Comparison of the inflorescence and shoot apical meristems reveals differences in gene expression and H3K27me3/H3K4me3 ratio and overexpression of a key methylase increases inflorescence meristem activity and panicle size.
Abstract
Rice inflorescence meristem (IM) activity is essential for panicle development and grain production. How chromatin and epigenetic mechanisms regulate IM activity remains unclear. Genome-wide analysis revealed that in addition to genes involved in the vegetative to reproductive transition, many metabolic and protein synthetic genes were activated in IM compared with shoot apical meristem and that a change in the H3K27me3/H3K4me3 ratio was an important factor for the differential expression of many genes. Thousands of genes gained or lost H3K27me3 in IM, and downregulation of the H3K27 methyltransferase gene SET DOMAIN GROUP 711 (SDG711) or mutation of the H3K4 demethylase gene JMJ703 eliminated the increase of H3K27me3 in many genes. SDG711-mediated H3K27me3 repressed several important genes involved in IM activity and many genes that are silent in the IM but activated during floral organogenesis or other developmental stages. SDG711 overexpression augmented IM activity and increased panicle size; suppression of SDG711 by RNA interference had the opposite effect. Double knockdown/knockout of SDG711 and JMJ703 further reduced panicle size. These results suggest that SDG711 and JMJ703 have agonistic functions in reprogramming the H3K27me3/H3K4me3 ratio and modulating gene expression in the IM.
INTRODUCTION
Rice (Oryza sativa) grain yield is mainly determined by number of panicles per plant, grain number per panicle, and grain weight. Among these factors, grain number per panicle contributes the most to grain yield (Sakamoto and Matsuoka, 2008; Xing and Zhang, 2010), and grain number, in turn, depends on inflorescence meristem (IM) activity to produce the primary branches and later spikelets during panicle development. The shift from the shoot apical meristem (SAM) to IM is accompanied by clear changes in a number of developmental and morphological traits. In rice, the first morphological change is a rapid increase in meristem size (Itoh et al., 2005). Genes involved in the vegetative to reproductive transition in rice include LEAFY-LIKE/ABERRANT PANICLE ORGANIZATION2 (RFL/APO2), SEPETALLA-LIKE MADS34/PANICLE PHYTOMER2 (MADS34/PAP2), and three APETALA1 (AP1)/ FRUITFULL-like genes (MADS14, MADS15, and MADS18); these genes are activated in the meristem during the transition and are required for panicle development (Fornara et al., 2004; Gao et al., 2010; Ikeda-Kawakatsu et al., 2012; Kobayashi et al., 2012). After the transition, the IM starts to initiate panicle branch meristem primordia. Several rice transcription factor genes have been shown to regulate IM activity, panicle branching, and grain production. For instance, increased expression of SPL14 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE14) promotes panicle branching and grain yield (Jiao et al., 2010; Miura et al., 2010). IM activity also requires cytokinins (Han et al., 2014) and rice genes involved in cytokinin accumulation in the IM regulate panicle branching and grain production. For instance, LONELY GUY (LOG), which encodes a novel enzyme that works in the final step of bioactive cytokinin synthesis, maintains rice IM activity and promotes panicle branching (Kurakawa et al., 2007). Reduced expression of the rice cytokinin oxidase gene GN1a/CKX2 causes accumulation of active cytokinins in the IM and increases the grain number (Ashikari et al., 2005). Therefore, regulation of IM size and activity is crucial to determining rice grain production. However, chromatin and epigenetic mechanisms implicated in regulating rice IM activity and panicle development remain unknown at the present time.
Important epigenetic regulators of developmental processes are the Polycomb-group (PcG) and Trithorax-group proteins, which catalyze histone H3 lysine 27 trimethylation (H3K27me3) or H3 lysine 4 trimethylation (H3K4me3), respectively (Pien and Grossniklaus, 2007; Steffen and Ringrose, 2014). The presence of H3K27me3 is largely correlated with gene silencing in animal and plant cells. In plants, the H3K27me3-marked genes have very low expression levels and often exhibit a high degree of tissue specificity (Zhang et al., 2007; Makarevitch et al., 2013), consistent with a function of H3K27me3 in maintaining gene repression during growth. H3K27me3 has been implicated in regulation of many plant developmental pathways, including seed development, flowering time, vernalization, and organ identity (reviewed in Köhler and Villar, 2008; Zheng and Chen, 2011). Whole-genome chromatin immunoprecipitation (ChIP) experiments have shown that ∼20% of Arabidopsis thaliana and rice genes are marked by H3K27me3 (He et al., 2010; Lafos et al., 2011). In both plants and animals, a remarkable number of transcription factors are targeted by H3K27me3 (Köhler and Villar, 2008; Zheng and Chen, 2011), further supporting the role of H3K27me3 in the regulation of developmental gene expression. Moreover, the levels of H3K27me3 were profiled in several plant tissues (Wang et al., 2009b; Lafos et al., 2011). These studies revealed large tissue-specific variation in H3K27me3 levels with many genes shown to gain or lose H3K27me3 during cell differentiation, demonstrating dynamic regulation of this epigenetic modification in responding to developmental signals in plants. In addition, H3K27me3 marks many stress-inducible genes; these marks are removed upon application of environmental signals or stresses (Charron et al., 2009; Li et al., 2013), suggesting that H3K27me3 is also dynamically regulated in response to environmental cues in plants.
About 40% of plant genes are marked by H3K4me3, which accumulates predominantly in 5′ of the genes (Zhang et al., 2009; He et al., 2010; Hu et al., 2012). Genes predominantly marked with H3K4me3 are actively transcribed in plants. H3K4me3 has been shown to play an important role in gene expression involved in plant development and stress adaptation (Avramova, 2009; Fromm and Avramova, 2014). Mutation of genes involved in H3K4 methylation and demethylation affected genome activity and plant growth (reviewed in Berr et al., 2011; Chen et al., 2011). For instance, mutation of JMJ703, an H3K4 demethylase gene, affects cell division in rice, resulting in reduced plant height with shorter internodes and smaller panicles (Chen et al., 2013).
H3K4me3 and H3K27me3 are considered to be mutually repulsive and antagonist marks for gene activity. However, many genes are simultaneously marked by both H3K4me3 and H3K27me3 (Zhang et al., 2009; He et al., 2010; Hu et al., 2012), referred to as “bivalent” domains (Young et al., 2011; Voigt et al., 2013). The bivalent modifications were identified on a subset of genes in Arabidopsis (Sequeira-Mendes et al., 2014). It is suggested the simultaneous presence of the active and the repressive modifications and associated complexes helps to maintain bivalent loci in a state that is both responsive to developmental cues and at the same time insensitive to subthreshold noise (Voigt et al., 2013).
