The ref5-1 mutant and its suppressors reveal that an intermediate in indole glucosinolate biosynthesis impacts the early steps of phenylpropanoid metabolism, an interaction that requires the Mediator complex.
Abstract
Plants produce an array of metabolites (including lignin monomers and soluble UV-protective metabolites) from phenylalanine through the phenylpropanoid biosynthetic pathway. A subset of plants, including many related to Arabidopsis thaliana, synthesizes glucosinolates, nitrogen- and sulfur-containing secondary metabolites that serve as components of a plant defense system that deters herbivores and pathogens. Here, we report that the Arabidopsis thaliana reduced epidermal fluorescence5 (ref5-1) mutant, identified in a screen for plants with defects in soluble phenylpropanoid accumulation, has a missense mutation in CYP83B1 and displays defects in glucosinolate biosynthesis and in phenylpropanoid accumulation. CYP79B2 and CYP79B3 are responsible for the production of the CYP83B1 substrate indole-3-acetaldoxime (IAOx), and we found that the phenylpropanoid content of cyp79b2 cyp79b3 and ref5-1 cyp79b2 cyp79b3 plants is increased compared with the wild type. These data suggest that levels of IAOx or a subsequent metabolite negatively influence phenylpropanoid accumulation in ref5 and more importantly that this crosstalk is relevant in the wild type. Additional biochemical and genetic evidence indicates that this inhibition impacts the early steps of the phenylpropanoid biosynthetic pathway and restoration of phenylpropanoid accumulation in a ref5-1 med5a/b triple mutant suggests that the function of the Mediator complex is required for the crosstalk.
INTRODUCTION
Plants produce a large number of low molecular weight compounds, such as terpenes, phenylpropanoids, alkaloids, and glucosinolates. Frequently called secondary or specialized metabolites, these compounds often play roles in responses to abiotic and biotic stresses. Phenylpropanoids are a group of secondary metabolites synthesized from the amino acid phenylalanine (Bonawitz and Chapple, 2010; Vanholme et al., 2012, 2013). One of the major phenylpropanoid pathway end-products is lignin, a heteropolymer important for the mechanical strength and hydrophobicity of the plant secondary cell wall. In addition to lignin, the pathway generates a wide range of soluble metabolites, such as flavonoids and hydroxycinnamate derivatives. The hydroxycinnamate ester sinapoylmalate is an example of one of these compounds that plays an important role in UV protection in Arabidopsis thaliana (Landry et al., 1995).
Glucosinolates, also known as mustard oil glucosides, are nitrogen- and sulfur-containing compounds found in the order Capparales, including Arabidopsis and oilseed rape (Brassica napus). In addition to their roles as defense molecules (Halkier and Gershenzon, 2006), glucosinolates are of great interest to humans because of the flavors they impart to various condiments and for their anticancer effects (Fahey et al., 2001; Mithen et al., 2003; Traka and Mithen, 2009). Over 100 glucosinolates are found in plants, and the Arabidopsis Columbia-0 (Col-0) ecotype alone contains ∼30 different types (Brown et al., 2003). Depending on their precursor amino acids, glucosinolates can be categorized into indole glucosinolates derived from Trp, aliphatic glucosinolates from Ala, Leu, Ile, Val, or Met, and aromatic glucosinolates from Phe and Tyr (Halkier and Gershenzon, 2006).
The content and composition of glucosinolates varies according to species, developmental stage, organ, and tissue (Brown et al., 2003). For example, indol-3-yl-methyl glucosinolate is predominant in Arabidopsis leaves, whereas 4-methoxyindol-3-yl-methyl glucosinolate and 1-methoxyindol-3-yl-methyl glucosinolate are abundant in Arabidopsis roots (Brown et al., 2003). The synthesis of indole glucosinolates starts with the conversion of tryptophan to indole-3-acetaldoxime (IAOx) by two cytochrome P450-dependent monooxygenases (P450s), CYP79B2 and CYP79B3 (Hull et al., 2000; Mikkelsen et al., 2000, 2004; Bak et al., 2001). Subsequently CYP83B1 catalyzes the conversion of IAOx into 1-aci-nitro-2-indolyl-ethane and then multiple further modifications result in the production of the various indole glucosinolates (Pfalz et al., 2009, 2011). IAOx is also a precursor for indole-3-acetic acid (IAA), a phytohormone, and camalexin, an important phytoalexin in Arabidopsis (Glawischnig et al., 2004; Nafisi et al., 2007). A recent study showed that indole-3-carbaldehyde and indole-3-carboxylic acid derivatives are made from IAOx as well (Böttcher et al., 2014). The conversion of IAOx to camalexin is thought to occur only under conditions of pathogen infection because both camalexin content and the expression of camalexin biosynthetic genes are low under normal growth conditions (Schuhegger et al., 2006; Nafisi et al., 2007).
Although glucosinolates and phenylpropanoids are synthesized through distinct biosynthetic pathways and have unique functions, a study of the ref2 mutant showed that there is crosstalk between the two pathways (Hemm et al., 2003). The ref2 mutant was originally identified from a forward genetic screen for mutants having a reduced epidermal fluorescence (ref) phenotype (Ruegger and Chapple, 2001). Wild-type Arabidopsis plants appear blue under UV light due to the presence of sinapoylmalate, a UV fluorescent secondary metabolite that is accumulated in the adaxial leaf epidermis. Sinapoylmalate is synthesized from phenylalanine via the phenylpropanoid pathway, and several biosynthetic enzymes and regulatory components with key roles in this biosynthetic pathway have been identified through the analysis of ref mutants (Ruegger and Chapple, 2001; Franke et al., 2002; Stout et al., 2008; Schilmiller et al., 2009). Surprisingly, despite the fact that it was first isolated based upon its phenylpropanoid metabolism defect, the identification of REF2 showed that the mutant is defective in the gene encoding CYP83A1, the enzyme that catalyzes the aldoxime-metabolizing step in aliphatic glucosinolate biosynthesis (Hemm et al., 2003). CYP83B1, a paralog of CYP83A1, which encodes the enzyme catalyzing the conversion of IAOx in indole glucosinolate biosynthesis, was originally isolated following a screen for high auxin phenotypes that identified the superroot2 (sur2) mutant (Delarue et al., 1998; Barlier et al., 2000). A global analysis of the sur2 mutant by the use of a combined transcriptomic and metabolomic approach revealed complex changes in pathways of secondary metabolism, hormone metabolism, and stress responses (Morant et al., 2010).
Here, we report the ref5-1 mutant as an allele of CYP83B1 and describe biochemical and genetic analyses that reveal crosstalk between indole glucosinolate biosynthesis and phenylpropanoid metabolism in both ref5 and wild-type plants. Our data suggest that alteration in the levels of IAOx or a subsequent metabolite affects early steps of the phenylpropanoid biosynthetic pathway through a mechanism that appears to require the Arabidopsis Mediator subunits MED5a and MED5b.
RESULTS
REF5 Encodes the Cytochrome P450 Monooxygenase CYP83B1
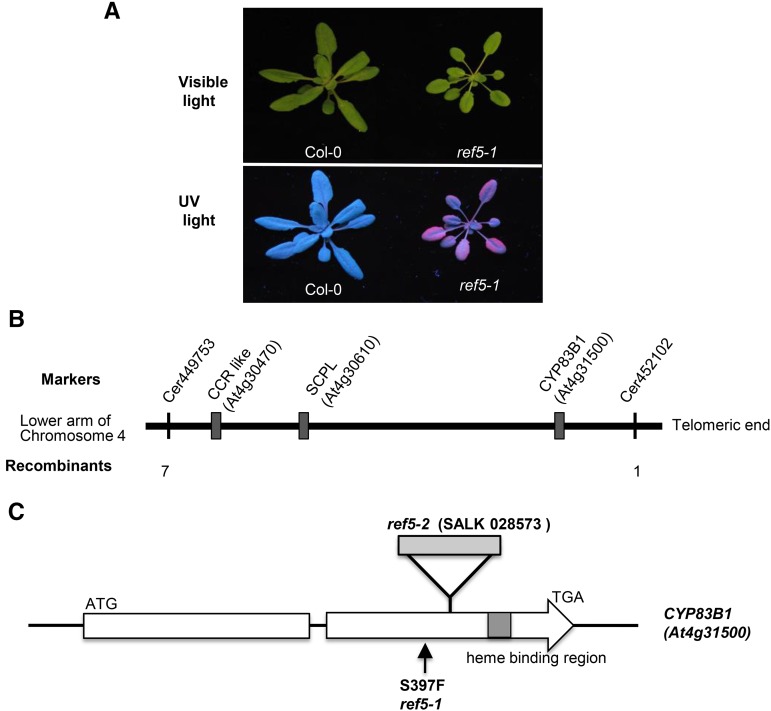
The ref5-1 mutant was isolated from a UV screen of pot-grown Arabidopsis plants evaluated at the rosette stage (Figure 1A; Ruegger and Chapple, 2001). Positional cloning was used to map the REF5 locus to a six-BAC interval at the bottom of chromosome 4 defined by markers Cer449753 and Cer452102. Three candidate genes, At4g30470, At4g30610, and At4g31500, which encode a cinnamoyl-CoA reductase-like protein, a serine carboxypeptidase-like protein, and a P450 (CYP83B1), respectively, were identified as REF5 candidates within this interval based upon their potential involvement in phenylpropanoid metabolism. Because there was only one recombination between the CYP83B1 locus and the marker Cer452102, CYP83B1 was pursued further as the putative REF5 gene (Figure 1B). Sequencing results showed that ref5-1 had a G-to-A missense mutation (TGTGTCACGAAAAACCGCCCA), which converts Ser-397 to Phe, 34 amino acids upstream of the heme binding motif (Figure 1C). It has already been shown that CYP83B1 catalyzes the conversion of IAOx to 1-aci-nitro-2-indolyl-ethane, which is subsequently converted to an S-alkylthiohydroximate in the presence of a thiol donor (Bak et al., 2001; Hansen et al., 2001) and that mutation of its paralog, CYP83A1, leads to a ref phenotype (Hemm et al., 2003).
Figure 1.
The Arabidopsis ref5-1 Mutant Is Defective in CYP83B1.
(A) Representative photographs of wild-type (Col-0) and ref5-1 plants under white light or UV light. Plants were compared 3 weeks after planting.
(B) Map-based cloning of the REF5 gene. The REF5 locus was mapped to a 6-BAC interval at the bottom of chromosome 4 defined by markers Cer449753 and Cer452102 (vertical bars). The annotation of three candidate genes within this interval, a cinnamoyl-CoA reductase-like protein (CCR like; At4g30470), a serine carboxypeptidase II-like protein (SCPL; At4g30610), and a cytochrome P450 monooxygenase 83B1 (CYP83B1; At4g31500), suggested that they might be involved in phenylpropanoid synthesis.
(C) Gene organization of REF5 and schematic representation of ref5-1 and ref5-2 mutants. White box indicates exons, gray box in second exon indicates the heme binding region, and the intervening line denotes an intron. The location of the T-DNA insertion in ref5-2 (SALK_028573) is shown above the genomic structure. The arrow indicates mutation position in ref5-1.
To evaluate whether REF5 encodes CYP83B1, we identified corresponding insertion mutants. SALK_028573 has a T-DNA insertion in the second exon of CYP83B1 and, when crossed to ref5-1, did not complement its biochemical or morphological phenotypes (Figures 1C, 2, and 3). As a result, SALK_028573 was designated ref5-2. Various other forward genetic screens have identified cyp83b1 alleles as summarized in Table 1, and the analysis of these mutants has elucidated the function of CYP83B1 as well as its regulation. For further study, we used the two null mutants ref5-2 and sur2-1, together with ref5-1.
