivation, which might be a strategy for cancer cells to survive under conditions of limited energy resources (51). In addition, we have also shown that YB-1, an oncogenic transcription factor, and Tip110 form a complex and reciprocally regulate each other's function (11). Hypoxia has been shown to induce phosphorylation of YB-1, which could induce the expression of proangiogenic factors that are important for tumor progression (29). Thus, these observations supported our findings that Tip110 plays an important role in host response to limited-oxygen conditions.
Upregulation of p53 by Tip110 expression (Fig. 5) is more likely through increased p53 protein stability, as there was no significant change in the p53 mRNA in the presence or absence of Tip110 under hypoxic condition (Fig. 7 and 8). On the other hand, p53 promoted Tip110 degradation under hypoxia and transactivated Tip110 transcription under normoxia (Fig. 4), which suggests that Tip110 and p53 regulate each other mainly through protein stabilization, whereas regulation of Tip110 transcription by p53 could be an additional mechanism that may regulate Tip110 protein homeostasis. Studies have shown that the increase in the p53 protein level during hypoxia is due to stabilization and is dependent on the presence of HIF-1α (52, 53). However, HIF-1α upregulation is not sufficient for p53 induction (54). Hypoxia-induced p53 fails to transactivate its target genes (55). Wild-type p53 has been shown to function as a molecular chaperone and to rescue mutant p53 in hypoxic tumors and subsequently cause tumor regression (56). There are multiple contradictory reports on reciprocal interaction of p53 with hypoxic signaling (39, 54, 57–60). These inconsistencies may likely be due to the types of cell lines used, the severity and the duration of hypoxia, and unknown cellular factors that under oxygen deficiency influence very differently the p53 protein level (34). The Tip110-p53 reciprocal regulation could contribute to the magnitude of Tip110 protein reduction in different cancer cell lines under similar hypoxic conditions (Fig. 1 to 5).
Tip110 overexpression resulted in the stabilization of HIF-α in U2OS cells under hypoxia (Fig. 5 and 8). However, in Saos-2 cells, the level of HIF-1α was highly increased when Tip110 was overexpressed under both normoxic and hypoxic conditions (Fig. 5C and D). p53 knockout mice have a higher HIF-1α protein level than wild-type mice following chronic hypoxia exposure (61). HIF-1α is overexpressed in metastatic osteosarcoma tumors (62) and plays important roles in bone repair (63). Amplification of HIF-1α-dependent responses to hypoxia via loss of p53 function contributes to the angiogenic switch during tumorigenesis (38). Furthermore, HIF-1α expression correlates with increasing tumor grade, invasion, and metastasis (64). Overexpression of c-Myc protein, which forms a reciprocal regulatory loop with Tip110 (19, 20), significantly stabilizes HIF-1α under normoxic conditions, enhances HIF-1α accumulation under hypoxic conditions, and is important for cancer development (65). Thus, there is a high probability that the level of Tip110 protein in response to limited-oxygen conditions contributes to the stability of p53 and/or HIF-1α during tumorigenesis.
We have shown that Tip110 upregulated p300 in Saos-2 under both normoxic and hypoxic conditions (Fig. 5C and D and Fig. 9A), and the expression of the p53(22/23) mutant, which is deficient in p300 binding, led to further reduction of Tip110 under hypoxia compared to the p53 wild type (Fig. 9B). p300 acts as transcriptional coactivator for several nuclear proteins involved in proliferation, cell cycle regulation, apoptosis, differentiation, and the DNA damage response (66). Among these factors are known oncoproteins, such as c-Jun and c-Fos, and tumor suppressor proteins, such as E2F, pRB, p53, and Smads (66). In addition, several studies have proposed a competition between p53 and HIF-1α for the limited amount of the shared transcriptional coactivator p300 (67, 68). Thus, depending on the relative amount of expression of either of those two factors in the context of Tip110 expression, this competition would explain how both p53 transcriptional activity and HIF-1 transcriptional activity are affected.
Although overexpression of Tip110 in either U2OS or Saos-2 cells has no effect on the cells' proliferation or apoptosis (data not shown), knockdown of Tip110 resulted in inhibition of the cells' proliferation and an increase in the number of apoptotic cells in U2OS only, yet no such effect was observed in Saos-2 cells (Fig. 10A and B). Interestingly, knockdown of Tip110 was more pronounced in U2OS cells than in Saos-2 cells cultured under hypoxia and under the same transfection conditions; however, the HIF-1α protein level was dramatically reduced in both cell types (Fig. 6A to D). Knockdown of HIF-1α enhances expression of HIF-2α, and vice versa, which results in increased cell viability and inhibits apoptosis (69). Thus, inhibition of cell proliferation by knockdown of Tip110 could be a consequence of the effect of Tip110 on the level of hypoxia-inducible factors in the presence of p53 under hypoxia, which merits further investigation.
