Abstract
T follicular helper (Tfh) cells are essential for germinal centers (GCs) and most long-term humoral immunity. Differentiation of Tfh cells depends on the transcriptional repressor B cell CLL/lymphoma 6 (Bcl6). Bcl6 mediates gene repression via the recruitment of corepressors. Currently, it is unknown how Bcl6 recruits corepressors to regulate gene expression of Tfh cells. In this article, we demonstrate, using a mutant form of Bcl6 with two BTB (bric-a-brac, tramtrack, broad-complex) mutations that abrogate corepressor binding, that the Bcl6 BTB domain is required for proper differentiation of Tfh and GC-Tfh cells in vivo. Importantly, we also observe a significant defect in GC B cell development. These results are consistent in multiple contexts, including a novel lymphocytic choriomeningitis virus nucleoprotein-specific TCR-transgenic mouse model. Taken together, these data suggest that the Bcl6 BTB domain is a key mediator of the differentiation of Tfh cells.
Introduction
The transcriptional repressor B cell CLL/lymphoma 6 (Bcl6) is essential for the differentiation of T follicular helper (Tfh) cells and germinal center (GC) B cells. Tfh cells are CD4 T cells specialized in providing help for B cells (1). The absence of Tfh cells results in the loss of GCs and, consequently, abrogated memory B cell, plasma cell, and neutralizing Ab responses. Thus, Tfh cells have critical roles in protective immune responses against pathogens, as well as deleterious roles in numerous autoimmune diseases (1, 2).
Bcl6 consists of a bric-a-brac, tramtrack, broad-complex (BTB/POZ) domain, a middle domain (also known as RDII), and a zinc finger domain consisting of six Kruppel-like zinc fingers (1). BTB domains are evolutionarily conserved protein-interaction domains that are widely present in transcription factors (3, 4). The BTB domain forms the interface of the obligate homodimer, and the corepressors BCOR, SMRT, and NCOR bind at the cleft formed by this interface (5–8). Although Bcl6 is required for Tfh cell differentiation (9–12), the contributions of its functional domains in CD4+ T cells are not well understood. In this study, we sought to examine the role of the Bcl6 BTB domain in Tfh cell differentiation and function.
Materials and Methods
Mice and vectors
C57BL/6J (B6) and CreCD4 mice were purchased from The Jackson Laboratory. Bcl6fl/fl (13), CD45.1-congenic, and Smarta TCR–transgenic (SM; specific for lymphocytic choriomeningitis virus [LCMV] gp66–77 on I-Ab) (14) mice were on a full B6 background and were bred at the La Jolla Institute for Allergy and Immunology. Bcl6BTBMUT mice, engineered to express the Bcl6 BTB domain mutant (BTBmut) from the endogenous Bcl6 locus, were generously provided by Dr. Ari Melnick (15). They were crossed to homozygosity at the La Jolla Institute for Allergy and Immunology for use in all experiments. NIP TCR-transgenic mice were generated as described below and in Supplemental Fig. 1. TCR hybridomas were generated (J. White and P. Marrack, unpublished observations), and TCR sequences were cloned and sequenced using cDNA isolated from LCMV-reactive clones. TCR sequences were expressed in 58α−β- T cell hybridomas and tested for reactivity against LCMV-infected dendritic cells. The TCRαβ pair showing the strongest reactivity (Vα1-Jα8 and Vβ6-Dβ1-Jβ2.3 rearrangements) was chosen and cloned into genomic TCR expression cassette vectors. Linearized DNA fragments were injected into fertilized C57BL/6 eggs at the University California, San Diego Transgenic Mouse Facility (La Jolla, CA). Pups were genotyped (Supplemental Fig. 1). A single α/β TCR-transgenic founder mouse (NIP) was selected and crossed to B6.SJL mice to generate CD45.1+ NIP mice. All animal experiments were conducted in accordance with approved animal protocols. The GFP-expressing retroviral expression vector pMIG was used. BTBmut Bcl6 retrovirus (BTBmut-RV) was generated by inducing two point mutations in the protein interaction domain that do not affect dimerization (16). RV particles were produced as previously described (9). Cell transfers into host mice were performed as described (9) by i.v. injection via the retro-orbital sinus. Transferred cells were allowed to rest in host mice for 3–5 d before infection or immunization. 5 × 105 transduced Smarta cells were transferred into each mouse for day 3 analysis, and 25 × 103 transduced Smarta cells were transferred into each mouse for day 7 analysis. For protein immunization, 5 × 105 cells were transferred into each mouse. 5 × 103 naive CD4 T cells from NIP TCR-transgenic or retrogenic mice were transferred into each mouse.
