Abstract
The highly conserved enzyme γ-glutamyltranspeptidase (GGT) plays an important role in metabolism of glutathione and glutamine. Yet, the regulation of ggt transcription in prokaryotes is poorly understood. In the human pathogen Campylobacter jejuni, GGT is important as it contributes to persistent colonization of the gut. Here we show that the GGT activity in C. jejuni is dependent on a functional RacRS (reduced ability to colonize) two-component system. Electrophoretic mobility shift and luciferase reporter assays indicate that the response regulator RacR binds to a promoter region ~80 bp upstream of the ggt transcriptional start site, which contains a recently identified RacR DNA binding consensus sequence. RacR needs to be phosphorylated to activate the transcription of the ggt gene, which is the case under low oxygen conditions in presence of alternative electron acceptors. A functional GGT and RacR are needed to allow C. jejuni to grow optimally on glutamine as sole carbon source under RacR inducing conditions. However, when additional carbon sources are present C. jejuni is capable of utilizing glutamine independently of GGT. RacR is the first prokaryotic transcription factor known to directly up-regulate both the cytoplasmic [glutamine-2-oxoglutarate aminotransferase (GOGAT)] as well as the periplasmic (GGT) production of glutamate.
Keywords: Campylobacter jejuni, gene regulation, two-component system, RacRS, γ-glutamyltranspeptidase, GGT, glutamine-2-oxoglutarate aminotransferase, GOGAT
Introduction
The enzyme γ-glutamyltranspeptidase (GGT, EC 2.3.2.2) is highly conserved among eukaryotic and prokaryotic organisms (Ong et al., 2008), where it has a key function in glutathione metabolism. In prokaryotes GGT is produced as a proenzyme in the cytoplasm and is then translocated into the periplasm where it undergoes autocatalytic cleavage. This proteolysis yields a mature dimer which transfers γ-glutamyl moieties from extracellular glutathione and related compounds to amino acids or peptides or catalyzes the hydrolysis of the glutamyl group to generate glutamate (Hanigan, 1998). GGT activity in Escherichia coli and Bacillus subtilis is maximal in stationary growth phase (Suzuki et al., 1986; Xu and Strauch, 1996). In B. subtilis GGT is indirectly transcriptionally regulated in response to low L-glutamate concentrations via the quorum sensing two-component system ComP/ComA (Kimura et al., 2004). In Helicobacter pylori expression of GGT is reported not to be growth phase dependent, but is up-regulated at low pH (Wachino et al., 2010) and to be involved in acid resistance and immune stimulation (Gong et al., 2010; Miller and Maier, 2014).
The bacterium C. jejuni is a major food borne pathogen in humans and colonizes the intestinal tract of many warm-blooded animals (Blaser, 1997). C. jejuni lacks the glycolytic enzyme phosphofructokinase and is therefore not able to use exogenous sugars as a carbon source, although some strains were shown to be able to metabolize fucose (Muraoka and Zhang, 2011; Stahl et al., 2011). Hence, amino acids (i.e., aspartate, glutamate, proline, and preferentially serine) are likely to sustain the growth of Campylobacter in the intestine (Guccione et al., 2008). Although the genome of C. jejuni encodes for a functional glutamine transporter (Lin et al., 2009), only isolates containing GGT are also able to utilize glutamine, and glutathione as sole carbon/energy source (Hofreuter et al., 2006). In the periplasm this enzyme converts glutamine and glutathione to glutamate, which is subsequently taken up via the aspartate/glutamate-binding protein PEB1 (Del Rocio Leon-Kempis et al., 2006). After glutathione cleavage by GGT the remaining dipeptide cys-gly is also imported by C. jejuni and used as sulfur source (Vorwerk et al., 2014). The expression of ggt is reported to be maximal in late log phase (Hyytiäinen et al., 2012). The presence of GGT allows C. jejuni strains to enhance their colonization persistence in the avian gut and to colonize the intestine of mice (Barnes et al., 2007; Hofreuter et al., 2008).
The regulator RacR (reduced ability to colonize) is like GGT needed to sustain the colonization of chickens (Brás et al., 1999) and is detected in nearly all C. jejuni isolates (Kordinas et al., 2005; Talukder et al., 2008; Quetz et al., 2012). Recently we showed that the RacRS two-component system of C. jejuni is active under low oxygen conditions in the presence of alternative electron acceptors (e.g., nitrate or TMAO; van der Stel et al., 2015). Under these conditions, RacR represses the transcription of several genes including the aspA gene and at the same time it activates the gltBD genes. The products of the gltBD genes form the glutamine-2-oxoglutarate aminotransferase (GOGAT) complex, which is responsible for cytosolic glutamate generation (Guccione et al., 2008). Glutamate is an important nitrogen source for bacteria as it functions as precursor for amino acid and nucleotide anabolism (Reitzer, 2003; Heeswijk et al., 2013; Hofreuter, 2014). Here we investigated whether the generation of periplasmic glutamate accomplished by GGT is also regulated by RacR.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. C. jejuni strains were cultured at 37°C or 42°C on Blood Agar Base No. 2 (BA) medium containing 5% horse blood or in Heart Infusion broth (HI; Oxoid), under microaerobic conditions (5% O2, 7.5% CO2, 7.5% H2, 80% N2), or under oxygen limited conditions (0.3% O2, 10% CO2, 10% H2, 80% N2). Kanamycin (25 μg ml-1) and/or chloramphenicol (15 μg ml-1) were added when appropriate. E. coli strains were routinely grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates supplemented with ampicillin (100 μg ml-1), kanamycin (30 μg ml-1), or chloramphenicol (30 μg ml-1).
Table 1.
Bacterial strains and plasmids used in this study.