A subset of the PcG proteins, including Enhancer of Zeste [E(z)], Extra sex combs (Esc), Su(z)12, and Nurf55 in Drosophila melanogaster form the Polycomb-repressive complex2 (PRC2), which is involved in catalyzing the addition of H3K27me3 (Schwartz and Pirrotta, 2008). Plants have clear homologs of the four core protein components of PRC2, often with multiple genes encoding each component. In Arabidopsis, CURLY LEAF (CLF), SWINGER (SWN), and MEDEA (MEA) encode E(z) homologs. MEA appears to function primarily in gametophyte and seed development, whereas CLF and SWN are broadly expressed and partially redundant in vegetative and reproductive development (Hennig and Derkacheva, 2009). The Arabidopsis clf swn double mutants show severe developmental defects (such as organ identity) and loss of H3K27me3 in vegetative tissues (Chanvivattana et al., 2004; Lafos et al., 2011), suggesting that PRC2 proteins also catalyze the addition of H3K27me3 in plants. The rice genome contains two genes encoding homologs of E(z) (CLF or SDG711 and EZ1 or SDG718), Su(Z)12 (EMF2a and EMF2b), and ESC (FIE1 and FIE2) (Luo et al., 2009; Liu et al., 2014).
In this work, we studied genome-wide levels of the H3K27me3 and H3K4me3 modifications and gene expression during the SAM-to-IM transition in rice and examined the function of SDG711 and JMJ703 in this process. The analysis revealed that change of H3K27me3/H3K4me3 ratio during the transition was critical for genome-wide gene expression reprogramming in IM and that both SDG711 and JMJ703 were required for acquisition of H3K27me3 on thousands of genes in the IM, including those known to be involved in IM activity and panicle development.
RESULTS
SDG711 Is Involved in the Control of Panicle Meristem Activity and Gene Expression
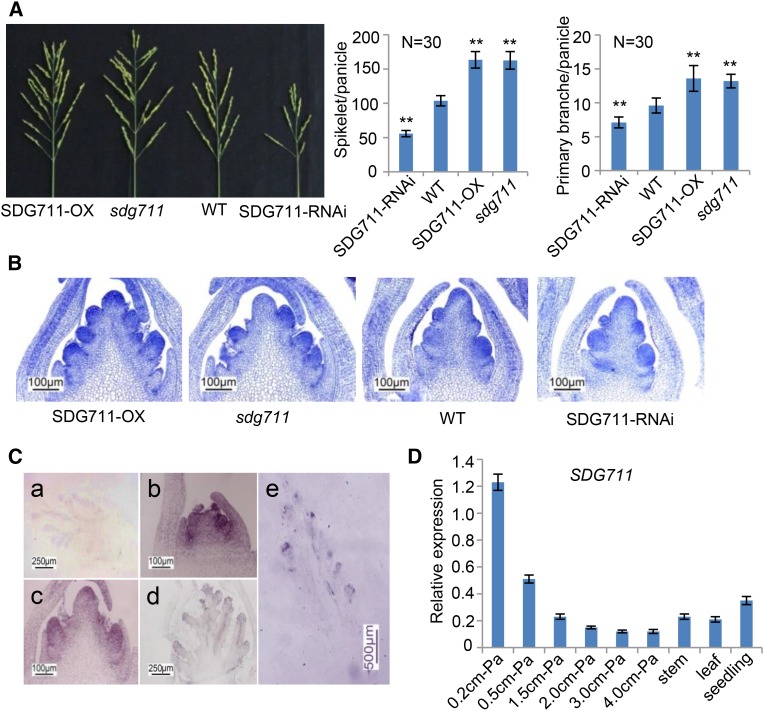
We recently characterized transgenic and mutant plants for the rice E(z) genes SDG711 and SDG718 (Liu et al., 2014). We observed that SDG711 expression levels affected panicle size: Overexpression of SDG711 led to the production of larger panicles with more primary branches and spikelets, and SDG711 RNA interference (RNAi) had the opposite effect (Figure 1A). Histological analysis of the IM revealed that the number of primary branch primordia increased and decreased in the overexpression and RNAi lines, respectively, compared with wild-type plants (Figure 1B), suggesting that SDG711 expression was important for IM activity. In situ hybridization experiments detected SDG711 transcripts throughout the IM with higher levels in the primordia of primary branches, spikelets, and florets (Figure 1C). The hybridization signals became weaker in more advanced stages of panicle development. Quantitative RT-PCR analysis indicated that SDG711 transcript level was relatively high in the IM at early stages and gradually decreased during IM growth (Figure 1D), confirming the in situ hybridization results. These observations suggested that the expression of SDG711 was dynamic in IM and might be involved in reprogramming the expression of genes involved in IM activity and panicle development.
Figure 1.
Up- and Downregulation of SDG711 Affected Panicle Development.
(A) Panicle phenotypes of SDG711 overexpression (OX), gain-of-function mutant (sdg711), and RNAi plants compared with the wild type (WT). Means ± sd of primary branches and spikelets per panicle (n = 30) are presented on the right. Significant differences between the transgenic or mutant plants and the wild type (Student’s t tests, P value < 0.01) are marked by double asterisks.
(B) Longitudinal section of inflorescence meristem from the indicated genotypes.
(C) In situ hybridization of SDG711 transcripts at different developmental stages of the inflorescence meristem. Length of the meristem was ∼1 mm in (a), 0.25 mm in (b), 0.5 mm in (c), 1.5 mm in (d), and 3 mm in (e). Probe used was sense in (a) and antisense in (b) to (e).
(D) Quantitative RT-PCR analysis of SDG711 transcripts in inflorescence meristem (at different developmental stages), stem, leaf, and seedling. Levels relative to ACTIN transcripts are presented (y axis).
