Abstract
Ligands for natural killer (NK) cell activating receptors can be released from tumor cells and are believed to promote tumor growth by acting as decoys for effector lymphocytes. In a recent paper published in Science, Deng et al. report another scenario in which a shed form of the MULT1 mouse NKG2D ligand boosts NK cell functions.
Natural killer (NK) cells are cytolytic and cytokine-producing lymphocytes of the innate immune system that participate in the control of tumor growth and microbial infections1. NK cell effector activities are tightly controlled by a fine balance of inhibitory and activating signals delivered by surface receptors. Activating receptors can recognize two types of ligands, self-molecules encoded by the host's own genome whose expression is upregulated upon cellular stress, or exogenous molecules produced by microbes during infection. NKG2D, one of the best characterized activating receptor expressed by NK and T cells, binds to several different ligands in human and mouse2. NKG2D ligands are poorly expressed on the vast majority of normal cell surfaces, but are upregulated on tumor and virus-infected cells. In addition, NKG2D ligands can be released by both surface cleavage and exosome excretion. It has been reported that shed ligands can block tumor cell recognition by effector cells by preventing NKG2D interaction with its ligands3. However, several reports do not correlate the presence of soluble ligands with decreased NKG2D expression nor functional activities.
Deng et al.4 focused their analysis on the NKG2D mouse ligand MULT1, which is commonly overexpressed on primary tumor cells. They first showed that MULT1-transduced fibroblast can cleave MULT1 from the plasma membrane, resulting in a released shed form in the supernatant. Shed MULT1 is of high affinity to NKG2D (∼13 nM) similar to recombinant MULT1. They further reveal the presence of shed MULT1 in the serum of mice developing spontaneous MULT1+ tumors. Interestingly, the authors detected a very high concentration of shed MULT1 in the sera of Apoe−/− mice exhibiting severe atherosclerosis and liver inflammation. Given that these autoimmune injuries observed in this mouse model depend on NKG2D activity5, it was unlikely that shed MULT1 exert an inhibitory effect on immunity.
Surprisingly, the authors further showed that mouse tumor cells engineered to release a secreted form of MULT1 (secMULT1) similar to the shed MULT1 were rejected when injected into syngenic mice. Tumor rejection is dependent on NK cells as cells grow in NK but not in CD8+ T cell-depleted host and requires NKG2D. Importantly, the controlled release of secMULT1 from tumors harboring inducible secMULT1 promotes tumor rejection. To rule out the possibility that tumor cell rejection was due to intrinsic modifications of tumor cells, the author monitored the rejection of a mixture of 9:1 secMULT1−: secMULT1+ tumor cells and showed an improved antitumoral effect on both secMULT1+ and, importantly, secMULT1− tumors. In addition, direct intratumoral injection of recombinant MULT1 promotes tumor rejection. These results suggested that soluble MULT1 mobilizes or activates anti-tumor effector cells. Deng et al. further reported increased frequencies of cytotoxic and IFN-γ-secreting NK cells associated with secMULT1+ tumors as compared to control tumor cells. Altogether, these data suggest that a shed NKG2D ligand can promote tumor rejection by boosting NK cell effector functions.