| Bacterial strains and plasmids | Relevant characteristics | Origin or reference |
| Strains | ||
| Escherichia coli TG1 | supE hsdΔ5 thiΔ(lac-proAB) F’ (traD36 proAB+ lacIq lacZΔM15); used for general cloning | Green and Sambrook (2012) |
| E. coli BL21(DE3) | F- ompT hsdSB (rB- mB-) gal dcm (DE3); used for protein overexpression | Novagen |
| E. coli S17 | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 (Tmpr Strr); used for conjugation | Parke (1990) |
| Campylobacter jejuni 81116 | Wildtype | Palmer et al. (1983) |
| C. jejuni ggt | 81116 derivative ggt::Cm | This study |
| C. jejuni racR | 81116 derivative racR::Cm | van der Stel et al. (2015) |
| C. jejuni racS | 81116 derivative racS::Cm | This study |
| C. jejuni racRS | 81116 derivative racRS::Cm | This study |
| C. jejuni racR+p | 81116 derivative racR::Cm + pMA1-1261-1263 | This study |
| C. jejuni racRS+p | 81116 derivative racRS::Cm + pMA1-1261-1263 | This study |
| C. jejuni 11168 | Wildtype | Palmer et al. (1983) |
| C. jejuni 81-176 ggt | 81-176 derivative ggt::Cm | This study |
| Plasmids | ||
| pBluescript II KS | ApR; 3.0 kb; LacZα | Stratagene |
| pGEM-T Easy | ApR; 3.0 kb; LacZα; TA cloning vector | Promega |
| pT7.7 | ApR; 2.5 kb; expression vector | Tabor and Richardson (1985) |
| pMA1 | KmR; 10 kb; E. coli/C. jejuni shuttle vector | van Mourik et al. (2008) |
| Plasmids constructed for mutagenesis | ||
| pUWM799 | ApR; 4.2 kb; pBluescript II KS/internal fragment of C. jejuni ggt | This study |
| pUWM804 | ApR CmR; 4.9 kb; pBluescript II KS/ggt::Cm | This study |
| pGEM-1261-1263 | ApR; 5.5 kb; pGEM-T Easy/C. jejuni 1261-1263 | This study |
| pGEM1261::Cm | ApR CmR; 6.3 kb; pGEM-T Easy/racR::Cm | This study |
| pGEM1262::Cm | ApR CmR; 6.3 kb; pGEM-T Easy/racS::Cm | This study |
| pGEM1261-62::Cm | ApR CmR; 5.3 kb; pGEM-T Easy/racRS::Cm | This study |
| Plasmids constructed for complementation | ||
| pMA1-1261-1263 | KmR;12.5 kb; pMA1/Cj1261-1263 | van der Stel et al. (2015) |
Construction of C. jejuni ggt, racS, and racRS Mutants
To inactivate the ggt gene, the ggt gene was amplified by PCR using the oligonucleotides Cjj67Sac and Cjj67Xba (Table 2). The obtained PCR product was digested with ClaI and PstI resulting in a 1.2-kb DNA fragment, which was ligated into pBluescript II KS to give plasmid pUWM799. Plasmid pUWM799 was digested with BglII to remove a 0.1-kb internal ggt fragment and was ligated to a 0.8-kb BamHI fragment containing the cat cassette (0.8 kb) of pRY109. The resulting ggt knockout construct pUWM804 contained the cat cassette in the same orientation as the ggt gene.
Table 2.
Oligonucleotides used in this study.
| Name | Sequence (5′-3′)a |
| Cjj67Sac | CGCGAGCTCGCTTTTTGCGGTGGTAGG |
| Cjj67Xba | AGTTCTAGAGGAGATCCTGTGCCTGTG |
| GGT204 | AAAGCATGCATTGCACTTTCAATAAATTTTAAATATTTTAGC |
| GGT104 | AAAGCATGCATAAAATGAGAATATTTGATAC |
| GGT69 | AAAGCATGCTTTAAATATTTTTATAAAAATATATC |
| GGT35 | AAAGCATGCAATTTATCAATACCCCTAGTTTTG |
| GGT28 | AAAGAGCTCCATTTTACTCCTTTTTAATGATATATAG |
| Primers for gel mobility shift assay | |
| GGTprR | GCTTCAAATTTCATATTGCACTT |
| GGTprFDIG | TTGAAATCGCAAATATAGCT |
| Cj200RDIG | GTTTTAGACTATCTGCAAAA |
| Cj201F | TTTCATCTTCAATATACTCTAA |
| CJ0145RDIG | TTAAAAACAATCTTCTTTCCAT |
| CJ0145F | TTTCTAGTACAGTAAGTGATATAGC |
| Primers used for real-time RT-PCR | |
| ggtftaq | TGCGAGTTATGGTTCAGGTG |
| ggtrtaq | TTAGCTTCTCCGCCTACAAG |
| gltBftaq2 | ACACGATGCCTGTGGTATCG |
| gltBrtaq2 | TCGGTGTTCAAGATTCATCAAAAT |
| aspAftaq | TATGGGATAAGCATAGTGAAGTTCAAG |
| aspArtaq | CGCTTTAATAATCGCATCTTGGA |
| rpoDftaq | GAACGAATTTGATTTAGCCAATGA |
| rpoDrtaq | TGTCCCATTTCTCTTAAATACATACGA |
aRestriction sites introduced for cloning purposes are underlined.
To construct the racS and racRS knockout constructs plasmid pGEM1261 (van der Stel et al., 2015), containing the genes Cj1261–Cj1263 was amplified by PCR using the primers RacSendBamHI/RacSstart2BamHI or Cj1261FBamHI/RacSendBamHI, respectively. The resulting PCR products were digested with BamHI and ligated to a 0.8-kb BamHI cat cassette from pAV35 (Vliet et al., 1998), resulting in plasmids pGEM1262::Cm and pGEM1261-1262::Cm, respectively.
The genes ggt, racS, and the racRS were disrupted in C. jejuni strain 81116 or 81-176 by natural transformation using the knock-out plasmids pUWM804, pJET1262::Cm and pGEM1261-1262::Cm, respectively. Double cross-over recombination events were confirmed by PCR.
Construction of racRS Complementation Strain
To complement the racRS mutant, the complementation plasmid pMA1–1261–1263 (van der Stel et al., 2015) was first transformed into E. coli S17 and then conjugated (Labigne-Roussel et al., 1987) to the racRS::Cm mutant strain.
Purification of Recombinant RacR and Cytoplasmic RacS
RacR (N-His) and RacScyto (N-His) were expressed and purified as described before (Wösten et al., 2004) using plasmids pT7.7-RacR(N-his) and pT7.7-RacScyto(N-his). Protein concentrations were determined using the BCA protein assay kit (Pierce).
Construction of ggt Promoter Luciferase Constructs
To localize where RacR binds on the ggt promoter, different lengths of the upstream region of the ggt gene were amplified by PCR. The PCRs were generated by fusion polymerase (Thermo) with Cj81116 genomic DNA as template and one of the primer pairs GGT28/GGT204, GGT28/GGT104, GGT28/GGT69, or GGT28/GGT35. The PCR products were digested with SacI and SphI (Thermo) and cloned into SacI, SphI digested plasmid pMA5-metK-luc (Bouwman et al., 2013) to replace the metK promoter. To obtain a promotorless luciferase vector, pMA5-metK-luc was digested with SacI and SphI, blunted with the blunting enzyme from the CloneJET PCR Cloning Kit (Thermo) and finally self-ligated. The plasmids were verified by sequencing (Macrogen). The obtained plasmids were transformed to E. coli S17 and subsequently conjugated to C. jejuni 81116 and the racRS mutant.
Real-Time RT-PCR
Total RNA was extracted from late logarithmic phase C. jejuni cultures grown in HI medium at 42°C under oxygen limiting conditions with the addition of 50 mM nitrate, using RNA-BeeTM kit (Tel-Test, Inc) according to the manufacturer’s specifications. Real-time RT-PCR analysis was performed as previously described (van der Stel et al., 2015). Primers used in this assay are listed in Table 2. Experiments were repeated with three independently grown cultures. Fold increase was calculated with the 2ΔΔCt method (Schmittgen and Livak, 2008) using rpoD as reference gene.
