Dear Editor,
Active DNA demethylation plays crucial roles in the regulation of gene expression and gene imprinting. In plants, active DNA demethylation is initiated by the ROS1/DME family of 5-methylcytosine-specific DNA glycosylases via a base excision repair mechanism1,2. ROS1 and DME are bifunctional DNA glycosylases that excise the 5-methylcytosine base and then cleave the DNA backbone at the abasic site, resulting in a gap with a 3′ phosphate or 3′ dRP (3′ α, β-unsaturated aldehyde) terminus. The DNA phosphatase ZDP and the apurinic/apyrimidinic endonuclease APE1L process the 3′ phosphate and 3′ dRP termini, respectively, to generate a 3′ OH group so that downstream polymerases and ligases can fill in the gap with an unmethylated cytosine3,4. Thus far, the DNA polymerases and ligases involved in this active DNA demethylation pathway are unclear. Here we show that Arabidopsis DNA LIGASE I (AtLIG1) co-localizes with ROS1, ZDP and APE1L in vivo. In addition, we found that AtLIG1 is essential for demethylation and activation of the maternally imprinted genes FWA and MEA in the endosperm. Our data suggest that AtLIG1 is the major DNA ligase that functions at the last step in active DNA demethylation in Arabidopsis.
The Arabidopsis genome encodes four DNA ligases: AtLIG1, AtLIG1a, AtLIG4 and AtLIG6 (Supplementary information, Figure S1A)5. AtLIG1 carries out the ligation reaction in DNA replication and base excision repair, whereas AtLIG4 is responsible for DNA ligation in the non-homologous end-joining pathway in DNA damage response6,7,8,9,10. There are no reported roles for AtLIG1a and AtLIG6. Transcriptome analyses revealed that AtLIG1a is probably not expressed, indicating that AtLIG1 may be the sole source of DNA ligase I activity in Arabidopsis8. In order to characterize the function of Arabidopsis DNA ligases (AtLIGs) in active DNA demethylation, T-DNA insertion lines for these genes were obtained (Supplementary information, Figure S1B). The homozygous mutant of atlig1 is embryonic lethal and we only obtained heterozygous mutants atlig1-1 (Col background) and atlig1-3 (C24 background), which produce small siliques and ∼50% aborted seeds (Supplementary information, Figure S2A). We also generated RNAi lines that have a reduced expression of AtLIG1 (Supplementary information, Figure S2B). Similar to previously reported AtLIG1 RNAi lines7, these RNAi lines exhibit a severe dwarf phenotype (Supplementary information, Figure S2C). The atlig1a-1, atlig1a-2, atlig4-5 and atlig6-1 mutants do not exhibit abnormal developmental phenotypes under normal growth conditions. To determine which of the ligases is involved in active DNA demethylation, we designed a chop-PCR assay to measure the DNA methylation level at the At1g26400 locus. Upon ROS1 dysfunction, the At1g26400 locus shows DNA hypermethylation and the DNA becomes resistant to cleavage by the methylation-sensitive restriction enzyme HhaI, and thereby can be amplified as a distinct band by PCR. AtLIG1 knockdown by RNAi, but not mutations of other AtLIGs, also led to DNA hypermethylation (Supplementary information, Figure S3A). Moreover, we performed bisulfite sequencing and confirmed that the AtLIG1 RNAi lines show DNA hypemethylation at this locus in the CG context. CHG and CHH methylation levels are also increased, although to a lesser extent (Supplementary information, Figure S3B). To further explore the roles of AtLIG4 and AtLIG6 in active DNA demethylation, we performed whole-genome bisulfite sequencing in the atlig4atlig6 double mutant but found that the double mutations did not affect DNA methylation levels when compared with ros1-4 and rdd mutants (Supplementary information, Table S1). Collectively, our data suggest that AtLIG1, but not other AtLIGs, is involved in the active DNA demethylation pathway.
