Abstract
Gelatinized starch-urea (Starea, SU) is an effective and economical source of urea for ruminants. Here we assessed the influence of dietary supplementation with gelatinized starch-urea on the diversity of intestinal bacteria in finishing cattle. Fifty steers were randomly allotted to five treatments with diets supplemented with different doses of Starea [0 % (SU0), 8 % (SU8), 16 % (SU16), 24 % (SU24), and 32 % (SU32) of urea-N in total nitrogen]. Denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes was used to examine the effect of dietary supplementation of Starea on intestinal bacterial flora. Shannon–Weaver and Simpson diversity indices consistently showed the lowest bacterial diversity in the SU0 treatment. Increasing doses of Starea increased the diversity up to SU24 after which, diversity decreased. Cluster analysis of 16S rRNA gene DGGE profiles indicates that the intestinal bacterial communities associated with cattle that were not supplemented with Starea in feed differed in composition and structure from those supplemented with Starea. The amount of Starea supplemented in cattle diets influenced the abundance of several key species affiliated with Lachnospiraceae, Ruminococcaceae, Peptostreptococcaceae, Comamonadaceae and Moraxellaceae. These results suggest that Starea influences the composition and structure of intestinal bacteria which may play a role in promoting ruminant health and production performance.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0526-8) contains supplementary material, which is available to authorized users.
Keywords: Intestinal bacteria, Cattle, Urea, Starea, Denaturing gradient gel electrophoresis
Introduction
The bacteria associated with the guts of cattle are diverse and abundant, with cell numbers often exceeding 1011 culture forming units (cfu) per gram of feces [1]. The primary function of intestinal bacteria is to catabolize carbohydrates not digested and absorbed in the upper gut, resulting in the production of organic acids, gases and short chain fatty acids [2]. There is strong epidemiological and experimental evidence that indicates a number of gut microorganisms have positive effects on gut health and the intestinal immune system (so called pro-biotics) [3]. Thus the bacterial composition in the intestine may have a great impact on the growth and health of cattle.
Dietary supplements can influence the composition of microbial communities. Urea is used as a common non-protein nitrogen supplement for ruminant diets for many years [4]. In the rumen, urea is rapidly hydrolyzed to NH3 that is synthesized into microbial protein [5]. Although many studies have demonstrated that the performance of urea supplements is usually poorer than that of natural protein feeds [6], urea has become a cost-effective protein replacement in consideration of increasing prices of protein feedstuff.
Gelatinized starch-urea (Starea), a mixture of gelatinized wheat starch and urea, is one of the most effective and economical slow-release urea products available today. As a nitrogen supplement Starea is comparable to the soybean meal diet fed to lactating cows to increase milk yield and that fed to young calves to maintain nitrogen balance [7, 8]. Treatments with different dietary doses of Starea have been reported to show similar growth promoting effects in growing-finishing beef cattle [9]. While it is clear that diet influences the community structure of intestinal microbiota [10, 11], information regarding the effects of urea on the composition of bacteria in cattle intestines is rare. There is only one report showing that urea content in the diet has no effect on fecal shedding of Escherichia coli O157:H7 [12]. Sequence analysis of the 16S rRNA genes has been used for decades to identify bacteria and construct phylogenic trees [13]. Methods based on sequence variations in the 16S rRNA genes, such as denaturing gradient gel electrophoresis (DGGE), can also be used to analyze bacterial diversity [14]. Recently, extensive analysis of 16S rRNA gene sequences has identified unique molecular features that can be used to achieve species level identification of bacteria, including in the genera Bacillus, Clostridium, Streptococcus, and Pseudomonas [15–18].
This study aimed to determine the effects of Starea on the diversity of intestinal bacteria using DGGE and 16S rDNA sequence analysis. The goal of the study was to provide new insight into the relationship between gut intestinal bacterial composition, treatment of feed with urea, and the potential role of these organisms in promoting ruminant growth and health.
Materials and Methods
Animals and Sampling
All procedures involving animals were approved by the China Agricultural University Institutional Animal Care and Use Committee. Fifty male crossbred steers (Limousin × Fuzhou, 18 mo old, bodyweight = 397.2 ± 19.5 kg) were allotted randomly to five dietary treatments (10 steers per treatment) with different doses of Starea. The basic diet for the animals consisted of corn silage, brewers grain, corn, cottonseed meal, corn starch, limestone, dicalcium phosphate and mineral premix. The diets of the five treatment groups were amended with 0 % (w/w) urea-N (control group, SU0), 8 % urea-N (SU8), 16 % urea-N (SU16), 24 % urea-N (SU24) and 32 % urea-N (SU32); these diets were formulated to meet the nutritional requirements of the steers. The animals were fed the aforementioned diets for a duration of 14 weeks. At the end of the 14 week period, three steers per group were selected randomly. Fecal samples were collected by hand wearing sterilized gloves and the samples were immediately stored in liquid nitrogen until used for DNA extraction.
