Abstract
In present study, several marine water samples collected from the North Goa Beaches, India for isolation of luminescent bacterial species. Isolates obtained labelled as DP1-5 and AB1-6. Molecular characterization including identification of a microbial culture using 16S rRNA gene based molecular technique and phylogenetic analysis confirmed that DP3 & AB1 isolates were Vibrio harveyi. All of the isolates demonstrated multiple metal resistances in terms of growth, with altered luminescence with variable metal concentration. Present investigations were an attempt towards exploring and reporting an updated diversity of bioluminescent bacterial species from various sites around the Goa, India which would be explored in future for constructing luminescence based biosensor for efficiently monitoring the level of hazardous metals in the environment.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0531-y) contains supplementary material, which is available to authorized users.
Keywords: Heavy metals pollution, Biosensor, Vibrio harveyi, Bioluminescent bacteria, 16S rRNA gene analysis, Bioassay, Monitoring
Introduction
Luminescent bacteria are the most abundant and widely distributed amongst the light-emitting organisms. The mechanism of the bioluminescence reaction is catalyzed by luciferase which produces light by the oxidation of a light-emitting molecule called the luciferin. The luciferase enzyme coded by the luxAB genes catalyses the unique light-producing bioluminescence reaction. For this light reaction, dissolved oxygen, reduced flavine mononucleotide and a long chain aldehyde coded by luxCDE genes are vital substrate requirements [1, 2]. The DNA sequences coding the proteins in the luminescent system are termed as the ‘lux genes’. The 2 polypeptide chains comprising the luciferase enzyme are encoded by luxAB genes. The aldehyde required for the reaction is synthesized by an enzymatic complex coded by three different genes present in all bioluminescent bacteria, luxCDE. This aldehyde is derived from the fatty acid pathway and can be recycled after each reaction. The three proteins that form the enzymatic complex comprises of a reductase, a synthetase and a transferase. Other proteins involved in the reaction and are only present in certain species of bacteria. For example, lumazine and yellow fluorescent protein (yfp) are two different proteins present in Photobacterium phosphoreum and in some strains of Vibrio fischeri respectively. Both are called accessory proteins because they are not crucial for the basic reaction completion [3]. Photobacterium phosphoreum is also known to contain LumP gene which codes for lumazine protein [4, 5]. The biological significance of luminescence to these bacterial species is still unclear; and therefore bioluminescence continues to be a subject of exploration. Presently most interesting theme of research is the prospective use of bacterial bioluminescence trait for deciphering various basic and applied problems. Hence understanding the biochemical and molecular basis of bioluminescence as well as effect of pollutants on it is of great importance. In view of the increasing ill-effects of environmental pollution, there is an utmost need for developing reliable tools for rapid assessment of the potential toxic and genotoxic effects of environmental samples. Bioluminescence can be used as a reliable reporter for the assessment or monitoring of various aquatic samples containing toxicants such as pesticides, PCBs, aromatic hydrocarbons, fuels, alkanes, alcohols and heavy metals because of its high degree of response, simple operation and low cost of performance [6–18]. Since marine luminous bacteria are ecologically resourceful, utilize several nutrients, occupy many niches in the marine environment and their bioluminescence being extremely sensitive to the toxicants, they are suitable bioassay candidate for detecting nano or pico molar concentrations of impurities in water bodies, pharmaceuticals and in the food industries [19]. These studies intend to investigate the physiological, biochemical as well as genetic diversity and relationships among bioluminescent bacterial strains isolated from various beaches of North Goa, India. Furthermore, current analysis is targeting to exemplify the potential use of newly isolated strains in construction of heavy metal bioassay or biosensor to monitoring the heavy metal contaminants in water samples.
Material and Methods
Water Sampling
Water samples collected from the several beaches of the Goa, India named as Dona Paulo, Miramar, Aguada in January 2013. The samples processed immediately in the laboratory for the bacterial isolation.