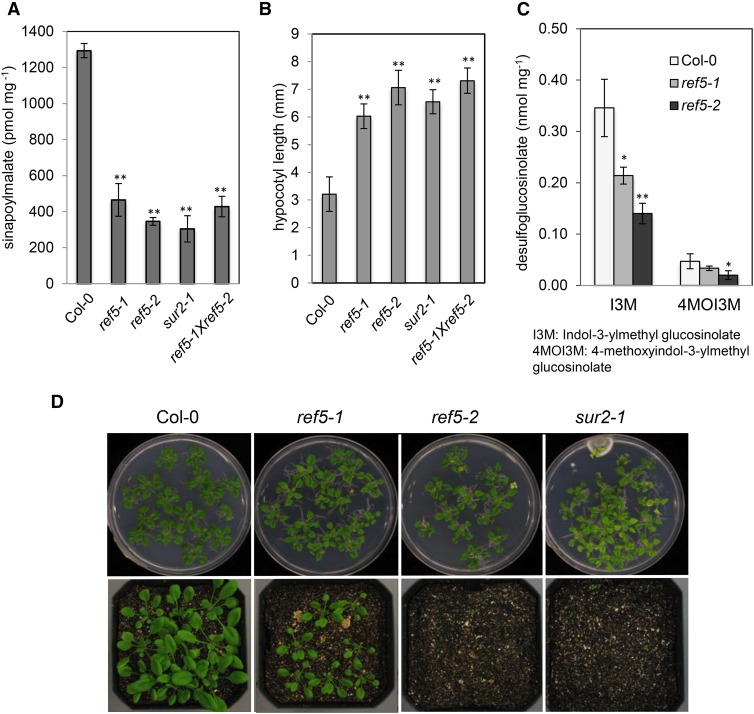
Figure 2.
The ref5-1 and ref5-2 Mutants Are Defective in Indole Glucosinolate Biosynthesis and Phenylpropanoid Biosynthesis.
(A) Sinapoylmalate content in wild-type, ref5 mutant, and ref5-1 × ref5-2 F1 plants. Data represent mean ± sd (n = 4).
(B) Hypocotyl length of 7-d-old wild-type, ref5 mutant, and ref5-1 × ref5-2 F1 plants. Seedlings were grown on MS plates for 7 d. Data represent mean ± sd (n = 10).
(C) Quantification of desulfoglucosinolate content in rosette leaves of ref5-1 and ref5-2 compared with the wild type. Glucosinolates are identified as follows: I3M, indol-3-methyl-glucosinolate; 4MOI3M, 4-methoxyindol-3-ylmethyl-glucosinolate. Data represent mean ± sd (n = 4). * and ** indicate P < 0.05 and P < 0.01 by a two-tailed Student’s t test compared with the wild type, respectively.
(D) Three-week-old ref5 mutants grown on MS plates or directly planted on soil compared with the wild type. When seeds were directly planted on soil, ref5-1 mutants display a less severe growth defect than ref5-2 and sur2-1.
Figure 3.
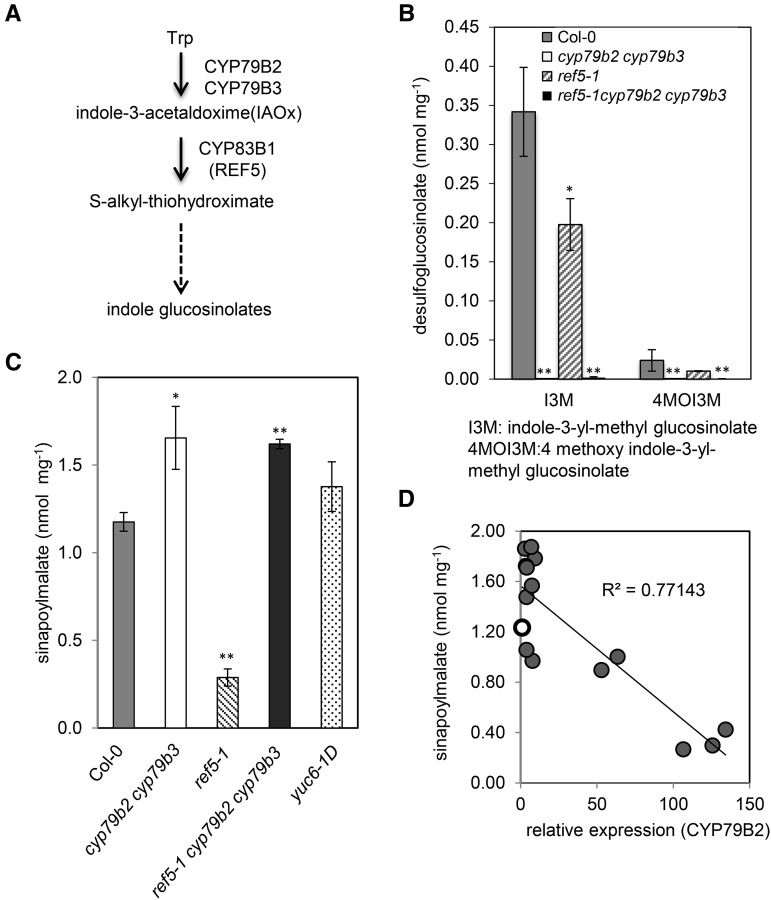
Phenylpropanoid Accumulation Is Limited by IAOx or an Aldoxime Derivative.
(A) Scheme of the indole glucosinolate biosynthesis pathway. The oxidation of Trp to IAOx is catalyzed by CYP79B2 and CYP79B3. CYP83B1/REF5 subsequently oxidizes IAOx to a product that spontaneously reacts to produce an S-alkylthiohydroximate. Further modifications, such as glucosylation and sulfation, give rise to the final indole glucosinolates.
(B) Desulfoglucosinolate content measured from rosette leaves of 3-week-old plants. Data represent mean ± sd (n = 4).
(C) Quantification by HPLC of sinapoylmalate in leaves of the wild type, ref5-1, cyp79b2 cyp79b3 double mutant, ref5-1 cyp79b2 cyp79b3 triple mutant, and yuc6-1D, an auxin accumulation mutant. Data represent mean ± sd (n = 5). * and ** indicate P < 0.05 and P < 0.01 compared with the wild type, respectively. * and ** indicate P < 0.05 and P < 0.01 by a two-tailed Student’s t test compared with the wild type, respectively.
(D) Impact of overexpression of CYP79B2 on levels of sinapoylmalate. Sinapoylmalate content in wild-type (open circle) and transgenic plants overexpressing CYP79B2 (closed circle) is shown.
Table 1. cyp83b1 Mutants Identified from Forward Genetic Screens.
| Mutant Name (Ecotype) | Mutation | Phenotype (Mutant Screen) | Reference |
|---|---|---|---|
| atr4-1 and atr4-2 (Col-0) | EMS (second mutagenesis in trp1-100) | Altered anthranilate synthase feedback inhibition by tryptophan, lesion-mimic | Smolen and Bender (2002) |
| gul1/sur2-7 (Ws-2) | EMS | Elongated hypocotyl when grown in 100 nm brassinazole | Maharjan et al. (2014) |
| red1 (No-0) | EMS (second mutagenesis in phyB overexpressor) | Elongated hypocotyl under continuous red light | Wagner et al. (1997) |
| Hoecker et al. (2004 | |||
| ref5-1 (Col-0) | EMS | Reduced epidermal fluorescence | This study |
| rnt1 (Ws) | T-DNA insertion (insertion in first exon) | Elongated hypocotyl and epinastic cotyledons | Winkler et al. (1998) |
| Bak and Feyereisen (2001) | |||
| sur2 (Ws) | T-DNA insertion | Adventitious roots, epinastic cotyledons | Delarue et al. (1998) |
| sur2 (Col-0) | Transposable element (En-1) insertion | Adventitious roots | Barlier et al. (2000) |
Ws, Wassilewskija.
The Defects in REF5/CYP83B1 Result in Alteration in Sinapoylmalate and Indole Glucosinolate Content
The ref phenotype of ref5-1 suggests that it is affected in leaf sinapoylmalate accumulation. HPLC analysis revealed that all cyp83b1 mutants as well as F1 plants of ref5-1 crossed with ref5-2 had reduced sinapoylmalate content compared with the wild type (Figure 2A). In cyp83b1 mutants, the accumulated IAOx is converted to IAA, which in turn causes high auxin phenotypes, such as adventitious rooting and long hypocotyls (Delarue et al., 1998; Barlier et al., 2000). Consistent with this and as shown in Figure 2B, ref5-1, ref5-2, and ref5-1 ref5-2 trans-heterozygotes have longer hypocotyls than the wild type. As we expected from the known role of CYP83B1 in indole glucosinolate biosynthesis, ref5-1 and ref5-2 leaves contain reduced levels of indole glucosinolates, such as indol-3-yl-methyl glucosinolate and 4-methoxyindol-3-yl-methyl glucosinolate (Figure 2C). It is notable that the reduction of indole glucosinolate content in the T-DNA insertion mutant ref5-2 was stronger than in the plants carrying the missense allele ref5-1. The sur2-1 and ref5-2 mutants showed strong growth defects on soil but grew substantially better on Murashige and Skoog (MS) media plates (Figure 2D); ref5-1 grew well under all conditions. Together, these data indicate that ref5-1 is a weak allele of CYP83B1.
To determine whether the ref5 mutation had an impact on lignin deposition like that observed in the ref2 mutant, lignin content and monomer composition were determined by both the Klason and the derivatization followed by reductive cleavage (DFRC) methods (Kaar and Brink, 1991; Lu and Ralph, 1997). Unlike the case with ref2 in which lignin syringyl monomer content is reduced, lignin content (wild type, 16.6% ± 0.3% dry weight; ref5-1, 17.1% ± 0.3% dry weight) and composition (Table 2) were not significantly affected.
Table 2. DFRC Lignin Monomer Composition and Klason Lignin Content of ref5 Mutants and Wild-Type Cell Walls (n = 3).
| Total DFRC (µmol g−1) | H-Lignin (µmol g−1) | G-Lignin (µmol g−1) | S-Lignin (µmol g−1) | Mol % G | Mol % S | |
|---|---|---|---|---|---|---|
| Col-0 | 64.2 ± 6.8 | 1.6 ± 0.1 | 42.9 ± 4.7 | 19.8 ± 2.2 | 66.8 ± 0.6 | 30.8 ± 0.7 |
| ref5-1 | 51.5 ± 0.1 | 1.1 ± 0.1 | 36.9 ± 0.3 | 13.5 ± 0.4 | 71.7 ± 0.6 | 26.3 ± 0.7 |
| ref5-2 | 50.8 ± 1.1 | 1.0 ± 0.1 | 35.7 ± 1.5 | 13.9 ± 0.5 | 70.5 ± 1.5 | 27.5 ± 1.6 |
Values shown are mean ± se. H, p-hydroxyphenyl; G, guaiacyl; S, syringyl.
The Accumulation of IAOx or an Aldoxime Derivative Perturbs Phenylpropanoid Biosynthesis
As mentioned above, both CYP79B2 and CYP79B3 function to produce the CYP83B1 substrate IAOx from tryptophan, and it has been shown that IAOx and indole glucosinolates are below the limits of detection in a cyp79b2 cyp79b3 double mutant, suggesting that no other enzymes are involved in IAOx synthesis (Figure 3A; Zhao et al., 2002; Sugawara et al., 2009). Consistent with previous reports, the indole glucosinolate content of cyp79b2 cyp79b3 and ref5-1 cyp79b2 cyp79b3 plants was at or below the limits of detection (Figure 3B). The ref5-1 mutant contained only reduced levels of indole glucosinolates, presumably because it is a weak allele and because of the partially redundant activity of the CYP83B1 paralog, CYP83A1 (REF2). Although ref5 mutants had decreased sinapoylmalate content as mentioned previously (Figure 2A), sinapoylmalate levels were instead increased in the cyp79b2 cyp79b3 double mutant and the ref5-1 cyp79b2 cyp79b3 triple mutant (Figure 3C). This result indicates that the perturbation of phenylpropanoids in ref5 mutants is not due to a deficiency in indole glucosinolates but is instead the result of the accumulation of IAOx or its derivatives. Furthermore, it shows that the crosstalk between the glucosinolate and phenylpropanoid pathways is relevant in wild-type plants and does not occur only in a mutant context. Thus, even in the wild type, cellular levels of IAOx or its derivatives impact the level of phenylpropanoids accumulated.