DNA Extraction
Frozen fecal samples were thawed at room temperature, and approximately 300 mg (wet weight) of samples was separately transferred into 2 mL sterile tubes. Total DNA was extracted according to a traditional method using a mini-bead beater (Biospec Products, Bartlesville, OK, USA) [19]. The tubes were bead-beaten at 5000 rpm for 3 min with 0.3 g of sterile zirconium beads (diameter, 0.1 mm) and then followed with phenol–chloroform extraction. DNA was precipitated with ethanol and suspended in 100 μL of nuclease-free TE solution. The integrity and concentration of DNA extracts were determined visually after electrophoresis on 1.2 % agarose gel (w/v) containing ethidium bromide.
PCR-DGGE Analysis
The V6 to V8 region of the bacterial 16S rRNA gene was amplified by PCR with the following primers: U968-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3′) and L1401 (5′-GCG TGT GTA CAA GAC CC-3′) [20]. PCR reactions contained 1 μL of template DNA, 1 μL of each primer (5 pmol/μL), 12.5 μL of Taq PCR Mastermix (Tiangen, China) and 9.5 μL of deionized distilled H2O. PCR was performed using the following conditions: initial denaturation for 5 min at 94 °C, 35 cycles of denaturation for 40 s at 94 °C, annealing for 40 s at 56 °C, and extension for 1 min at 72 °C, and a final extension for 5 min at 72 °C.
PCR amplicons were subjected to sequence-specific separation using a DCode DGGE system (Bio-Rad Laboratories, Hercules, CA, USA) using previously described methods [19, 21]. Briefly, the amplicons were separated in 8 % (w/v) polyacrylamide gel containing a linear gradient (43–58 %) of urea and formamide. Electrophoresis was initiated by pre-running in 0.5× TAE buffer for 10 min at 200 V and subsequently performed at 85 V for 16 h at a temperature of 60 °C. Following electrophoresis, the polyacrylamide gel was stained for 20 min with 1 μg/mL ethidium bromide, illuminated by ultraviolet light, and photographed. Dominant bands in the DGGE gel were excised with sterilized scalpels and incubated in nuclease-free TE solution overnight at 4 °C to allow the DNA in the bands to diffuse to the liquid [21].
Cloning and Sequencing
DNA that eluted from the excised gel fragments was re-amplified by PCR with the primer pair U968 (5′-AA CGC GAA GAA CCT TAC-3′) and L1401. PCR reaction and cycling conditions were the same as described above. PCR products were verified by electrophoresis on a 1.2 % agarose gel. PCR products were purified using a Gel Purification Kit (Bioteke, Beijing, China) and ligated into a pMD™ 19-T Vector (Takara, Dalian, Liaoning, China) in accordance with the manufacturer’s instructions. Recombinant clones were grown in Luria–Bertani broth with ampicillin (100 μg/mL) as the selection agent. Recombinant bacterial cultures was directly prepared for PCR amplification with the universal primers M13(-47) (5′-CGC CAG GGT TTT CCC AGT CAC GAC-3′) and M13(-48) (5′-AGC GGA TAA CAA TTT CAC ACA GGA-3′). Agarose gel electrophoresis was performed to determine the presence of amplification products and the PCR products were reanalyzed using DGGE, as described above, to verify that the band position matched the original position in the DGGE electropherogram. Recombinant bacterial cell pellets were directly sent to Invitrogen Corporation (Shanghai, China), and the sequencing of plasmid insert was completed.
Diversity and Cluster Analyses
The QuantityOne software package (Bio-Rad Laboratories) was used to quantitatively compare patterns in the 16S rRNA gene DGGE for use in calculating bacterial community diversity and to generate matrices for use in the generation of dendrograms describing community similarity. Dendrograms were constructed using the unweighted pair group mean average method [22]. Bacterial community diversity was measured using Shannon–Weaver (H) and Simpson diversity (D = 1 − λ) indices [23]. Data were analyzed by one-way ANOVA using GLM procedure of SAS 9.13 software (SAS Inst. Inc., Cary, NC, USA). Multiple comparisons were tested with the PDIFF option to determine differences among treatments. A correlation analysis between diversity index and dosage of Starea was performed using conic fitting method by Microsoft Office Excel Software.
