Abstract
Human infections with non-typhoidal Salmonella (NTS) serovars are increasingly becoming a threat to human health globally. While all motile Salmonellae have zoonotic potential, Salmonella Enteritidis and Salmonella Typhimurium are most commonly associated with human disease, for which poultry are a major source. Despite the increasing number of human NTS infections, the epidemiology of NTS in poultry in India has not been fully understood. Hence, as a first step, we carried out epidemiological analysis to establish the incidence of NTS in poultry to evaluate the risk to human health. A total of 1215 samples (including poultry meat, tissues, egg and environmental samples) were collected from 154 commercial layer farms from southern India and screened for NTS. Following identification by cultural and biochemical methods, Salmonella isolates were further characterized by multiplex PCR, allele-specific PCR, enterobacterial repetitive intergenic consensus (ERIC) PCR and pulse field gel electrophoresis (PFGE). In the present study, 21/1215 (1.73 %) samples tested positive for NTS. We found 12/392 (3.06 %) of tissue samples, 7/460 (1.52 %) of poultry products, and 2/363 (0.55 %) of environmental samples tested positive for NTS. All the Salmonella isolates were resistant to oxytetracycline, which is routinely used as poultry feed additive. The multiplex PCR results allowed 16/21 isolates to be classified as S. Typhimurium, and five isolates as S. Enteritidis. Of the five S. Enteritidis isolates, four were identified as group D Salmonella by allele-specific PCR. All of the isolates produced different banding patterns in ERIC PCR. Of the thirteen macro restriction profiles (MRPs) obtained by PFGE, MRP 6 was predominant which included 6 (21 %) isolates. In conclusion, the findings of the study revealed higher incidence of contamination of NTS Salmonella in poultry tissue and animal protein sources used for poultry. The results of the study warrants further investigation on different type of animal feed sources, food market chains, processing plants, live bird markets etc., to evaluate the risk factors, transmission and effective control measures of human Salmonella infection from poultry products.
Keywords: Isolation, Identification, NTS, Zoonotic Salmonella, Genetic diversity, Poultry products, India
Introduction
Salmonellosis continues to be a major public health problem globally. Salmonellosis is caused by two species of Salmonella (Salmonella enterica and Salmonella bongori), which are etiological agents for diarrhoeal and systemic infection in humans. Human infections with non-typhoidal Salmonella (NTS) are frequently associated with the consumption of contaminated food [1]. This can result in diarrhoeal disease, bacteraemia and extraintestinal focal infection in infants such as meningitis, and may also result in more serious complications among the elderly and immunocompromised patients [2]. S. enterica serovar Enteritidis (S. Enteritidis) is a major public health problem globally and is the most common serotype among the NTS in the US [3]. Poultry and their products are the most frequently implicated reservoirs of NTS in the human food chain. India is one of the largest producers of poultry in the world. Among the foods of animal origin, chicken meat and eggs are widely accepted, are not subject to a religious taboo, and are the only available animal protein source to a vast majority of poor people in India. Increased trade and globalization of the modern poultry industry has resulted in new and more complex opportunities for the spread of Salmonella [4, 6]. More than 2500 serotypes of Salmonella have been reported to date [5], but only about 10 % of these have been isolated from poultry.
The distribution of Salmonella serotypes from poultry varies geographically and temporally, although several serotypes are consistently found at a high incidence [6]. In recent years, concern about poultry eggs and meats contaminated with Salmonella has gained significant attention because of the high occurrence of antimicrobial-resistant bacteria connected with human illness [7–9]. A high prevalence of antimicrobial resistant Salmonella in poultry and foods of animal origin has been reported earlier from India [10, 11]. During the last few years, the National Egg Co-ordination Committee (NECC), Govt. of India has taken steps to promote egg as a source of good quality protein and the consumption of egg has increased noticeably. There are mass poultry production regions in several parts of the country especially in southern states, Tamil Nadu and Andhra Pradesh. Though the consumption has been promoted, no effective measures are taken to monitor the quality of the poultry meat and egg reaching the market. Molecular subtyping of Salmonella isolates is an invaluable epidemiological tool that can be used to track the source of infection and to determine the epidemiological link between isolates from poultry and human sources. Pulsed-field gel electrophoresis (PFGE) and other DNA based characterization techniques like ERIC PCR, Multiplex PCR and Allele specific PCR provides information that can be used to evaluate epidemiological associations with a high degree of confidence. In India, other than some limited studies on the incidence of salmonellosis, no systematic studies have been conducted to establish the epidemiology and characterize NTS in poultry [12–14]. This study describes the isolation, characterization and epidemiology of NTS in poultry and their products in Southern India using conventional culture methods and molecular techniques.