Infections and immunizations
LCMV Armstrong stocks were prepared and quantified as previously described (9). Infections were performed by i.p. injection of 0.5–2 × 105 PFU LCMV Armstrong/mouse. gp61-keyhole limpet hemocyanin (KLH) was prepared in alum and injected as described previously (17). A total of 20 μg gp61-KLH was resuspended in alum for bilateral footpad injections.
Flow cytometry
Flow cytometry was done with mAbs against SLAM (CD150; BioLegend) and CD4, CD8, CD44, CD62L, CD25, B220, Fas, and GL7 (all from eBioscience). Stains were done for 30 min at 4°C in PBS supplemented with 0.5% BSA and 0.1% sodium azide, unless specified otherwise. CXCR5 staining was done as described (9, 18). Intracellular staining for Bcl6 was performed with an Alexa Fluor 647–conjugated mAb to Bcl6 (clone K112-91; BD Pharmingen) using the Foxp3 intracellular staining kit buffers and protocol (eBioscience).
Statistical analysis
Statistical tests were performed using Prism 5.0 (GraphPad). The p values were calculated by two-tailed unpaired Student t tests with a 95% confidence interval. Error bars depict the SEM.
Results and Discussion
The Bcl6 BTB domain regulates Tfh cell differentiation
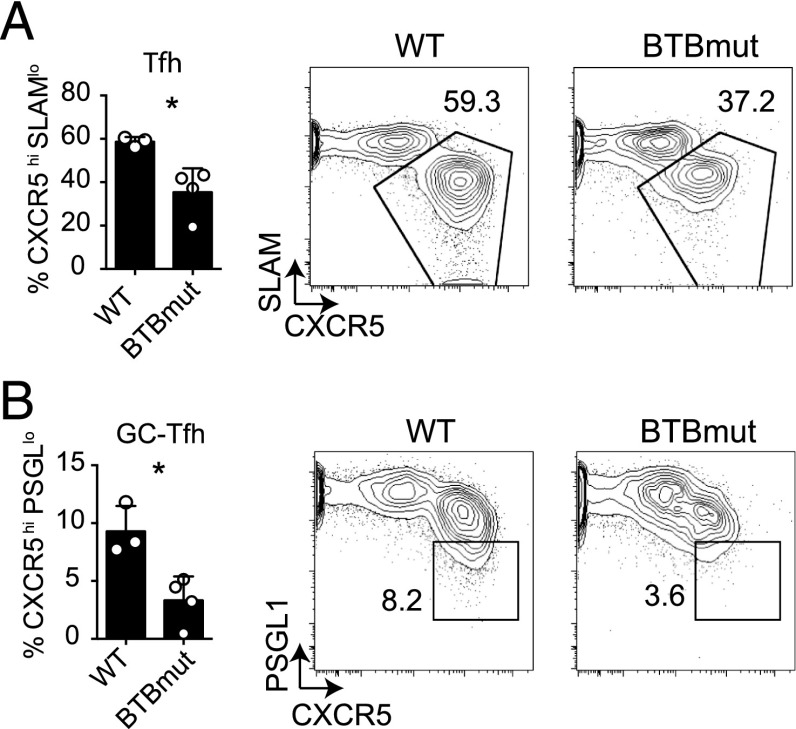
Given that the Bcl6 BTB domain is known to interact with corepressors (such as BCOR) in B cells that are also expressed in CD4 T cells, we sought to determine the role of the Bcl6 BTB domain in Tfh cell differentiation. To this end, we generated N21K and H116A mutations in the Bcl6 BTB domain. These point mutations together were shown to prevent corepressor binding to the BTB domain without affecting the ability of Bcl6 to dimerize (16). Similar levels of Bcl6 protein expression were observed in CD4 T cells transduced with an RV expressing either wild-type (WT) Bcl6 or BTBmut, and the amount of Bcl6 expression was comparable to that of GC-Tfh cells (Fig. 1A). To examine the role of the Bcl6 BTB domain in vivo, we determined whether expression of the Bcl6 BTBmut protein in Bcl6-deficient cells could rescue Tfh cell development in response to acute LCMV infection. LCMV gp-specific Bcl6fl/fl CreCD4 SM CD45.1 cells were transduced with Bcl6 WT, BTBmut, or an empty GFP vector (GFP) and transferred to Bcl6fl/fl CreCD4 hosts. We first examined early Tfh cell development 3 d following an acute LCMV infection. Cells transduced with the empty vector (GFP+) did not differentiate into Tfh cells, confirming the requirement of Bcl6 for Tfh cell differentiation. Ectopic expression of Bcl6 WT was sufficient to rescue Tfh cell development (Fig. 1B). We observed a defect in early Tfh cell differentiation in BTBmut+ SM cells compared with Bcl6-WT+ SM cells (p < 0.0001, Fig. 1B). We