Phylogenetic Tree Construction
16S rRNA gene sequences obtained from DGGE bands were aligned to sequences available in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Ribosomal Database Project databases to determine the closest matches and phylogenetic relationships [24]. 16S rRNA genes from DGGE bands and from closely related sequences were aligned using ClustalX (version 2.1) [25]. A matrix describing the evolutionary distance between sequences was computed from the sequence alignment using MEGA 5.05 software [26]. This matrix was used to generate a phylogenetic tree using the neighbor-joining algorithm [27].
Results
DGGE Profiles and Community Diversity
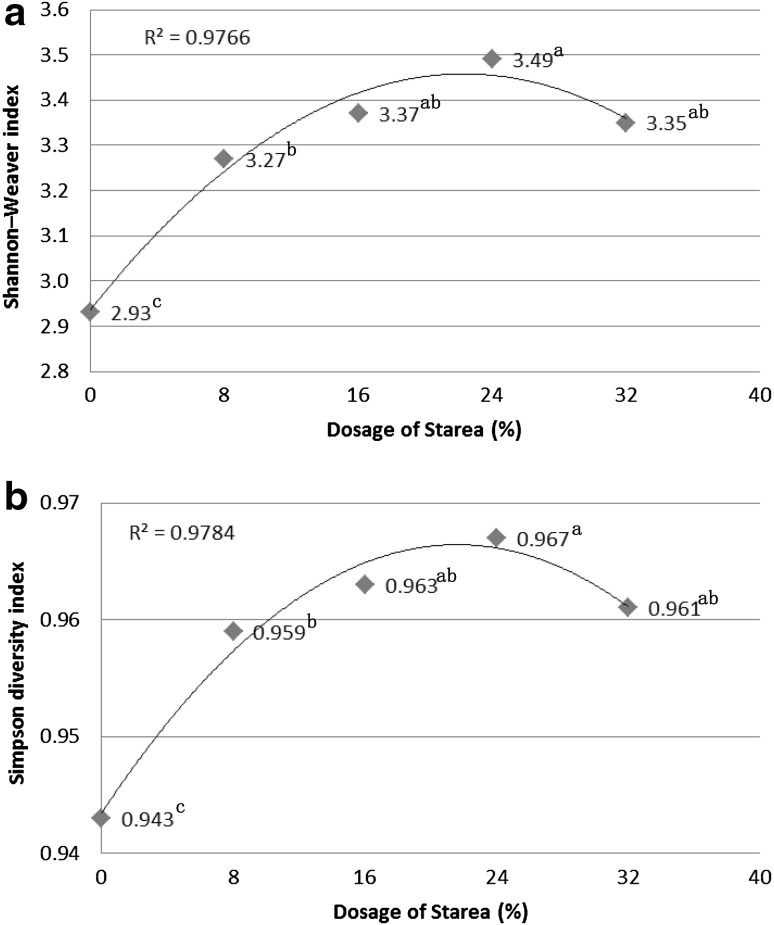
Each DGGE profile was characterized by the presence of dominant bands, or OTUs, with a background of distinct lower intensity bands from less abundant OTUs (Fig. 1). The correlation analysis (Fig. 2) consistently and significantly showed SU0 harbored the lowest diversity among the treatment groups. In addition, increasing doses of Starea up to 24 % w/w resulted in systematic increases in bacterial 16S rRNA gene diversity. Lower 16S rRNA gene diversity associated with the SU32 treatment which may indicate that Starea becomes toxic when administered at this concentration.
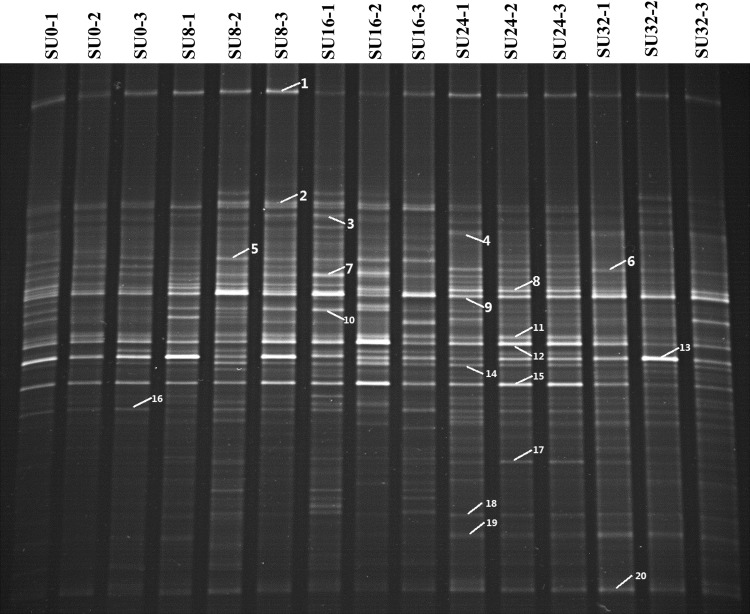
Fig. 1.