Materials and Methods
Sample Collection
A total of 1215 samples were collected from 154 commercial layer farms from Southern India and screened for the presence of Salmonella. The samples comprised: poultry tissue samples (liver, ovary, intestinal contents, spleen and yolk from dead birds), poultry products (meat, egg, feed, fishmeal, meat and bone meal) and environmental samples (water, drag swab, boot swab, muconium and fecal samples).
Isolation
The drag swab, boot swab, muconium, fecal, egg, feed, fishmeal and meat and bone meal were subjected to pre- enrichment with buffered peptone water (25 g in 225 ml of buffered peptone water) and incubated at 37 °C for 24 h. Tissue samples were directly inoculated into selective enrichment broth. One ml of pre-enrichment culture was transferred to 10 ml of tetrathionate broth for selective enrichment and incubated at 37 °C for 24 h. The main purpose of selective enrichment broth was selective inhibition of bacteria other than Salmonella, so that Salmonella can be isolated in the selective plating media. A loopful of inoculum from enrichment broth was streaked onto brilliant green agar (BGA) and incubated at 37 °C for 24 h and observed for the development of characteristic colonies. The highly Salmonella suggestive pink colour colonies were picked out from BGA and streaked onto desoxycholate citrate agar, MacConkey agar and Salmonella differential agar and incubated at 37 °C for 24 h and the colony character was observed. The colonies which were highly suggestive of Salmonella were subjected to further identification and characterization. Suspected colonies of Salmonella from MacConkey agar plates were inoculated into brain heart infusion broth and incubated at 37 °C for 6 h. One to two drops of actively grown broth culture was subjected for motility test by hanging drop method under compound microscope. The colonies from BHI agar plates were stabbed into TSI slants and incubated at 37 °C for 24 h and observed for acid, gas and H2S production. The first step in eliminating non-Salmonella organism was done by urease test. The isolates which produced acid butt and alkaline slant with or without H2S in TSI slants were selected and stabbed into urease agar slant incubated 37 °C for overnight and observed. The isolates also tested for indole production, MR–VP test, and utilization carbohydrates namely lactose, arabinose, maltose, sorbitol and dulcitol. The culture which was identified as Salmonella by the above tests was further confirmed by Hi-Salmonella identification kit procured from Hi-Media, Mumbai and the results were interpreted according to the interpretation chart supplied by the manufacturer.
Antibiogram (Kirby–Bauer Method)
The antibiotic sensitivity of all isolates was carried out using a previously described method [15]. The commonly used antimicrobial discs namely amoxicillin (Am) 30 mcg/disc, chloramphenicol (C) 30 mcg/disc, ciprofloxacin (Cf) 30 mcg/disc, co-trimoxazole (Co) 25 mcg/disc, enrofloxacin (Ex) 10 mcg/disc, gentamicin (G) 30 mcg/disc, norfloxacin (Nx), 10 mcg/disc and oxytetracycline (O) 30 mcg/disc were used. The sensitivity pattern of all the Salmonella isolates was determined using Muller Hinton Agar plates (Himedia Pvt., Ltd., Mumbai) incubated at 37 °C for 16 h. The zone of the inhibition of the isolates were measured using high sensitivity zone scale (Hi-Media Pvt., Ltd., Mumbai). The pattern of sensitivity/resistance determined as per the disc manufacturers guidelines (Hi-Media Pvt., Ltd., Mumbai).
Molecular Characterization
The isolates identified at genus level as Salmonella by biochemical tests were subsequently characterized further using the following tests: multiplex PCR, allele-specific PCR, enterobacterial repetitive intergenic consensus (ERIC) PCR, and pulse field gel electrophoresis (PFGE).
Genomic DNA was extracted from isolates using the Ready Template Genomic DNA purification kit (Heleni Biomolecules, Chennai). A multiplex PCR method developed by [16] was used, with modifications in the primer sets. The sets of primers used in the multiplex PCR assay were Pef A-Fwd-5′-TTC CATTATTGCACTGGGTG-3′, PefA-Rev-5′-AAGCCACTGCGAAAGATGCC-3′, that were selected based on the 5′–3′ conserved region of the fimbrial virulence gene (pefA). The pefA gene amplifies both in S. Typhimurium and S. Enteritidis. However, these two serovars were differentiated using an additional primer (F-5′-AAGTTGTTCAGCTGGGTACC-3′) targeting the gene kpnl which is present in S. Typhimurium but not in S. Enteritidis.