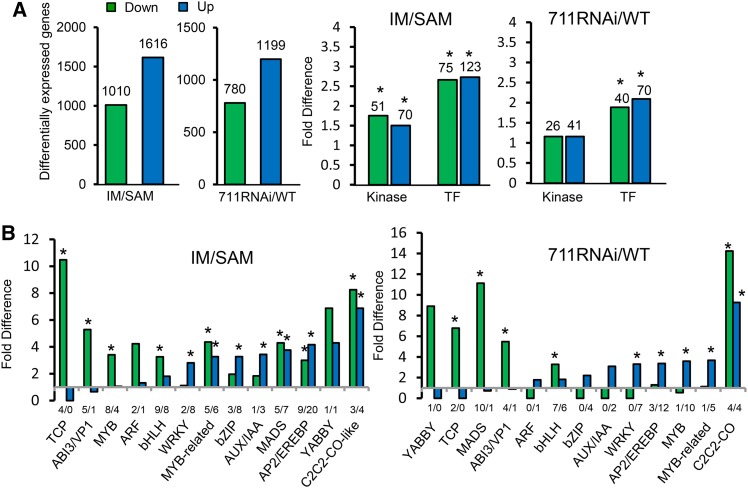
To study gene expression changes during the SAM-to-IM transition and to examine the function of SDG711 in the process, we analyzed the transcriptomes of wild-type SAM and IM and SDG711 RNAi IM (lines 3, 7, and 9 combined) (Liu et al., 2014). The developmental stages of SAM and IM were determined as previously described (Itoh et al., 2005). Precisely, SAMs were harvested from 4-week-old plants at the 6- to 7-leaf stage, and IMs (∼1 to 2 mm) were collected from 6- to 7-week-old SDG711 RNAi plants and 7- to 8-week-old wild-type grown in the field during the May to June period in the Wuhan area (see Methods). To avoid effects due to tissue culture for the transgenic plants, wild-type plants used in this study were regenerated from callus culture. Two biological replicates were sequenced with the Illumina HiSeq-2000 sequencing system and analyzed by using TopHat (version 2.0.13) to align the clean tags filtered by Trimmomatic (version 0.32) to the reference genome of Nipponbare (version 7.0). Cufflinks (version 2.2.1) was used for identification of differentially expressed genes (see Methods). About 21 to 34 million clean reads per sample were obtained. More than 95% of the clean reads for each sample were mapped to the reference genome (Supplemental Figure 1A). The two replicates displayed a high repeatability (R2 > 0.96) (Supplemental Figure 1B). A total of 15,491 and 15,949 genes were expressed in wild-type SAM and IM, respectively (Supplemental Figure 1C). A number of transposable element (TE) genes were found to be expressed, but the transcript levels were relative low (Supplemental Figures 1C and 1D). In IM, 1616 and 1010 genes were respectively up- and downregulated more than 2-fold (P value < 0.05) compared with SAM (Figure 2A). When we analyzed the differentially regulated genes, we found that many metabolic pathways were enriched (false discovery rate [FDR] < 0.01) for upregulation (Supplemental Table 1). Most of the enriched pathways were related to photosynthesis, glycolysis, amino acid biosynthesis, and phenylpropanoid biosynthesis, indicating that general metabolic and protein synthetic activities were higher in IM. In addition, genes involved in hormone signal transduction were also found to be enriched (FDR < 0.01) among the upregulated genes (Supplemental Table 1). A similar number of expressed genes (15,922) were detected in SDG711 RNAi IM (Supplemental Figure 1A). Compared with the wild type, 1199 genes were upregulated and 780 were downregulated (>2-fold, P value < 0.05) (Figure 2A). About 42.5% (518/1199 genes) of the upregulated genes overlapped with those induced in IM, while only 4% (50/1199) corresponded to those repressed in IM (Supplemental Figure 2). In addition, genes of some metabolic pathways upregulated in wild-type IM were further induced by SDG711 RNAi (Supplemental Table 1). These observations suggested that SDG711 might have a function to repress genes upregulated during the SAM-to-IM transition.
Figure 2.
Differential Gene Expression between IM and SAM and between SDG711 RNAi IM and the Wild Type.
(A) Left histograms: numbers of up- and downregulated genes (>2-fold, P value < 0.05) between IM and SAM and between SDG711 RNAi IM and the wild type (WT). Right histograms: numbers of protein kinase and transcription factor (TF) genes differentially regulated in IM compared with SAM or in SDG711 RNAi compared with the wild type. Significant enrichments (Fisher’s tests, P value <0.05) are indicated by asterisks.
(B) Relative enrichments of transcription factor families. Significant enrichments (Fisher’s tests, P value <0.05) are indicated by asterisks. Down/upregulated genes numbers are indicated below the bars. Fold changes are differences relative to the observed frequencies of specific subsets within the genome.
When we analyzed the categories of the differentially expressed genes, we found that those encoding protein kinases (RGAP version 7.0 annotation) and transcription factors (http://drtf.cbi.pku.edu.cn) were significantly enriched (Fisher’s tests, P value < 0.05), with 123 and 75 transcription factor genes and 70 and 51 protein kinase genes being up- and downregulated, respectively, in IM compared with SAM (Figure 2A). Differentially regulated transcription factors genes found in SDG711 RNAi IM were also significantly enriched (Fisher’s tests, P value < 0.05), with 70 and 40 being up- and downregulated, respectively, compared with the wild type (Figure 2B). Several specific transcription factor families were enriched among the differentially expressed genes (Fisher’s tests, P value < 0.05) (Figure 2B).
Among the differentially regulated transcription factor genes, the three AP1-like genes (MADS15, MADS18, and MADS14) and the SEP-like gene (MADS34/PAP2), which are IM mark genes and have overlapping function to specify the panicle meristem identity (Kobayashi et al., 2012), were upregulated by 5- to 355-fold (P value <0.05) in wild-type IM compared with SAM (Table 1). In addition, RFL/APO2, the rice ortholog of LEAFY (another marker gene of the vegetative to reproductive transition) (Rao et al., 2008; Ikeda-Kawakatsu et al., 2012), was also upregulated in IM. These data confirmed the developmental stages of the collected meristem samples. Other transcription factor genes previously shown to be involved in panicle development were found to be upregulated, including MADS56 (Ryu et al., 2009), SPL14 (Jiao et al., 2010; Miura et al., 2010), EATB (Ethylene-responsive factor Associated with Tillering and panicle Branching) (Qi et al., 2011), AP2-39 (Yaish et al., 2010), and the GATA C2C2 zinc finger transcription factor gene (Os05g50270) (Wang et al., 2009a). Conversely, other transcription factor genes, such as MADS23 and MADS27 (which are AGL17 homologs), and a few AP2 genes (AP22-106, AP2-125, and AP2-33/IDS1 [INDETERMINATE SPIKELET1]) (Lee and An, 2012) were found to be among the downregulated genes in IM (Table 1).