DGGE profiles of amplified V6–V8 regions of bacterial 16S rRNA genes. Labeled bands were excised for cloning and sequencing and correspond with those presented in Table 1
Fig. 2.
The correlation analysis between diversity index and dosage of Starea. Values with different superscript letters were significantly different (P < 0.05) on the basis of statistical analysis by SAS software
Cluster Analysis of DGGE 16S rRNA Gene Profiles
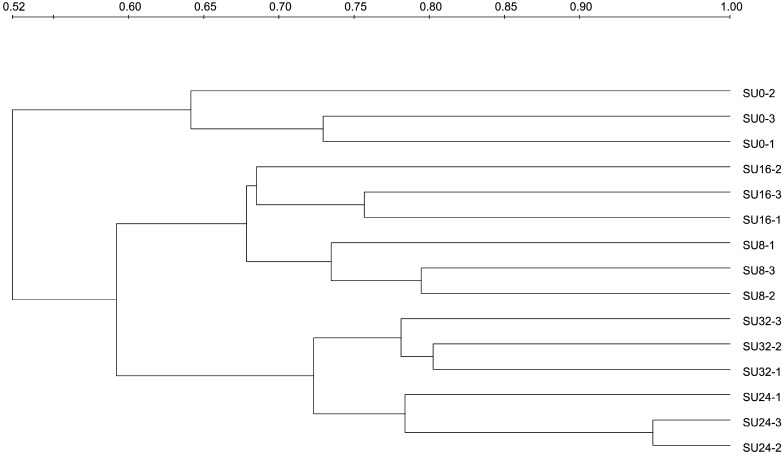
Bacterial 16S rRNA genes recovered from animals administered under the same Starea treatment regime formed clusters in the dendrogram analysis (Fig. 3), indicating that they harbored similar community structures. All of the samples were divided into two distinct clusters. One of these clusters contained SU0, whereas the other cluster consisted of all Starea treatments. In Starea treatments, SU8 and SU16 were grouped together, whereas SU24 and SU32 were grouped in one cluster. This indicates that Starea treatment and the amount of Starea administered both influenced the composition of bacterial communities in cattle intestines.
Fig. 3.
Cluster analysis of 16S rRNA gene DGGE profiles associated with five Starea treatment groups
16S rRNA Gene Analysis
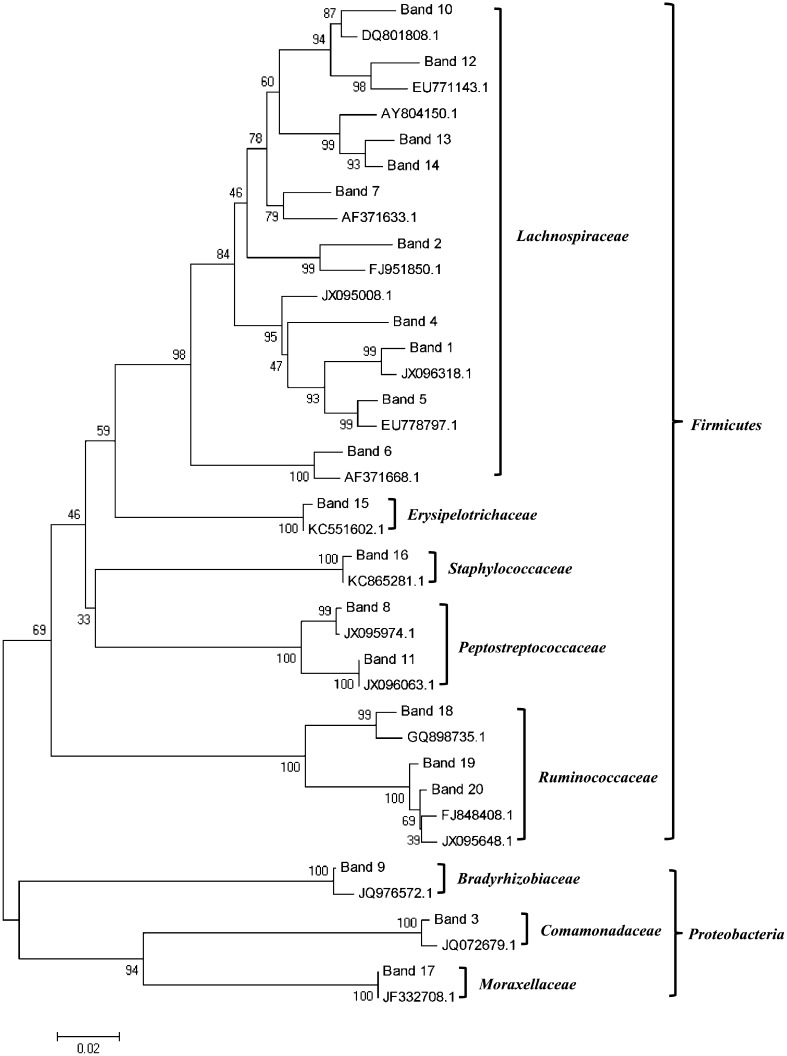
A total of 20 16S rRNA gene fragments were excised from DGGE gels and were sequenced (Table 1). The majority of the 16S rRNA gene sequences recovered from animals were affiliated with uncultured bacteria. Phylogenetic analysis results were shown in Fig. 4. All of the sequences could be assigned to two phyla, three of which belonged to the Proteobacteria and the 17 of which were assigned to Firmicutes. Among the 17 sequences that were affiliated with the phylum Firmicutes, 10 (58.8 %) were related to the family Lachnospiraceae, 3 (17.6 %) were related to the family Ruminococcaceae, and 2 (11.8 %) were affiliated with the family Peptostreptococcaceae.