then examined Tfh and GC B cell development at 7 d following acute LCMV infection. Again, there was a significant impairment in Tfh cell differentiation in BTBmut+ cells (p < 0.0001, Fig. 1C) compared with WT. This was coupled with an even larger reduction in GC-Tfh cells (p = 0.006, Fig. 1D). GC-Tfh cells are the more polarized Tfh cells that are present in the GCs and are identifiable as CXCR5hiPD1hiPSGL1loBcl6hi cells (1). In parallel, we observed a significant defect in GC B cell development (p = 0.005, Fig. 1E. Thus, Bcl6 BTB domain functions appeared to be necessary for the majority of Tfh differentiation and B cell help.
FIGURE 1.
The Bcl6 BTB domain is necessary for proper Tfh cell differentiation. (A) Similar levels of Bcl6 expression in Bcl6-RV, BTBmut-RV, and GC-Tfh cells. Bcl6 mean fluorescence intensity of Bcl6fl/fl CreCD4 CD4 T cells transduced with either BTBmut-RV or Bcl6-RV compared with ex vivo GC-Tfh cells isolated from mice at 7 d following LCMV infection. (B–E) Bcl6fl/fl CreCD4 SM cells were retrovirally transduced with empty GFP vector, Bcl6 WT, or BTBmut; transferred to Bcl6fl/fl CreCD4 mice; and analyzed at day 3 or 7 following an acute LCMV infection. (B) Early Tfh cell differentiation (CXCR5hiSLAMlo) by transduced Bcl6fl/fl CreCD4 SM cells at day 3. (C) Tfh cell differentiation (CXCR5hiSLAMlo) by transduced Bcl6fl/fl CreCD4 SM cells at day 7. (D) GC-Tfh cell differentiation (CXCR5hiPD1hi) by transduced Bcl6fl/fl CreCD4 SM cells at day 7. (E) GC B cells (PNAhiFashi) at day 7. At least three mice were used for each condition. Experiments were repeated at least three times. **p < 0.01, ****p < 0.0001.
Impaired Tfh differentiation by germline Bcl6BTBMUT CD4 T cells
These results are different from a recent report using mixed bone marrow chimeric mice with Bcl6BTBMUT and Tcrb−/− donor cells, in which no difference in total Tfh cell frequencies was observed upon immunization with SRBCs, although Ag-specific cells were not directly assessed in that study (19). Therefore, in light of those data, it was incumbent upon us to explore the role of the Bcl6 BTB domain in Tfh cell differentiation in multiple contexts, directly examining Ag-specific Tfh cells in each case. Thus, we obtained the germline knock-in Bcl6BTBMUT mice for further studies. CD4 T cells were isolated from Bcl6BTBMUT SM CD45.1+ or WT SM CD45.1+ mice and transferred to Bcl6fl/fl CreCD4 hosts. Ag-specific Tfh and GC-Tfh cell differentiation was examined at 7 d following acute LCMV infection. We observed a significant defect in both Tfh cell (p = 0.017, Fig. 2A) and GC-Tfh cell (p = 0.014, Fig. 2B) differentiation, confirming the requirement for the Bcl6 BTB domain in the generation of Tfh cells.