The allele-specific PCR was used to differentiate group D Salmonella from group B Salmonella as well as from non-Salmonella organisms based on amplification of the rfbS gene which is present only in group D Salmonella. Allele-Specific PCR analysis of Salmonella isolates was performed according to the method of [17] using the following primers, Fwd-5′-TCACGACTTACATCCTAC-3′ and Rev-5′-CTGCTATATCAGCACAAC-3′. ERIC PCR analysis for intra-serotype typing of Salmonella isolates in this study was performed according to the method of [18] using the following primer set ERIC1-5′-ATGTAAGCTCCTGGGGATTCA C-3′, ERIC2-5′-AAGTAAGTGACTGGGGTGAGC G-3′. PFGE was performed to study the genetic relatedness of the Salmonella isolates using the XbaI-restriction enzyme as described by [19]. The normalization, recognition and assignment of the bands were performed with the software programme FINGERPRINTING II (Bio-Rad) using Dice-UPGMA algorithm.
Results
Incidence of NTS in Poultry and Their Products
In the present study 21/1215 (1.73 %) of samples were confirmed as positive for Salmonella (Table 1). Tissue samples showed highest isolation of NTS (12/392), followed by poultry products (7/460), and environmental samples (2/363). Among the tissue samples, the recovery of NTS was highest in liver samples (5/104, 4.80 %), and among poultry products viz., poultry feed, meat and bone meal (7/73, 9.60 %). The isolation was high in layer chicks below 3 weeks of age (4/55, 7.27 %) followed by layer chickens below 40 weeks of age (4/111, 3.60 %) and layer chickens above 40 weeks of age (4/226, 1.77 %). Tissue samples collected from affected birds revealed the high incidence of NTS in winter 17/607 (2.80 %) than summer 4/608 (0.65 %).
Table 1.
Non-typhoidal Salmonella (NTS) isolates obtained from poultry, poultry products and environmental samples
| Type of sample | No. of samples | No. of isolates obtained | Sample wise isolation percentage |
|---|---|---|---|
| Poultry tissue samples | |||
| Yolk | 21 | 01 | 4.76 |
| Liver | 104 | 05 | 4.80 |
| Ovary | 96 | 02 | 2.08 |
| Intestinal content | 103 | 02 | 1.94 |
| Spleen | 11 | – | |
| Pooled organs | 57 | 02 | 3.5 |
| Total tissue samples | 392 | 12 | 3.06 |
| Poultry products | |||
| Egg | 213 | – | – |
| Feed | 67 | 05 | 7.4 |
| Meat sample | 174 | – | – |
| Meat and bone meal | 06 | 02 | 33.3 |
| Total poultry products | 460 | 07 | 1.52 |
| Environmental samples | |||
| Muconium | 12 | – | – |
| Water | 73 | – | – |
| Drag swab | 58 | 01 | 1.72 |
| Boot swab | 61 | – | – |
| Fecal sample | 159 | 01 | 0.62 |
| Total environmental samples | 363 | 02 | 0.55 |
| Total | 1215 | 21 | 1.73 |
Antibiogram of Salmonella Isolates
The antibiotic sensitivity of 21 salmonella isolates was determined using a selection of most commonly used antibiotics in poultry industry in India. The percentage of isolates regarded as sensitive to each antibiotic was as follows: ciprofloxacin (100 %), enrofloxacin (90 %), norfloxacin (81 %), chloramphenicol (62 %), amoxicillin (62 %), co-trimoxazole (38 %), and gentamicin (9 %). Notably, all the isolates were resistant to oxytetracycline.
Molecular Characterization
Multiplex PCR
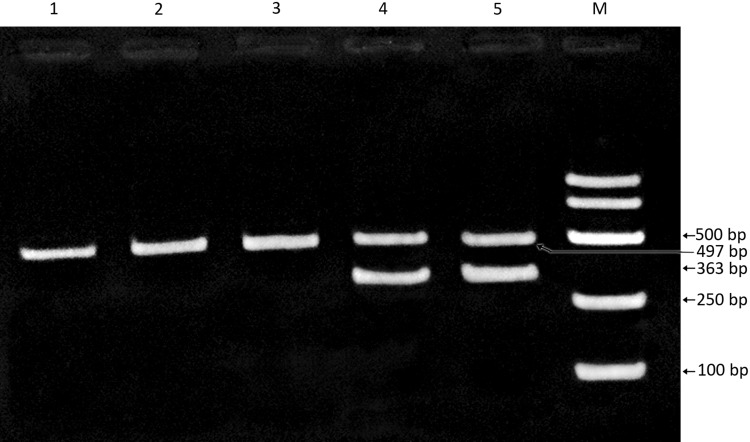
Based on the results of multiplex PCR, 16 isolates that yielded two distinct products of 363 and 497 bp length were categorized as Salmonella Typhimurium (isolates no. S2, S44, S87 S99, S100, S102, S184, S185, S186, S255, S380, S381, S711, S714, S931 and S932). Five isolates (S585, S586, S759, S791 and S828) which yielded a single product of 497 bp were categorized as S. Enteritidis (Fig. 1).