Table 1.
Taxonomic affiliation of predominant 16S rRNA genes excised from DGGE gels
| Band | Closest relative (accession no.)a | Similaritya (%) | Family or genusb | Treatment groupc |
|---|---|---|---|---|
| 1 | Uncultured bacterium clone Hmb2-20 (JX096318.1) | 99 | Lachnospiraceae | All |
| 2 | Uncultured bacterium clone N19 (FJ951850.1) | 96 | Lachnospiraceae | All |
| 3 | Uncultured bacterium clone NBBPI0308_82 (JQ072679.1) | 99 | Comamonadaceae | All except SU32 |
| 4 | Uncultured bacterium clone Fa1-57 (JX095008.1) | 95 | Lachnospiraceae | SU24 and SU32 |
| 5 | Uncultured bacterium clone SBSD_aaa03e05_1 (EU778797.1) | 98 | Lachnospiraceae | All except SU32 |
| 6 | Uncultured bacterium clone p-3487-9F3 (AF371668.1) | 98 | Lachnospiraceae | SU24 and SU32 |
| 7 | Uncultured bacterium clone p-2186-s959-3 (AF371633.1) | 96 | Lachnospiraceae | All |
| 8 | Uncultured bacterium clone E2-38 (JX095974.1) | 99 | Clostridium | All except SU16 |
| 9 | Uncultured bacterium clone 4040RB24 (JQ976572.1) | 99 | Bosea | All |
| 10 | Uncultured bacterium clone RL201_aai47a11 (DQ801808.1) | 97 | Lachnospiraceae | All except SU32 |
| 11 | Uncultured bacterium clone Hma1-37 (JX096063.1) | 100 | Clostridium | All |
| 12 | Uncultured bacterium clone AE3_aaa02f01 (EU771143.1) | 97 | Lachnospiraceae | All |
| 13 | Roseburia faecis strain M88/1 (AY804150.1) | 95 | Lachnospiraceae | All |
| 14 | Roseburia faecis strain M88/1 (AY804150.1) | 97 | Lachnospiraceae | All |
| 15 | Uncultured bacterium clone GD 1-79 (KC551602.1) | 99 | Turicibacter | All |
| 16 | Staphylococcus cohnii strain 3BP (KC865281.1) | 99 | Staphylococcus | All |
| 17 | Uncultured bacterium clone N30.4 (JF332708.1) | 100 | Acinetobacter | SU24 and SU32 |
| 18 | Uncultured bacterium clone S4-69 (GQ898735.1) | 99 | Ruminococcaceae | SU24 and SU32 |
| 19 | Uncultured bacterium clone Hdb2-73 (JX095648.1) | 99 | Ruminococcaceae | SU24 and SU32 |
| 20 | Uncultured bacterium clone DLN-96 (FJ848408.1) | 99 | Ruminococcaceae | All |
Band numbers correspond with those presented in Fig. 1
aThe information was obtained from the National Center for Biotechnology Information (NCBI) database
bThe information was obtained from the Ribosomal Database Project (RDP) database
cA DGGE band that only existed in one lane of a treatment group was not counted
Fig. 4.