GGT Activity Assay
To assay the GGT activity the production of 3-carboxy-4-nitroaniline was followed by measuring the absorbance at 405 nm according to a modified procedure described by (Chevalier et al., 1999). Briefly, 1 mL of bacterial culture was pelleted and stored at −80°C for at least 1 h. The pellet was resuspended in 250 μL buffer A (50 mM Tris/HCl (pH 7.6), 1 μg/mL lysozyme), and incubated for 30 min on ice. Next, the bacteria were disrupted by sonication followed by centrifugation (10 min at 12000 × g at 4°C). From the cell free bacterial lysate 20 μl was mixed together with 180 μl of a reagent containing 2.9 mM L-γ-glutamyl-3-carboxy-4-nitroanilide, 100 mM glycylglycine and 100 mM Tris-HCl (pH 8.2). Samples were measured every 60 s during an incubation period of 30 min at 37°C. From these graphs the slope of all values in a linear range was calculated. Protein concentration was determined using the BCA method (Pierce). GGT activity is expressed as nmol min-1 mg protein-1. The data shown represents at least three independent experiments.
Luciferase Assay
Expression of the luciferase in C. jejuni 81116 and racRS mutant strain harboring the pMA5-ggtprom-luc plasmids was measured as previously described (Bouwman et al., 2013). Briefly, overnight cultures were diluted to an OD550 nm of 0.05 and grown for 7.5 h in HI with 50 mM KNO3 in an oxygen limiting atmosphere at 37°C. One milliliter of each culture was pelleted (8000 ×g, 5 min, 4°C) and suspended in 100 μL RLB buffer (Promega) supplemented with 0.5% Triton-X100. Suspensions were stored at −80°C for at least 30 min to disrupt the bacteria. Bacterial lysate (20 μL) was mixed with 50 μL of luciferase reagent (Promega) and RLU’s were measured immediately on a luminometer (TD20/20, Turner Designs). The data shown represents at least three independent experiments.
Gel Mobility Shift Assay
The promoter regions upstream of ggt, phoX (Cj0145), and Cj0200c were amplified by PCR using Dreamtaq polymerase (Thermo) and one of the primers sets GGTpromDIG/GGT204, GGTpromDIG/GGT104, GGTpromDIG/GGT69, GGTpromDIG/GGT35, Cj145F/CJ0145RDIG, and Cj200F/Cj200RDIG, respectively, (Table 2) and Cj81116 chromosomal DNA as template. Primers GGTpromDIG, Cj0145RDIG, and Cj200RDIG were ordered with a digoxigenin (DIG) label (Eurofins genomics). Approximately, 50 fmol DIG-labeled PCR fragments was incubated with His-tagged RacR, RacScyto, and 2 mM ATP for 30 min on ice. RacR, RacScyto, and ATP were preincubated for 5 min at 37°C to allow phosphorylation. The binding buffer used for protein-DNA incubations was 20 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 100 mM KCl, 100 μg/ml bovine serum albumin, 1 μg/ml poly-(dI-dC) and 10% glycerol. Samples (10 μl) were run on a 4% non-denaturing Tris glycine polyacrylamide gel at 4°C. After electrophoresis the DNA was blotted on a hybond-N+ membrane (Amersham) and PCR fragments were visualized using α-DIG-AP, Fab fragments, and CSPD substrate (Roche).
Growth Curves
Campylobacter jejuni precultures were grown for 7 h in HI medium with the addition of 50 mM nitrate at oxygen limiting conditions at 42°C. The precultures were diluted 50 times in 300 μL DMEM (without glucose, glutamine, pyruvate, bicarbonate, and phenol red, D5030, Sigma) with the addition of 10 mM TMAO and 10 mM glutamate, glutamine or aspartate, and growth curves were generated at 42°C in a 100 well honeycomb plate which was continuously shaking in a Bioscreen C MRB (Oy Growth Curves Ab) computer-controlled incubator. The incubator was placed inside an anaerobic chamber (Coy Labs, Grass Lake, MI, USA), due to suboptimal gas exchange in the honeycomb plate the oxygen concentration was set to 1%, which yielded comparable growth as when bacteria were grown in rectangular flasks inside an anaerobic jar containing 0.3% O2. The OD600nm of cultures was recorded every 15 min during 45 h. For clarity reasons only point at 2.5 h, or 5 h intervals are shown. Experiments were repeated three times in duplicate.
Statistical Analysis
Prism software (GraphPad, San Diego, CA, USA) was used for statistical analysis. Results are shown as mean ± SEM. Data was analyzed by one-way ANOVA, followed with Bonferroni post hoc tests; P < 0.05 was considered statistically significant.
Results
RacR Regulates ggt Transcription and Activity
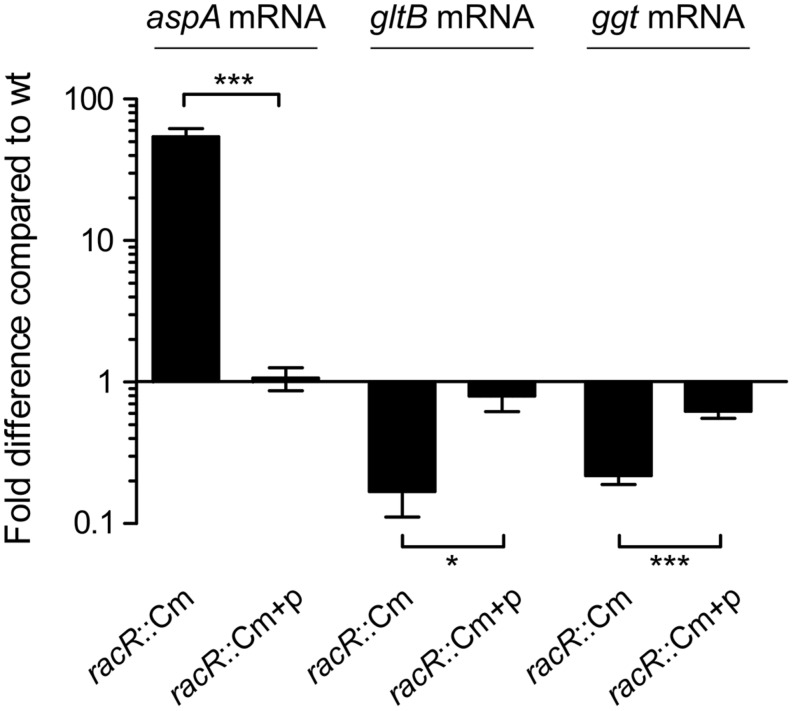
As the C. jejuni RacRS two-component system regulates the cytoplasmic glutamate production by activating the gltBD genes (van der Stel et al., 2015), we wondered whether GGT, responsible for the periplasmic glutamate production, is also regulated by the RacRS system. To investigate this we measured the ggt transcripts in the C. jejuni 81116 wildtype strains, the isogenic racR mutant, and in the complemented racR mutant, grown until late log phase under RacRS inducing conditions, i.e., 0.3% O2 with 50 mM nitrate. To verify that we used RNA that was isolated under RacR inducing conditions, we also measured the transcripts of aspA and gltB genes. Using real time RT-PCR we observed that inactivation of racR resulted in a 55-fold increase of aspA mRNA, and a sixfold decrease of the gltB consistent with our previous results (van der Stel et al., 2015) and confirming that RacR is induced under these conditions. A significant fivefold decrease was observed for the ggt mRNA transcripts. The differences between the wt and the racR mutant were almost restored to wt levels by introducing complementation plasmid harboring the RacRS operon (Figure 1). These results suggest that RacR has a strong influence on the production of glutamate as it not only activates the transcription of the genes required for the cytoplasmic glutamate production (gltB), but also the periplasmic production of glutamate (ggt).