Isolation of Bioluminescent Bacteria
Collected samples were then spread plated on modified nutrient agar media (contains 25 % sea salt with 3 % glycerol) [20] and incubated in dark at 21 °C for 24 h as isolates shows enhanced luminescence when grown in dark. The luminescent colonies were picked maintained on modified nutrient agar at an interval of 4 days for continued luminosity & the isolates were stored at −20 °C with addition of 50 % glycerol. Gram and flagella staining were performed as per the standard procedure [21]. The morphology of the bacterial isolates was studied and confirmed using scanning electron microscopy. Motility was observed by using ‘hanging drop preparation method’ as per the standard protocol [22].
Biochemical Characterisation
In order to characterize the isolates for biochemical parameters a total of 14 biochemical assays [23] such as: Oxidase, Nitrate reduction, Gelatinase, Simmons citrate, Indole, Methyl Red, Voges-proskauer, Catalase, Urease, Cytochrome oxidase, Amylase, Gelatin liquefaction and Hydrogen sulphide production test were performed. For carbohydrate utilization test various sugars were used- glucose, lactose, mannitol, sucrose, maltose and pyruvate. All the controls (positive and negative) and the blank used for different biochemical test were incubated at respective optimum growth temperature (37 °C).
Bacterial Identification
In order to confirm the identity of the isolates genomic DNA was extracted using an Axyprep Bacterial Genomic DNA Miniprep Kit (Axygen). Further, fragment of 16S rRNA gene was amplified by Polymerase Chain Reaction (PCR) with the aid of universal primers for bacteria (27F, 5-AGAGTTTGATCMTGGCTCAG and 1492R, 5-TACGGYTACCTTGTTACGACT-3). Amplification parameters for bacterial 16S rRNA gene were as follow: an initial denaturation of 94 °C for 3 min., 30 cycles of (94 °C 30 s, 52.7 °C 30 s, and 72 °C 1.30 min). DNA sequencing reaction of PCR amplicon was carried out with 8F and 1492R primers using BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyzer. The 16S rRNA gene sequence was used to carry out BLAST with the nrdatabase of NCBI genbank database. Based on maximum identity score first ten sequences were selected and aligned using multiple alignment software program Clustal W. Distance matrix was generated using RDP database and the phylogenetic tree was constructed using MEGA 4 [24].
Antibiotic and Heavy Metal Sensitivity Profiling
4–5 colonies of isolated strains were inoculated with a loop to 5 ml Nutrient Broth aided with 50 % (v/v) sea salt & incubated at 21 °C for overnight to achieve light to moderate turbidity. By using standardized inoculum the entire agar surface of the plate was streaked with the swab & dried for 30 min. To study the antibiotic sensitivity profiling the Himedia made Icosa G-I-Minus antibiotic susceptibility testing discs was applied using aseptic technique and incubated at 21 °C for 16–18 h. The diameters of zones (millimetre) indicating growth inhibition was record using a calibrated zone scales. Heavy metal sensitivity profile was developed by standard well diffusion assay. In each of the inoculated plates well was created by using 6 mm cork borer and 100 µl of various concentrations of heavy metals ranging between 0.05 and 0.50 mg/L was added. The diameters of zones (millimetre) showing growth inhibition was record using a calibrated zone scales.
Qualitative Estimation of Luminescence
Bacterial culture streaked on modified nutrient agar plates was partitioned in four compartments streaked & incubated at 21 °C. Partitioned culture plates were sprayed with three heavy metal solutions of 0.05, 0.25, 0.50 mg/L and incubated for 30 min at 21 °C. Measurement of qualitative (Visual) alterations in luminescence due to presence of respective heavy metal concentrations was made by observing these plates in completely dark room.
Results and Discussion
Isolation of Bioluminescent Bacteria
These bioluminescent isolates differ both in light emitting intensities and morphologies. Fine isolated bluish-green glowing colonies of the bacterial isolates were obtained.
Morphological Characterization
The characterisation with regard to size, shape, arrangement of cells and motile features, colonial characteristics viz., form, elevation, margin, colour, surface, density, consistency and presence of flagella have been proved to be fruitful in better understanding of the morphology of bacterial isolates using different staining procedures. The presence of flagella was also confirmed by the flaring outgrowth of the inoculated bacterial culture around the line of stab in SIM (Sulfite Indole Motility) medium. The SEM analysis provided an additional insight to the external morphology and appearance of the bacterial cells (Fig. 1a, b).