It is clear that in Arabidopsis IAOx is a precursor for IAA because cyp79b2 cyp79b3 mutants contain reduced levels of IAA, whereas CYP79B2 overexpression plants and cyp83b1 mutants have an increased IAA content (Delarue et al., 1998; Barlier et al., 2000; Zhao et al., 2002; Sugawara et al., 2009; Maharjan et al., 2014). To test whether the increased IAA content in ref5 mutants might be what influences phenylpropanoid metabolism, we examined sinapoylmalate content in the yuc6-1D mutant, a mutant that exhibits high auxin phenotypes due to the activation of an auxin biosynthesis enzyme that functions independently of the IAOx pathway (Kim et al., 2007; Sugawara et al., 2009; Mashiguchi et al., 2011; Ross et al., 2011; Stepanova et al., 2011; Won et al., 2011). As shown in Figure 3C, yuc6-1D mutants contain wild-type levels of sinapoylmalate, suggesting that the IAOx-dependent perturbation of phenylpropanoids in ref5 is not due to elevated levels of IAA.
To independently evaluate the link between IAOx and phenylpropanoid metabolism, CYP79B2 overexpression lines were generated by transforming a cauliflower mosaic virus 35S promoter-driven CYP79B2 construct into the wild type. CYP79B2 overexpression plants displayed characteristic high auxin phenotypes such as elongated hypocotyls and narrow downward-curled leaves as previously reported (Zhao et al., 2002). As shown in Figure 3D, transgenic plants expressing higher levels of CYP79B2 transcripts contained less sinapoylmalate in their leaves. The negative correlation between CYP79B2 expression and sinapoylmalate content confirms that the phenylpropanoid defect in ref5 mutants is not dependent on the CYP83B1 mutation per se, but instead on the accumulation of IAOx or a derivative thereof.
The ref2 ref5 Double Mutant Displays Synergistic Phenotypes
The aliphatic oximes derived from chain-elongated homologs of methionine are efficiently metabolized by CYP83A1/REF2, whereas CYP83B1/REF5 metabolizes these substrates with very low efficiency. In contrast, CYP83B1 has a higher affinity for aromatic oximes, particularly in the case of IAOx, where there is a 50-fold difference in Km values between CYP83A1 and CYP83B1 (Naur et al., 2003). Because CYP83A1/REF2 and CYP83B1/REF5 have similar functions in glucosinolate biosynthesis and both mutants show phenylpropanoid defect phenotypes, we tested the hypothesis that the ref2 ref5 double mutants would exhibit additive phenylpropanoid alteration phenotypes.
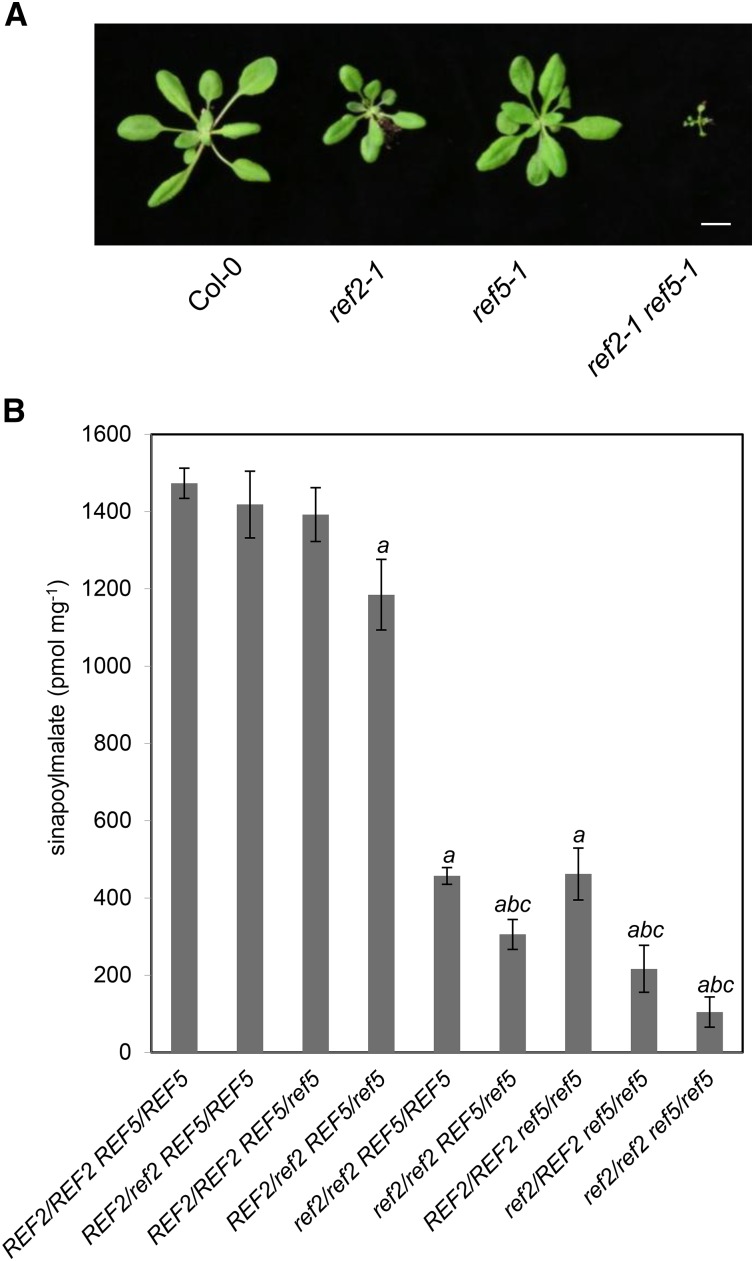
As shown in Figure 4A, the ref2 ref5 double mutant grows more slowly than the wild type and the single mutants and ultimately fails to bolt and flower. To determine whether the double mutant has a more extreme phenylpropanoid metabolic phenotype, sinapoylmalate levels in the segregating F2 population from a ref2 × ref5 cross were analyzed. Both ref2 and ref5 single mutants showed ∼70% reduction in wild-type sinapoylmalate content but ref2 ref5 double mutants exhibited a 96% reduction (Figure 4B). A further reduction in sinapoylmalate content was observed in ref2/ref2 REF5/ref5 and ref2/REF2 ref5/ref5 plants compared with either of the single mutants, indicating that in the context of homozygous loss-of-function alleles of one aldoxime-oxidizing P450, wild-type alleles of the other P450 become haploinsufficient. It is also notable that ref2/REF2 ref/5REF5 trans-heterozygous plants contain less sinapoylmalate than the wild type even though both ref2 and ref5 are recessive (Figure 4B). These data suggest that in ref2/REF2 ref5/REF5 plants, one nonfunctional allele of each P450 leads to the accumulation of sufficient aldoxime(s) that phenylpropanoid content is affected.
Figure 4.
The ref2-1 ref5-1 Double Mutants Display Severe Growth Defects and a Reduction of Sinapoylmalate Contents.
(A) A representative ref2-1 ref5-1 double mutant compared with Col-0, ref2-1, and ref5-1. Four-week-old wild-type, ref2-1, ref5-1, and ref2-1 ref5-1 plants were grown under long-day conditions (16 h light/8 h dark). Bar = 1 cm.
(B) Quantification of sinapoylmalate content in leaves of genotyped members of a ref2-1 × ref5-1 F2 segregating population. Four-week-old F2 individual lines were analyzed for sinapoylmalate content and genotyped for the ref2-1 and ref5-1 mutations. Data represent mean ± sd (n = 4). a, b, and c indicate P < 0.05 compared with the wild type, ref2-1, and ref5-1, respectively, by a Student’s t test.
O-Methyltransferases Are Not the Target for IAOx-Dependent Phenylpropanoid Pathway Inhibition
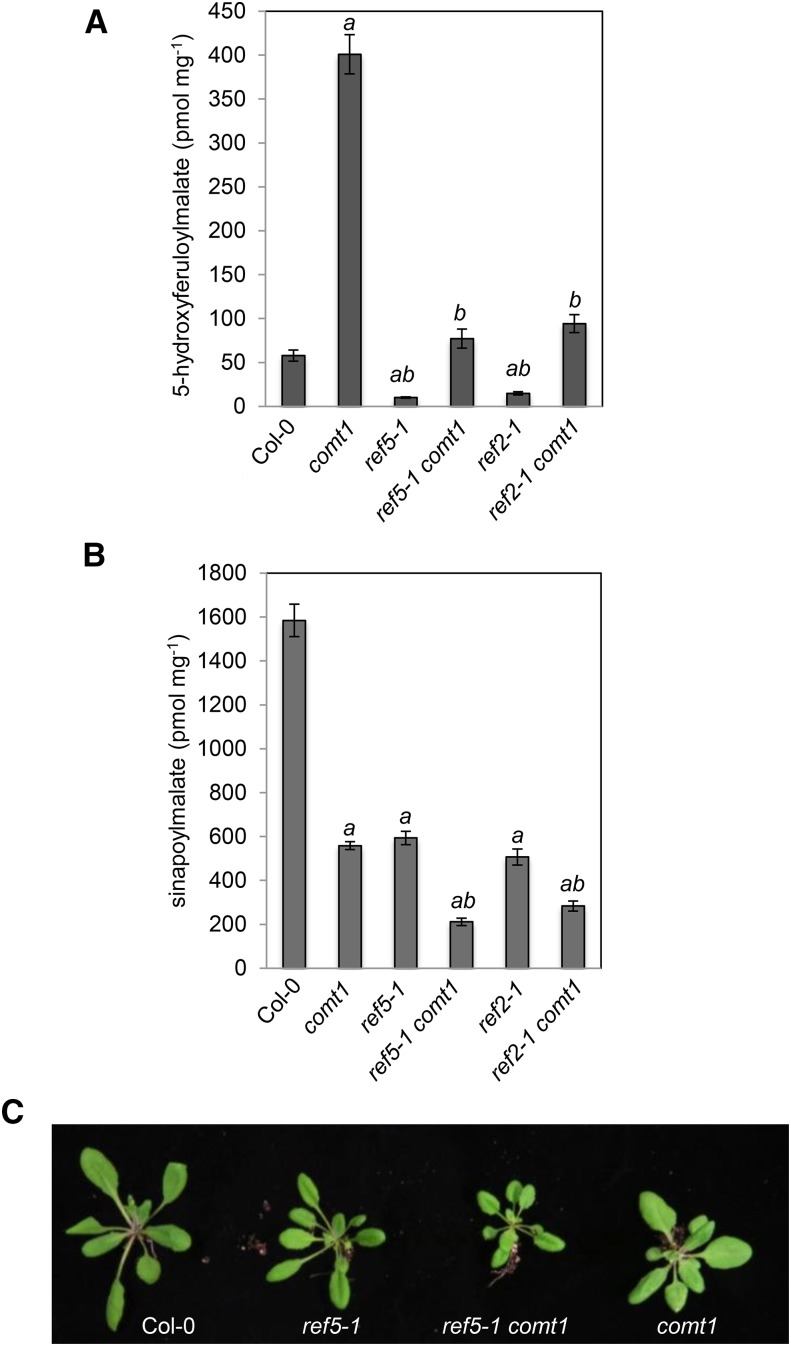
It was proposed that the inhibition of caffeic acid O-methyltransferase (COMT) activity by methylthioalkyl aldoximes in ref2 leaves may account for the ref2 phenylpropanoid phenotype (Hemm et al., 2003). It has recently been shown that the Arabidopsis COMT null mutant, comt1, accumulates 5-hydroxy-feruloylmalate (5-OH-feruloylmalate), which is found only at very low levels in the wild type (Goujon et al., 2003; Do et al., 2007), suggesting that if the aldoximes that accumulate in ref2 and ref5 inhibit COMT, these mutants should also accumulate 5-OH-feruloylmalate. Instead, we found that its levels in ref5-1 and ref2-1 mutants were even lower than in the wild type (Figure 5A). Furthermore, the content of both 5-OH-feruloylmalate and sinapoylmalate in ref5-1 comt1 and ref2-1 comt1 double mutants was lower than in the comt1 single mutant (Figures 5A and 5B), indicating that COMT is not the site of phenylpropanoid pathway inhibition in ref2 and ref5 mutants.
Figure 5.
Analysis of Soluble Metabolites in ref5-1 comt1 and ref2-1 comt1 Double Mutants.