Table 1. Relative Transcript Levels and H3K27me3 Changes of Panicle Development and Hormone-Related Genes in IM Compared to SAM and in SDG711RNAi IM Compared to the Wild Type.
| Gene Name | Locus Number | IM/SAM (H3K27me3) | SDG711RNAi IM/Wild Type (H3K27me3) |
|---|---|---|---|
| MADS15/AP1 | LOC_Os07g01820 | 355.60 (↓) | 0.51 (−) |
| MADS34/PAP2 | LOC_Os03g54170 | 200.86 (↑) | 0.36 (↓) |
| MADS14/AP1 | LOC_Os03g54160 | 15.24 (−) | 0.72 (↓) |
| MADS18/AP1 | LOC_Os07g41370 | 5.06 (−) | 1.05 (↓) |
| MADS56 | LOC_Os10g39130 | 3.43 (−) | 1.06 (↓) |
| MADS23 | LOC_Os08g33488 | 0.41 (↑) | 0.80 (↓) |
| MADS27 | LOC_Os02g36924 | 0.33 (↑) | 1.01 (↓) |
| AP2-33/IDS1 | LOC_Os03g60430 | 0.43 (↑) | 0.56 (↓) |
| AP2-106 | LOC_Os07g22730 | 0.06 (↑) | 0.85 (↓) |
| AP2-121 | LOC_Os03g09170 | 3.59 (−) | 3.08 (↓) |
| AP2-123 | LOC_Os09g28440 | 3.62 (−) | 2.22 (↓) |
| AP2-125 | LOC_Os03g08470 | 0.30 (↑) | 1.05 (↓) |
| AP2-163 | LOC_Os03g64260 | 2.05 (↓) | 2.09 (−) |
| C2C2-GATA | LOC_Os05g50270 | 3.77 (↓) | 0.85 (−) |
| JAZ1/EG2 | LOC_Os04g55920 | 1.56 (↑) | 2.65 (↓) |
| EATB | LOC_Os09g28440 | 3.62 (−) | 2.22 (↓) |
| RFL/APO2 | LOC_Os04g51000 | 4.24 (−) | 1.20 (↓) |
| LOG | LOC_Os01g40630 | 4.14 (−) | 2.03 (↓) |
| SPL14 | LOC_Os08g39890 | 4.54 (−) | 1.11 (−) |
| TDC2 | LOC_Os08g04540 | 2.62 (↓) | 5.32 (−) |
| YUC9 | LOC_Os01g16714 | 0.87 (↓) | 2.02 (−) |
| PIN9 | LOC_Os01g58860 | 11.75 (−) | 1.88 (↓) |
| IAA8 | LOC_Os02g49160 | 1.62 (−) | 3.43 (−) |
| IAA3 | LOC_Os12g40900 | 1.49 (↑) | 2.25 (↓) |
| GH3-2 | LOC_Os01g55940 | 11.21 (−) | 2.96 (↓) |
| GH3-6 | LOC_Os05g05180 | 1.99 (−) | 2.06 (↓) |
| SAUR23 | LOC_Os04g56690 | 24.39 (−) | 5.85 (↓) |
| SAUR11 | LOC_Os02g42990 | 4.77 (−) | 2.61 (−) |
| CKX4 | LOC_Os01g71310 | 1.52 (−) | 2.05 (↓) |
| CKX2/GNA1 | LOC_Os01g10110 | 1.37 (↑) | 2.12 (↓) |
| RR11 | LOC_Os02g42060 | 3.27 (−) | 0.66 (−) |
| RR3 | LOC_Os02g58350 | 0.79 (−) | 0.16 (−) |
In SDG711 RNAi plants, we observed that the expression of MADS34/PAP2 was decreased and that of EATB and JAZ1/EG2 (EXTRA GLUME2; which encodes a zinc finger protein involved in spikelet development) (Cai et al., 2014) was upregulated by more than 2-fold (Table 1). In addition, many genes involved in auxin and cytokinin biosynthesis and signaling were differentially expressed in wild-type IM versus SAM or in SDG711 RNAi IM compared with the wild type (Table 1). Remarkably, several auxin biosynthesis (TDC2 and YUC9) and responsive genes (GH3-2, GH3-6, SAUR23, SAUR11, IAA3, and IAA8) and cytokinin oxidase genes (i.e., CKX2/Gn1a and CKX4) were upregulated by more than 2-fold in SDG711 RNAi IM (Table 1). Conversely, the cytokinin A-type responsive regulator gene RR3 was repressed by more than 6-fold (Table 1).
To confirm the RNA-seq data, we performed RT-PCR analysis of CKX gene family in IM of wild-type, SDG711 RNAi, and overexpression (lines 2, 4, and 5 combined) plants and a gain-of-function mutant (sdg711) (Liu et al., 2014). The analysis revealed that the expression of several members (i.e., CKX2/Gn1a, CKX3, CKX4, CKX5, CKX10, and CKX11) of this gene family was increased in the RNAi lines but repressed in the overexpression plants and the gain-of-function mutant (Supplemental Figure 3A). Gn1a/CKX2 is expressed in IM and its expression level affects panicle size (Ashikari et al., 2005). We performed in situ hybridization to analyze the transcript levels of Gn1a/CKX2 in IM of different genotypes. The experiments revealed lower transcript levels of the gene in the overexpression and higher levels in the RNAi compared with wild-type plants (Supplemental Figure 3B). Dosage of cytokinins in IM revealed that different types of active cytokinins were increased in the overexpression and decreased in the RNAi plants compared with the wild type (Supplemental Figure 3C). These data suggested that the panicle phenotype of SDG711 transgenic plants may be partly related to CKX genes expression and cytokinin levels in the meristem. In addition, quantitative RT-PCR was performed to validate the RNA-seq data of 18 genes with or without differential expression between SAM and IM or between the wild type and SDG711 RNAi IM (Supplemental Figure 4).
Genome-Wide H3K27me3 and H3K4me3 Changes during the SAM-to-IM Transition
To study H3K27me3 changes during the vegetative-to-reproduction transition in rice and examine the function of SDG711, we analyzed genome-wide H3K27me3 in SAM and IM of callus-regenerated wild-type and SDG711 RNAi plants by ChIP-seq. H3K4me3 is generally thought to be antagonistic to H3K27me3 in the regulation of gene expression. In addition, previous data showed that mutation of H3K4me3 demethylase gene JMJ703 affected panicle size (Chen et al., 2013). To study the functional relationship between H3K27me3 and H3K4me3 during the transition, we investigated genome-wide H3K4me3 levels in wild-type SAM and IM and H3K4me3 and H3K27me3 in jmj703 mutant IM (from 7- to 8-week-old plants). SAM and IM were harvested at the same stages as for the RNA-seq sample collection. The precipitated DNAs were sequenced by the Illumina HiSeq-2000 sequencing system. The analyzed materials and the ChIP-seq data are listed in Supplemental Table 2. Clean reads were aligned with the reference genome (The Institute for Genomic Research, TIGR version 7.0) using Bowtie2 (version 2.1.0) to determine the frequency of reads matching each genomic base pair position. Differential H3K27me3 levels between different samples were observed at specific genomic locations, while most regions were at about similar levels (Supplemental Figure 5). More than 10,000 H3K27me3 and 20,000 H3K4me3 peaks were identified (P value <1e-5) using MACS (Version 1.4) (Supplemental Table 2). From 65 to 74% of H3K27me3 peaks were found to match with genes (non-TE), 10 to 12% to TEs, and 16 to 23% in the intergenic region. By contrast, more than 84% of H3K4me3 peaks mapped to non-TE genes (Supplemental Figure 6).