FIGURE 1.
RacR activates ggt transcription. Real-time RT-PCR data showing the transcript fold difference of the aspA, gltB, and ggt mRNA in the Campylobacter jejuni racR mutant (racR::Cm) and complemented strain (racR::Cm+p) compared to the wt. Cultures were grown in Heart Infusion broth (HI) medium with addition of nitrate until end-log phase under oxygen limiting conditions. Fold increase is calculated using the 2ΔΔCt method using rpoD as reference gene. Data represent the mean values and SE of three independent experiments. *p < 0.05, ***p < 0.001.
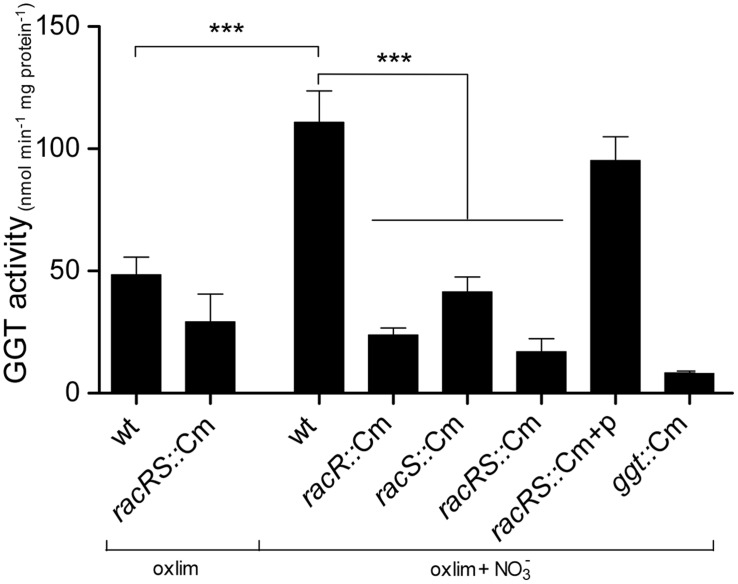
To verify that the two-component system RacR/RacS also influences the GGT enzyme activity, we measured the GGT activity in C. jejuni stationary phase cultures grown at 0.3% O2 with or without the addition of nitrate (Figure 2). Only background levels of GGT activity were observed in the ggt mutant indicating that this enzyme is solely responsible for the production of 3-carboxy-4-nitroaniline in the GGT assay. Maximum GGT activity of bacteria grown in HI liquid medium was observed in stationary phase (data not shown). A low GGT activity was measured in wt and racR mutant when the strains were grown at 0.3% O2; however, the GGT activity increased threefold in the wt bacteria when nitrate was present. This induction was not observed in the racRS double mutant or the single racR, or racS mutant strains. When the racRS, racR, or racS mutants were complemented with a plasmid harboring the RacRS operon the GGT activity was restored to wildtype levels. These results indicate that GGT activity largely depends on a functional and activated RacR and RacS, as exists under limited oxygen condition in the presence of an alternative electron acceptor.
FIGURE 2.
RacR influences the γ-glutamyltranspeptidase (GGT) activity. GGT activity was measured in cell lysate from stationary phase (20 h) cultures of wildtype 81116, the racR, racS, and racRS mutant strains, as well as the complemented racRS mutant, grown in HI medium at oxygen limiting conditions with or without the addition of nitrate. Mean and SE of at least three independent experiments are shown. ***p < 0.001.
RacR Protein Binds to the Promoter Region of the ggt Gene
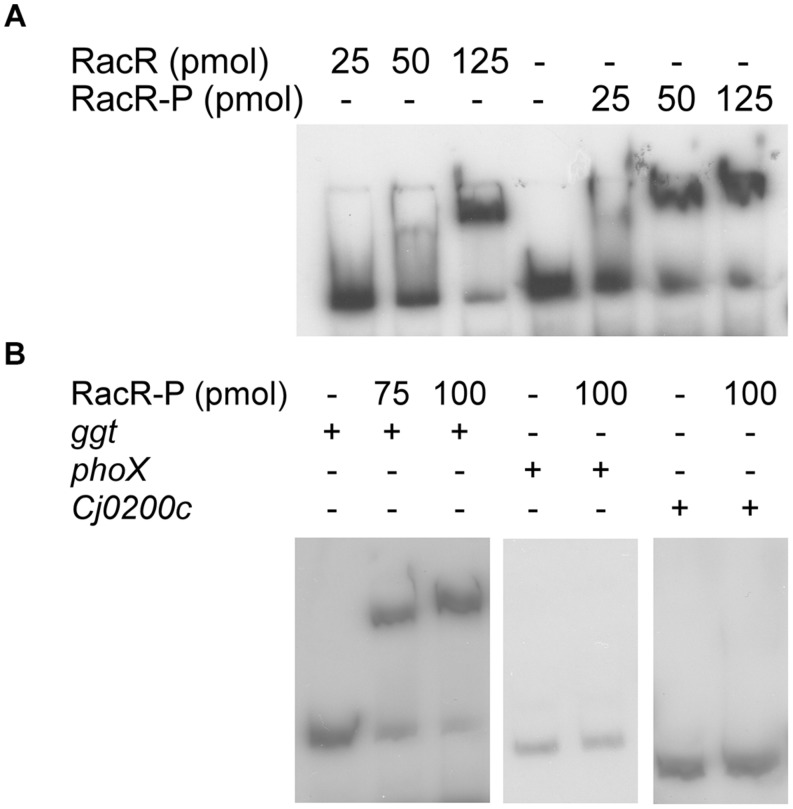
To investigate whether RacR activates the ggt gene directly by binding to the ggt promoter region, electrophoretic mobility shift assays (EMSA) were performed. Hereto the RacR response regulator and the cytoplasmic region of RacS were isolated as His-tagged recombinant proteins and together with ATP incubated with DIG-labeled DNA fragments containing the promoter region of the ggt gene or, as a negative control, the promoter regions of the phoX and Cj0200c genes. Incubation of RacR and the cytosolic region of RacS in the presence of ATP led to rapid phosphorylation of RacR (van der Stel et al., 2015). Unphosphorylated RacR bound to the ggt promoter but less RacR was needed when it was phosphorylated by the cytoplasmic part of RacS (Figure 3A). Phosphorylated RacR did not bind to the phoX and Cj0200c promoter fragments as no band shifts were observed, while a clear band shift was seen for the ggt promoter fragment (Figure 3B). These results indicate that RacR specifically binds to the ggt promoter and that phosphorylation of RacR enhances the binding affinity.
FIGURE 3.
RacR binds to the ggt promoter region as shown by electrophoretic mobility shift assays (EMSA). DIG-labeled PCR fragments (~50 fmol) containing the ggt, phoX, or Cj0200c promoter regions were incubated with RacR as indicated. (A) Influence of the phosphorylation of RacR on the binding to the ggt promoter. RacR was phosphorylated by RacScyto in the presence of ATP. (B) EMSA of the ggt, phoX, and Cj0200c promoter regions with phosphorylated RacR protein. The phoX and Cj0200c promoter regions were used as negative controls. RacR-P, phosphorylated RacR.