Fig. 1.
Multiplex PCR assay of non typhoid salmonella (NTS) isolates targeting pefA and kpnl genes. NTS isolates obtained from poultry and their products were subjected to multiplex PCR targeting a conserved region of the fimbrial virulence gene (pefA) and kpnl gene to distinguish between S. Typhimurium and S. Enteritidis. Lanes 1–3 Salmonella isolates (S759, S 791 and S828) showing a single PCR product of 497 bp was confirmed as S. Enteritidis. Lanes 4, 5 Salmonella isolates (S931 and S932) showing two distinct bands of 363 and 497 bp were confirmed as S. Typhimurium. Lane M 250 bp DNA ladder
Allele-Specific PCR
Five isolates of S. Enteritidis confirmed by multiplex PCR were further characterized by allele-specific PCR, which showed that 4/5 S. Enteritidis isolates belonged to group D Salmonella (S586, S759 S791 and S828) with a single 720 bp product.
ERIC PCR
ERIC PCR was carried out in order to assess the level of variation between the isolates. All of the isolates produced eight different banding patterns (ERIC 1–8) ranging from two to six bands with the fragments ranging from 150 to 2000 bp. Six isolates showed ERIC profile 1 (S2, S44, S87, S184, S381, and S759) and only one isolate each showed ERIC profile seven and eight (S828 and S185 respectively). The remaining profiles had two to four isolates (S99, S100, S102, S186, S255, S380, S585, S586, S711, S714, S791, S931 and S932).
PFGE
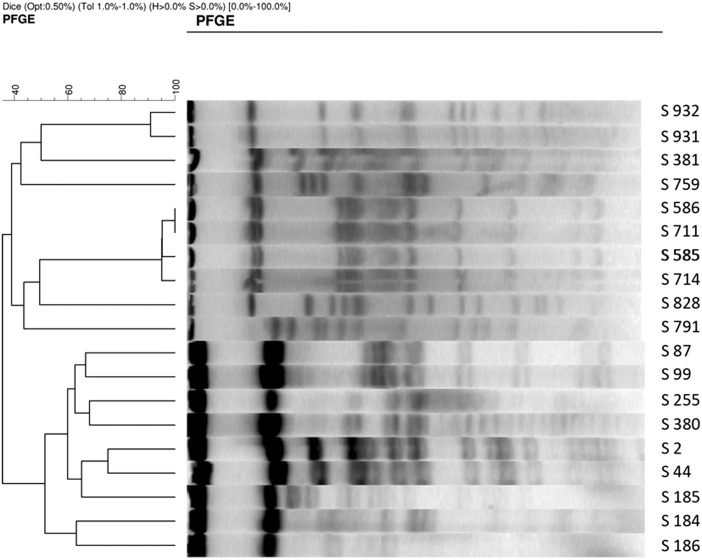
PFGE was used to study the degree of genetic relatedness of the Salmonella strains by XbaI digestion of genomic DNA. The 21 Salmonella isolates could be ascribed to one of 11 macrorestriction profiles (MRPs), each of which contained up to 13 restriction fragments with a molecular size ranging from 40 to 700 kb. Five of the MRPs (MRP1, MRP2, MRP3, MRP4 and MRP5) contained two isolates each (10.5 %), MRP 6 contained four isolates (21 %) and remaining 6 profiles contained a single isolate. The dendrogram of the 19 isolates produced four clusters based on 40–60 % similarity (Fig. 2). Cluster I contained four isolates comprising two MRPs; Cluster II, three isolates (two MRPs); Cluster III, two isolates (two MRPs) and cluster IV consisted of ten isolates comprising six MRPs.
Fig. 2.