It is known that tumor tissues experiencing hypoxia will be exposed to oxygen levels that vary in amplitude and duration (acute or chronic hypoxia) (70). Hypoxia and transforming growth factor β (TGF-β) signaling drive tumor bone metastases in a bone metastasis mouse model (71). Using the same mouse model, we found little Tip110 protein expression in the central hypoxic region of the tumor and significantly reduced levels of HIF-1α and p53 in the same regions (Fig. 11A and B). c-Myc protein expression has been detected in the tumor area proximal to the blood vessels (moderate hypoxia), but no c-Myc was detected in the tumor area distant from blood vessels (severe hypoxia or anoxia) (51). Similarly to c-Myc, the Tip110 expression level was O2 concentration dependent (Fig. 3), and knockdown of Tip110 resulted in reduction of p53 and HIF-1α under hypoxic conditions (Fig. 6A and B). Although the Tip110 protein level is highly elevated in a number of malignant tumor cell lines and cancerous tissues (16–25), it is also possible that the Tip110 expression level is affected by the tumor microenvironment, such as under low oxygen tension. Taken together, these results indicate that the control of Tip110 protein level may have important functional consequences on the cross talk with key signaling pathways controlling cell survival, which is likely to have an impact on p53 and HIF targeting strategies for hypoxia-associated diseases.
ACKNOWLEDGMENTS
We thank Mark Hannink for providing UB-HA plasmid and Jiayuh Lin for providing p53 and p53(22/23) plasmids. We thank Xiangle Sun for her assistance with flow cytometry.
REFERENCES
- 1.Wang GL, Jiang BH, Rue EA, Semenza GL. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. 2003. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 3.Green DR, Kroemer G. 2009. Cytoplasmic functions of the tumour suppressor p53. Nature 458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Bayle JH, Olson D, Levine AJ. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 6.Novotny I, Blazikova M, Stanek D, Herman P, Malinsky J. 2011. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 22:513–523. doi: 10.1091/mbc.E10-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanek D, Rader SD, Klingauf M, Neugebauer KM. 2003. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada K, Yamada A, Yang D, Itoh K, Shichijo S. 2001. Binding of a SART3 tumor-rejection antigen to a pre-mRNA splicing factor RNPS1: a possible regulation of splicing by a complex formation. Int J Cancer 93:623–628. doi: 10.1002/ijc.1391. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Kim BO, Kao C, Jung C, Dalton JT, He JJ. 2004. Tip110, the human immunodeficiency virus type 1 (HIV-1) Tat-interacting protein of 110 kDa as a negative regulator of androgen receptor (AR) transcriptional activation. J Biol Chem 279:21766–21773. doi: 10.1074/jbc.M314321200. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Li J, Kim BO, Pace BS, He JJ. 2002. HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J Biol Chem 277:23854–23863. doi: 10.1074/jbc.M200773200. [DOI] [PubMed] [Google Scholar]
- 11.Timani KA, Liu Y, He JJ. 2013. Tip110 interacts with YB-1 and regulates each other's function. BMC Mol Biol 14:14. doi: 10.1186/1471-2199-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W, Liu Y, Timani KA, He JJ. 2014. Tip110 protein binds to unphosphorylated RNA polymerase II and promotes its phosphorylation and HIV-1 long terminal repeat transcription. J Biol Chem 289:190–202. doi: 10.1074/jbc.M113.529784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trede NS, Medenbach J, Damianov A, Hung LH, Weber GJ, Paw BH, Zhou Y, Hersey C, Zapata A, Keefe M, Barut BA, Stuart AB, Katz T, Amemiya CT, Zon LI, Bindereif A. 2007. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci U S A 104:6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timani KA, Liu Y, Suvannasankha A, He JJ. 2014. Regulation of ubiquitin-proteasome system-mediated Tip110 protein degradation by USP15. Int J Biochem Cell Biol 54:10–19. doi: 10.1016/j.biocel.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Nakao M, Shichijo S, Sasatomi T, Takasu H, Matsumoto H, Mori K, Hayashi A, Yamana H, Shirouzu K, Itoh K. 1999. Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res 59:4056–4063. [PubMed] [Google Scholar]