(A) and (B) The level of 5-OH-feruloylmalate (A) and sinapoylmalate (B) in the wild type, ref5-1, comt1, ref5-1 comt1, ref2-1, and ref2-1 comt1. Data represent mean ± sd (n = 5). a and b indicate P < 0.01 compared with the wild type and comt1, respectively, from a Student’s t test.
(C) Morphology of 4-week-old ref5-1 comt1 plants compared with the respective single mutants.
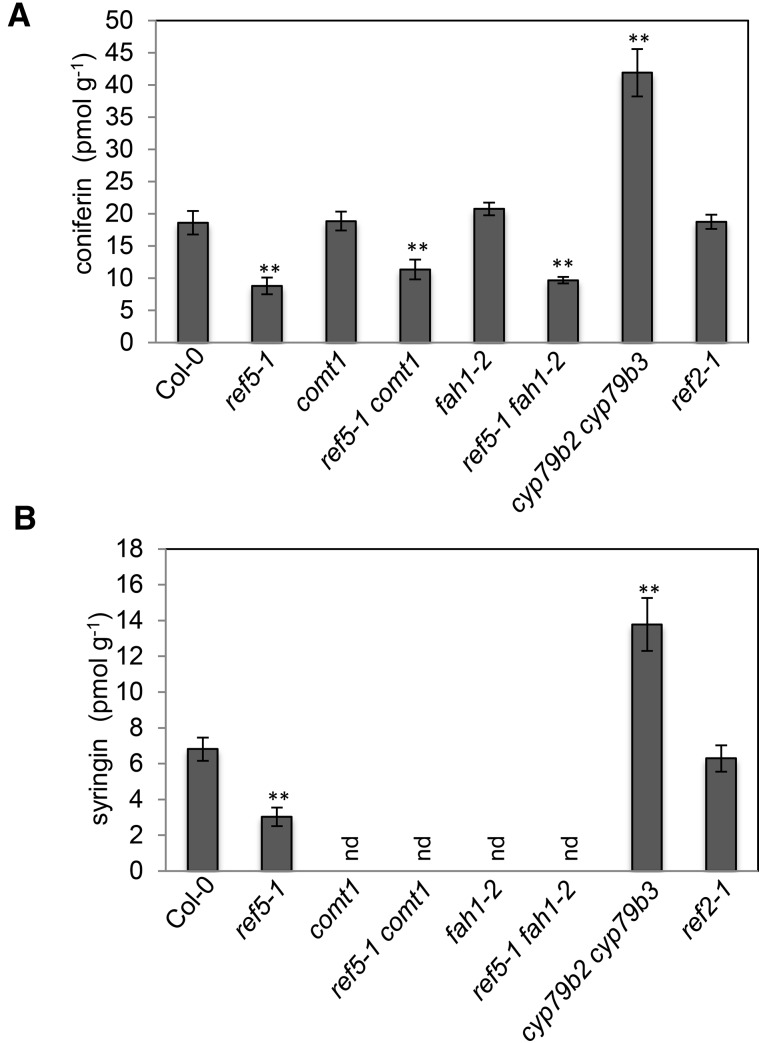
Knowing that indole glucosinolate biosynthesis is active in Arabidopsis roots (Brown et al., 2003), we extended our analyses to light-grown Arabidopsis seedling roots, which are known to accumulate coniferin and syringin, the 4-O-glucosides of the monolignols coniferyl alcohol and sinapyl alcohol, respectively (Hemm et al., 2004). HPLC analysis revealed that coniferin and syringin were reduced by 50% in ref5-1 roots but increased in the cyp79b2 cyp79b3 double mutant (Figures 6A and 6B). In comt1 and the ferulate 5-hydroxylase (F5H)-deficient fah1-2 mutant (Meyer et al., 1996), coniferin content was similar to the wild type, but ref5-1 comt1 and ref5-1 fah1-2 double mutants contained reduced levels, suggesting that the inhibition occurs earlier in the pathway (Figure 6A). It is notable that syringin was absent in comt1 roots, being replaced by a major, earlier eluting compound that is presumably 5-hydroxyconiferin, indicating that COMT is required for syringin biosynthesis (Figure 6B).
Figure 6.
The Phenylpropanoid Content of ref5 Mutant Roots Is Reduced.
Coniferin (A) and syringin (B) content in seedling roots as determined by HPLC. Data represent mean ± sd (n = 3). Syringin content in comt1, fah1-2, ref5-1 comt1, and ref5-1 fah1-2 mutants was below the limit of detection. nd, not detected. Asterisks indicate P < 0.01 by a two-tailed Student’s t test compared with the wild type.
It was previously suggested that COMT1 and caffeoyl-CoA O-methyltransferase (CCoAOMT1) act redundantly to methylate the 3-hydroxy position of the phenolic ring of monolignols (Do et al., 2007). The comt1 ccoaomt1 double mutants do not develop further than the seedling stage and the production of soluble metabolites such as 5-OH-feruloylmalate and sinapoylmalate is significantly reduced (Do et al., 2007). We reasoned that if CCoAOMT1 activity was affected in ref5 mutants, a ref5-1 comt1 double mutant should contain less coniferin than ref5-1 and might show the severe growth defects observed in comt1 ccoaomt1 double mutants. In contrast, the ref5-1 comt1 double mutant grew normally and the level of coniferin was similar to ref5-1, suggesting that the inhibition occurs upstream of COMT, F5H, and CCoAOMT (Figures 5C and 6A).
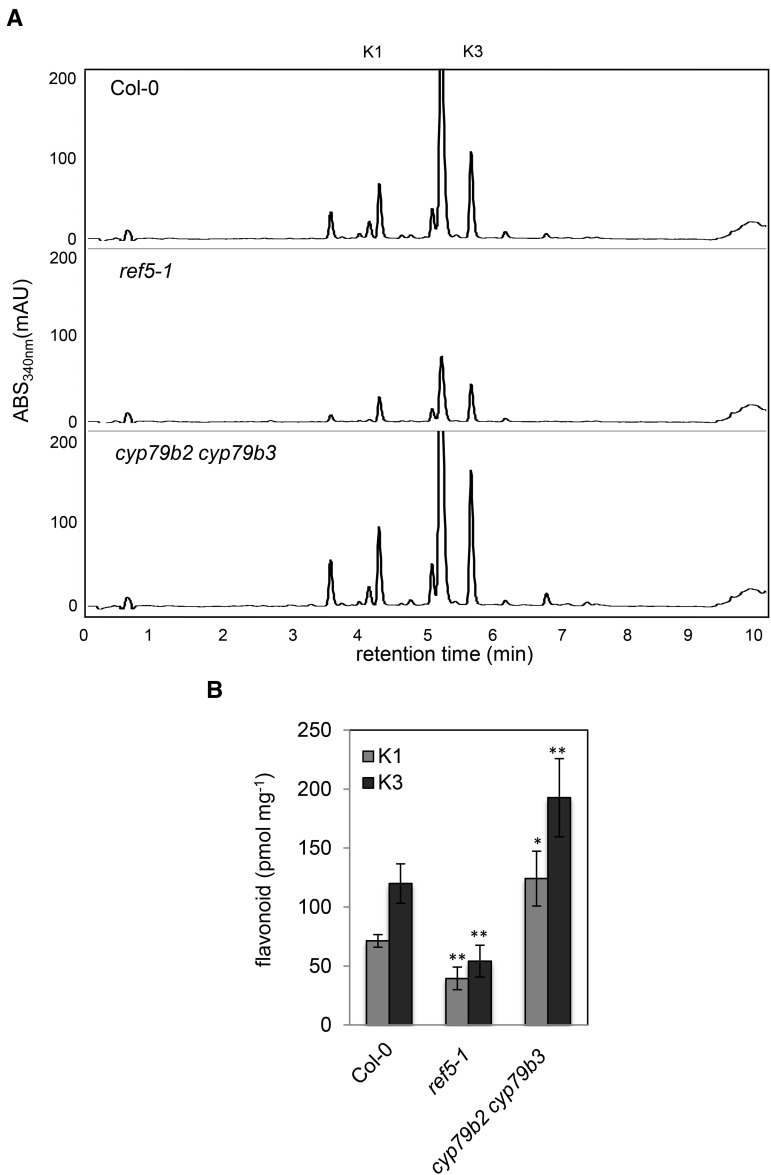
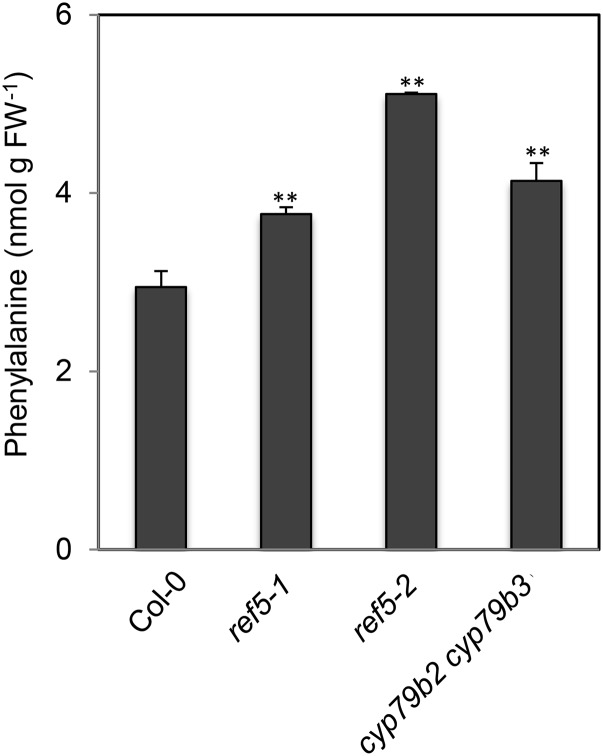
In the course of the previous analyses, we noticed that the level of flavonoids was modestly but consistently reduced in ref5 and enhanced in cyp79b2 cyp79b3 mutants (Figures 7A and 7B). To determine whether phenylalanine supply or its subsequent metabolism is affected in ref5 plants, we measured the phenylalanine content of both ref5 and cyp79b2 cyp79b3 plants and found that it was increased compared with the wild type (Figure 8). The fact that cyp79b2 cyp79b3 and ref5 mutants display opposite phenylpropanoid phenotypes indicates that at least in these mutants, there is not a strong correlation between phenylpropanoid metabolism and steady state phenylalanine pools. Taken together, these results suggest that inhibition or downregulation of phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate CoA ligase (4CL), the three steps common to sinapoylmalate and flavonoid biosynthesis, may be the cause of phenylpropanoid perturbation in ref5 mutants.
Figure 7.
Flavonoid Content in ref5 Mutants and cyp79b2 cyp79b3 Double Mutants Compared with the Wild Type.
HPLC profiles of UV absorbing metabolites (A) and quantification of flavonoids (B) from leaves of 4-week-old wild type, ref5-1, and cyp79b2 cyp79b3 double mutants. Two flavonoid peaks are shown as K1 and K3. K1, kaempferol 3-O-[6’’-O-(rhamnosyl)glucoside] 7-O-rhamnoside; K3, kaempferol 3-O-rhamnoside 7-O-rhamnoside. Data represent mean ± sd (n = 4). * and ** indicate P < 0.05 and P < 0.01 by a two-tailed Student’s t test compared with the wild type, respectively.
Figure 8.
Phenylalanine Content in ref5 Mutants and cyp79b2 cyp79b3 Double Mutants.
Data represent mean ± sd (n = 3). Asterisks indicate P < 0.01 by a two-tailed Student’s t test compared with the wild type.