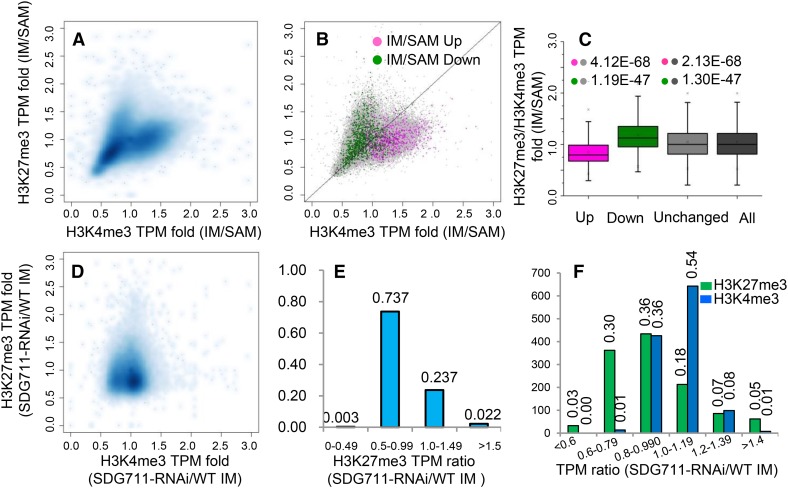
To analyze the function of H3K27me3 and H3K4me3 in gene activity, we examined the quantitative relationships between the ChIP-seq and the RNA-seq reads using contour plots. As expected, we observed a positive correlation between levels of H3K4me3 (tags per million [TPM]), but not H3K27me3, and transcript abundance (reads per kilobase per million reads [RPKM]) in both SAM (R2 =0.219) and IM (R2 = 0.362) (Supplemental Figures 7A and 7B). Interestingly, H3K27me3 and H3K4me3 in both SAM (R2 = 0.471) and IM (R2 = 0.282) displayed a positive correlation in the contour plots, in which a bimodal distribution pattern was observed (Supplemental Figure 7C). This bimodal distribution pattern of the two epigenetic marks was also observed in rice seedlings (He et al., 2010), suggesting that there might be a concurrence of H3K27me3 and H3K4me3 in many genes. Next, we analyzed the change of H3K27me3 and H3K4me3 levels between IM and SAM. We found a bimodal distribution of the changes of both epigenetic marks between the two meristems (Figure 3A), indicating that the amounts (TPM) of H3K27me3 and H3K4me3 on many genes increased in IM, but genes with higher increases of H3K27me3 showed lower levels of H3K4me3, and vice versa. Comparison between the ChIP-seq and RNA-seq data revealed that upregulated genes in IM were mostly distributed in genes with lower H3K27me3/H3K4me3 ratios and downregulated genes were found to have higher H3K27me3/H3K4me3 ratios (Figures 3B and 3C). These observations suggested that change of H3K27me3/H3K4me3 ratio was implicated in overall gene expression changes during the SAM-to-IM transition.
Figure 3.
Quantitative Changes (TPM) of H3K27me3 and H3K4me3 and Correlation with Differential Gene Expression between IM and SAM and between SDG711 and the Wild Type.
(A) Contour plots of H3K4me3 and H3K27me3 changes between IM and SAM.
(B) Distribution of up- and downregulated genes (>2-fold, P value <0.05) in IM compared with SAM.
(C) Box plots of up- and downregulated, unchanged, and all rice genes relative to H3K27me3/H3K4me3 ratio. P values (t tests) from comparison between differentially regulated gene and unchanged or all genes are indicated.
(D) Contour plots of H3K4me3 and H3K27me3 changes between SDG711 RNAi and the wild type.
(E) Distribution of different fractions of H3K27me3 TPM ratio between SDG711 RNAi and the wild type (WT). The ratio of ∼74% of genes was below 1. y axis, percentage of genes.
(F) Distribution of upregulated genes (>2-fold, P value <0.05) on TMP ratio fractions of H3K27me3 or H3K4me3 between SDG711 RNAi and the wild type. y axis, gene numbers.
Differential H3K27me3 between Shoot and Panicle Meristems
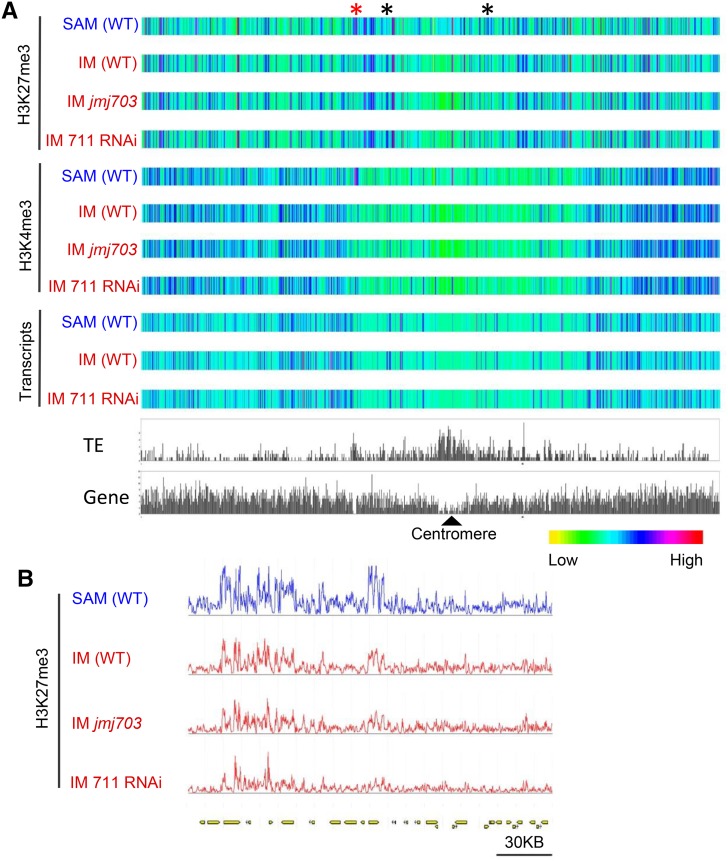
Globally, H3K27me3 or H3K4me3 in the different meristem samples displayed about the same profiles along chromosomes (Figure 4A). However, clear differences of H3K27me3 in discrete chromosomal locations could be observed between SAM and IM (Figures 4A and 4B), suggesting that changes in contiguous H3K27me3-marked genomic regions might occur during the SAM-to-IM transition (Figure 4B). We identified a total of 15,247 genes marked by H3K27me3 in SAM and 13,954 in IM, representing 25 to 27% of total genes (Figure 5A). For H3K4me3, we identified 26,562 genes in SAM and 26,474 in IM, corresponding to ∼47% of total rice genes (Figure 5B). The percentages of both types of marked genes were comparable to previous data in rice and Arabidopsis (Cui et al., 2010; He et al., 2010; Lafos et al., 2011). The H3K27me3-marked genes displayed a higher tissue specificity score compared with unmarked genes (Supplemental Figure 8), confirming the notion that H3K27me3 preferentially marks developmental genes (Kinoshita et al., 2001; Chanvivattana et al., 2004; Schubert et al., 2005). There were 10,233 genes (representing 67% of those marked in SAM and 73% in IM) marked commonly in both meristems. Compared with SAM, IM showed a gain of 3721 and a loss of 5014 H3K27me3-marked genes (Table 2). By contrast, most of the H3K4me3-marked genes (97%) were common in both the SAM and IM (Figure 5B).