- 16.Fukuda K. 2001. Expression of the SART3 antigens in oral cancers. Kurume Med J 48:55–58. doi: 10.2739/kurumemedj.48.55. [DOI] [PubMed] [Google Scholar]
- 17.Kawagoe N, Shintaku I, Yutani S, Etoh H, Matuoka K, Noda S, Itoh K. 2000. Expression of the SART3 tumor rejection antigen in renal cell carcinoma. J Urol 164:2090–2095. doi: 10.1016/S0022-5347(05)66975-3. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Lee MR, Timani K, He JJ, Broxmeyer HE. 2012. Tip110 maintains expression of pluripotent factors in and pluripotency of human embryonic stem cells. Stem Cells Dev 21:829–833. doi: 10.1089/scd.2011.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Timani K, Mantel C, Fan Y, Hangoc G, Cooper S, He JJ, Broxmeyer HE. 2011. TIP110/p110nrb/SART3/p110 regulation of hematopoiesis through CMYC. Blood 117:5643–5651. doi: 10.1182/blood-2010-12-325332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Timani K, Ou X, Broxmeyer HE, He JJ. 2013. C-MYC controlled TIP110 protein expression regulates OCT4 mRNA splicing in human embryonic stem cells. Stem Cells Dev 22:689–694. doi: 10.1089/scd.2012.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murayama K, Kobayashi T, Imaizumi T, Matsunaga K, Kuramoto T, Shigemori M, Shichijo S, Itoh K. 2000. Expression of the SART3 tumor-rejection antigen in brain tumors and induction of cytotoxic T lymphocytes by its peptides. J Immunother 23:511–518. doi: 10.1097/00002371-200009000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Niiya F, Nishizaka S, Matsunaga K, Koufuji K, Mori M, Katai H, Yamana H, Itoh K. 2000. Expression of SART3 tumor-rejection antigen in gastric cancers. Jpn J Cancer Res 91:337–342. doi: 10.1111/j.1349-7006.2000.tb00950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suefuji Y, Sasatomi T, Shichijo S, Nakagawa S, Deguchi H, Koga T, Kameyama T, Itoh K. 2001. Expression of SART3 antigen and induction of CTLs by SART3-derived peptides in breast cancer patients. Br J Cancer 84:915–919. doi: 10.1054/bjoc.2000.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka S, Tsuda N, Kawano K, Sakamoto M, Nishida T, Hashimoto T, Shichijo S, Kamura T, Itoh K. 2000. Expression of tumor-rejection antigens in gynecologic cancers. Jpn J Cancer Res 91:1177–1184. doi: 10.1111/j.1349-7006.2000.tb00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuda N, Murayama K, Ishida H, Matsunaga K, Komiya S, Itoh K, Yamada A. 2001. Expression of a newly defined tumor-rejection antigen SART3 in musculoskeletal tumors and induction of HLA class I-restricted cytotoxic T lymphocytes by SART3-derived peptides. J Orthop Res 19:346–351. doi: 10.1016/S0736-0266(00)90031-7. [DOI] [PubMed] [Google Scholar]
- 26.Iseki K, Matsunaga H, Komatsu N, Suekane S, Noguchi M, Itoh K, Yamada A. 2010. Evaluation of a new oil adjuvant for use in peptide-based cancer vaccination. Cancer Sci 101:2110–2114. doi: 10.1111/j.1349-7006.2010.01653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komohara Y, Harada M, Arima Y, Suekane S, Noguchi M, Yamada A, Itoh K, Matsuoka K. 2006. Anti-cancer vaccine candidates in specific immunotherapy for bladder carcinoma. Int J Oncol 29:1555–1560. doi: 10.3892/ijo.29.6.1555. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki N, Maeda Y, Tanaka S, Hida N, Mine T, Yamamoto K, Oka M, Itoh K. 2002. Detection of peptide-specific cytotoxic T-lymphocyte precursors used for specific immunotherapy of pancreatic cancer. Int J Cancer 98:45–50. doi: 10.1002/ijc.10145. [DOI] [PubMed] [Google Scholar]
- 29.Coles LS, Lambrusco L, Burrows J, Hunter J, Diamond P, Bert AG, Vadas MA, Goodall GJ. 2005. Phosphorylation of cold shock domain/Y-box proteins by ERK2 and GSK3beta and repression of the human VEGF promoter. FEBS Lett 579:5372–5378. doi: 10.1016/j.febslet.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 30.Gordan JD, Thompson CB, Simon MC. 2007. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, Duong V, Dunn LK, Mauviel A, Guise TA. 2011. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res 71:175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem 280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 33.Lin J, Chen J, Elenbaas B, Levine AJ. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev 8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 34.Sermeus A, Michiels C. 2011. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis 2:e164. doi: 10.1038/cddis.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin-Lee YC, Tatebe S, Savaraj N, Ishikawa T, Tien Kuo M. 2001. Differential sensitivities of the MRP gene family and gamma-glutamylcysteine synthetase to prooxidants in human colorectal carcinoma cell lines with different p53 status. Biochem Pharmacol 61:555–563. doi: 10.1016/S0006-2952(00)00592-X. [DOI] [PubMed] [Google Scholar]