PFGE profiling of non-typhoid salmonella (NTS) isolates NTS isolates obtained from poultry and their products were subjected to PFGE profiling to study the degree of genetic relatedness. Isolates generated restriction fragments with molecular size ranging from 40 to 700 kb fragments, representing 11 distinct macro restriction profiles. Dendrogram showing similarities of Xba1 digested macro restriction profiles of NTS isolates generated by bio-rod finger printing with the band matching coefficient of Dice and the UPGMA clustering
Discussion
Infection with bacteria of the genus Salmonella is responsible for a variety of acute and chronic diseases in poultry. Infected poultry flocks are also among the most frequently implicated reservoirs of Salmonellae that can be transmitted through the food chain to humans. Poultry producers are facing with intensifying pressures from public health authorities, elected officials and consumers regarding food safety issues. The isolation of Salmonella is reported more often in poultry products than from any other animal source [20]. In earlier years, the marketing of eggs, meat and its products in India was mainly depending upon domestic market. In the current scenario, the population of both layer and broilers are increasing rapidly and hence the poultry industry needs to export these products to other countries. The importing countries also showing interest to import poultry products from countries like India, since it is economical. But those importing countries willing to import egg, meat and its product from India have certain norms and one of the important criteria is that the product should be free from Salmonella and the flock from which the meat or egg is obtained should also be free from Salmonella. For the creation of Salmonella free conditions, surveillance of Salmonella and rapid detection of Salmonella is absolutely mandatory. The existence of various serovars and molecular epidemiological pattern of Salmonella infection need to be studied.
This study tested poultry tissue samples, products and environment samples collected from 154 commercial layer farms in South India and found 1.73 % of the samples tested to be positive for NTS. Previous studies conducted in south India in 1996 and 2008 reported 2.7 and 2.6 % incidence respectively [13, 14]. The exact reason for this difference is not clear; it is possible that a difference in the sampling, choice of farms, and methodology employed may be responsible. It is also possible that the low incidence of NTS in the present study compared with earlier studies could be due to improved biosecurity measures and periodical surveillance. However, further detailed studies on the correlation of levels of biosecurity and incidence of NTS are needed.
The present study found higher levels of NTS isolation from tissue samples and poultry products compared with the environmental samples. Similar observations of higher incidence of Salmonella from poultry and poultry products compared to other sources were also made in the UK [20, 21]. Among the tissue samples tested in the present study, the recovery of NTS was higher in liver. Chicken liver continues to be a popular organ meat and dishes made with chicken liver are a delicacy in south India. Chicken livers have previously been implicated as a source for human Salmonella infections. For example, a multistate outbreak of human Salmonella Heidelberg infections in USA was linked to “kosher broiled chicken livers” [22].
The present study found a higher incidence of Salmonella in chicks than older birds. The incidence of Salmonella was found to be much higher in chicks below 3 weeks of age (7.27 %) than layer chicken below 40 weeks (3.6 %) and above 40 weeks of age (1.78 %). It is well known that younger chicken are relatively more susceptible to Salmonella than older birds [6]. The higher level of NTS recovery from chicks could be due to their higher susceptibility. The study found higher incidence of NTS in winter (2.80 %) than summer (0.66 %). A potential possibility for the lower NTS isolation in summer could be due to the dis-infecting ability of higher temperatures in summer. Average summer temperatures in south India typically are between 35 and 40 °C. In contrast, increased contamination and prolonged persistence of Salmonella in feed may have contributed to higher NTS recovery during winter season. Similar observations were reported in a previous study [23] in which higher incidence of Salmonella in 0–21 days age group of birds (5.47 %) than birds older than 3 weeks was found. The study also reported higher incidence of Salmonellosis in rainy and winter season.
Several factors such as ingredients used in preparing feed have been implicated to be the major source of contamination and depending upon the nature of feed, the incidence could vary [24]. The incidence of Salmonella in poultry feed and feed ingredients were found to be 9.6 % in this study. The incidence of Salmonella in poultry feed and feed ingredients are known to be highly variable ranging from 0 to 78 % [25, 26]. Various factors including source and quality of feed ingredients, storage conditions could contribute to this variation. The recovery of Salmonella in meat and bone meal alone was 33.33 % whereas the incidence in the compound feed was 7.5 %. Higher incidence of Salmonella in meat and bone meal could be due to improper sterilization of these ingredients and suggest that they may be the major source of Salmonella in compound feed.
The isolation of Salmonella from environmental samples such as drag swab, boot swab was 0.55 %. This number is relatively low compared to an earlier study which reported that the frequency of isolating Salmonella in the environmental samples ranged widely from 7.9 to 95.7 % [27]. Various factors including methods of sample collection, number of samples tested and level of bio-security in poultry farms could contribute to the differences in the levels of Salmonella detection in the environmental samples. Out of the 154 poultry farms screened in the present study, 11 farms were found to be positive for NTS incidence that had overcrowding, poor farm hygiene, lack of adequate biosecurity measures and infestation of rodents and insects. Mice, wild birds, ants and snakes are known to play important role in the transmission of Salmonella among birds, flocks and farms [28–30]. Hence, there is an association between the farm hygiene and incidence of NTS in poultry farms.