RacR Binds to a RacR Binding Consensus Sequence in Front of the ggt Promoter
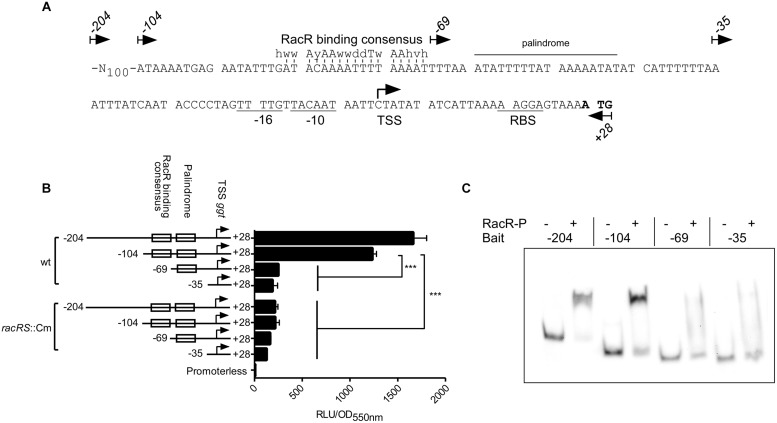
To investigate where RacR binds on the ggt promoter, an in silico analysis of the promoter region was performed (Figure 4A). Based on this analysis, primers were designed to amplify DNA fragments in order to study the different elements on the ggt promoter region. Besides the full intergenic region between the ggt and c8j_0034 gene (-204 nt fragment) three truncated ggt promoter elements were generated; (1) a −104 nt fragment containing a putative RacR binding consensus sequence, a palindromic sequence and the putative −35, −16 and −10 region; (2) a −69 nt fragment lacking the putative RacR binding consensus sequence, and (3) a -35 nt fragment only containing the putative −35, −16 and −10 regions. These promoter elements were cloned in front of the luciferase reporter gene, replacing the metK promoter located on plasmid pMA5-metK-luc (Bouwman et al., 2013). Luciferase activity was measured in the wt and racRS mutant strain under RacR inducing conditions. Because of poor stability of the luciferase enzyme at 42°C (data not shown), all luciferase reporter assay experiments were performed at 37°C. In wt bacteria high luciferase activity was measured only from the promoter elements −204 and −104, both containing the predicted RacR nucleotide binding site (Figure 4B). All promoter fragments resulted in a low luciferase activity in the racRS mutant, which was, however, still higher than the luciferase activity of the strain carrying a promoterless luciferase plasmid. These results indicate that the region upstream of the ggt gene containing the predicted RacR consensus sequence is important for enhancing of the ggt transcription in a RacRS dependent manner.
FIGURE 4.
RacR binds to a specific region on the ggt promoter. (A) Nucleotide sequence and features of the ggt promoter. The start codon ATG is indicated in bold face, the putative -10 and -16 regions and ribosomal binding site (RBS) are underlined. A palindromic sequence is indicated with a horizontal bar. The previously identified RacR binding consensus sequence (van der Stel et al., 2015) is indicated above the predicted RacR binding site, vertical lines indicate matching nucleotides. Arrows indicate the 5′ termini and direction of the primers used to generate the ggt promoter elements for the luciferase reporter plasmids and EMSA bait DNA. The transcriptional start site of the ggt gene identified by (Dugar et al., 2013) is indicated with a hooked arrow. (B) Luciferase activities using different lengths of the region upstream of the ggt gene are determined in wt and racRS::Cm mutant bacteria. Cultures were grown until late-log phase at oxygen limiting conditions with the addition of nitrate. Data represents the mean and SE of three independent experiments. (C) EMSA experiments using the different ggt promoter elements. DIG-labeled PCR fragments (~50 fmol) were mixed with or without 50 pmol RacR and 25 pmol RacScyto in the presence of ATP. RacR-P phosphorylated RacR.
In order to verify the luciferase reporter results, the different ggt promoter elements were subjected to EMSA experiments (Figure 4C). The -204 and -104 ggt promoter elements, harboring the predicted RacR binding site showed a distinct bandshift when phosphorylated RacR was present. In accordance with the luciferase assay results RacR did not bind to the two shorter fragments. These results prove that phosphorylated RacR binds to the upstream region of the ggt promoter region containing the predicted RacR binding consensus nucleotide site.
RacR is Important for C. jejuni to Generate More Biomass out of Glutamine, under RacRS Inducing Conditions
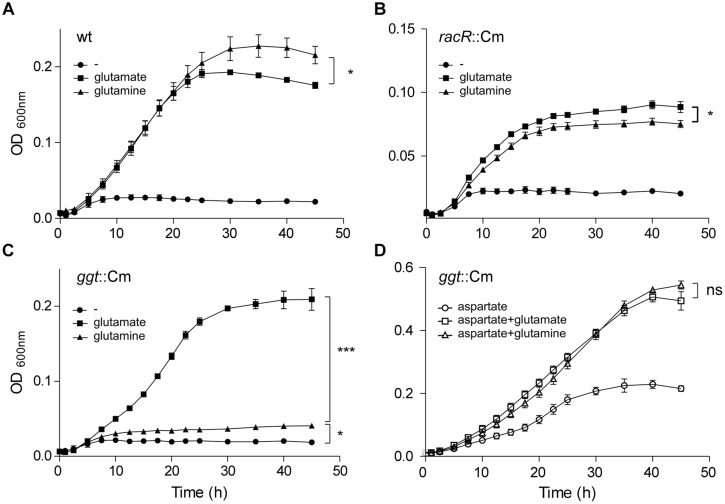
To investigate whether the GGT activity contributes to an increased bacterial fitness, growth curves in DMEM medium with or without 10 mM glutamate or 10 mM glutamine were recorded for the wt, the racR and ggt mutant strains under RacR inducing conditions. To facilitate growth TMAO was used as electron acceptor, because nitrate proved to be detrimental for growth at these nutrient poor conditions. The growth rates of the wt grown with either glutamate or glutamine as sole carbon source were very similar, although the maximum OD600 nm was higher when glutamine was present (Figure 5A). The racR mutant strain on the other hand showed a reduced growth rate when grown on glutamine compared to glutamate and consistently reached a slightly lower OD600 nm (Figure 5B). Furthermore compared to the wt, the racR mutant shows a reduced growth rate and a lower maximum growth yield. The ggt mutant strain grew comparable to the wt on glutamate, but hardly grew on glutamine (Figure 5C), verifying that GGT is needed to utilize glutamine as sole carbon/energy source (Hofreuter et al., 2006). These results prove that under low oxygen conditions in the presence of alternative electron acceptors the RacRS system is important for the conversion of glutamine to glutamate.
FIGURE 5.