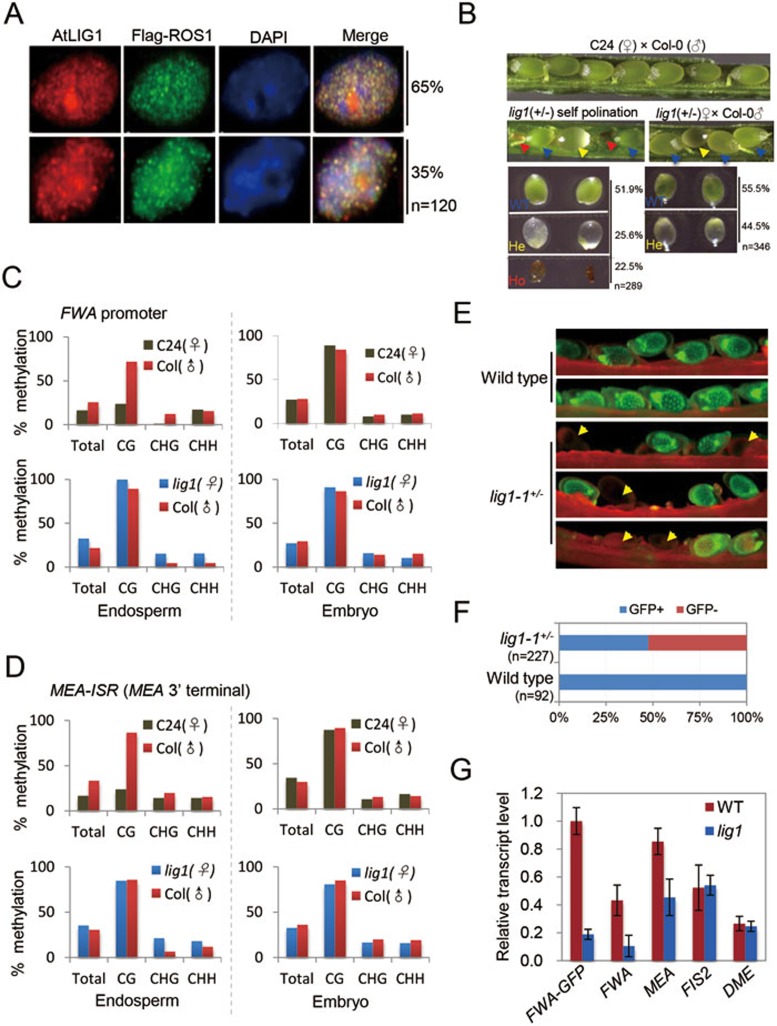
Our previous data show that ZDP and APE1L co-localize with ROS1 in subnuclear foci3,4. To test whether AtLIG1 may co-localize with other enzymes in the active DNA demethylation pathway, we performed co-immunolocalization assays. The expression of FLAG-tagged ROS1, driven by its native promoter in ros1-1 mutant plants, is visualized by an antibody against the FLAG tag. AtLIG1 is stained by a custom-made primary antibody and a fluorescence-tagged secondary antibody. AtLIG1 co-localizes with ROS1 in nucleoplasmic foci, as shown by the strong yellow signals (Figure 1A). In 89% of the cells, AtLIG1 co-localizes with APE1L in the nucleolus and in nucleoplasmic foci, whereas in 11% of the cells, AtLIG1 and APE1L substantially co-localize in nucleoplasmic foci but not in the nucleolus (Supplementary information, Figure S4A). AtLIG1 and ZDP also co-localize in nucleoplasmic foci in ∼87% of the cells (Supplementary information, Figure S4B). Thus, AtLIG1 co-localizes with known components of the active DNA demethylation machinery in distinct subnuclear foci.
Figure 1.
AtLIG1 is required for genomic imprinting in Arabidopsis. (A) Dual immunolocalization using anti-AtLIG1 (red) in transgenic lines expressing Flag-ROS1 (green). In all panels the DNA was stained with DAPI (blue). The frequency of nuclei displaying each interphase pattern is shown on the right. (B) Phenotype of developing F1 seeds at 7 days after pollination (DAP) in a cross between wild type (WT) and mutant. The percentages of normal, early aborting (homozygous) and late aborting (heterozygous) seeds are shown on the right. (C-D) Methylation percentage of the 5′ SINE-related repeat of FWA (C) or MEA 3′ terminal (D) in the WT embryo and endosperm or mutant embryo and endosperm with maternal atlig1. Methylation levels for each fraction were determined by bisulfite sequencing. (E) Fluorescence images of pFWA::ΔFWA-GFP seeds at 4 DAP. pFWA::ΔFWA-GFP was introduced into atlig1(+/−) background by genetic crosses, and homozygous pFWA::ΔFWA-GFP transgenic plants were selected by genotyping. Yellow arrowheads show seeds with a reduced GFP signal presumably because of their maternal inheritance of atlig1. (F) The percentages of GFP-positive and GFP-negative seeds in WT and atlig1 mutant with pFWA::ΔFWA-GFP reporter. (G) Expression levels of imprinted genes in WT and atlig1 mutant seeds at 3 DAP. ACT11 was used as an internal control. Two biological replicates were performed, and very similar results were obtained. Standard errors were calculated from three technical repeats (n = 3).