Industrialization of poultry production and the widespread use of nontherapeutic antimicrobial growth promoters have increased the risk of emergence of antibiotic resistance strains. As a consequence, a reduced effectiveness of several classes of antibiotics for treating infections in humans and livestock is becoming a major problem [31]. High resistance of NTS isolates to oxytetracycline observed in this study could due to its extensive and indiscriminate usage in poultry feed as a growth promoter. In contrast NTS isolates showed higher sensitivity to less commonly used antibiotics such as ciprofloxacin, enrofloxacin and norfloxacin.
The molecular techniques to genotype Salmonella isolates are often more discriminatory and rapid and have largely replaced phenotypic methods, such as serotyping and phage typing for epidemiological investigations in many laboratories [32–34]. Genotyping of Salmonella is a rapidly expanding field and many new methods have been developed in recent years. However, it is increasingly becoming evident that a single method might not work for all isolates and it is necessary to find out a method or combination of methods capable of differentiating clones of a particular serovar or phage type.
Differentiation of S. Typhimurium and S. Enteritidis could be made based on the presence in the former and absence in the latter of a restriction site for the enzyme Kpnl in the pefA gene [16]. To differentiate S. Typhimurium and S. Enteritidis, we have used PCR with a primer set targeting Kpnl enzyme target sequence in pefA gene.
Allele specific PCR has been widely used to differentiate the group D Salmonella from other groups and non Salmonella [17, 35, 36]. By allele specific PCR, four isolates were identified as S. Enteritidis and one isolate (S585) could not be identified possibly due to the loss or absence of the specific sites for primer binding in the chromosomal DNA.
ERIC PCR uses ERIC sequences as PCR primer binding sites to study the distribution of repetitive sequences [37] and is a useful tool for fingerprinting Salmonella genomes [37]. In this study eight ERIC profiles were identified and different isolates obtained from the same flock showed similar banding pattern. Similarly an earlier study [38] studied strain differentiation of Indian Salmonella isolates by ERIC PCR and identified 21 molecular types from 24 strains of the four serovars (S. Dublin, S. Abortusequi, S. Choleraesuis and S. Bareilly). EIC PCR profiling showed the genetic diversity among NTS isolates in south India. Salmonella with different genotypes could vary in their pathogenicity and it would be useful to map association between genotype and pathogenicity.
PFGE has been shown to be an accurate and reproducible method for genotyping of S. enterica subsp. enterica serovar Abortusovis (SAO) and S. Gallinarum and provides useful information in support of traditional epidemiological investigations [39]. We found identical PFGE profiles among S. Enteritidis strains isolated from one region which were considerably different from the profiles of isolates obtained from a different location. These findings indicate that PFGE profiling could be used in epidemiological investigation to establish genetic diversity of NTS isolates which may be useful as an epidemiological tool to track human salmonellosis outbreaks.
In conclusion this study isolated NTS from poultry and associated products and environmental substrates in southern India. Multiplex PCR with Salmonella serovar specific primers could be used for better characterization of NTS in poultry and related products. The poultry industry employs many people in India, and provides a source of low cost animal protein which plays an important role in the social and cultural lives of millions of people in India. People who work and live closely with the poultry are conceivably at a higher risk of acquiring the NTS infection. In addition to the organized poultry sector, small scale family poultry production is practiced by some rural households where each family keeps 5–20 hens which are mostly looked after by women and children. Therefore it is also essential to study the incidence of NTS in backyard poultry to evaluate the risk of infection of children and women. Hence, there is an urgent need to systematically evaluate risk factors of human infections with NTS in India in order to develop intervention strategies to control this important food borne zoonosis.
Acknowledgments
The authors are thankful to the Tamil Nadu Veterinary and Animal Sciences University for financial support and The University of Nottingham (S.V.K. and R.A.) for technical input into this research work.
Conflict of interest
None.