Phenylpropanoid/Glucosinolate Crosstalk Occurs Early in the Phenylpropanoid Pathway
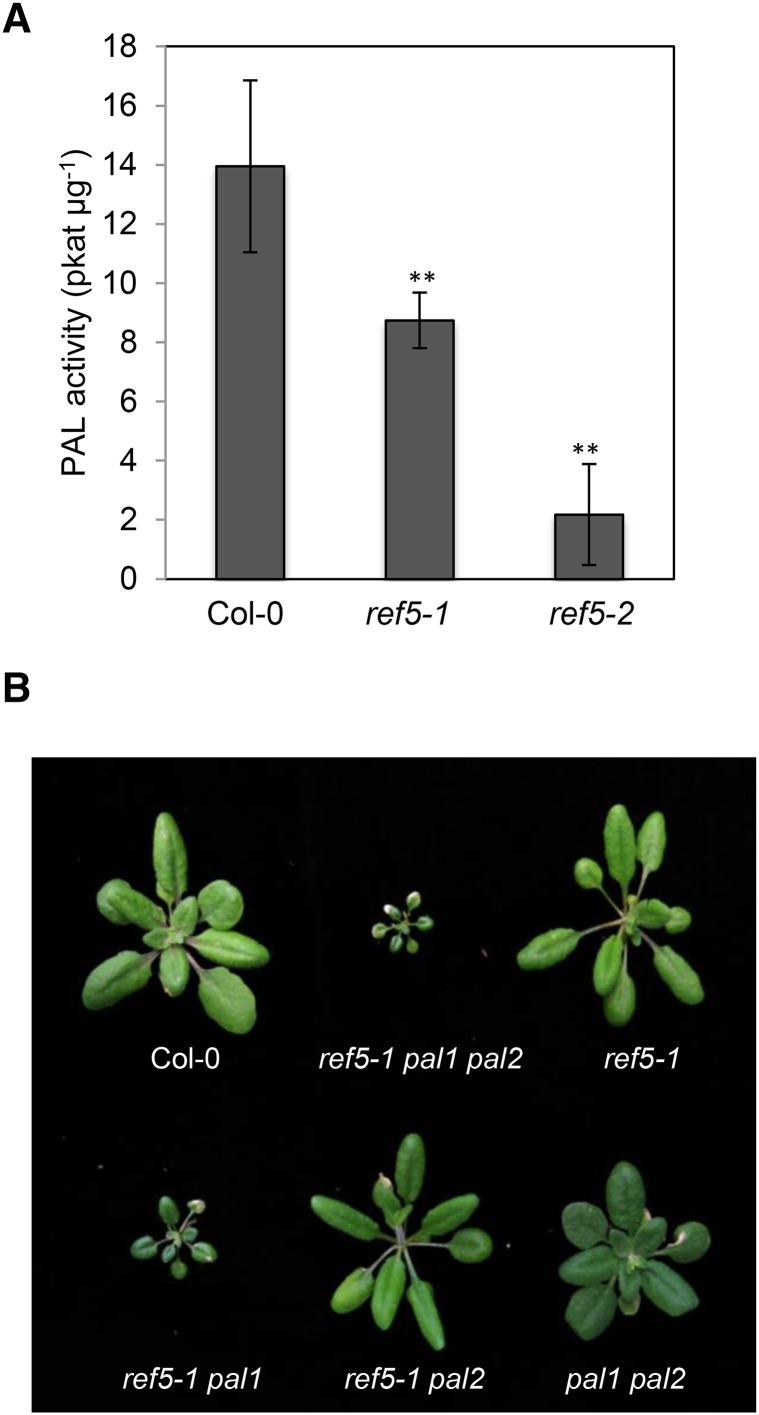
Finally, we measured PAL activity in desalted crude protein extracts of seedlings and found that ref5-1 and ref5-2 mutants contain ∼70 and 25% of wild-type PAL activity, respectively (Figure 9A). There are four PAL genes (PAL1-PAL4) in Arabidopsis. Although none of the single pal mutants has obvious visible phenotypes, pal1 pal2 double mutants lack anthocyanins in various organs and have only 25 to 30% of wild-type PAL activity, suggesting that PAL1 and PAL2 are major, functionally redundant enzymes (Raes et al., 2003; Rohde et al., 2004; Huang et al., 2010). To determine whether either of these PALs is a preferential target for IAOx-mediated suppression, we generated ref5-1 pal1 and ref5-1 pal2 double mutants and a ref5-1 pal1 pal2 triple mutant. Interestingly, although the ref5-1 pal2 double mutant is indistinguishable from ref5-1, the ref5-1 pal1 double mutant displayed growth defects (Figure 9B), indicating that IAOx accumulation in ref5-1 represses PAL activity mainly through impacting PAL2. Furthermore, the ref5-1 pal1 pal2 triple mutant displayed more severe growth defects than the ref5-1 pal1 double mutant, developing small dark-green rosette leaves but failing to bolt (Figure 9B; Supplemental Figure 1). Considering that the pal1/2/3/4 quadruple mutants are less affected developmentally than ref5-1 pal1 pal2 plants, it is likely that accumulation of IAOx or its derivative affects a step or steps in the phenylpropanoid pathway in addition to PAL (Huang et al., 2010; Supplemental Figure 1). To test this hypothesis, we crossed ref5-1 to the leaky C4H mutant ref3-2 (Schilmiller et al., 2009). These double mutants show severe growth defects similar to the C4H null mutant (Figure 10). These data suggest that C4H activity may also be repressed in ref5-1.
Figure 9.
PAL Activity in ref5 Mutants Compared with the Wild Type and Growth Phenotypes of the ref5-1 pal1 pal2 Triple Mutant and the ref5-1 pal1 and ref5-1 pal2 Double Mutants.
(A) PAL activity in crude extracts of 2-week-old wild-type and ref5 seedlings. PAL activity is expressed as average PAL activity in pkat µg−1 protein ± sd. Asterisks indicate P < 0.01 by a two-tailed Student’s t test compared with the wild type.
(B) Four-week-old soil-grown ref5-1 pal1 pal2 triple mutant (ref5-1 pal1/2), ref5-1 pal1, and ref5 pal2 double mutants compared with the wild type, ref5-1, and the pal1 pal2 double mutant.
Figure 10.
Growth Phenotypes of ref5-1 ref3-2 Double Mutants.
MED5, a Repressor of Phenylpropanoid Metabolism, Is Required for Glucosinolate/Phenylpropanoid Crosstalk
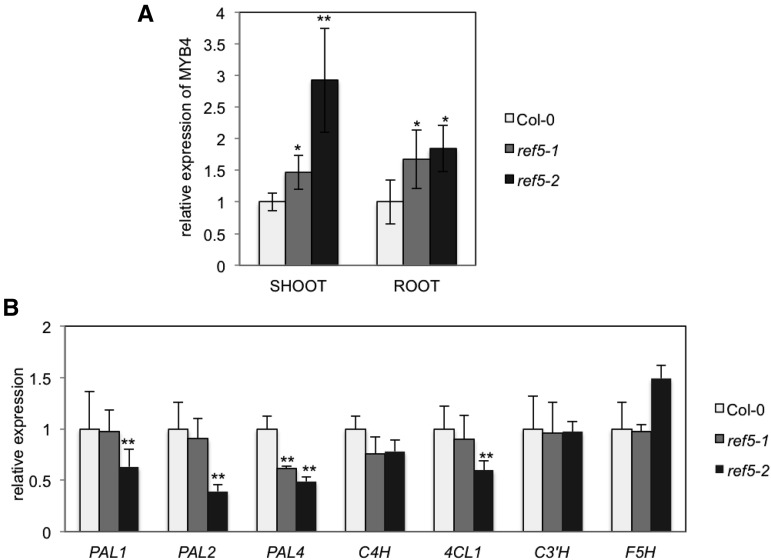
Biosynthesis of phenylpropanoids is tightly controlled by an array of transcriptional regulatory mechanisms (Zhong and Ye, 2009). Among several transcription factors, MYB4 has been shown to function as a transcriptional repressor for the expression of early phenylpropanoid genes (Jin et al., 2000). In a previously published data set (Zhao et al., 2002), we found that MYB4 expression was increased ∼12-fold exclusively in plants overexpressing CYP79B2 (CYP79B2ox), whereas other high auxin conditions did not show significant changes in MYB4 expression. Based on this finding, we tested the hypothesis that the accumulation of aldoxime or aldoxime derivatives in ref5 enhances the expression of MYB4, which in turn represses the expression of phenylpropanoid biosynthetic enzymes. As shown in Figure 11, MYB4 expression was increased in ref5 mutants. The transcript levels of PAL, C4H, and 4CL were not affected in the ref5-1 mutant, although their expression was slightly reduced in ref5 null mutants compared with the wild type (Figure 11B).
Figure 11.
Relative Expression of MYB4 and Phenylpropanoid Biosynthetic Genes in ref5 Mutants.
At1g13320 was used for internal control. The relative expression was calculated with 2−ΔΔCT. Data represent mean ± sd (n = 3). * and ** indicate P < 0.05 and P < 0.01, respectively, compared with the wild type.
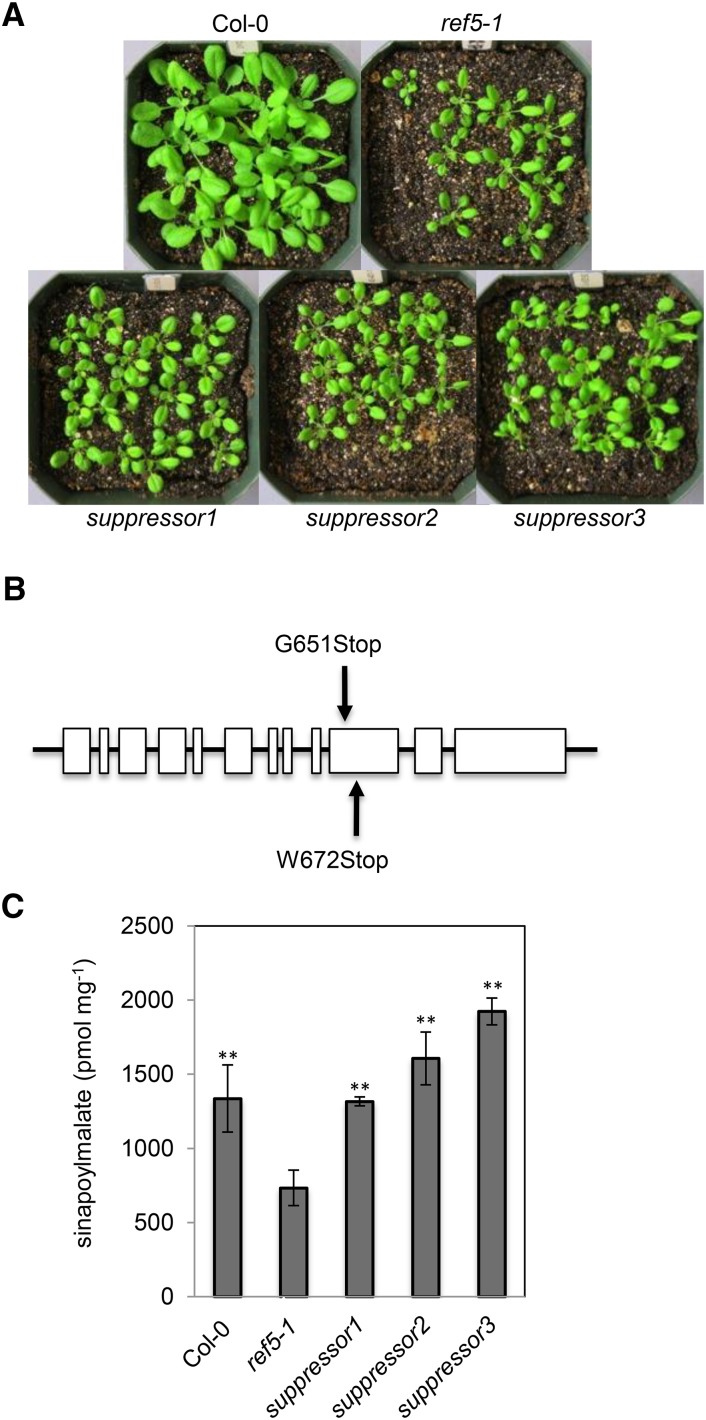
To identify genes required for the crosstalk between glucosinolate biosynthesis and phenylpropanoid metabolism, we performed a ref5 suppressor screen and isolated three independent lines that had a restored fluorescence phenotype when observed under UV light. As shown in Figure 12A, suppressors displayed the high auxin morphological phenotypes typical of ref5, such as elongated hypocotyls and narrow downward-curled leaves, but HPLC analyses revealed that sinapoylmalate content was restored to wild-type levels (Figure 12C). The high auxin morphology and restored sinapoylmalate content in the suppressors suggest that they continue to synthesize IAA in an IAOx-dependent manner but are defective in the crosstalk between IAOx and phenylpropanoid metabolism.