Figure 4.
Heat Map of H3K27me3 and H3K4me3 Levels and Transcript Abundance on Chromosome 3.
(A) Comparison between SAM and IM from different genotypes. Regions with clear difference between SAM and IM are indicated by asterisks. Relative abundance of TEs and genes of the chromosome are indicated.
(B) Close-up view of the H3K27me3-marked region indicated by the red asterisk.
Figure 5.
Venn Diagrams of Differentially Marked Genes by H3K27me3 and H3K4me3, between SAM and IM, and between SDG711 RNAi or jmj703 IM and the Wild Type.
Differentially marked genes by H3K27me3 ([A], [C], and [E]) and H3K4me3 ([B], [D], and [F]) between SAM and IM ([A] and [B]) and between SDG711 RNAi ([C] and [D]) or jmj703 IM and the wild type ([E] and [F]). Black circle represents overlap between genes that lost H3K27me3 in SDG711 RNAi or in both SDG711 RNAi and jmj703 mutant IM and those gained H3K27me3 in IM relative to SAM.
Table 2. Numbers of Genes with Differential H3K27me3 in IM Compared to SAM and in SDG711 RNAi IM Compared to the Wild Type.
|
IM/SAM | |||
|---|---|---|---|
| H3K27me3 Loss | H3K27me3 Gain | Genome | |
| Total | 5,014 (191↑140↓) | 3,721 (122↑72↓) | 55,546 |
| Protein kinases | 168* (7↑5↓) | 155* (6↑2↓) | 1,600 |
| Transcription factors | 219* (17↑13↓) | 133* (9↑6↓) | 1,549 |
| SDG711 RNAi IM/wild type | |||
| K27me3 Loss |
H3K27me3 Gain |
Genome |
|
| Total | 5,406 (134↑65↓) | 1,378 (28↑16↓) | 55,546 |
| Protein kinases | 248* (4↑2↓) | 39 (0↑0↓) | 1,600 |
| Transcript factors | 120 (11↑4↓) | 36 (2↑0↓) | 1,549 |
Numbers of genes that showed upregulation (indicated by upwards arrows) or downregulation (indicated by downwards arrows) by more than 2-fold (P value < 0.05) are in parentheses. Significant enrichments (Fisher’s tests, P value < 0.05) are indicated by asterisks.
Analysis of the differentially marked genes revealed that several cellular pathways were enriched for gain of H3K27me3 in IM (Supplemental Table 3). In addition, transcription factor and protein kinase genes were enriched for gain or loss of H3K27me3 in IM (Fisher’s tests, P value < 0.05) (Table 2). Further analysis revealed that some of transcription factor subfamilies (such as ARF, Aux/IAA, MADS, AP2, TCP, etc.) were enriched for differential H3K27me3 in IM (Supplemental Figure 9).
When compared with the RNA-seq data, we found no general correlation between loss or gain of H3K27me3 and up- or downregulation of gene expression (>2-fold) (Table 2). Nevertheless, we observed that the expression of a number of genes encoding transcription factors, protein kinases, and metabolic enzymes was correlated with the gain or loss of H3K27me3 during the SAM-to-IM transition (Supplemental Data Set 1). Among the upregulated IM and panicle development regulatory genes, MADS15 (AP1-like), MADS5 (SEP1-like), and the GATA C2C2 zinc finger gene (Os05g50270) were found to lose H3K27me3 (Table 1). Conversely, downregulated transcription factor genes such as MADS23, MADS27, AP22-106, AP2-125, and AP2-33/IDS1 gained H3K27me3 in IM (Table 1). Several highly upregulated genes involved in photosynthesis (e.g., light-harvesting complex I chlorophyll a/b binding protein 1, Os06g21590), glycolysis (e.g., pyruvate decarboxylase, Os03g18220), cytokinin synthesis (e.g., cytokinin dehydrogenase, Os01g56810), and linoleic acid metabolism (e.g., lipoxygenase Os12g37260) lost H3K27me3 (Supplemental Data Set 1). Quantitative ChIP-PCR was performed to validate the ChIP-seq data of 14 genes with or without differential H3K27me3 between SAM and IM (Supplemental Figure 4A).
Effect of SDG711 RNAi on Genome-Wide H3K27me3 in the Inflorescence Meristem
Compared with wild-type IM, SDG711 RNAi reduced the amount of H3K27me3 (TPM) of a majority of the marked genes (RNAi/wild type < 1) (Figures 3D and 3E). Consequently, the changes of the two marks in SDG711 RNAi IM were not correlated and a water drop-like, instead of a bimodal, pattern was observed (Figure 3D). Comparison with the RNA-seq data revealed that upregulated genes (>2-fold, P value < 0.05) in SDG711 RNAi IM were mostly distributed in genes with reduced H3K27me3 (RNAi/wild type < 1) and increased H3K4me3 (RNAi/wild type > 1) levels (Figure 3F), supporting that the change of H3K27me3/H3K4me3 ratio was important for gene activity.