References
- 1.Bakeri SA, Yasin RM, Koh YT, et al. Genetic diversity of human isolates of Salmonella enterica serovar Enteritidis in Malaysia. J Appl Microbiol. 2003;95:773–780. doi: 10.1046/j.1365-2672.2003.02033.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhowmick PP, Srikumar S, Devegowda D, et al. Serotyping and molecular characterization for study of genetic diversity among seafood associated nontyphoidal Salmonella serovars. Indian J Med Res. 2012;135:371–381. [PMC free article] [PubMed] [Google Scholar]
- 3.Varga C, Pearl DL, McEwen SA et al (2013) Incidence, distribution, seasonality, and demographic risk factors of Salmonella Enteritidis human infections in Ontario, Canada, 2007–2009. BMC Infect Dis 13:212 [DOI] [PMC free article] [PubMed]
- 4.Gast RK, Guraya R, Guard J. Salmonella enteritidis deposition in eggs after experimental infection of laying hens with different oral doses. J Food Prot. 2013;76:108–113. doi: 10.4315/0362-028X.JFP-12-268. [DOI] [PubMed] [Google Scholar]
- 5.Grimont PA, Weill F. Antigenic formulae of the Salmonella serovars. 9. Paris: World Health Organization Collaborating Centre for Reference and Research on Salmonella, Institute Pasteur; 2007. [Google Scholar]
- 6.Gast R (2003) Paratyphoid infections. In: Saif M, Barnes HJ, Fadly AM, Glisson JR, McDougald LR, Swayne DE (eds) Poult, 11th edn. Iowa State University Press, Ames, pp 583–613
- 7.Breuil J, Brisabois A, Casin I, et al. Antibiotic resistance in salmonellae isolated from humans and animals in France: comparative data from 1994 and 1997. J Antimicrob Chemother. 2000;46:965–971. doi: 10.1093/jac/46.6.965. [DOI] [PubMed] [Google Scholar]
- 8.CDC Multidrug resistant Salmonella serovar Typhimurium—United States, 1996. Morb Mort Wkly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 9.Davis MA, Hancock DD, Besser TE, et al. Changes in antimicrobial resistance among Salmonella enterica Serovar typhimurium isolates from humans and cattle in the Northwestern United States, 1982–1997. Emerg Infect Dis. 1999;5:802–806. doi: 10.3201/eid0506.990610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatha AAM, Lakshmanaperumalsamy P. Prevalence of Salmonella in fish and crustaceans in Coimbatore, South India. Food Microbiol. 1997;14:111–116. doi: 10.1006/fmic.1996.0070. [DOI] [Google Scholar]
- 11.Suresh T, Srinivasan D, Hatha AAM, Lakshmanaperumalsamy P. A study on the incidence, antimicrobial resistance and survival of Salmonella and E. coli isolated from broiler chicken retail outlets. Microbes Environ. 2000;15:173–181. doi: 10.1264/jsme2.2000.173. [DOI] [Google Scholar]
- 12.Gopala Krishna Murthy T, Srinivasan P, Saravanan S, Mohan B. Salmonella contamination in poultry. Indian Vet J. 2011;88:147–148. [Google Scholar]
- 13.Murugadas V (2008) Molecular characterization of Salmonella enterica subsp. enterica from poultry and its products. M.V.Sc. thesis, Tamil Nadu Veterinary and Animal Sciences University
- 14.Purushothaman V, David P, Venkatesan R. Comparison of plasmid profile analysis, serotyping, resistotyping, biotyping and antimicrobial susceptibility testing as epidemiological tools in the strain identification of Salmonella isolates from avian source. Indian J Anim Sci. 1996;66:419–430. [Google Scholar]
- 15.Bauer R, Kirby MDK, Sherris JC, Turck M. Antibiotic susceptibility testing by standard single disc diffusion method. Am J Clin Pathol. 1996;14:493–496. [PubMed] [Google Scholar]
- 16.Cortez ALL, Carvalho ACFB, Ikuno AA, et al. Identification of Salmonella spp. isolates from chicken abattoirs by multiplex-PCR. Res Vet Sci. 2006;81:340–344. doi: 10.1016/j.rvsc.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Shah DH, Park J-H, Cho M-R, et al. Allele-specific PCR method based on rfbS sequence for distinguishing Salmonella gallinarum from Salmonella pullorum: serotype-specific rfbS sequence polymorphism. J Microbiol Methods. 2005;60:169–177. doi: 10.1016/j.mimet.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Cao S-Y, Wang M-S, Cheng A-C, et al. Comparative analysis of intestinal microbial community diversity between healthy and orally infected ducklings with Salmonella enteritidis by ERIC-PCR. World J Gastroenterol (WJG) 2008;14:1120–1125. doi: 10.3748/wjg.14.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo Y-S, Lee S-H, Shin E-K, et al. Pulsed-field gel electrophoresis genotyping of Salmonella gallinarum and comparison with random amplified polymorphic DNA. Vet Microbiol. 2006;115:349–357. doi: 10.1016/j.vetmic.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Myint M (2004) Epidemiology of Salmonella contamination of poultry products: knowledge gaps in the farm to store products. Discussion submitted to Faculty of the Graduate School of University of Maryland, College Park