Phylogenetic tree based on 16S rRNA gene sequences from the dominant species in DGGE profiles. The close relatives were represented by accession number from NCBI database. The topology of the tree was estimated by parsimony from 1000 bootstrap replications. Numbers at the nodes were percentages supported by bootstrap evaluation
Numbers and Percentages of Predominant Bacterial Species in Different Groups
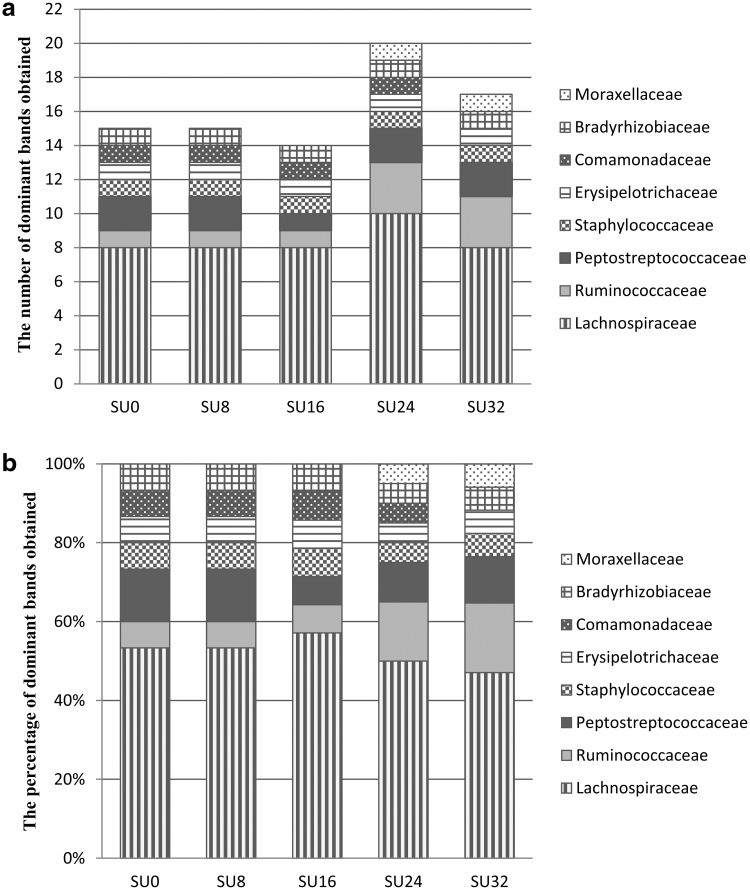
In each group, over 80 % of the dominant 16S rRNA gene bands were from sequences that represented phylum Firmicutes, with the majority of those exhibiting close affiliation with the family Lachnospiraceae (Fig. 5). The members of dominant bacterial species in different treatment groups were different with some 16S rRNA gene bands only being present in specific treatment group. For example, SU16 contained the least number of 16S rRNA gene sequences affiliated with Clostridium and 16S rRNA gene sequences affiliated with Acinetobacter only occurred in SU24 and SU32.
Fig. 5.
The number (a) and percentage (b) of predominant bands in each treatment group. Specific band only existed in one lane of a treatment group was not counted in the figure
Discussion
Supplementation of ruminant diets with Starea increased bacterial community diversity and increases in the dose of Starea systematically increased diversity up to 24 % w/w; diversity was lower in SU32 when compared with SU24 treatment. The decline of diversity in SU32 might be due to toxicity of urea when applied at that concentration. Treatment of animals with 24 % urea-N resulted in the highest diversity. Cluster analysis (Fig. 3) showed that bacterial community in the gut might be divided into three types based on the dose of Starea in the diet, and the degree of similarity of bacterial community from the same treatment differed among the three types.
The majority of the intestinal bacteria have not been cultured [28]. Consistent with this, the majority of the 16S rRNA gene sequences excised from DGGE profiles were affiliated with uncultured bacteria (Table 1) and almost all of the bacterial families obtained here were reported to be found in gut bacteria [29]. In agreement with the view that bacterial species within the gastrointestinal tract are structured by diet [30], the composition of intestinal bacteria differed on the basis of the dose of Starea. Treatment group SU24 contained the most members of Lachnospiraceae and this has previously been shown to be the most abundant member in the proximal large intestine of cattle [31]. Sequences affiliated with Ruminococcaceae were only present in treatment groups SU24 and SU32. Cultivated representatives of Lachnospiraceae and Ruminococcaceae are carbohydrate utilizing, butyrate-producing bacteria [32]. They are common intestinal bacteria and sensitive to the condition of the host health.