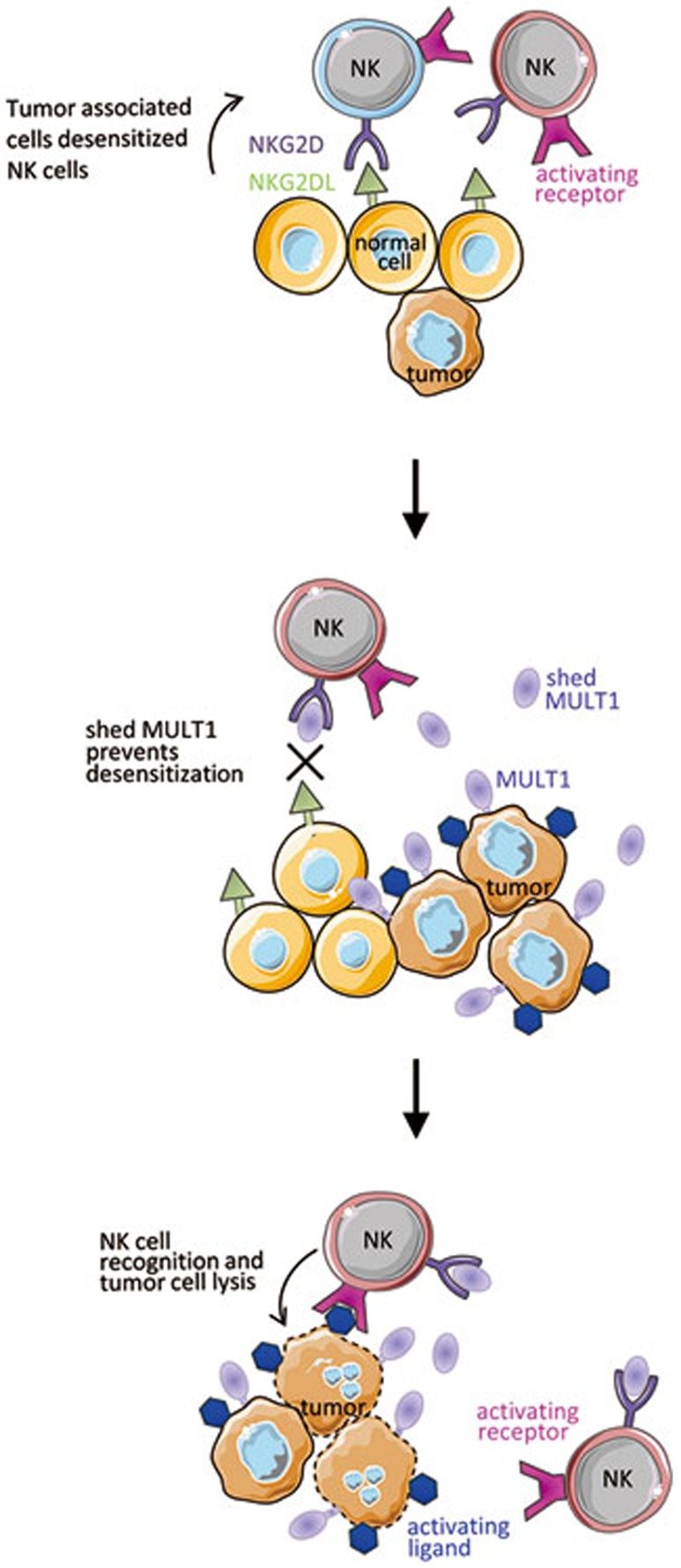
Shed MULT1 could crosslink NKG2D and thus activate NK cells. However, shed and secMULT1 are monomeric molecules similar to the recombinant MULT1 which fails to activate NK cells in vitro. Formation of multivalent structures in vivo was not detected. In addition, whereas the transmembrane form of MULT1 can activate NK cells by crosslinking NKG2D and induces NKG2D downregulation, soluble MULT1 upregulates NKG2D on the NK cell surface. This upregulation is probably due do a decreased downregulation of NKG2D surface expression because no increase in NKG2D mRNA or protein was observed. Based on these findings, the authors hypothesized that NKG2D ligands expressed on non-tumor host cell membrane continuously engage NKG2D on NK cells, leading to NKG2D downregulation and NK cell desensitization, whereas soluble MULT1 blocks these interactions to increase NK cell responsiveness (Figure 1). Along this line, NK cells from mutant mice genetically deficient for the NKG2D ligand expressed by tumor-associated myeloid cells are not desensitized.
Figure 1.
Tumor-associated cells express NKG2DL which can desensitize NK cells. Tumor shedding of MULT1 delivers soluble MULT1 that outcompetes for NKG2D binding and prevents NK cell desensitization. Boosted NK cell functions lead to improved tumor cell rejection by other activating receptors.