The RacRS system is important for the generation of glutamate out of glutamine. Growth curves were generated of the wt 81116 (A), the racR (B), and ggt (C) mutants grown in DMEM (circles) or with the addition of 10 mM glutamate (squares) or 10 mM glutamine (triangles) at 1% O2 with the addition of 10 mM TMAO as electron acceptor. (D) The same as (C) but with the extra addition of 10 mM aspartate to the medium. Data represent the mean and standard error of three independent experiments. Significance was calculated using the maximally obtained OD600nm values. ns, not significant; *p < 0.05, ***p < 0.001.
Campylobacter jejuni is Capable of Utilizing Glutamine in a GGT Independent Manner
Although GGT is required for growth with glutamine as sole carbon source, we observed that the ggt mutant reached a higher OD600nm when grown on DMEM plus glutamine compared to DMEM alone (Figure 5C). C. jejuni possess a glutamine transporter PaqPQ that has been shown to be functional under nutrient rich conditions (Lin et al., 2009). To investigate whether the ggt mutant is able to grow on glutamine under nutrient rich conditions, aspartate was added to the DMEM medium. Both the growth rate and bacterial yield of the ggt mutant increased with glutamate and glutamine compared to aspartate alone (Figure 5D). Similar results were obtained when serine was used instead of aspartate (data not shown). This GGT independent glutamine utilization phenotype was also seen for the C. jejuni 81-176 ggt mutant strain and the C. jejuni 11168 strain, which naturally lacks the ggt gene (data not shown). These results indicate that GGT is important when other energy sources than glutamines are not available.
Discussion
The highly conserved enzyme GGT in prokaryotes plays a key role in the glutamine and glutathione metabolism. However, the regulation of the transcription of this gene is poorly understood. Here we identified the response regulator RacR as the first prokaryotic transcription factor that directly regulates ggt gene transcription. The C. jejuni RacR activates ggt gene transcription under low oxygen conditions in the presence of alternative electron acceptors. The RacRS system therefore not only activates the cytoplasmic production of glutamate by upregulating the GOGAT system (van der Stel et al., 2015), but also influences the periplasmic production of glutamate by upregulating the GGT enzyme, and thus ensuring the use of extracellular glutamine as energy source under RacR inducing conditions.
Previously, the ggt gene was not identified as part of the RacR regulon as the micro-arrays used in that study were based on a C. jejuni strain that lacks the ggt gene (van der Stel et al., 2015). Only 31% of C. jejuni strains contain a ggt gene (de Haan et al., 2012). Because both RacR and GGT have been shown to be important for host colonization (Brás et al., 1999; Barnes et al., 2007; Hofreuter et al., 2008), we assumed that the RacRS system might also regulate the ggt gene. Real-time RT-PCR and GGT enzyme activity assays clearly showed that RacR activates the transcription of the ggt gene and GGT enzyme activity under limited oxygen conditions in the presence of alternative electron acceptors (Figures 1 and 2), the same result was observed in a C. jejuni 81-176 racR mutant (data not shown). Similar results were obtained in the racRS and the racS mutants, showing that both RacR and RacS are needed to activate ggt transcription.
Unequivocal evidence that ggt is a member of the RacRS regulon was obtained from EMSA and luciferase reporter assays (Figures 3 and 4). EMSA results showed that phosphorylated RacR strongly interacts with the ggt promoter, indicating direct regulation of ggt by RacR upon phosphorylation. The C. jejuni 81116 ggt promoter contains a conserved −10 and −16 regions but no −35 region (Petersen et al., 2003; Dugar et al., 2013). Apparently a strongly conserved −10 and −16 regions are not sufficient to activate the ggt transcription without additional transcription factors, as seen by real-time RT-PCR and luciferase assay (Figures 1 and 4). These results suggest that although a conserved consensus sequence for the −35 region of sigma 70 regulated promoters in C. jejuni is lacking the region upstream of the −16 region is essential to activate sigma 70 regulated promoters as has been observed already (Dugar et al., 2013; Salamasznska-Guz et al., 2013). Using the previously obtained RacR binding consensus (van der Stel et al., 2015), a potential RacR binding site was found ~80-bp upstream of the transcriptional start site (TSS) (Dugar et al., 2013), besides that, a palindromic sequence was found ~60-bp upstream of the TSS, which could be a potential regulatory element. Different lengths of the ggt promoter cloned in front of a promoterless luciferase reporter gene and EMSA experiments showed that only the fragments containing the predicted RacR binding nucleotide sequence were activated in the luciferase assay and band shifted in the EMSA experiment. This result clearly shows that the ggt gene belongs to the RacRS regulon.
Although present in many bacterial species, knowledge regarding the regulation of the GGT is limited. In E. coli and B. subtilis GGT activity is maximal in stationary growth phase (Tabor and Richardson, 1985; Xu and Strauch, 1996), while in H. pylori, an organism closely related to C. jejuni, ggt transcription is growth phase independent (Wachino et al., 2010). In C. jejuni the highest GGT activity measured at microaerophilic conditions is seen on plates in logarithmic growth phase (Barnes et al., 2007), however, when we used HI liquid medium the highest GGT activity was observed in stationary growth phase, independent of the oxygen concentration (data not shown). Nutrient availability might explain this difference as nitrogen-limiting conditions has been shown to activate the B. subtilis GGT (Kimura et al., 2004). So far, only in B. subtilis a transcription factor ComA has been identified to indirectly regulate the ggt gene. ComA of the quorum sensing two-component system ComP/ComA activates the ggt transcription, but has no influence on the gltBD genes (Ogura et al., 2001). The C. jejuni RacRS system is therefore unique that it directly regulates the periplasmic as well as cytoplasmic glutamate production in response to the available electron acceptors.
The GOGAT activity in other bacteria is strictly regulated, based on carbon, nitrogen, and energy status of the cell (Ninfa and Jiang, 2005), also because it requires the high energetic cofactor NADPH to generate glutamate. GGT does, however, not require high energetic co-factors; this could be a reason why it is expressed by C. jejuni in late log/stationary phase to scavenge for nutrients when energy levels are low.
The role of GGT varies among organisms, in animals the enzyme is used to recycle glutathione, while in yeast cells GGT is used as metabolic enzyme to utilize nitrogen sources (Mehdi and Penninckx, 1997). In H. pylori it was shown that an ammonia generating cycle consisting of periplasmic GGT and AnsA is present that aids in acidic resistance (Miller and Maier, 2014) and is essential for colonization and pathogenicity. The role of GGT in C. jejuni has been proposed to be metabolic, necessary for the acquisition of energy, carbon, and nitrogen, by deamination of glutamine, or acquisition of additional sulfur by metabolizing glutathione (Hofreuter, 2014; Vorwerk et al., 2014). Furthermore, GGT has been shown to be important for the pathogenesis of C. jejuni, by inhibiting cell proliferation and causing apoptosis (Barnes et al., 2007; Floch et al., 2014). Here we show that ggt is co-regulated with other metabolic genes in C. jejuni by RacR (Figure 1). In the periplasm GGT converts glutamine and glutathione to glutamate (Figure 6), which is subsequently transported to the cytoplasm via glutamate transporter PEB1 (Del Rocio Leon-Kempis et al., 2006). Up-regulation of ggt yields a growth advantage when grown on glutamine, confirming a metabolic role for GGT. This growth advantage is, however, only observed when other more favorable nutrients are less available (Figure 5D). When other carbon sources are present, the transporter PaqPQ (Lin et al., 2009) probably imports glutamine, which is converted to glutamate by the GOGAT system. Recently it has been reported that most Campylobacter cow isolates lack the ggt gene, while ggt is common in poultry and human isolates (de Haan et al., 2012). C. jejuni clades that lack ggt often harbor the fucose utilization gene cluster, while co-occurrence is rarely observed (de Haan et al., 2012), which could explain why strains lacking ggt are able to colonize chickens while a constructed ggt mutant can not. As humans and poultry are both omnivores, they consume a diet richer in protein than cattle. This results in a higher glutamine concentration in the gut (Stella et al., 2006), which is in favor of GGT possessing C. jejuni strains.