Peak analysis revealed a loss of 5406 and a gain of 1378 H3K27me3-marked genes and a relative smaller change of H3K4me3-marked genes (with a gain of 1513 and a loss of 576 genes) in SDG711 RNAi IM (Figures 5C and 5D). There was no enrichment between loss of H3K27me3 and gain of H3K4m3, while enrichment between loss of H3K4me3 and loss of H3K27me3 was observed (Supplemental Table 4). Interestingly, 56% (2083 out of 3721) of the genes that gained H3K27me3 in wild-type IM (compared with SAM) were found to lose the mark in SDG711 RNAi (Figure 5A). In addition, most of the cellular pathways that were enriched for gain of H3K27me3 in wild-type IM were found to be enriched for loss of H3K27me3 in SDG711 RNAi IM (Supplemental Table 3). These observations suggested that SDG711 was required for acquisition of H3K27me3 during SAM-to-IM transition. We found that 68 genes that showed both upregulation and loss of H3K27me3 in SDG711 RNAi IM were also upregulated (>2-fold) in wild-type IM (Supplemental Figure 2 and Supplemental Data Set 2), suggesting that these IM-induced genes were repressed by SDG711-mediated H3K27me3. Seven genes that showed both upregulation and H3K27me3 loss in SDG711 RNAi were downregulated (>2-fold) in wild-type IM compared with SAM (Supplemental Figure 2 and Supplemental Table 6), suggesting that SDG711-mediated H3K27me3 was involved in the repression of these “SAM-induced” genes in IM.
We found that 248 kinase and 120 transcription factor genes lost H3K27me3, while relatively fewer genes of these categories gained H3K27me3 in the RNAi IM (Table 2). A majority of the differentially regulated genes involved in IM activity and panicle development and in auxin and cytokinin synthesis or signaling were found to lose H3K27me3 in the RNAi IM (Table1), among which the panicle developmental genes (EBTA and JAZ1/EG2) (Qi et al., 2011; Cai et al., 2014), auxin-responsive genes (IAA3, GH3-2, and SAUR23), and CKX genes (CKX2/Gn1a and CKX4) were upregulated. In addition, many upregulated metabolic genes were found to lose H3K27me3 (Supplemental Data Set 1). These genes might be targets for repression by SDG711-mediated H3K27me3 in IM. Quantitative ChIP-PCR was performed to validate the ChIP-seq data of four genes with or without differential H3K27me3 between SDG711 RNAi and wild-type IM (Supplemental Figure 4B).
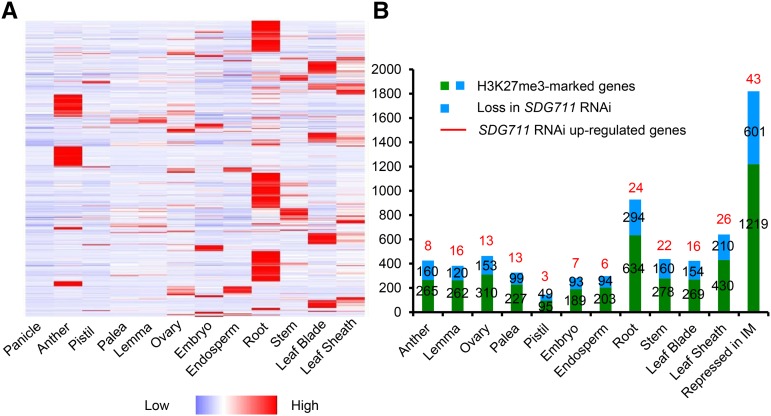
Comparison of the present RNA-seq and H3K27me3 ChIP data with rice transcriptomic data (http://ricexpro.dna.affrc.go.jp) revealed that 1820 H3K27me3-marked genes repressed in IM are expressed during floral organogenesis or other developmental stages (Figure 6). About one-third (601/1820) of the genes lost H3K27me3 in SDG711 RNAi IM (Figure 6), some of which were derepressed (Figure 6; Supplemental Data Set 3). This analysis suggested that SDG711 was involved in H3K27me3 of genes repressed in IM.
Figure 6.
Involvement of SDG711-Mediated H3K27me3 in Maintaining Repression in IM of Genes That Are Expressed during Floral Organogenesis and Other Developmental Stages.
(A) Heat map of transcripts in the indicated organ/tissues of 1820 genes that were marked by H3K27me3 and repressed in IM.
(B) Number of H3K27me3-marked genes in IM that are expressed in the indicated organs (red and blue bars). SDG711 RNAi reduced about one-third of the marked genes (red bars). Numbers of genes that were derepressed in SDG711 RNAi IM are indicated on the top of the bars.
Functional Relationship between SDG711 and JMJ703 in H3K27me3 and Panicle Development
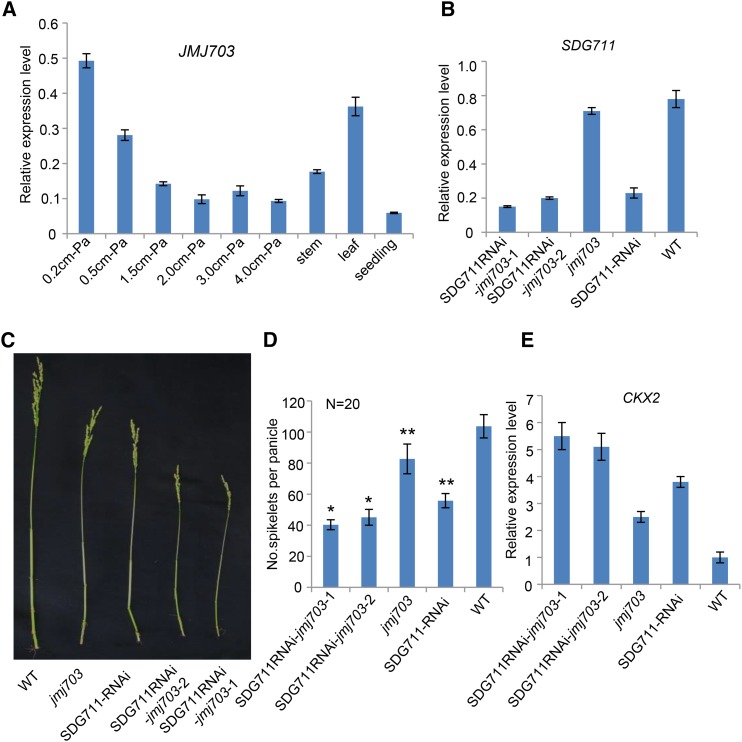
The jmj703 mutation resulted in a change of a relatively small portion of genome-wide H3K4me3 (Figure 5F), which confirmed previously published data (Cui et al., 2013), but led to a loss of 2957 and a gain of 2447 H3K27me3-marked genes in IM (Figure 5E). Comparison of differentially marked genes in jmj703 and SDG711 RNAi plants revealed that 2307 out of 2952 (i.e., 2307 + 645) (78%) genes that lost H3K27me3 in jmj703 overlapped with those that lost the mark in SDG711 RNAi plants, 50% of which (1172/2307) corresponded to the genes that gained the mark in wild-type IM (Figures 5A and 5E). Conversely, 607 out of 1378 (607+771) (44%) genes that gained H3K27me3 in SDG711 RNAi were found to overlap with those that gained the mark in jmj703 (Figure 5E). These observations suggested that JMJ703 might have a function in SDG711-mediated H3K27me3 in IM. Like SDG711, JMJ703 expression in IM gradually decreased during panicle development (Figure 7A). To study the functional relationship between the two genes, we constructed the double knockdown/knockout plants by introducing the SDG711 RNAi construct into the jmj703 mutant. These plants displayed a more severe panicle phenotype (Figures 7B to 7D). The expression of CKX2/Gn1a in double knockdown/knockout plants was higher than in SDG711 RNAi or jmj703 mutant (Figure 7E), suggesting that the two genes had an additive function to repress the expression of CKX2/Gn1a. H3K27me3 ChIP analysis of the CKX2/Gn1a locus revealed that H3K27me3 levels decreased in SDG711RNAi but increased in overexpression and gain-of-function mutant plants, whereas H3K4me3 levels (mostly at the 5′end) of the gene were reversely regulated (Figures 