FIGURE 2.
Impaired Tfh cell differentiation by Bcl6BTBMUT CD4 T cells. Bcl6BTBMUT SM or WT SM cells were transferred to Bcl6fl/fl CreCD4 mice and analyzed 7 d following acute LCMV infection. (A) Tfh (CXCR5hiSLAMlo) Bcl6BTBMUT SM cells. (B) GC-Tfh (CXCR5hiPSGL1lo) Bcl6BTBMUT SM cells. At least three mice were used for each condition. Experiments were repeated at least three times. *p < 0.05.
BTB-dependent Tfh cell differentiation in NIP-transgenic Bcl6BTBMUT CD4 T cells
One possibility is that this observed phenotype may be selective to the SM CD4 T cell response, although SM CD4 T cells have been very informative in numerous contexts (9, 14, 17, 20). To this end, we developed a new LCMV-specific TCR-transgenic mouse. The LCMV nucleoprotein (NP) is the major target of the Ab response during LCMV infection, and the currently used gp-specific SM CD4 T cells do not allow for the study of the immunodominant Ab response, because gp-specific CD4 T cells do not provide help to NP-specific B cells (14). Epitope mapping defined a specific peptide (NP 311–325) of the NP that is efficiently recognized by CD4 T cells during LCMV infection (21, 22). For this reason, NP 311–325 was chosen as a target to generate a novel TCR-transgenic mouse model (NIP, Supplemental Fig. 1). NIP-transgenic CD4 T cells transferred into B6 mice exhibited robust expansion and differentiated into Tfh and GC-Tfh cells following LCMV infection (Supplemental Fig. 1B–E). NIP CD4 T cells transferred into B6 hosts were able to induce robust B cell responses, including the generation of GC B cells (Supplemental Fig. 1F), plasma cells (Supplemental Fig. 1G), and LCMV-specific Abs (Supplemental Fig. 1H).
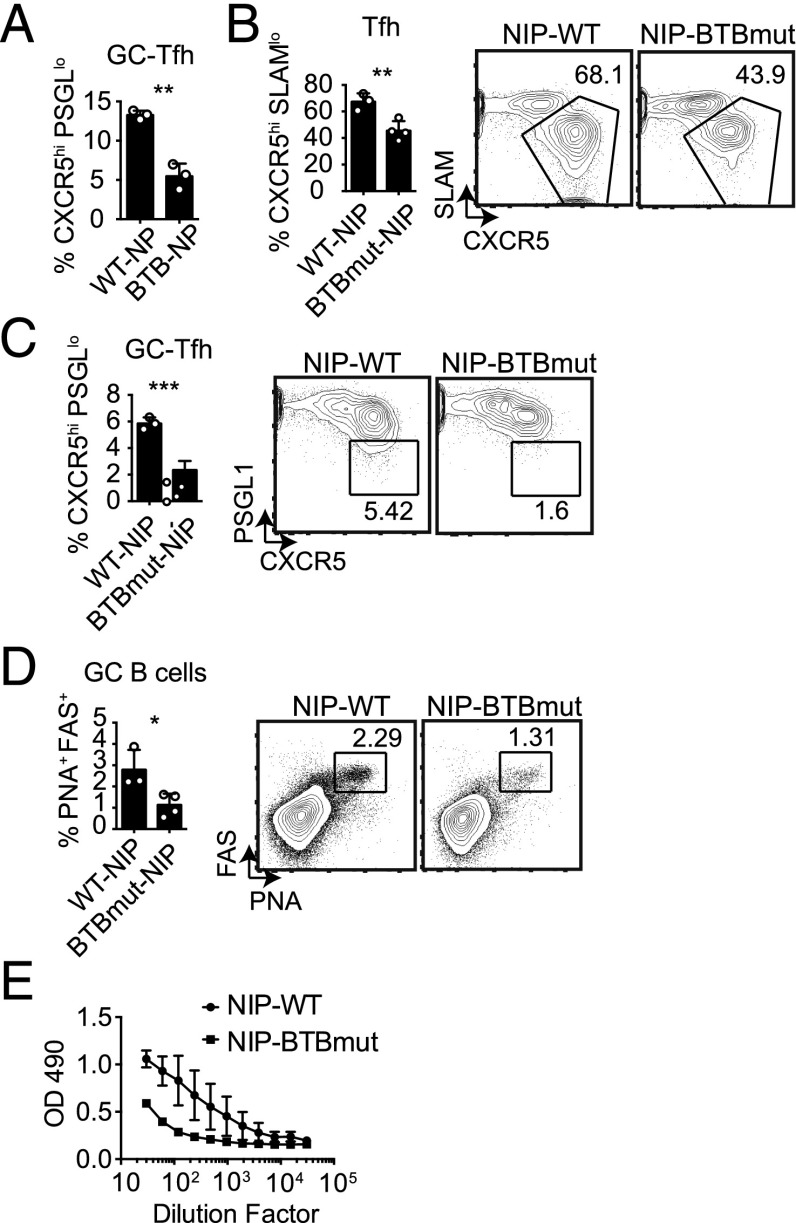