Fig. 1.

SEM photograph of the bacterium isolated from, a Aguadha Beach Sample; b Dona Paulo Beach Sample. Sample a exhibit a basic morphology of elongated bacilli with 1–1.44 µm size, in contrast samples b shows a shorter bacilli morphology of 1 µm size
Biochemical Characterization
In the biochemical characterization samples differ in their ability to metabolize carbohydrates such as Maltose could be utilized by both of the isolates while Lactose is not utilized by any of the bioluminescent isolates. Vibrio harveyi strain DP1 was unable to utilize citrate, while both isolates were found to be Indole positive. Enzymes such as catalase, urease, amylase were found to be absent for AB isolate while in contrast to this gelatinase was actively present for Vibrio harveyi strain AB1 isolate (Table. I in ESM). All strains gave a positive reaction to the oxidase test and dependent on NaCl for growth as well as luminescence production.
16S rRNA and Phylogenetic Analysis
The total genomic DNA was isolated by Axyprep Bacterial Genomic DNA miniprep kit (Genaxy). Fragment of 16S rRNA gene was amplified by PCR with 8F and 1492R primers from the above isolated DNA samples. A single discrete PCR amplicon band of 1438 and 1500 bp was resolved on Agarose Gel for AB & DP isolate respectively.
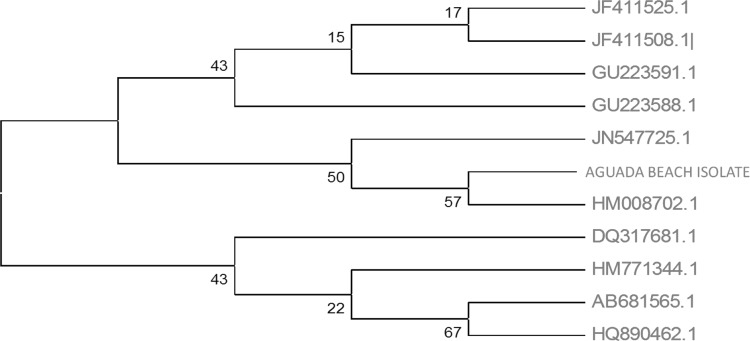
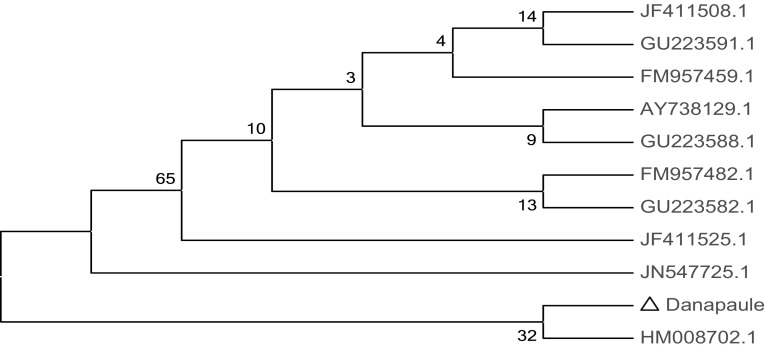
The PCR amplicon was purified to remove contaminants by QIAquick Gel Extraction Kit (QIAGEN) and respective consensus sequences of 1438 bp and 1500 bp 16S rDNA genes was generated by BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyzer. Based on BLAST and MEGA 4 the isolates regonised, AB as Vibrio harveyi [KF607036] and DP as Vibrio harveyi [KF607037] (Figs. 2, 3).
Fig. 2.
Phylogenetic analysis of 16S rDNA gene amplified from Aguada Beach (AB) isolates and identified as Vibrio harveyi strain NB0901 with close homologs
Fig. 3.