- 36.Rochette PJ, Bastien N, Lavoie J, Guerin SL, Drouin R. 2005. SW480, a p53 double-mutant cell line retains proficiency for some p53 functions. J Mol Biol 352:44–57. doi: 10.1016/j.jmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa R, Kusunoki M, Yanagi H, Noda M, Furuyama JI, Yamamura T, Hashimoto-Tamaoki T. 2001. Dual antitumor effects of 5-fluorouracil on the cell cycle in colorectal carcinoma cells: a novel target mechanism concept for pharmacokinetic modulating chemotherapy. Cancer Res 61:1029–1037. [PubMed] [Google Scholar]
- 38.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. 2000. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev 14:34–44. doi: 10.1101/gad.14.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graeber TG, Peterson JF, Tsai M, Monica K, Fornace AJ Jr, Giaccia AJ. 1994. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol 14:6264–6277. doi: 10.1128/MCB.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Royds JA, Dower SK, Qwarnstrom EE, Lewis CE. 1998. Response of tumour cells to hypoxia: role of p53 and NFkB. Mol Pathol 51:55–61. doi: 10.1136/mp.51.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groulx I, Lee S. 2002. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol 22:5319–5336. doi: 10.1128/MCB.22.15.5319-5336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang LE, Gu J, Schau M, Bunn HF. 1998. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A 95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim BM, Choi JY, Kim YJ, Woo HD, Chung HW. 2007. Reoxygenation following hypoxia activates DNA-damage checkpoint signaling pathways that suppress cell-cycle progression in cultured human lymphocytes. FEBS Lett 581:3005–3012. doi: 10.1016/j.febslet.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 44.Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. 1998. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem 273:11995–11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- 45.Gu W, Shi XL, Roeder RG. 1997. Synergistic activation of transcription by CBP and p53. Nature 387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 46.Somasundaram K, El-Deiry WS. 1997. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene 14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 47.Yu J, de Belle I, Liang H, Adamson ED. 2004. Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol Cell 15:83–94. doi: 10.1016/j.molcel.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 48.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. 1995. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev 9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 49.Juarez P, Mohammad KS, Yin JJ, Fournier PG, McKenna RC, Davis HW, Peng XH, Niewolna M, Javelaud D, Chirgwin JM, Mauviel A, Guise TA. 2012. Halofuginone inhibits the establishment and progression of melanoma bone metastases. Cancer Res 72:6247–6256. doi: 10.1158/0008-5472.CAN-12-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirao M, Hashimoto J, Yamasaki N, Ando W, Tsuboi H, Myoui A, Yoshikawa H. 2007. Oxygen tension is an important mediator of the transformation of osteoblasts to osteocytes. J Bone Miner Metab 25:266–276. doi: 10.1007/s00774-007-0765-9. [DOI] [PubMed] [Google Scholar]
- 51.Okuyama H, Endo H, Akashika T, Kato K, Inoue M. 2010. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res 70:10213–10223. doi: 10.1158/0008-5472.CAN-10-2720. [DOI] [PubMed] [Google Scholar]