Figure 12.
Three MED5b Mutants Were Isolated from a ref5 Suppressor Screen.
(A) Morphological phenotype of the three ref5 suppressors. Three-week-old suppressors are compared with the wild type and the ref5-1 mutant.
(B) The location of mutations in ref5 suppressors is shown above the schematic of the MED5b (At3g23590) genomic locus. The two mutations in the three suppressors result in early stops.
(C) Sinapoylmalate content measured in whole rosette leaves of 3-week-old plants. Data represent mean ± sd (n = 4). Asterisks indicate P < 0.01 by a two-tailed Student’s t test compared with ref5-1.
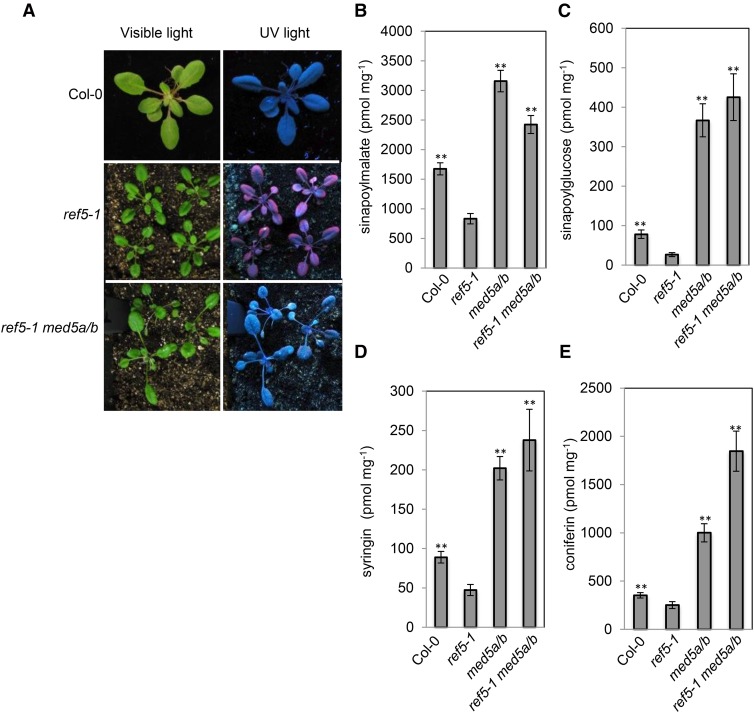
Whole-genome sequencing analyses revealed that all three of the suppressors have mutations in the same gene, At3g23590, which encodes the Arabidopsis Mediator subunit MED5b/RFR1. The first has a C-to-T nonsense mutation in the 10th exon of MED5b (TAGGTTCCGTAACTTGCTTG), which terminates the protein after Pro-650 (Figure 12B), and the other two have the same G-to-A nonsense mutation in the 10th exon of MED5b (AGTGTATCATGAACTCTTCCTT), which results in an early stop after Ser-671 (Figure 12B). It has been shown previously that the Arabidopsis Mediator subunit MED5b/RFR1 and its paralog, MED5a/REF4, function semiredundantly in the repression of phenylpropanoid biosynthesis and med5a med5b double mutants contain increased levels of phenylpropanoids (Bonawitz et al., 2012, 2014). Consistent with the identification of MED5b as a ref5 suppressor, a ref5-1 med5a med5b triple mutant generated by crossing ref5-1 with two T-DNA insertion mutants in MED5a and MED5b did not display a ref phenotype, although the characteristic ref5 high auxin morphology was maintained (Bonawitz et al., 2012; Figure 13A). HPLC analyses showed increased phenylpropanoids in leaves and seedling roots (Figures 13B to 13E). The observation that perturbation of a major transcriptional coregulatory complex restores phenylpropanoid metabolism in the ref5 mutant background strongly suggests that the crosstalk between the glucosinolate and phenylpropanoid biosynthetic pathways occurs at the transcriptional level. Based on data we present here, we propose a model for the crosstalk between two secondary metabolite biosynthesis pathways in which the level of IAOx or its derivative impacts PAL activity, mainly through PAL2, and other early steps of the phenylpropanoid pathway including C4H. The repression occurs at the transcriptional level through Arabidopsis Mediator Subunit 5a/b (MED5A and MED5B). MYB4, a repressor of phenylpropanoid biosynthesis gene expression, may play a role in the crosstalk.
Figure 13.
The Disruption of MED5a/b Restores Phenylpropanoid Content in ref5-1.
(A) Morphology and ref phenotype in 3-week-old ref5-1 med5a/b triple mutant compared with its single mutants. ref5-1 med5a/b plants exhibit the high auxin phenotypes similar to ref5-1 but the ref phenotype is suppressed.
(B) to (E) Soluble metabolite analysis in mature leaves and seedling roots. Sinapoylmalate and sinapoylglucose content was measured in 3-week-old whole rosette leaves ([B] and [C]). Syringin and coniferin content was measured in 2-week-old seedling roots ([D] and [E]). Asterisks indicate P < 0.01 by a two-tailed Student’s t test compared with ref5-1.
DISCUSSION
As sessile organisms, plants have acquired various mechanisms that allow them to survive deleterious environmental changes. One of these strategies is the production of specialized secondary metabolites. Although many of these molecules have specific biological roles and are produced through unique biosynthetic pathways, plants often orchestrate the synthesis of members of multiple groups of specialized metabolites simultaneously to draw the most advantageous outcome. For example, under microbial invasion or elicitation conditions, plants induce the production of defense molecules such as phytoalexins or glucosinolates as a chemical barrier and activate genes involving the biosynthesis of cell wall components, which may function as a physical barrier (Rinaldi et al., 2007; Tronchet et al., 2010). Although the biosynthesis and regulation of many specialized metabolites has been extensively studied, how plants coordinate their production is not yet well understood.
In this study, we demonstrated that IAOx, an intermediate in tryptophan-derived glucosinolate biosynthesis, or a derivative thereof, limits phenylpropanoid accumulation. The biochemical and genetic data presented here suggest that the crosstalk between two specialized metabolite pathways involves the repression of an early step(s) of the phenylpropanoid pathway results in decreased PAL activity and requires the Arabidopsis Mediator complex component MED5b. Most importantly, this process was found to occur not only in a mutant context but also in wild-type plants.
ref5 Is an Allele of CYP83B1
Several cyp83b1 mutants have been isolated from a variety of forward genetic mutant screens, and their phenotypes have shed light on the regulation of the indole glucosinolate pathway as well as the function of CYP83B1 itself (Table 1). sur2, rnt1, red1, and gul1 mutants were isolated based upon their characteristic morphology (Wagner et al., 1997; Delarue et al., 1998; Winkler et al., 1998; Barlier et al., 2000; Bak and Feyereisen 2001; Hoecker et al., 2004; Maharjan et al., 2014). Their phenotypes, such as elongated hypocotyls, down-curled leaves, and adventitious roots, are often observed in high auxin mutants, suggesting a connection between indole glucosinolate metabolism and auxin biosynthesis (Zhao et al., 2002; Kim et al., 2007). Indeed, cyp83b1 mutants and transgenic plants overexpressing CYP79B2 contain higher amounts of free IAA (Delarue et al., 1998; Zhao et al., 2002). IAOx is believed to be a precursor of IAA, although the enzymes involved in this transformation are still unknown (Barlier et al., 2000; Zhao et al., 2002). atr4-1 and atr4-2 mutants were identified from a forward genetic screen for plants with altered tryptophan feedback resistance phenotypes. atr4 mutants exhibit upregulation of MYB34/ATR1, ASA1, and CYP79B2, and the study revealed a molecular mechanism for feedback inhibition in tryptophan biosynthesis (Smolen and Bender, 2002; Celenza et al., 2005).
Although most cyp83b1 mutants were isolated based on their growth phenotypes under specific conditions, ref5-1 was isolated due to an apparently unrelated biochemical phenotype. As a new cyp83b1 mutant, ref5-1 provides additional evidence for crosstalk between the phenylpropanoid and indole glucosinolate biosynthetic pathways. This crosstalk had been suggested from two previous independent investigations: characterization of ref2/cyp83a1 mutants and metabolomic analysis of sur2 mutants (Hemm et al., 2003; Morant et al., 2010). Morant et al. (2010) reported that the accumulation of various indolic and phenolic compounds was reduced in sur2 seedlings but reported increased coniferin and syringin levels, whereas our results show the opposite in ref5-1 (Morant et al., 2010; Figures 6A, 6B, and 13B to 13E). Considering that the sur2 results represent metabolite amount per whole seedling and ours are calculated on a root fresh weight basis, it is possible that the severe morphological phenotypes of sur2 mutants may explain this apparent contradiction. The ability to analyze cyp83b1-induced phenotypes that occur as a result of a weak allele that does not generate severe developmental phenotypes is a significant advantage provided by the ref5-1 allele.
ref2 and ref5 Mutants Display Overlapping but Unique Phenotypes
Although both ref2 and ref5 mutants are altered in phenylpropanoid metabolism, they have distinct phenotypes. Both ref2 and ref5 mutants contain reduced levels of sinapoylmalate but coniferin and syringin levels in ref2 roots are not altered, whereas they are reduced in ref5 (Figures 6A and 6B; Hemm et al., 2003). Also, in ref2 stems, syringyl lignin monomer deposition, which occurs primarily in interfasicular fibers, is 25 to 50% of that found in the wild type, whereas syringyl lignin content in ref5 mutants is unaffected (Table 2; Hemm et al., 2003). The expression of both CYP83A1 (REF2) and CYP79F1/BUS1 is low in seedling roots, whereas the expression of CYP83B1 (REF5) and CYP79B2/CYP9B3 is relatively high (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al., 2007; Supplemental Figure 2). Similarly, CYP79B2/CYP79B3 expression is low in stems, whereas both CYP83A1 (REF2) and CYP79F1/BUS1 are expressed at relatively high levels (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Hemm et al., 2003; Winter et al., 2007; Supplemental Figure 2). Given these observations, it seems likely that the relative expression of these genes at the organ and even cell type level results in the unique monolignol glucoside and lignin phenotypes of the ref2 and ref5 mutants.
Alteration of Aldoxime Levels Limits Phenylpropanoid Metabolism
The data presented here show that the alteration of IAOx or a subsequent metabolite in ref5 mutants reduces phenylpropanoid accumulation, whereas the opposite is true in cyp79b2 cyp79b3 mutants in which phenylpropanoid compound accumulation is increased. The reduced phenylpropanoid phenotypes in IAOx accumulation mutants would be of interest but not of wide significance if it only occurred in ref5 mutants; however, the increased levels of coniferin, syringin, flavonoids, and sinapoylmalate in the cyp79b2 cyp79b3 double mutant indicate that the presence of aldoximes or their derivatives in the wild type limits phenylpropanoid production. The levels of IAOx and IAOx derivatives in plants, including cyp79b2 cyp79b3 mutants, have been determined previously (Sugawara et al., 2009). IAOx is present in very low level in Arabidopsis at 1.7 ng per g fresh weight. These levels are approximately one-tenth of the concentration of IAA. IAOx is absent in cyp79b2 cyp79b3 mutants and is present at 4.2 ng per g fresh weight in the sur1 mutant, which shows a high auxin phenotype similar to ref5/sur2/cyp83b1, suggesting that in ref5, the levels of IAOx might be very similar.