We examined the role of the Bcl6 BTB domain in the context of a retrogenic mouse model in which bone marrow from Bcl6BTBMUT or B6 (WT) mice was transduced with an NP-specific TCRα and TCRβ RV construct (NIP-RV), and the transduced bone marrow was transferred into irradiated WT recipients (23). WT or Bcl6BTBMUT NP-specific naive CD4 T cells were isolated from these mice after reconstitution and transferred to Bcl6fl/fl CreCD4 hosts. The NIP+ WT and NIP+ Bcl6BTBMUT CD4 T cells expanded robustly in response to an acute LCMV infection. At 7 d following LCMV infection, we observed a significant defect in Tfh cell differentiation (Supplemental Fig. 2) and GC-Tfh cell differentiation (Fig. 3A, Supplemental Fig. 2) for Bcl6BTBMUT NIP-RV+ mice compared with WT NIP-RV+ mice (p = 0.01 and p = 0.0014). We then made use of germline NIP TCR-transgenic mice crossed with Bcl6BTBMUT mice. Bcl6BTBMUT NIP or WT NIP CD4 T cells were transferred into Bcl6fl/fl CreCD4 recipients followed by LCMV infection. We observed severe defects in Tfh cell (p = 0.0088, Fig. 3B) and GC-Tfh cell (p = 0.0006, Fig. 3C) differentiation in Bcl6BTBMUT NIP CD4 T cells. The consequence of the substantial loss of GC Tfh cells was a significant decrease in GC B cell development (Fig. 3D) and a reduced anti-LCMV IgG response (Fig. 3E). Taken together, these data demonstrate that the Bcl6 BTB domain is important for Tfh cell differentiation and function in response to at least two Ags.
FIGURE 3.
Bcl6BTBMUT LCMV-specific NIP-transgenic CD4 T cells are defective in Tfh cell differentiation and function. CD4 T cells from Bcl6BTBMUT NIP-TCR+ retrogenic mice were transferred to Bcl6fl/flCreCD4 mice and analyzed at day 7 following acute LCMV infection. (A) GC-Tfh cells (CXCR5hiPSGL1lo) among Bcl6BTBMUT NIP-TCR+ cells. (B–E) Bcl6BTBMUT NIP or WT NIP cells were transferred to Bcl6fl/fl CreCD4 mice and analyzed following acute LCMV infection. (B) Tfh (CXCR5hiSLAMlo) Bcl6BTBMUT NIP cells. (C) GC-Tfh (CXCR5hiPSGL1lo) Bcl6BTBMUT NIP cells. (D) GC B cells (PNAhiFashi). (E) LCMV-specific IgG. At least three mice were used for each condition. Experiments in (B)–(D) were repeated at least three times. *p < 0.05, **p < 0.01, ***p < 0.001.