Intestinal microbiota is associated with host health, public health, and environmental quality [2, 3, 33]. In the present study, the results demonstrated Starea increases the diversity of cattle intestinal bacterial communities. The amount of Starea supplemented in cattle diets was shown to shift the composition of bacterial community communities and influenced the abundance of several key species. Treatment group SU16 contained the least representation of 16S rRNA gene sequences related to Clostridium. Clostridium contains around 100 species that include common free-living bacteria, as well as pathogens [34]. Comamonadaceae, a family of the Betaproteobacteria, was not dominant in treatment group SU32. Comamonadaceae strains are involved in the oxidative deamination of aniline [35]. Acinetobacter, which belong to the class Gammaproteobacteria, only occurred in treatment groups SU24 and SU32. The species are a key source of infection in debilitated human patients, in particular the species Acinetobacter baumannii that causes nosocomial infections including meningitis, bacteraemia and pneumonia [36]. The results of this cultivation independent analysis suggest that the positive growth promoting effects associated with Starea treatment in cattle may be due to differences in the bacterial communities that are selected for in their intestines. Application of next generation sequencing techniques, in particular the use of metagenomics and metatranscriptomics, will provide additional new perspective into the functional traits that are selected for in the intestinal bacteria in cattle fed differing amounts of Starea. In addition, future studies using the newly developed methods to identify intestinal bacteria at the species level [15, 17, 18, 37] will provide further information on the positive effects of dietary amendment with Starea on cattle growth and health.
Electronic supplementary material
Acknowledgments
This study was supported by a Grant from the National Natural Science Foundation of China (No. 31172231) and the Earmarked Fund for Modern Agro-Industry Technology Research System.
References
- 1.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr. 1997;21:357–365. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- 3.Hooper LV, Gordon JI. Commensal host–bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 4.Mapato C, Wanapat M, Cherdthong A. Effects of urea treatment of straw and dietary level of vegetable oil on lactating dairy cows. Trop Anim Health Prod. 2010;42:1635–1642. doi: 10.1007/s11250-010-9613-3. [DOI] [PubMed] [Google Scholar]
- 5.Taylor-Edwards CC, Hibbard G, Kitts SE, McLeod KR, Axe DE, Vanzant ES, Kristensen NB, Harmon DL. Effects of slow-release urea on ruminal digesta characteristics and growth performance in beef steers. J Anim Sci. 2009;87:200–208. doi: 10.2527/jas.2008-0912. [DOI] [PubMed] [Google Scholar]
- 6.Velloso L, Perry TW, Peterson RC, Beeson WM. Effect of dehydrated alfalfa meal and of fish solubles on growth and nitrogen and energy balance of lambs and beef cattle fed a high urea liquid supplement. J Anim Sci. 1971;32:764–768. doi: 10.2527/jas1971.324764x. [DOI] [PubMed] [Google Scholar]
- 7.Helmer LG, Bartley EE, Deyoe CW. Feed processing. VI. Comparison of starea, urea, and soybean meal as protein sources for lactating dairy cows. J Dairy Sci. 1970;53:883–887. doi: 10.3168/jds.S0022-0302(70)86312-3. [DOI] [Google Scholar]
- 8.Morrill JL, Dayton AD. Soybean meal versus starea at two concentrations for young calves. J Dairy Sci. 1974;57:427–429. doi: 10.3168/jds.S0022-0302(74)84908-8. [DOI] [Google Scholar]
- 9.Ma W, Ren L, Wang L, Ding J, Zhao J, Meng Q. Effect of supplemental levels of gelatinized starch-urea on growth performance and plasma biochemical indices of growing-finishing beef cattle. Chin J Anim Nutr. 2011;23:1710–1715. [Google Scholar]
- 10.Kim M, Kim J, Kuehn LA, Bono JL, Berry ED, Kalchayanand N, Freetly HC, Benson AK, Wells JE. Investigation of bacterial diversity in the feces of cattle fed different diets. J Anim Sci. 2014;92:683–694. doi: 10.2527/jas.2013-6841. [DOI] [PubMed] [Google Scholar]
- 11.Castillo-Lopez E, Ramirez Ramirez HA, Klopfenstein TJ, Anderson CL, Aluthge ND, Fernando SC, Jenkins T, Kononoff PJ. Effect of feeding dried distillers grains with solubles on ruminal biohydrogenation, intestinal fatty acid profile, and gut microbial diversity evaluated through DNA pyro-sequencing. J Anim Sci. 2014;92:733–743. doi: 10.2527/jas.2013-7223. [DOI] [PubMed] [Google Scholar]
- 12.Paddock ZD, Walker CE, Drouillard JS, Nagaraja TG. Dietary monensin level, supplemental urea, and ractopamine on fecal shedding of Escherichia coli O157:H7 in feedlot cattle. J Anim Sci. 2011;89:2829–2835. doi: 10.2527/jas.2010-3793. [DOI] [PubMed] [Google Scholar]