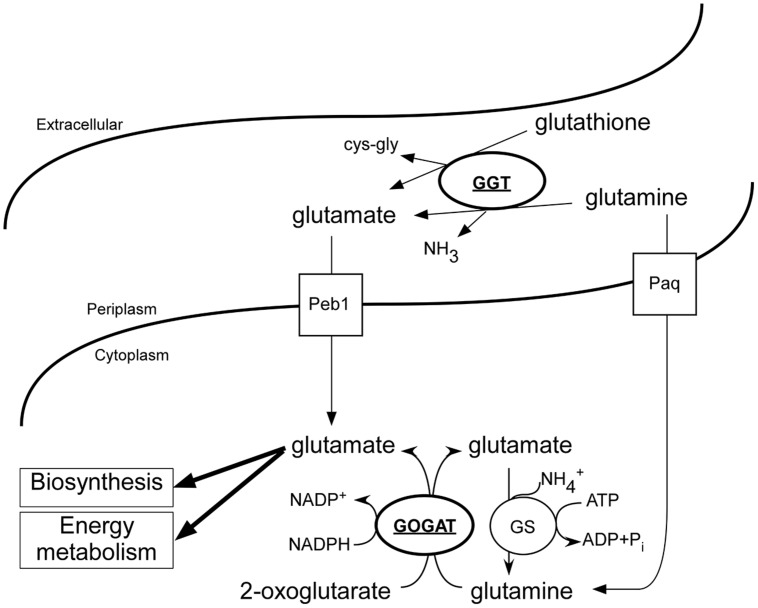
FIGURE 6.
Overview of the C. jejuni glutamate generating enzymes and involved transport systems. Under low oxygen conditions in the presence of additional electron acceptors, RacR is activated and enhances the production of GGT and GOGAT (bold and underlined). Glutamine and glutathione are converted to glutamate in the periplasm by GGT; glutamate is consequently imported and used as carbon/energy source and anabolic precursor. Independent of GGT, glutamine can be imported and converted to glutamate by GOGAT, however, this process is dependent on the presence of additional carbon/energy sources.
Overall, we show that the C. jejuni RacRS two-component system directly regulates the ggt gene transcription under limited oxygen conditions when alternative electron acceptors are present. The RacRS system is the first identified system that directly regulates both the periplasmic glutamate production (GGT) as well as the cytoplasmic glutamate production (GOGAT) and plays an important role in the metabolism of C. jejuni.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the fellowship of the Royal Netherlands Academy of Arts and Science, NWO-VIDI grant 917.66.330 and NWO-ECHO grant 711.012.007 to MW. As well as by the Polish Ministry of Science and Higher Education, grant N401 183 31/3968.
References
- Barnes I. H., Bagnall M. C., Browning D. D., Thompson S. A., Manning G., Newell D. G. (2007). γ-Glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 43 198–207. 10.1016/j.micpath.2007.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M. J. (1997). Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176 S103–S105. 10.1086/513780 [DOI] [PubMed] [Google Scholar]
- Bouwman L. I., Niewold P., van Putten J. P. M. (2013). Basolateral invasion and trafficking of Campylobacter jejuni in polarized epithelial cells. PLoS ONE 8:e54759 10.1371/journal.pone.0054759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brás A. M., Chatterjee S., Wren B. W., Newell D. G., Ketley J. M. (1999). A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J. Bacteriol. 181 3298–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Thiberge J.-M., Ferrero R. L., Labigne A. (1999). Essential role of Helicobacter pylori γ-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol. Microbiol. 31 1359–1372. 10.1046/j.1365-2958.1999.01271.x [DOI] [PubMed] [Google Scholar]
- de Haan C. P., Llarena A.-K., Revez J., Hänninen M.-L. (2012). Association of Campylobacter jejuni metabolic traits with multilocus sequence types. Appl. Environ. Microbiol. 78 5550–5554. 10.1128/AEM.01023-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rocio Leon-Kempis M., Guccione E., Mulholland F., Williamson M. P., Kelly D. J. (2006). The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60 1262–1275. 10.1111/j.1365-2958.2006.05168.x [DOI] [PubMed] [Google Scholar]
- Dugar G., Herbig A., Förstner K. U., Heidrich N., Reinhardt R., Nieselt K., et al. (2013). High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 9:e1003495 10.1371/journal.pgen.1003495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch P., Pey V., Castroviejo M., Dupuy J. W., Bonneu M., de La Guardia A. H., et al. (2014). Role of Campylobacter jejuni gamma-glutamyl transpeptidase on epithelial cell apoptosis and lymphocyte proliferation. Gut Pathog. 6 20 10.1186/1757-4749-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M., Ling S. S. M., Lui S. Y., Yeoh K. G., Ho B. (2010). Helicobacter pylori γ-glutamyl transpeptidase is a pathogenic factor in the development of peptic ulcer disease. Gastroenterology 139 564–573. 10.1053/j.gastro.2010.03.050 [DOI] [PubMed] [Google Scholar]
- Green M. R., Sambrook J. (eds) (2012). “Molecular cloning: a laboratory manual,” Cold Spring Harbor, Berlin, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Guccione E., Del Rocio Leon-Kempis M., Pearson B. M., Hitchin E., Mulholland F., Van Diemen P. M., et al. (2008). Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69 77–93. 10.1111/j.1365-2958.2008.06263.x [DOI] [PubMed] [Google Scholar]
- Hanigan M. H. (1998). γ-Glutamyl transpeptidase, a glutathionase: its expression and function in carcinogenesis. Chem. Biol. Interact. 111–112, 333–342. 10.1016/S0009-2797(97)00170-1 [DOI] [PubMed] [Google Scholar]
- Heeswijk W. C., van Westerhoff H. V., Boogerd F. C. (2013). Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol. Mol. Biol. Rev. 77 628–695. 10.1128/MMBR.00025-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D. (2014). Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni. Front. Cell. Infect. Microbiol. 4:137 10.3389/fcimb.2014.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D., Novik V., Galán J. E. (2008). Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4 425–433. 10.1016/j.chom.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Hofreuter D., Tsai J., Watson R. O., Novik V., Altman B., Benitez M., et al. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74 4694–4707. 10.1128/IAI.00210-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytiäinen H., Juntunen P., Akrenius N., Hänninen M.-L. (2012). Importance of RNA stabilization: evaluation of ansB, ggt, and rpoA transcripts in microaerophilic Campylobacter jejuni 81–176. Arch. Microbiol. 194 803–808. 10.1007/s00203-012-0820-3 [DOI] [PubMed] [Google Scholar]
- Kimura K., Tran L.-S. P., Uchida I., Itoh Y. (2004). Characterization of Bacillus subtilis γ-glutamyltransferase and its involvement in the degradation of capsule poly-γ-glutamate. Microbiology 150 4115–4123. 10.1099/mic.0.27467-0 [DOI] [PubMed] [Google Scholar]