- 21.Davies R, Breslin M. Observations on Salmonella contamination of commercial laying farms before and after cleaning and disinfection. Vet Rec. 2003;152:283–287. doi: 10.1136/vr.152.10.283. [DOI] [PubMed] [Google Scholar]
- 22.CDC (2011) Multistate outbreak of human salmonella heidelberg infections linked to “kosher broiled chicken livers” from Schreiber processing corporation. http://www.cdc.gov/salmonella/heidelberg-chickenlivers/111011/index.html?s_cid=ccu112111_015
- 23.Islam M, Das B, Hossain K, et al. A study of the occurrence of poultry diseases in sylhet region of Bangladesh. Int J PoultSci. 2003;2:354–356. [Google Scholar]
- 24.Okoli I, Endujihe G, Ogbuewu I. Frequency of isolation of Salmonella from commercial poultry feeds and their anti microbial resistance profiles. Online J Health Allied Sci. 2006;5:2–3. [Google Scholar]
- 25.Veldman A, Vahl HA, Borggreve GJ, Fuller DC. A survey of the incidence of Salmonella species and Enterobacteriaceae in poultry feeds and feed components. Vet Rec. 1995;136:169–172. doi: 10.1136/vr.136.7.169. [DOI] [PubMed] [Google Scholar]
- 26.Ward J, Griffin M, Egan J (1996) Evaluation of some rapid methods for the detection of Salmonella in poultry carcasses, feed and environmental samples. In: Proceedings of the monit. proced. rapid. detect. methods tech., Newbury, UK, pp 123–127
- 27.Wales A, Breslin M, Davies R. Semiquantitative assessment of the distribution of Salmonella in the environment of caged layer flocks. J Appl Microbiol. 2006;101:309–318. doi: 10.1111/j.1365-2672.2006.02916.x. [DOI] [PubMed] [Google Scholar]
- 28.Angen O, Skov M, Chriel M, et al. A retrospective study on Salmonella infection in Danish broiler flocks. Prev Vet Med. 1996;26:223–237. doi: 10.1016/0167-5877(95)00549-8. [DOI] [Google Scholar]
- 29.Davies RH, Nicholas RA, McLaren IM, et al. Bacteriological and serological investigation of persistent Salmonella enteritidis infection in an integrated poultry organisation. Vet Microbiol. 1997;58:277–293. doi: 10.1016/S0378-1135(97)00157-0. [DOI] [PubMed] [Google Scholar]
- 30.Carrique-Mas JJ, Breslin M, Snow L, et al. Persistence and clearance of different Salmonella serovars in buildings housing laying hens. Epidemiol Infect. 2009;137:837–846. doi: 10.1017/S0950268808001568. [DOI] [PubMed] [Google Scholar]
- 31.Gilchrist MJ, Greko C, Wallinga DB, et al. The potential role of concentrated animal feeding operations in infectious disease epidemics and antibiotic resistance. Environ Health Perspect. 2007;115:313–316. doi: 10.1289/ehp.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Threlfall EJ, Frost JA. The identification, typing and fingerprinting of Salmonella: laboratory aspects and epidemiological applications. J Appl Bacteriol. 1990;68:5–16. doi: 10.1111/j.1365-2672.1990.tb02542.x. [DOI] [PubMed] [Google Scholar]
- 33.Boxrud D, Pederson-Gulrud K, Wotton J, et al. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J Clin Microbiol. 2007;45:536–543. doi: 10.1128/JCM.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torpdahl M, Sørensen G, Lindstedt B-A, Nielsen EM. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg Infect Dis. 2007;13:388–395. doi: 10.3201/eid1303.060460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verma N, Reeves P. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989;171:5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai AR, Shah DH, Shringi S, et al. An allele-specific PCR assay for the rapid and serotype-specific detection of Salmonella pullorum. Avian Dis. 2005;49:558–561. doi: 10.1637/7385-052205R.1. [DOI] [PubMed] [Google Scholar]
- 37.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxena MK, Singh VP, Lakhcharua BD, et al. Strain differentiation of Indian isolates of Salmonella by ERIC-PCR. Res Vet Sci. 2002;73:313–314. doi: 10.1016/S0034-5288(02)00030-9. [DOI] [PubMed] [Google Scholar]
- 39.Dionisi A, Carattoli A, Luzzi I, et al. Molecular genotyping of Salmonella abortionist by pulse field gel electrophoresis. Vet Microbiol. 2006;116:217–223. doi: 10.1016/j.vetmic.2006.03.008. [DOI] [PubMed] [Google Scholar]