The Bcl6 BTB domain regulates Tfh cell differentiation and GC B cell development following protein immunization
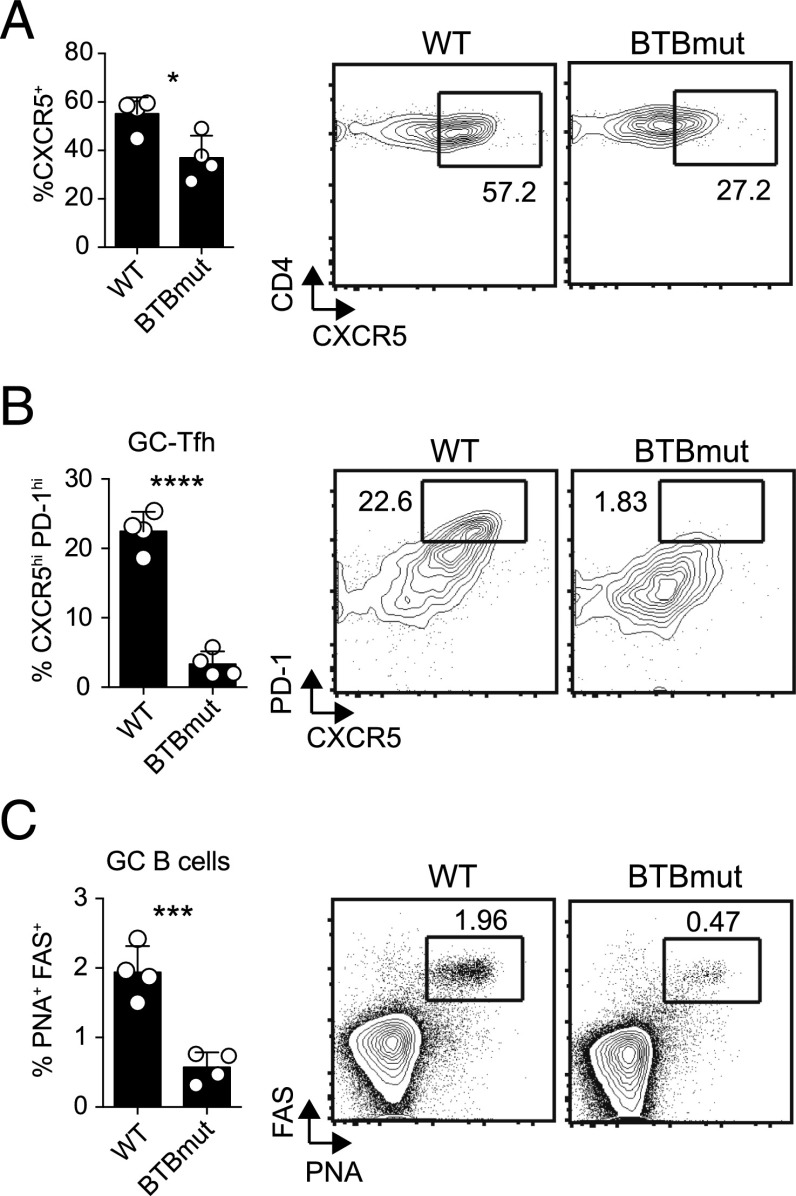
To examine the role of the Bcl6 BTB domain in another experimental context, we used a protein immunization model: KLH conjugated with gp61–80 peptide in alum (17). We transferred Bcl6BTBMUT SM or WT SM CD4 T cells into Bcl6fl/fl CreCD4 hosts, followed by immunization with gp61-KLH in alum. At 10 d following immunization, lymph nodes were harvested and examined for Ag-specific Tfh cell differentiation and function. We observed a significant reduction in CXCR5 expression by Bcl6BTBMUT CD4 T cells (p = 0.0195, Fig. 4A) and a severe defect in GC-Tfh cell differentiation (p < 0.0001, Fig. 4B). Bcl6BTBMUT CD4 T cells were functionally deficient because they were unable to promote GC B cell responses (p = 0.0008, Fig. 4C). We also observed defects in Tfh and GC-Tfh cell differentiation in Bcl6fl/fl CreCD4 mice receiving BTBmut-RV+ SM CD4 T cells compared with Bcl6-WT-RV+ SM CD4 T cells at 10 d following immunization (data not shown). Thus, the Bcl6 BTB domain is needed for optimal Tfh cell differentiation and function in both the context of protein immunizations and an acute viral infection.