Phylogenetic analysis of 16S rDNA gene amplified from Dona Paulo Beach (DP) isolates and identified as Vibrio harveyi strain NB0901 with close homologs
Antimicrobial Susceptibility of the Bacterial Isolate
Antibiotics susceptibility tests for each isolates were performed using disc diffusion method. Antibiotic impregnated paper discs (Himedia made Icosa G-I-Minus) were applied onto the agar surface, incubated overnight at 21 °C and then diameter of zone of inhibition (mm) around antibiotic discs to the nearest whole millimeter was recorded. Both bacterial isolate was observed to be susceptible to Kanamycin (30 µg), Gentamicin (10 µg) and Streptomycin (10 µg). For Sparfloxacin (5 µg) and Ciprofloxacin (5 µg) DP isolate showed resistance while AB isolate was found to be sensitive and on other hand DP isolate indicated sensitivity towards Amikacin (30 µg) for which Vibrio harveyi strain AB1 isolate shown intermediate susceptibility. Few antibiotics such as Augmentin (30 µg), Cefpodoxime (10 µg) & Ticarcillin (75 µg) showed no zone formation for both of the isolates (Table-II in ESM).
Effect of Different Heavy Metals on Bacterial Isolate
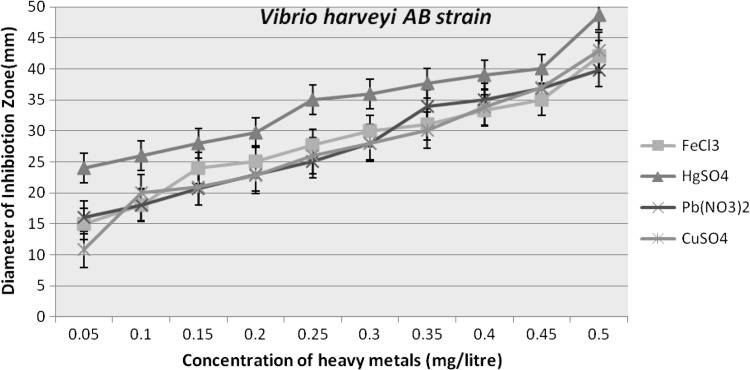
The effect of various concentrations of compounds containing heavy metals as Pb(NO3)2,CuSO4, FeCl3 and HgSO4 was also seen on the growth and luminescence of V. harveyi. The incubated plates were observed for the diameter of the zone of growth and luminescence inhibition around the heavy metal, in terms of millimetre (mm) (Table-III in ESM; Figs. 4, 5).
Fig. 4.
Graph indicating heavy metal sensitivity profile of isolated Vibrio harveyi DP strain against various concentrations of heavy metals (0.05–0.50 mg/l)
Fig. 5.
Graph indicating heavy metal sensitivity profile of isolated Vibrio harveyi AB strain against various concentrations of heavy metals (0.05–0.50 mg/l)
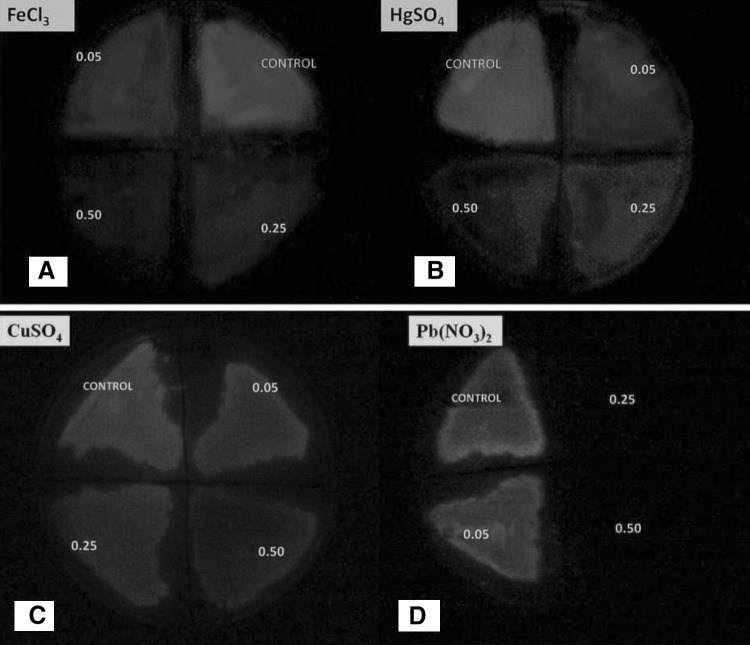
Visual Evidences for Luminescence Alteration due to Heavy Metal Presence
Both the strains were examined for luminescence alteration against various heavy metal concentrations and based on results Vibrio harveyi AB strain was considered for further analysis. The change in the luminescence intensity of Vibrio harveyi AB strain were spray analyzed for the 0.05,0.25 and 0.50 mg/litre of FeCl3, HgSO4, CuSO4, Pb(NO3)2 (Fig. 6a–d). The results suggested that define change in the luminescence was recorded in Pb (NO3)2 at the 0.25 onwards with complete loss of luminescence. While at 0.05 mg/litre the luminescence remain unchanged compared to control. For FeCl3 and HgSO4 similar trend was apparent where luminescence decreased with increased metal concentration as compared to control. In contrast CuSO4 treated sample showed very slight change in luminescence. Luminescent biosensor strain explored during present investigation is highly responsive to wide range of concentrations of FeCl3, HgSO4, CuSO4 ions, except for Pb(NO3)2 and can be used to quantify their bioavailability in polluted water samples. Few recent studies reported recombinant GFP based bacterial biosensor responded to Pb2+ in the range of 50–400 μM [25]. Collectively, previous and present studies indicates the need for development of lead specific luminescence based recombinant bacterial strain which could provide a wide range for lead metal sensing in terms of luminescence changes.