- 52.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. 1998. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 53.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E. 1998. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 54.Wenger RH, Camenisch G, Desbaillets I, Chilov D, Gassmann M. 1998. Up-regulation of hypoxia-inducible factor-1alpha is not sufficient for hypoxic/anoxic p53 induction. Cancer Res 58:5678–5680. [PubMed] [Google Scholar]
- 55.Shenberger JS, Dixon PS. 1999. Oxygen induces S-phase growth arrest and increases p53 and p21(WAF1/CIP1) expression in human bronchial smooth-muscle cells. Am J Respir Cell Mol Biol 21:395–402. doi: 10.1165/ajrcmb.21.3.3604. [DOI] [PubMed] [Google Scholar]
- 56.Gogna R, Madan E, Kuppusamy P, Pati U. 2012. Chaperoning of mutant p53 protein by wild-type p53 protein causes hypoxic tumor regression. J Biol Chem 287:2907–2914. doi: 10.1074/jbc.M111.317354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. 2002. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol 22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hubert A, Paris S, Piret JP, Ninane N, Raes M, Michiels C. 2006. Casein kinase 2 inhibition decreases hypoxia-inducible factor-1 activity under hypoxia through elevated p53 protein level. J Cell Sci 119:3351–3362. doi: 10.1242/jcs.03069. [DOI] [PubMed] [Google Scholar]
- 59.Wouters A, Pauwels B, Lambrechts HA, Pattyn GG, Ides J, Baay M, Meijnders P, Dewilde S, Vermorken JB, Lardon F. 2009. Chemoradiation interactions under reduced oxygen conditions: cellular characteristics of an in vitro model. Cancer Lett 286:180–188. doi: 10.1016/j.canlet.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, Hill RP. 2004. Hypoxia enhances metastatic efficiency by up-regulating Mdm2 in KHT cells and increasing resistance to apoptosis. Cancer Res 64:4180–4189. doi: 10.1158/0008-5472.CAN-03-3038. [DOI] [PubMed] [Google Scholar]
- 61.Mizuno S, Bogaard HJ, Kraskauskas D, Alhussaini A, Gomez-Arroyo J, Voelkel NF, Ishizaki T. 2011. p53 gene deficiency promotes hypoxia-induced pulmonary hypertension and vascular remodeling in mice. Am J Physiol Lung Cell Mol Physiol 300:L753–761. doi: 10.1152/ajplung.00286.2010. [DOI] [PubMed] [Google Scholar]
- 62.Mizobuchi H, Garcia-Castellano JM, Philip S, Healey JH, Gorlick R. 2008. Hypoxia markers in human osteosarcoma: an exploratory study. Clin Orthop Relat Res 466:2052–2059. doi: 10.1007/s11999-008-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wan C, Gilbert SR, Wang Y, Cao X, Shen X, Ramaswamy G, Jacobsen KA, Alaql ZS, Eberhardt AW, Gerstenfeld LC, Einhorn TA, Deng L, Clemens TL. 2008. Activation of the hypoxia-inducible factor-1alpha pathway accelerates bone regeneration. Proc Natl Acad Sci U S A 105:686–691. doi: 10.1073/pnas.0708474105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. 1999. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res 59:5830–5835. [PubMed] [Google Scholar]
- 65.Doe MR, Ascano JM, Kaur M, Cole MD. 2012. Myc posttranscriptionally induces HIF1 protein and target gene expression in normal and cancer cells. Cancer Res 72:949–957. doi: 10.1158/0008-5472.CAN-11-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyer NG, Ozdag H, Caldas C. 2004. p300/CBP and cancer. Oncogene 23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 67.Schmid T, Zhou J, Brune B. 2004. HIF-1 and p53: communication of transcription factors under hypoxia. J Cell Mol Med 8:423–431. doi: 10.1111/j.1582-4934.2004.tb00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmid T, Zhou J, Kohl R, Brune B. 2004. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1). Biochem J 380:289–295. doi: 10.1042/BJ20031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menrad H, Werno C, Schmid T, Copanaki E, Deller T, Dehne N, Brune B. 2010. Roles of hypoxia-inducible factor-1alpha (HIF-1alpha) versus HIF-2alpha in the survival of hepatocellular tumor spheroids. Hepatology 51:2183–2192. doi: 10.1002/hep.23597. [DOI] [PubMed] [Google Scholar]
- 70.Hockel M, Vaupel P. 2001. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst 93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 71.Dunn LK, Mohammad KS, Fournier PG, McKenna CR, Davis HW, Niewolna M, Peng XH, Chirgwin JM, Guise TA. 2009. Hypoxia and TGF-beta drive breast cancer bone metastases through parallel signaling pathways in tumor cells and the bone microenvironment. PLoS One 4:e6896. doi: 10.1371/journal.pone.0006896. [DOI] [PMC free article] [PubMed] [Google Scholar]