Our data show that PAL and possibly other early steps in phenylpropanoid biosynthesis are affected by elevated IAOx levels in ref5. Considering that nearly 30% of organic carbon in plant biomass is channeled through the phenylpropanoid pathway, control of phenylpropanoid production as it enters the pathway is likely to be an important way to regulate carbon flux. PAL is the first committed enzyme in the phenylpropanoid pathway, and many studies have identified mechanisms that control PAL activity, including transcriptional, translational, and posttranslational regulation as well as product inhibition (reviewed in Zhang and Liu, 2015). Arabidopsis PAL is encoded by four genes, PAL1 to PAL4, and it is thought that PAL isoforms may have distinct but overlapping functions in plant growth and development. For example, studies with pal mutants showed that PAL1, PAL2, and PAL4 are associated with lignin biosynthesis, whereas PAL1 and PAL2 are responsible for soluble metabolite biosynthesis (Wanner et al., 1995; Raes et al., 2003; Rohde et al., 2004; Huang et al., 2010). PAL1 and PAL2 function redundantly in flavonoid biosynthesis and growth because neither the pal1 nor the pal2 single mutant shows the flavonoid deficiency or growth phenotypes that pal1 pal2 double mutants do (Rohde et al., 2004; Huang et al., 2010). The difference of growth phenotype between ref5-1 pal1 and ref5-1 pal2 indicates that the IAOx accumulation in ref5 mutant primarily impacts PAL2 (Figure 9B). The fact that the ref5-1 pal1 pal2 triple mutant grows more poorly than the pal1/2/3/4 quadruple mutant suggests that the IAOx accumulation may repress an additional step(s) in the phenylpropanoid pathway besides PAL. Consistent with this model, the growth phenotype of the ref5-1 ref3-2 mutant suggests that the IAOx accumulation may impact C4H activity as well (Figure 10).
How the altered IAOx level influences multiple steps of phenylpropanoid pathway simultaneously is not clear. Biosynthesis of phenylpropanoids is tightly controlled through multiple levels of regulation. At the transcriptional level, an array of transcription factors from MYB, NAC, WRKY, and KNOX families have been shown to function as transcriptional activators or repressors of lignin biosynthesis (reviewed in Zhao and Dixon, 2011). Among them, MYB4 has been shown to function as a transcriptional repressor of phenylpropanoid biosynthetic genes, specifically those in the early steps of the pathway (Jin et al., 2000). It was also shown that any condition with an enhanced level of IAOx or its derivatives may directly or indirectly activate the expression of MYB4 (Zhao et al., 2002). Morant et al. (2010) reported changes in expression of multiple transcription factors in the sur2 mutant, and among them, MYB4 was upregulated 1.7-fold. Consistent with this report, MYB4 expression was increased in ref5 seedlings compared with the wild type (Figure 11A). The expression of PAL, C4H, and 4CL was slightly downregulated in the ref5 null mutant but surprisingly was not changed in plants carrying the ref5-1 weak allele even though both mutants had significantly reduced PAL activity (Figure 9). These findings suggest that other mechanisms of PAL downregulation, possibly including ubiquitin-mediated turnover (Zhang et al., 2013a), may be involved in the reduction of its enzyme activity (Figure 9A).
Phenylpropanoid-Glucosinolate Crosstalk Requires the Mediator Subunit MED5b
Our forward genetic mutant screen for ref5-1 suppressors isolated three MED5b alleles. Mediator is a large multisubunit transcriptional coregulatory complex, and it has been shown that it plays essential roles in transcriptionally regulated processes in many organisms (reviewed in Kornberg, 2007; Conaway and Conaway, 2011). Recent progress has revealed that in plants, Mediator functions in diverse biological processes include plant immunity (Kidd et al., 2009; Wathugala et al., 2012; Zhang et al., 2012; Zhang et al., 2013b; Lai et al., 2014), freezing tolerance (Knight et al., 2009; Hemsley et al., 2014), flowering (Imura et al., 2012; Zheng et al., 2013; Lai et al., 2014), hormone responses (Chen et al., 2012; Lai et al., 2014), and phenylpropanoid homeostasis (Bonawitz et al., 2012, 2014). A pair of recent studies with loss-of-function mutants showed that disruption of both MED5a and its paralog, MED5b, results in enhanced expression of phenylpropanoid biosynthetic genes and hyperaccumulation of various phenylpropanoids (Bonawitz et al., 2012, 2014). Three med5b alleles were identified from the ref5 suppressor screen, and ref5-1 med5a/b triple mutants accumulate elevated levels of phenylpropanoids as observed in the med5a/b double mutant (Figure 13). These genetic results suggest that at some level, transcriptional regulation of gene expression is involved in the crosstalk between glucosinolate and phenylpropanoid biosynthesis, potentially including a role for MYB4. On the other hand, we cannot rule out the formal possibility that elimination of MED5a/b function simply overrides the impact of IAOx or its derivatives given that the absence of MED5a/b results in enhanced phenylpropanoid gene expression and the hyperaccumulation of various phenylpropanoids (Bonawitz et al., 2012, 2014). Further studies with more ref5 suppressors will elucidate the molecular mechanism(s) behind the glucosinolate/phenylpropanoid metabolic crosstalk, including how the accumulation of IAOX is sensed in vivo, how the derived signal is transmitted to the nucleus, and whether any additional Mediator subunits are involved.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type. Plants were cultivated at a light intensity of 100 μE m−2 s−1 at 22°C under a photoperiod of 16 h light/8 h dark unless otherwise specified. For seedlings, seeds were surface sterilized and plated on modified MS ammonium-free medium containing 1% sucrose (Murashige and Skoog, 1962). For root samples, seedlings were vertically grown on ammonium-free MS media containing 1% sucrose and 1.2% agar.
T-DNA insertion mutants ref5-2 (SALK_028573), pal1-3 (SALK_096474C), and pal2-3 (GABI-692H09-025071) were obtained from the ABRC (The Ohio State University, Columbus, OH). Primers CC3405 (left primer), CC3406 (right primer), and CC3407 (border primer) were used to genotype for the presence of the T-DNA insertion in ref5-2. Primers CC4010 (left primer), CC4011 (right primer), and CC2449 (border primer) for pal1-3 and primers CC4061 (left primer), CC4062 (right primer), and CC2501 (border primer) for pal2-3 were used for genotyping. The pal1/2/3/4 quadruple mutant was pal1-3 pal2-3 pal3-2 pal4-2, and the pal1pal2 double mutant was pal1-3 pal2-3 (Huang et al., 2010). Primer sequences are shown in Supplemental Table 1.
Cloning of the REF5 Gene
A ref5 mapping population was generated by crossing the ref5-1 mutant with Landsberg erecta. F1 individuals were allowed to self-pollinate, and F2 plants were screened under UV light for the ref5 phenotype. A collection of 22 simple sequence length polymorphism markers (http://carnegiedpb.stanford.edu/publications/methods/ppsuppl.html), which span the Arabidopsis genome, were used to genotype the F2 plants. After a marker linked to the REF5 locus was determined, plants carrying recombinant chromosomes in the REF5 region were used to define a mapping interval using other further designed simple sequence length polymorphism or cleaved amplified polymorphic sequence (CAPS) markers.
To identify the single nucleotide polymorphism (SNP) in the putative ref5 mutant, a pair of flanking primers, CC1352 and CC1357, located at the 5′ and 3′ untranslated region of the CYP83B1 genomic DNA were designed to amplify a sequencing template from the putative ref5 mutant genomic DNA, which was then sequenced employing primers CC1353, CC1354, CC1355, CC1356, CC1358, CC1359, and CC1360 at the Purdue University Genomics Facility (Supplemental Table 1).
The PCR-based derived CAPS method was employed (Neff et al., 1998) to verify the mutation identified by sequence analysis. Primers were designed using the derived CAPS Finder 2.0 program available online (http://helix.wustl.edu/dcaps/dcaps.html). A normal forward primer CC1691 was used with a mismatched reverse primer CC1548 to amplify 300-bp PCR products for wild-type and ref5-1 DNA templates, respectively (Supplemental Table 1). This primer pair creates a XhoI site specifically in the wild-type REF5 allele, which results in a 25-bp difference between wild-type and ref5-1 PCR products after digestion.
Lignin Analysis
For cell wall preparation, Arabidopsis stem tissue was ground to a fine powder in liquid nitrogen and extracted sequentially with 100 mM sodium phosphate buffer (pH 7.2), 70% ethanol, and acetone. To measure lignin content, cell wall samples were analyzed using the microscale Klason method (Kaar and Brink, 1991). Lignin monomer composition was determined by the DFRC method (Lu and Ralph, 1997) as described previously (Li et al., 2010).
HPLC Analysis of Secondary Metabolites
For soluble metabolites analyses, samples were frozen in liquid nitrogen immediately and stored in the freezer until ready for extraction. Frozen tissues were extracted with 50% methanol (v/v) at 65°C for 1 h at a tissue concentration of 100 mg mL−1. Samples were centrifuged at 16,000g for 10 min before HPLC analysis. Ten microliters of extraction was loaded on a Shim-pack XR-ODS column (Shimadzu) and separated at an increasing concentration of acetonitrile from 2 to 25% for 30 min in 0.1% formic acid at a flow rate of 1 mL min−1. Compounds were identified based on their retention times and UV spectra. Sinapate esters and flavonoids were quantified using sinapic acid and kaempferol as their standards, respectively.
For analysis of glucosinolate contents in plant tissues, desulfoglucosinolate extracts were prepared according to the protocol previously described (Lee et al., 2012). Desulfoglucosinolates were identified based on their retention times and UV spectra from published results and quantified from peak areas at 229 nm relative to the internal standard after adjusting HPLC response factors as suggested by Brown et al. (2003) (Kiddle et al., 2001; Reintanz et al., 2001; Hemm et al., 2003).
Generation of Transgenic Plants Overexpressing CYP79B2
To generate a CYP79B2 overexpression construct, the CYP79B2 open reading frame was amplified by PCR using the primer pair CC3582 and CC3583 (Supplemental Table 1). The amplified PCR product was cloned into a Gateway entry vector that was modified from pENTR1A (Life Technologies) by replacing the kanamycin selection marker with an ampicillin resistance gene. The resulting CYP79B2 entry clone was subsequently recombined with destination vector, pCC0995 (Weng et al., 2010), in which expression of the open reading frame was driven by the 35S promoter to generate the 35S:CYP79B2 construct. The 35S:CYP79B2 construct was introduced into wild-type plants via Agrobacterium tumefaciens (strain C58 pGV3850)-mediated transformation. To select for herbicide-resistant transformants, 1-week-old T1 plants were sprayed with 0.2% Finale solution (glufosinate ammonium; Farnam Companies). Third and fourth rosette leaves from 4-week-old surviving T1 plants were harvested for RNA extraction and sinapoylmalate quantification.
PAL Activity Assay
For crude protein extraction, 1 g of 2-week-old seedlings was harvested and ground in liquid nitrogen. Five milliliters of extraction buffer containing 100 mM Tris-HCl, pH 8.3, 2 mM DTT, and 10% (v/v) glycerol was added, and the mixture was stirred at 4°C for 30 min. After centrifugation at 10,000g for 10 min, the supernatant was collected, desalted into extraction buffer on a PD-10 column (GE Healthcare Life Science) following the manufacturer’s protocol, and assayed for PAL activity. Total protein content was measured using the BCA Protein Assay Kit (Pierce) using BSA as a standard. To measure the activity of PAL, 100-μL aliquots of the protein extracts was incubated with 4 mM Phe in reaction buffer (100 mM Tris-HCl, pH 8.3, 2 mM DTT, and 10% [v/v] glycerol) in a total volume of 500 µL. Reactions were incubated at 37°C for 90 min and then stopped by addition of 50 μL of glacial acetic acid. Reaction products were extracted with 750 μL ethyl acetate, 500 μL of which was removed, dried in a speed vac, and redissolved in 50 μL of 50% methanol. Ten microliters of the final extract was analyzed by HPLC to quantify the reaction product, cinnamic acid.
RNA Extraction and Quantitative RT-PCR
RNA extraction and quantitative real-time PCR experiments were performed as previously described (Li et al., 2010) using the following primers: PAL1, CC3280 and CC3281; PAL2, CC3282 and CC3283; PAL4, CC2016 and CC2017; C4H, CC3258 and CC3259; 4CL1, CC3256 and CC3257; C3′H, CC2839 and CC2840; F5H, CC3272 and CC3273; CCoAOMT, CC3844 and CC3845; MYB4, CC3813 and CC3814; CYP79B2, CC3663 and CC3664; reference gene (At1g13320), CC2558 and CC2559 (Supplemental Table 1). RNA was isolated from 10-d-old seedlings for measurement of phenylpropanoid gene and MYB4 expression. For CYP79B2 overexpression analysis, 4-week-old rosette leaves were used for RNA extraction.
ref5-1 Suppressor Screen
Approximately 150,000 ref5-1 seeds were mutagenized in a 0.3% ethyl methanesulfonate (EMS) solution for 16 h. After extensive rinsing with running water, the seeds were planted and grown in the greenhouse. M2 seeds from 48 seed pools were collected and ∼30,000 M2 plants were screened for mutant identification. Three weeks after planting, plants were examined under a UV transilluminator with a peak wavelength of 302 nm (Model TM-36; UVP), and mutants with ref5-1 morphology having restored leaf epidermal fluorescence were selected as suppressors. Out of five putative suppressors, the genomes of three displaying strong phenotypes were sequenced.