Fig. 6.
a–d Photographic evidences indicating effect of increasing toxic concentrations (0.05, 0.25, 0.50 mg/L) of various heavy metals on test organism-Vibrio harveyi AB strain
Electronic supplementary material
Acknowledgments
The financial support under the Major research project sponsored by University Grant of Commission, Govt. of India, New Delhi is gratefully acknowledged.
Conflict of interest
All authors have none to declare.
Contributor Information
Neha A. Thakre, Phone: + 91- 9823018963, Email: thakre.neha5@gmail.com
Arti S. Shanware, Phone: + 91 (0712)2552080, Email: biotechneha23@gmail.com, Email: artishanware@gmail.com
References
- 1.Charrier T, Durand MJ, Affi M,Jouanneau S, Gezekel H, Thouand G (2010) Bacterial bioluminescent biosensor characterisation for on-line monitoring of heavy metals pollutions in waste water treatment plant effluents. “Biosensors” book, Chapter 11, ISBN 978-953-7619-99-2. doi: 10.5772/7210
- 2.Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell ZT, Baldwin TO. Fre is the major flavin reductase supporting bioluminescence from Vibrioharveyi luciferase in Escherichiacoli. J Biol Chem. 2009;284:8322–8328. doi: 10.1074/jbc.M808977200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato Y, Shimizu S, Ohtaki A, Noguchi K, Miyatake H, Dohmae N, Sasaki S, Odaka M, Yohda M. Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl- 8-(1′-D-ribityl) lumazine, and its analogues, riboflavin and flavin mononucleotide, at high resolution. J Bacteriol. 2010;192:127–133. doi: 10.1128/JB.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Small ED, Koka P, Lee J. Lumazine protein from the bioluminescent bacterium photobacterium phosphoreum. Purification and characterization. J Biol Chem. 1980;255:8804–8810. [PubMed] [Google Scholar]
- 6.Abd-El-Haleem D, Moawad H, Zaki E, Zaki S. A luxCDABE-based bioluminescent bioreporter for the detection of phenol. J Ind Microbiol Biotechnol. 2002;29:233–237. doi: 10.1038/sj.jim.7000309. [DOI] [PubMed] [Google Scholar]
- 7.Backhaus T, Grimme LH. The toxicity of antibiotic agents to the luminescent bacterium Vibriofischeri. Chemosphere. 1999;38:3291–3301. doi: 10.1016/S0045-6535(98)00560-8. [DOI] [PubMed] [Google Scholar]
- 8.Choi SH, Gu MB. A portable toxicity biosensor using freeze-dried recombinant bioluminescent bacteria. Biosens Bioelectron. 2002;17:433–440. doi: 10.1016/S0956-5663(01)00303-7. [DOI] [PubMed] [Google Scholar]
- 9.Cho JC, Park KJ, Ihm HS, Park JE, Kim SY, Kang I, Lee KH, Jahng D, Lee DH, Kim SJ. A novel continuous toxicity test system using a luminously modified freshwater bacterium. Biosens Bioelectron. 2004;20:338–344. doi: 10.1016/j.bios.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 10.King JMH, DiGrazia PM, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler GS. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 11.Minak-Bernero V, Bare RE, Haith CE, Grossman MJ. Detection of alkanes, alcohols, and aldehydes using bioluminescence. Biotechnol Bioeng. 2004;87:170–177. doi: 10.1002/bit.20089. [DOI] [PubMed] [Google Scholar]