The homozygous atlig1 mutant has been reported to be maternally lethal11. Consistent with the previous report, we also observed brown early aborting seeds and white late aborting seeds, which may correspond to homozygous atlig1 seeds and heterozygous seeds carrying a maternal allele of atlig1 mutation, respectively (Figure 1B). Maternal lethality phenotypes can be caused by aberrant expression of maternally imprinted genes and defects in the central cell or the endosperm2,12,13. FWA and MEA are two well-known maternally imprinted genes, and their maternal expression in the endosperm requires active DNA demethylation initiated by the ROS1 paralog DME12,14. To investigate whether the methylation of FWA promoter and MEA ISR (intergenic subtelomeric repeat) in endosperm tissues is affected by the atlig1 mutation, we dissected the seeds of eight DAP (days after pollination) and collected the endosperm tissues from wild-type (WT) and white translucent mutant seeds. Then we carried out bisulfite sequencing analyses of the FWA promoter and MEA ISR region. Our data show that the maternal CG, CHG and CHH methylation levels of the 5′ direct repeats in the FWA promoter in the WT C24 endosperm are 23.81%, 0.8% and 17.14%, respectively (Figure 1C). In the endosperm of late aborting seeds (presumably of the atlig1-1+/− genotype with a maternal atlig1 mutation), the maternal methylation levels of the FWA promoter in CG, CHG and CHH contexts are changed to 100%, 15.35% and 15.71%, respectively (Figure 1C). There is a less increase in paternal CG methylation levels in the FWA promoter (from 71.85% to 91.11%), and we did not observe any increase in paternal CHG and CHH methylation levels (Figure 1C). The DNA methylation levels in all sequence contexts are similar between WT and late aborting seeds in the embryo (Figure 1C). The maternal CG, CHG and CHH methylation levels of MEA ISR in the WT C24 endosperm are 24%, 14.29% and 14.29%, respectively. They are increased to 84.92%, 21.43% and 18.15% in the endosperm of late aborting seeds (Figure 1D). Similarly, the paternal MEA ISR methylation levels remain unchanged (Figure 1D). These results suggest that the FWA promoter and MEA ISR are hypermethylated in the endosperm of mutant seeds harboring a maternal allele of atlig1. To examine whether the abnormal seeds in the atlig1-1+/− mutant have aberrant expression of FWA, we introduced a pFWA::ΔFWA-GFP reporter into the atlig1-1+/− mutant14. We found that atlig1-1+/− plants produced about 52.4% seeds that were defective in pFWA::ΔFWA-GFP expression (Figure 1E and 1F). The ratio of GFP-positive and GFP-negative seeds is in accordance with that of normal seeds and aborting seeds. To further measure the mRNA levels of FWA and MEA in the endosperm of WT and late aborting seeds, we performed real-time PCR and found that the expression levels of FWA-GFP, FWA and MEA, but not DME and FIS2, are downregulated in the atlig1 mutant endosperm (Figure 1G). Taken together, our data suggest that the defects in the atlig1 endosperm are associated with DNA hypermethylation and impaired expression of imprinted genes.
Our findings are consistent with previous data showing that mutations of components in the active DNA demethylation pathway, e.g., DME, SSRP1, ZDP and APE1L, result in defective activation of maternally imprinted genes and cause aberrant seed development2,4,13. A previous study reported normal expression of FWA and a limited, but not significantly altered, expression of MEA in the atlig1 mutant8. One possibility for this disparity is that materials of different ecotypes and different developmental stages were used. The previous study used the atlig1-3 mutant (C24 background) and examined the FWA-GFP and MEA-GUS signals in the central cell, whereas we used the atlig1-1 mutant (Col background) and examined the GFP signal in the endosperm.
In conclusion, our study reveals that AtLIG1 is an important component of the active DNA demethylation machinery. This allows us to have a more complete picture of the active DNA demethylation pathway in Arabidopsis (Supplementary information, Figure S5).
Acknowledgments
The work was supported by US National Institutes of Health Grants R01GM070795 and R01GM059138 (to J-K Z), and by the Chinese Academy of Sciences and by Peking-Tsinghua Center for Life Science.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Supplementary information Figures S1–S5, Materials and Methods and Tables S1 and S2
References
- Gong Z, Morales-Ruiz T, Ariza RR, et al. Cell. 2002. pp. 803–814. [DOI] [PubMed]
- Choi Y, Gehring M, Johnson L, et al. Cell. 2002. pp. 33–42. [DOI] [PubMed]
- Martinez-Macias MI, Qian W, Miki D, et al. Mol Cell. 2012. pp. 357–370. [DOI] [PMC free article] [PubMed]
- Li Y, Cordoba-Canero D, Qian W, et al. PLoS Genet. 2015. p. e1004905. [DOI] [PMC free article] [PubMed]
- Sunderland PA, West CE, Waterworth WM, et al. Plant J. 2006. pp. 356–367. [DOI] [PubMed]
- Taylor RM, Hamer MJ, Rosamond J, et al. Plant J. 1998. pp. 75–81. [DOI] [PubMed]
- Waterworth WM, Kozak J, Provost CM, et al. BMC Plant Biol. 2009. p. 79. [DOI] [PMC free article] [PubMed]
- Cordoba-Canero D, Roldan-Arjona T, Ariza RR. Plant J. 2011. pp. 693–702. [DOI] [PubMed]
- West CE, Waterworth WM, Jiang Q, et al. Plant J. 2000. pp. 67–78. [DOI] [PubMed]
- van Attikum H, Bundock P, Overmeer RM, et al. Nucleic Acids Res. 2003. pp. 4247–4255. [DOI] [PMC free article] [PubMed]
- Andreuzza S, Li J, Guitton AE, et al. Development. 2010. pp. 73–81. [DOI] [PubMed]
- Gehring M, Huh JH, Hsieh TF, et al. Cell. 2006. pp. 495–506. [DOI] [PMC free article] [PubMed]
- Ikeda Y, Kinoshita Y, Susaki D, et al. Dev Cell. 2011. pp. 589–596. [DOI] [PubMed]
- Kinoshita T, Miura A, Choi Y, et al. Science. 2004. pp. 521–523. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information Figures S1–S5, Materials and Methods and Tables S1 and S2