Whole-Genome Sequencing of ref5 Suppressors
Whole-genome sequencing was performed by the Purdue University Genomics Core Facility using an Illumina HiSeq2500 instrument. Approximately 29 to 34 million 100-bp paired-end reads resulted in 14- to 19-fold genomic coverage when aligned to the TAIR10 reference genome sequence. SNPs were called using Bowtie2 (Langmead and Salzberg, 2012). The SNPs in MED5b/RFR1 were identified by screening candidate genes using the Integrated Genome Viewer (Robinson et al., 2011; Thorvaldsdóttir et al., 2013). The PCR-based cut/CAPS method was performed to verify the mutations identified by sequence analysis. CC1770 and CC1775 were used to amplify 950-bp PCR products for wild-type and suppressor DNA templates. To confirm the C-to-T mutation in one of the putative ref5-1 suppressors, PCR product was digested with AciI, which cuts the wild-type amplicon but not the one from the suppressor. For the G-to-A mutation in the other suppressors, PCR products were digested with HinfI, which cuts the wild-type amplicon but not the one from the suppressor.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: REF5/CYP83B1 (At4g31500), REF2/CYP83A1 (At4g13770), CYP79B2 (At4g39950), CYP79B3 (At2g22330), CYP79F1 (At1g16410), PAL1 (At2g37040), PAL2 (At3g53260), PAL4 (At4b10340), C4H (At2g30490), 4CL1 (At1g51680), C3′H (At2g40890), F5H (At4g36220), CCoAOMT (At4g34050), MYB4 (At4g38620), COMT (At5g54160), MED5a/REF4 (At2g48110), and MED5b/RFR1 (At3g23590).
Supplemental Data
Supplemental Figure 1. Growth phenotypes of ref5-1 pal1 pal2 plants grown on soil.
Supplemental Figure 2. Expression pattern of CYP83B1, CYP83A1, CYP79B2, CYP79B3, and CYP79F1.
Supplemental Table 1. A list of primer sequences used.
Supplementary Material
Acknowledgments
This work was supported as part of the Center for Direct Catalytic Conversion of Biomass to Biofuels, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences under Award DE-SC0000997. This material is also based upon work supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under Award DE-FG02-07ER15905. We acknowledge Yu Han for initiating this study and identifying the position of the ref5 mutation. We thank Zhixiang Chen for providing seeds of the pal mutants and Bruce Cooper for help with mass spectrometry analyses.
AUTHOR CONTRIBUTIONS
J.I.K. and C.C. designed the experiments. J.I.K., N.A.A., and W.L.D. performed research. J.I.K. and C.C. wrote the article.
Glossary
- Col-0
Columbia-0
- IAOx
indole-3 acetaldoxime
- IAA
indole-3-acetic acid
- DFRC
derivatization followed by reductive cleavage
- MS
Murashige and Skoog
- COMT
caffeic acid O-methyltransferase
- 5-OH-feruloylmalate
5-hydroxy-feruloylmalate
- PAL
phenylalanine ammonia lyase
- C4H
cinnamate 4-hydroxylase
- CAPS
cleaved amplified polymorphic sequence
- SNP
single nucleotide polymorphism
- EMS
ethyl methanesulfonate
References
- Bak S., Feyereisen R. (2001). The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Tax F.E., Feldmann K.A., Galbraith D.W., Feyereisen R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I., Kowalczyk M., Marchant A., Ljung K., Bhalerao R., Bennett M., Sandberg G., Bellini C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97: 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N.D., Chapple C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44: 337–363. [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Soltau W.L., Blatchley M.R., Powers B.L., Hurlock A.K., Seals L.A., Weng J.-K., Stout J., Chapple C. (2012). REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 287: 5434–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N.D., Kim J.I., Tobimatsu Y., Ciesielski P.N., Anderson N.A., Ximenes E., Maeda J., Ralph J., Donohoe B.S., Ladisch M., Chapple C. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380. [DOI] [PubMed] [Google Scholar]
- Böttcher C., Chapman A., Fellermeier F., Choudhary M., Scheel D., Glawischnig E. (2014). The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis thalinana. Plant Physiol. 165: 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.D., Tokuhisa J.G., Reichelt M., Gershenzon J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481. [DOI] [PubMed] [Google Scholar]
- Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R.C., Conaway J.W. (2011). Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Prinsen E., Onckelen H.V., Caboche M., Bellini C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14: 603–611. [DOI] [PubMed] [Google Scholar]
- Do C.T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., Lapierre C., Jouanin L. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129. [DOI] [PubMed] [Google Scholar]
- Fahey J.W., Zalcmann A.T., Talalay P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56: 5–51. [DOI] [PubMed] [Google Scholar]
- Franke R., Humphreys J.M., Hemm M.R., Denault J.W., Ruegger M.O., Cusumano J.C., Chapple C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30: 33–45. [DOI] [PubMed] [Google Scholar]
- Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon T., Sibout R., Pollet B., Maba B., Nussaume L., Bechtold N., Lu F., Ralph J., Mila I., Barrière Y., Lapierre C., Jouanin L. (2003). A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51: 973–989. [DOI] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Hansen C.H., Du L., Naur P., Olsen C.E., Axelsen K.B., Hick A.J., Pickett J.A., Halkier B.A. (2001). CYP83b1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 276: 24790–24796. [DOI] [PubMed] [Google Scholar]
- Hemm M.R., Ruegger M.O., Chapple C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm M.R., Rider S.D., Ogas J., Murry D.J., Chapple C. (2004). Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38: 765–778. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Hurst C.H., Kaliyadasa E., Lamb R., Knight M.R., De Cothi E.A., Steele J.F., Knight H. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Toledo-Ortiz G., Bender J., Quail P.H. (2004). The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219: 195–200. [DOI] [PubMed] [Google Scholar]
- Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y.H., Yu J.Q., Chen Z. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153: 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97: 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura Y., Kobayashi Y., Yamamoto S., Furutani M., Tasaka M., Abe M., Araki T. (2012). CRYPTIC PRECOCIOUS/MED12 is a novel flowering regulator with multiple target steps in Arabidopsis. Plant Cell Physiol. 53: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19: 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaar W.E., Brink D.L. (1991). Simplified analysis of acid-soluble lignin. J. Wood Chem. Technol. 11: 465–477. [Google Scholar]
- Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle G., Bennett R.N., Botting N.P., Davidson N.E., Robertson A.A.B., Wallsgrove R.M. (2001). High-performance liquid chromatographic separation of natural and synthetic desulphoglucosinolates and their chemical validation by UV, NMR and chemical ionisation-MS methods. Phytochem. Anal. 12: 226–242. [DOI] [PubMed] [Google Scholar]
- Kim J.I., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145: 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Mugford S.G., Ulker B., Gao D., Thorlby G., Knight M.R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58: 97–108. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D. (2007). The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 104: 12955–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Schluttenhofer C.M., Bhide K., Shreve J., Thimmapuram J., Lee S.Y., Yun D.-J., Mengiste T. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5: 3064. [DOI] [PubMed] [Google Scholar]
- Landry L.G., Chapple C.C., Last R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109: 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kaminaga Y., Cooper B., Pichersky E., Dudareva N., Chapple C. (2012). Benzoylation and sinapoylation of glucosinolate R-groups in Arabidopsis. Plant J. 72: 411–422. [DOI] [PubMed] [Google Scholar]
- Li X., Bonawitz N.D., Weng J.-K., Chapple C. (2010). The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F.C., Ralph J. (1997). Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: Protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45: 2590–2592. [Google Scholar]
- Maharjan P.M., Dilkes B.P., Fujioka S., Pěnčík A., Ljung K., Burow M., Halkier B.A., Choe S. (2014). Arabidopsis gulliver1/SUPERROOT2-7 identifies a metabolic basis for auxin and brassinosteroid synergy. Plant J. 80: 797–808. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Cusumano J.C., Somerville C., Chapple C.C. (1996). Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc. Natl. Acad. Sci. USA 93: 6869–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Hansen C.H., Wittstock U., Halkier B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275: 33712–33717. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M.D., Naur P., Halkier B.A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37: 770–777. [DOI] [PubMed] [Google Scholar]
- Mithen R., Faulkner K., Magrath R., Rose P., Williamson G., Marquez J. (2003). Development of isothiocyanate-enriched broccoli, and its enhanced ability to induce phase 2 detoxification enzymes in mammalian cells. Theor. Appl. Genet. 106: 727–734. [DOI] [PubMed] [Google Scholar]
- Morant M., Ekstrøm C., Ulvskov P., Kristensen C., Rudemo M., Olsen C.E., Hansen J., Jørgensen K., Jørgensen B., Møller B.L., Bak S. (2010). Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2. Mol. Plant 3: 192–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P., Petersen B.L., Mikkelsen M.D., Bak S., Rasmussen H., Olsen C.E., Halkier B.A. (2003). CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Neff J.D., Chory J., Pepper A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392. [DOI] [PubMed] [Google Scholar]
- Pfalz M., Vogel H., Kroymann J. (2009). The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21: 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M., Mikkelsen M.D., Bednarek P., Olsen C.E., Halkier B.A., Kroymann J. (2011). Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23: 716–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133: 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., Godde M., Uhl R., Palme K. (2001). Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13: 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi C., Kohler A., Frey P., Duchaussoy F., Ningre N., Couloux A., Wincker P., Le Thiec D., Fluch S., Martin F., Duplessis S. (2007). Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol. 144: 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J.J., Tivendale N.D., Reid J.B., Davies N.W., Molesworth P.P., Lowe E.K., Smith J.A., Davidson S.E. (2011). Reassessing the role of YUCCAs in auxin biosynthesis. Plant Signal. Behav. 6: 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Chapple C. (2001). Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159: 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.-K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782. [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B.L., Olsen C.E., Svatoš A., Halkier B.A., Glawischnig E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141: 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen G., Bender J. (2002). Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J., Romero-Severson E., Ruegger M.O., Chapple C. (2008). Semidominant mutations in reduced epidermal fluorescence 4 reduce phenylpropanoid content in Arabidopsis. Genetics 178: 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M., Mithen R. (2009). Glucosinolates, isothiocyanates and human health. Phytochem. Rev. 8: 269–282. [Google Scholar]
- Tronchet M., Balagué C., Kroj T., Jouanin L., Roby D. (2010). Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J.H., Goeminne G., Halpin C., Rohde A., Morreel K., Boerjan W. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Wagner D., Hoecker U., Quail P.H. (1997). RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell 9: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner L.A., Li G., Ware D., Somssich I.E., Davis K.R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27: 327–338. [DOI] [PubMed] [Google Scholar]
- Wathugala D.L., Hemsley P.A., Moffat C.S., Cremelie P., Knight M.R., Knight H. (2012). The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195: 217–230. [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Mo H., Chapple C. (2010). Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64: 898–911. [DOI] [PubMed] [Google Scholar]
- Winkler R.G., Frank M.R., Galbraith D.W., Feyereisen R., Feldmann K.A. (1998). Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Isolation of mutations in the cytochrome p450 gene superfamily. Plant Physiol. 118: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gou M., Liu C.J. (2013a). Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25: 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yao J., Zhang Y., Sun Y., Mou Z. (2013b). The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75: 484–497. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu C.J. (2015). Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 8: 17–27. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Dixon R.A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16: 227–233. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guan H., Leal F., Grey P.H., Oppenheimer D.G. (2013). Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLoS ONE 8: e53924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (2009). Transcriptional regulation of lignin biosynthesis. Plant Signal. Behav. 4: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.