- 12.Ren S, Frymier PD, Schultz TW. An exploratory study of the use of multivariate techniques to determine mechanisms of toxic action. Ecotoxicol Environ Saf. 2003;55:86–97. doi: 10.1016/S0147-6513(02)00132-X. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz MJ, Lopez-Jaramillo L, Redondo MJ, Font G. Toxicity assessment of pesticides using the microtox test: application to environmental samples. Bull Environ Contam Toxicol. 1997;59:619–625. doi: 10.1007/s001289900524. [DOI] [PubMed] [Google Scholar]
- 14.Stocker J, Balluch D, Gsell M, Harms H, Feliciano J, Daunert S, Malik KA, van der Meer JR. Development of a set of simple bacterial biosensors for quantitative and rapid measure-ments of arsenite and arsenate in potable water. Environ Sci Technol. 2003;37:4743–4750. doi: 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- 15.Tauriainen S, Karp M, Chang W, Virta M. Luminescent bacterial sensor for cadmium and lead. Biosens Bioelectron. 1998;13:931–938. doi: 10.1016/S0956-5663(98)00027-X. [DOI] [PubMed] [Google Scholar]
- 16.Unitzur S, Lahav T, Ulitzur N. A novel and sensitive test for rapid determination of water toxicity. Environ Toxicol. 2002;17:291–296. doi: 10.1002/tox.10060. [DOI] [PubMed] [Google Scholar]
- 17.Van Dyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, LaRossa RA. Rapid and sensitive pollutant detection by induction of heat shock gene bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weitz HJ, Campbell CD, Killham K. Development of a novel, bioluminescence-based fungal bioassay for toxicity testing. Environ Microbiol. 2002;4:422–429. doi: 10.1046/j.1462-2920.2002.00315.x. [DOI] [PubMed] [Google Scholar]
- 19.Ramaiah N, Chandramohan I. Ecological and Laboratory Studies on the Role of Luminous Bacteria and their Luminescence in Coastal Pollution Surveillance. Mar Pollut Bull. 1993;26:190–201. doi: 10.1016/0025-326X(93)90621-P. [DOI] [Google Scholar]
- 20.Shanware AS, Thakre NA, Pande SS. Isolation and characterization of novel marine luminescent bacteria from Diu beach, India. J Pharm Res. 2013;7:529–533. doi: 10.1016/j.jopr.2013.05.019. [DOI] [Google Scholar]
- 21.Koneman EW, et al. Color Atlas and Textbook of Diagnostic Microbiology. 5. Philadelphia: J.B. Lippincott Company; 1997. [Google Scholar]
- 22.Mac Faddin, JF (1980) Media for isolation, cultivation, identification, maintenance of medical bacteria. Vol I, Baltimore, Williams and Wilkins
- 23.Baumann P, Schubert RHW (1984) Family II. Genetic diversity of luminous Vibrio Vibrionaceae. In Bergey’s manual of systematic Krieg, N. R. and J.G. Holt (Eds.), 1:516–550
- 24.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty T, Gireesh Babu P, Alam A, Chaudhari A. GFP expressing bacterial biosensor to measure lead contamination in aquatic environment. Curr Sci. 2008;94:800–805. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.