FIGURE 4.
Defective Tfh cell differentiation and function in the absence of a functional Bcl6 BTB domain following protein immunization. Bcl6BTBMUT SM or WT SM cells were transferred to Bcl6fl/fl CreCD4 mice and analyzed following gp61-KLH (alum) immunization on day 10. (A) Tfh (CXCR5hiSLAMlo) Bcl6BTBMUT SM cells. (B) GC-Tfh (CXCR5hiPD1hi) Bcl6BTBMUT SM cells. (C) GC B cells (PNAhiFashi). At least three mice were used for each condition. Experiments were repeated at least three times. *p < 0.05, ***p < 0.001, ****p < 0.0001.
Our results demonstrate the importance of the Bcl6 BTB repressor domain in CD4 T cells in the differentiation of Tfh cells and in the ability of Tfh cells to provide help to B cells. A previous study did not observe a Tfh cell defect in Bcl6BTBMUT CD4 T cells (19), possibly because Ag-specific Tfh cells were not analyzed directly or possibly as the result of a confounding negative-feedback Bcl6 autoregulatory mechanism involving cullin3, whereby cullin3 is most likely recruited via the Bcl6 BTB domain (24). Alternatively, the BTB domain of Bcl6 has more prominent roles for Bcl6 activity in some in vivo contexts than others. Bcl6 functions in Tfh cells via binding to thousands of genes, demonstrating that Bcl6 is deeply integrated into Tfh cell biology (25). In this study, Tfh cell defects in the absence of BTB domain function were consistent when assessing Ag-specific Tfh cells using two Ag specificities, as well as in the context of both acute viral infection and protein immunization. The defect in Tfh cell differentiation and function that was observed is likely due to the inability of Bcl6 to recruit corepressors (e.g., SMRT, NCOR, and BCOR), and, thus, repress target genes. Indeed, it was found that BCOR-deficient CD4 T cells are defective for GC-Tfh cell differentiation and B cell help functions (26). Thus, BCOR may be the main corepressor interacting with the Bcl6 BTB domain in Tfh cells. SMRT, NCOR, and BCOR are all expressed in CD4 T cells, and further investigation of the comparative roles of each of these Bcl6 BTB domain–interacting transcription factors is needed. This report highlights the importance of the repressive ability of the Bcl6 BTB domain in Tfh cells for successful GC reactions and robust Ab responses.
Supplementary Material
Acknowledgments
We thank Marc Jenkins, Jessica Yang, Vivian Bardwell, and Noah Tubo for sharing unpublished data; Ari Melnick for sharing Bcl6BTBMUT mice; and Janice White and Philippa Marrack for generating T cell hybridomas for LCMV NP 311–325.
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants R01 AI063107 and U19 AI109976 (to S.C.).
The online version of this article contains supplemental material.