- Kordinas V., Nicolaou C., Ioannidis A., Papavasileiou E., John Legakis N., Chatzipanagiotou S. (2005). Prevalence of four virulence genes in Campylobacter jejuni determined by PCR and sequence analysis. Mol. Diagn. J. Devoted Underst. Hum. Dis. Clin. Appl. Mol. Biol. 9 211–215. 10.2165/00066982-200509040-00008 [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A., Harel J., Tompkins L. (1987). Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 169 5320–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A. E., Krastel K., Hobb R. I., Thompson S. A., Cvitkovitch D. G., Gaynor E. C. (2009). Atypical roles for Campylobacter jejuni amino acid ATP binding cassette transporter components PaqP and PaqQ in Bbcterial stress tolerance and pathogen-host cell dynamics. Infect. Immun. 77 4912–4924. 10.1128/IAI.00571-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi K., Penninckx M. J. (1997). An important role for glutathione and γ-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology 143 1885–1889. 10.1099/00221287-143-6-1885 [DOI] [PubMed] [Google Scholar]
- Miller E. F., Maier R. J. (2014). Ammonium metabolism enzymes aid Helicobacter pylori acid resistance. J. Bacteriol. 196 3074–3081. 10.1128/JB.01423-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka W. T., Zhang Q. (2011). Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193 1065–1075. 10.1128/JB.01252-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Jiang P. (2005). PII signal transduction proteins: sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8 168–173. 10.1016/j.mib.2005.02.011 [DOI] [PubMed] [Google Scholar]
- Ogura M., Yamaguchi H., Yoshida K., Fujita Y., Tanaka T. (2001). DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29 3804–3813. 10.1093/nar/29.18.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong P.-L., Yao Y.-F., Weng Y.-M., Hsu W.-H., Lin L.-L. (2008). Residues Arg114 and Arg337 are critical for the proper function of Escherichia coli γ-glutamyltranspeptidase. Biochem. Biophys. Res. Commun. 366 294–300. 10.1016/j.bbrc.2007.11.063 [DOI] [PubMed] [Google Scholar]
- Palmer S. R., Gully P. R., White J. M., Pearson A. D., Suckling W. G., Jones D. M., et al. (1983). Water-borne outbreak of Campylobacter gastroenteritis. Lancet 321 287–290. 10.1016/S0140-6736(83)91698-7 [DOI] [PubMed] [Google Scholar]
- Parke D. (1990). Construction of mobilizable vectors derived from plasmids RP4 pUC18 and pUC19. Gene 93 135–137. 10.1016/0378-1119(90)90147-J [DOI] [PubMed] [Google Scholar]
- Petersen L., Larsen T. S., Ussery D. W., On S. L. W., Krogh A. (2003). RpoD promoters in Campylobacter jejuni exhibit a strong periodic signal instead of a -35 box. J. Mol. Biol. 326 1361–1372. 10.1016/S0022-2836(03)00034-2 [DOI] [PubMed] [Google Scholar]
- Quetz J., da S., Lima I. F. N., Havt A., Prata M. M. G., Cavalcante P. A., et al. (2012). Campylobacter jejuni infection and virulence-associated genes in children with moderate to severe diarrhoea admitted to emergency rooms in northeastern Brazil. J. Med. Microbiol. 61 507–513. 10.1099/jmm.0.040600-0 [DOI] [PubMed] [Google Scholar]
- Reitzer L. (2003). Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57 155–176. 10.1146/annurev.micro.57.030502.090820 [DOI] [PubMed] [Google Scholar]
- Salamasznska-Guz A., Grodzik M., Klimuszko D. (2013). Mutational analysis of cj0183 Campylobacter jejuni promoter. Curr. Microbiol. 67 696–702. 10.1007/s00284-013-0420-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Stahl M., Friis L. M., Nothaft H., Liu X., Li J., Szymanski C. M., et al. (2011). L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U.S.A. 108 7194–7199. 10.1073/pnas.1014125108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella C., Beckwith-Hall B., Cloarec O., Holmes E., Lindon J. C., Powell J., et al. (2006). Susceptibility of human metabolic phenotypes to dietary modulation. J. Proteome Res. 5 2780–2788. 10.1021/pr060265y [DOI] [PubMed] [Google Scholar]
- Suzuki H., Kumagai H., Tochikura T. (1986). gamma-Glutamyltranspeptidase from Escherichia coli K-12: formation and localization. J. Bacteriol. 168 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. (1985). A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U.S.A. 82 1074–1078. 10.1073/pnas.82.4.1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder K. A., Aslam M., Islam Z., Azmi I. J., Dutta D. K., Hossain S., et al. (2008). Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 46 1485–1488. 10.1128/JCM.01912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vliet A. H. M., van Wooldridge K. G., Ketley J. M. (1998). Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180 5291–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Stel A.-X., van Mourik A., Heijmen-van Dijk L., Parker C. T., Kelly D. J., van de Lest C. H. A., et al. (2015). The Campylobacter jejuni RacRS system regulates fumarate utilization in a low oxygen environment. Environ. Microbiol. 17 1049–1064. 10.1111/1462-2920.12476 [DOI] [PubMed] [Google Scholar]
- van Mourik A., Bleumink-Pluym N. M., van Dijk L., van Putten J. P., Wösten M. M. (2008). Functional analysis of a Campylobacter jejuni alkaline phosphatase secreted via the Tat export machinery. Microbiology 154 584–592. 10.1099/mic.0.2007/012120-0 [DOI] [PubMed] [Google Scholar]
- Vorwerk H., Mohr J., Huber C., Wensel O., Schmidt-Hohagen K., Gripp E., et al. (2014). Utilization of host-derived cysteine-containing peptides overcomes the restricted sulphur metabolism of Campylobacter jejuni. Mol. Microbiol. 93 1224–1245. 10.1111/mmi.12732 [DOI] [PubMed] [Google Scholar]
- Wachino J.-I., Shibayama K., Suzuki S., Yamane K., Mori S., Arakawa Y. (2010). Profile of expression of Helicobacter pylori γ-Glutamyltranspeptidase. Helicobacter 15 184–192. 10.1111/j.1523-5378.2010.00755.x [DOI] [PubMed] [Google Scholar]
- Wösten M. M., Wagenaar J. A., van Putten J. P. (2004). The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279 16214–16222. 10.1074/jbc.M400357200 [DOI] [PubMed] [Google Scholar]
- Xu K., Strauch M. A. (1996). Identification, sequence, and expression of the gene encoding gamma-glutamyltranspeptidase in Bacillus subtilis. J. Bacteriol. 178 4319–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]