- 13.Rajendhran J, Gunasekaran P. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res. 2011;166:99–110. doi: 10.1016/j.micres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhu W-Y, Williams BA, Konstantinov SR, Tamminga S, De Vos WM, Akkermans ADL. Analysis of 16S rDNA reveals bacterial shift during in vitro fermentation of fermentable carbohydrate using piglet faeces as inoculum. Anaerobe. 2003;9:175–180. doi: 10.1016/S1075-9964(03)00083-0. [DOI] [PubMed] [Google Scholar]
- 15.Porwal S, Lal S, Cheema S, Kalia VC. Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus. PLoS ONE. 2009;4:e4438. doi: 10.1371/journal.pone.0004438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Mehariya S, Ray S, Mishra A, Kalia VC. Biodiesel industry waste: a potential source of bioenergy and biopolymers. Indian J Microbiol. 2014;55:1–7. doi: 10.1007/s12088-014-0509-1. [DOI] [Google Scholar]
- 17.Kalia VC, Mukherjee T, Bhushan A, Joshi J, Shankar P, Huma N. Analysis of the unexplored features of rrs (16S rDNA) of the Genus Clostridium. BMC Genomics. 2011;12:18. doi: 10.1186/1471-2164-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal D, Verma M, Lal R. Exploring internal features of 16S rRNA gene for identification of clinically relevant species of the genus Streptococcus. Ann Clin Microbiol Antimicrob. 2011;10:28. doi: 10.1186/1476-0711-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe T, Asakawa S, Nakamura A, Nagaoka K, Kimura M. DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol Lett. 2004;232:153–163. doi: 10.1016/S0378-1097(04)00045-X. [DOI] [PubMed] [Google Scholar]
- 22.Wang JQ, Yin FG, Zhu C, Yu H, Niven SJ, de Lange CFM, Gong J. Evaluation of probiotic bacteria for their effects on the growth performance and intestinal microbiota of newly-weaned pigs fed fermented high-moisture maize. Livestock Sci. 2012;145:79–86. doi: 10.1016/j.livsci.2011.12.024. [DOI] [Google Scholar]
- 23.Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LH, Ravelonandro P, Mavingui P. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol. 2011;75:377–389. doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Sun L, Wu H, Hu Z, Liu W, Li Y, Wen X. The intestinal microbial diversity in mud crab (Scylla paramamosain) as determined by PCR-DGGE and clone library analysis. J Appl Microbiol. 2012;113:1341–1351. doi: 10.1111/jam.12008. [DOI] [PubMed] [Google Scholar]
- 28.Haverson K, Rehakova Z, Sinkora J, Sver L, Bailey M. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol. 2007;119:243–253. doi: 10.1016/j.vetimm.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Rudi K, Moen B, Sekelja M, Frisli T, Lee MR. An eight-year investigation of bovine livestock fecal microbiota. Vet Microbiol. 2012;160:369–377. doi: 10.1016/j.vetmic.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Murphy P, Bello FD, O’Doherty JV, Arendt EK, Sweeney T, Coffey A. Effects of cereal beta-glucans and enzyme inclusion on the porcine gastrointestinal tract microbiota. Anaerobe. 2012;18:557–565. doi: 10.1016/j.anaerobe.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Dougal K, de la Fuente G, Harris PA, Girdwood SE, Pinloche E, Newbold CJ. Identification of a core bacterial community within the large intestine of the horse. PLoS ONE. 2013;8:e77660. doi: 10.1371/journal.pone.0077660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol. 2014;87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 33.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 2010;44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 34.Costa MC, Reid-Smith R, Gow S, Hannon SJ, Booker C, Rousseau J, Benedict KM, Morley PS, Weese JS. Prevalence and molecular characterization of Clostridium difficile isolated from feedlot beef cattle upon arrival and mid-feeding period. BMC Vet Res. 2012;8:38. doi: 10.1186/1746-6148-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boon N, Goris J, De Vos P, Verstraete W, Top EM. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl Environ Microbiol. 2001;67:1107–1115. doi: 10.1128/AEM.67.3.1107-1115.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamouda A, Findlay J, Al Hassan L, Amyes SG. Epidemiology of Acinetobacter baumannii of animal origin. Int J Antimicrob Agents. 2011;38:314–318. doi: 10.1016/j.ijantimicag.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Bhushan A, Joshi J, Shankar P, Kushwah J, Raju SC, Purohit HJ, Kalia VC. Development of genomic tools for the identification of certain pseudomonas up to species level. Indian J Microbiol. 2013;53:253–263. doi: 10.1007/s12088-013-0412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.