The induction of cell desensitization by a frequent or even constant stimulation is a very common mechanism across living objects. Regarding NK cells, another example of tuning via desensitization resides in the impact of the long lasting absence of MHC class I molecules in their environment. Indeed, NK cells are hyporesponsive in a MHC-I-deficient host6. There are accumulating data indicating that in the absence of engagement of inhibitory receptors for MHC class I molecules, NK cells get desensitized due to their chronic interaction with endogenous stimulating ligands7. Indeed, in the absence of engagement of this inhibitory pathway, NK cell activation would be unleashed8. This scenario is supported by a series of in vitro and in vivo experiments in which NK cells are desensitized following chronic exposure to stimulatory molecules expressed at the surface of interacting cells9,10. Thus, the induction of MHC class I downregulation or NKG2D ligand upregulation boosts NK cell function, whereas the sustained lack of MHC class I or expression of NKG2D ligands impairs NK cell reactivity. This tuning of immune response as a function of the speed of change of the stimuli detected by lymphocytes is at the center of the recently proposed Discontinuity Theory11.
Finally, consistent with their findings with secMULT1 but somewhat counter-intuitively, Deng et al. also show that NKG2D receptor deficiency or blockade using anti-NKG2D monoclonal antibodies mimics the effect of soluble MULT1. Indeed, in both conditions, NK cell effector functions are boosted, resulting in improved tumor rejection. Similarly, blocking other NK activating receptors, such as NKp46, may also lead to NK cell desensitization12. Checkpoint inhibitory receptors are revolutionizing the treatment of cancers by inhibiting the inhibitory receptors. The findings reported by Deng et al. together with earlier results propose alternative strategies of cancer treatment using antibodies that are directed against activating receptors. In the case of NKG2D, the chronic engagement of NK cells with membrane-bound NKG2D ligand affects not only NKG2D-dependent but also NKG2D-independent signaling pathways9. The blockade of NKG2D desensitization by antibodies directed against NKG2D should thus also boost NK cell activation via other pathways, such as antibody-dependent cell cytotoxicity. However, the precise identification of the ligand-receptor pair involved in the control of tumors by NK cells will be a limiting factor to these innovative therapeutic approaches. Indeed, antibodies against activating receptors should be designed to boost NK cell reactivity but should not block the recognition of the tumors by NK cells. Finally, as the tuning of NK cell reactivity by soluble NKG2D ligands depends on their affinity for NKG2D, the pre-clinical development of this new class of drug candidates might reveal novel pharmacokinetics and the pharmacodynamics guidelines.
References
- Vivier E, Raulet DH, Moretta A, et al. Science. 2011. pp. 44–49. [DOI] [PMC free article] [PubMed]
- Raulet DH, Guerra N. Nat Rev Immunol. 2009. pp. 568–580. [DOI] [PMC free article] [PubMed]
- Groh V, Wu J, Yee C, et al. Nature. 2002. pp. 734–738. [DOI] [PubMed]
- Deng W, Gowen BG, Zhang L, et al. Science. 2015. pp. 136–139. [DOI] [PMC free article] [PubMed]
- Xia M, Guerra N, Sukhova GK, et al. Circulation. 2011. pp. 2933–2943. [DOI] [PMC free article] [PubMed]
- Hoglund P, Brodin P. Nat Rev Immunol. 2010. pp. 724–734. [DOI] [PubMed]
- Viant C, Fenis A, Chicanne G, et al. Nat Commun. 2014. pp. 5108–5118. [DOI] [PubMed]
- Raulet DH, Vance RE. Nat Rev Immunol. 2006. pp. 520–531. [DOI] [PubMed]
- Coudert JD, Scarpellino L, Gros F, et al. Blood. 2008. pp. 3571–3578. [DOI] [PubMed]
- Oppenheim DE, Roberts SJ, Clarke SL, et al. Nat Immunol. 2005. pp. 928–937. [DOI] [PubMed]
- Pradeu T, Jaeger S, Vivier E. Nat Rev Immunol. 2013. pp. 764–769. [DOI] [PubMed]
- Narni-Mancinelli E, Jaeger BN, Bernat C, et al. Science. 2012. pp. 344–348. [DOI] [PubMed]