- B6
- C57BL/6J
- Bcl6
- B cell CLL/lymphoma 6
- BTBmut
- BTB domain mutant; BTBmut-RV, BTBmut Bcl6 retrovirus
- GC
- germinal center
- KLH
- keyhole limpet hemocyanin
- LCMV
- lymphocytic choriomeningitis virus
- NP
- nucleoprotein
- RV
- retrovirus
- SM
- Smarta TCR transgenic
- Tfh
- T follicular helper
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueno H., Banchereau J., Vinuesa C. G. 2015. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 16: 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell V. J., Treisman R. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8: 1664–1677. [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu A. M., Sant’Angelo D. B. 2011. The BTB-ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. J. Immunol. 187: 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh K. D., Bardwell V. J. 1998. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17: 2473–2484. [DOI] [PubMed] [Google Scholar]
- 6.Huynh K. D., Fischle W., Verdin E., Bardwell V. J. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14: 1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 7.Dhordain P., Albagli O., Lin R. J., Ansieau S., Quief S., Leutz A., Kerckaert J. P., Evans R. M., Leprince D. 1997. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 94: 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghetu A. F., Corcoran C. M., Cerchietti L., Bardwell V. J., Melnick A., Privé G. G. 2008. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell 29: 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston R. J., Poholek A. C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A. L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroenke M. A., Eto D., Locci M., Cho M., Davidson T., Haddad E. K., Crotty S. 2012. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 188: 3734–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurieva R. I., Chung Y., Martinez G. J., Yang X. O., Tanaka S., Matskevitch T. D., Wang Y.-H., Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325: 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D., Rao S., Tsai L. M., Lee S. K., He Y., Sutcliffe E. L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31: 457–468. [DOI] [PubMed] [Google Scholar]
- 13.Kaji T., Ishige A., Hikida M., Taka J., Hijikata A., Kubo M., Nagashima T., Takahashi Y., Kurosaki T., Okada M., et al. 2012. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 209: 2079–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oxenius A., Bachmann M. F., Zinkernagel R. M., Hengartner H. 1998. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28: 390–400. [DOI] [PubMed] [Google Scholar]
- 15.Huang C., Gonzalez D. G., Cote C. M., Jiang Y., Hatzi K., Teater M., Dai K., Hla T., Haberman A. M., Melnick A. 2014. The BCL6 RD2 domain governs commitment of activated B cells to form germinal centers. Cell Reports 8: 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad K. F., Melnick A., Lax S., Bouchard D., Liu J., Kiang C.-L., Mayer S., Takahashi S., Licht J. D., Privé G. G. 2003. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 12: 1551–1564. [DOI] [PubMed] [Google Scholar]
- 17.Choi Y. S., Yang J. A., Yusuf I., Johnston R. J., Greenbaum J., Peters B., Crotty S. 2013. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 190: 4014–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi Y. S., Kageyama R., Eto D., Escobar T. C., Johnston R. J., Monticelli L., Lao C., Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34: 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C., Hatzi K., Melnick A. 2013. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat. Immunol. 14: 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harker J. A., Lewis G. M., Mack L., Zuniga E. I. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 334: 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow C., Oseroff C., Peters B., Nance-Sotelo C., Sidney J., Buchmeier M., Sette A., Mothé B. R. 2008. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. J. Virol. 82: 11734–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homann D., Lewicki H., Brooks D., Eberlein J., Mallet-Designé V., Teyton L., Oldstone M. B. A. 2007. Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II. Virology 363: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holst J., Szymczak-Workman A. L., Vignali K. M., Burton A. R., Workman C. J., Vignali D. A. A. 2006. Generation of T-cell receptor retrogenic mice. Nat. Protoc. 1: 406–417. [DOI] [PubMed] [Google Scholar]
- 24.Mathew R., Mao A.-P., Chiang A. H., Bertozzi-Villa C., Bunker J. J., Scanlon S. T., McDonald B. D., Constantinides M. G., Hollister K., Singer J. D., et al. 2014. A negative feedback loop mediated by the Bcl6-cullin 3 complex limits Tfh cell differentiation. J. Exp. Med. 211: 1137–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzi K., Nance J. P., Kroenke M. A., Bothwell M., Haddad E. K., Melnick A., Crotty S. 2015. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 212: 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J. A., Tubo N. J., Gearhart M. D., Bardwell V. J., Jenkins M. K. 2015. BCL6-interacting corepressor contributes to germinal center T follicular helper cell formation and B cell helper function. J. Immunol. 194: 5604–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.