8A and 8B). In jmj703 mutant, increases of H3K4me3 and decreases of H3K27me3 were detected on the locus. In the double knockdown/knockout plants, we observed a further increase of H3K4me3 and a further decrease of H3K27me3 (Figure 8C). In addition, overexpression of JMJ703 (under the control of the maize ubiquitin gene promoter) reduced H3K4me3 but augmented H3K27me3 on the locus (Figure 8C). These data suggested a mutually repulsive relationship between H3K4me3 and H3K27me3 on CXK2/Gn1a and revealed an agonistic relationship between JMJ703 and SDG711 in regulating H3K27me3 and H3K4me3 on the locus. ChIP analysis of wild-type plants with anti-SDG711 (Liu et al., 2014) and Ubi-JMJ703-Flag transgenic plants with anti-Flag suggested that both SDG711 and JMJ703 directly target to the CKX2 locus (Figure 8D).
Figure 7.
SDG711 RNAi in jmj703 Mutant Background Further Reduced Panicle Size.
(A) Quantitative RT-PCR analysis of JMJ703 transcript levels (relative to ACTIN transcripts) in IM (at different stages), stem, leaf, and seedling.
(B) Relative transcript levels (to ACTIN) of SDG711 in jmj703, SDG711-RNAi, and the double knockdown/knockout lines compared with the wild type (WT).
(C) Panicle phenotype of the indicated genotypes.
(D) Spikelet numbers per panicle of the indicated genotypes. Bars = means ± sd from 20 panicles per genotype. Significant differences (Student’s t tests, P value <0.05) compared with the wild type are indicated by double asterisks (P value < 0.01). Significant difference between the double knockdown/knockout and SDG711 RNAi is indicated by single asterisks (P value < 0.05).
(E) Relative transcript levels (to ACTIN) of CKX2/Gn1a in IM of the indicated genotypes. For the RT-PCR data, bar = means ± sd from three biological repeats.
Figure 8.
SDG711 and JMJ703 Are Involved in Histone Modification and Repression of OsCKX2/Gn1a.
(A) Diagram of the CKX2/Gn1a locus. The transcribed region is represented by the thick line. Relative positions of the transcription start site (TSS) and the primer sets (P1 to P4) used in ChIP experiments are indicated.
(B) ChIP assay with anti H3K27me3 (left) and anti H3K4me3 (right) of IM chromatin of wild-type (DJ), SDG711 RNAi, gain-of-function mutant (sdg711), and overexpression (OX) plants.
(C) ChIP assay with anti H3K27me3 (left) and anti H3K4me3 (right) of IM chromatin of SDG711 RNAi, jmj703 mutant, SDG711 and JMJ703 double knockdown/knockout, JMJ703 overexpression plants, and their respective wild-type plants (DJ and ZH11).
(D) SDG711 and JMJ703 were associated directly with CKX2/Gn1a locus. ChIP assays were performed with the same samples as in (B) using anti-SDG711 (left) and with the indicated genotypes using anti-FLAG (right). Anti-IgG was used as controls in the ChIP assays.
DISCUSSION
H3K27me3 Is Dynamically Regulated during the SAM-to-IM Transition
Our data revealed differential gene expression and histone methylation patterns in rice IM compared with SAM from plants grown in the paddy field in Wuhan. The weather conditions during the growth season may influence rice growth and flowering time to some extent, which may vary slightly year to year but are very constant in this major rice production area in China. We cannot exclude that some of the differential gene expression and histone methylation between SAM and IM is related to growth conditions. However, because the meristem size was the criterion for SAM and IM sampling and the different genotypes were grown under the same conditions, the high throughput data should reflect the main difference between SAM and IM and between the different genotypes. In addition to genes regulating the SAM-to-IM transition, genes of metabolic pathways such as photosynthesis, secondary metabolism, glycolysis, and amino acid synthesis were highly induced, which may represent a characteristic molecular feature that distinguishes IM from SAM in rice (Supplemental Table 1). Possibly, the increased metabolic activity may be required to support the rapid growth of the inflorescence. A recent article also showed that metabolic activity increased in rice young panicle (Sharma et al., 2012).
Furthermore, the high-throughput data analysis revealed that many TE-related genes were expressed in both SAM and IM, although their expression levels were relatively very low (RPKM of most TE is < 1; Supplemental Figure 1D). This is consistent with previous data showing that many TE genes are transcriptionally active in maize shoot meristems (Ohtsu et al., 2007; Li et al., 2010; Martínez and Slotkin, 2012). In particular, a recent work showed that many TEs are transcribed in rice vegetative and reproductive shoot apex (Tamaki et al., 2015). The mechanism of release of the TE silencing in meristems is not clear. However, the ChIP-seq data indicated that ∼10 to 12% of H3K27me3 peaks corresponded to TEs in rice SAM and IM. This is consistent with previous data showing that there is a high number of TEs marked by H3K27me3 in Arabidopsis SAM (Lafos et al., 2011). It is suggested that there may be an unexpected role for PcG proteins in the regulation of TEs specifically in stem cell harboring meristems (Lafos et al., 2011). It is unclear whether there is a causal link between TE expression and differential H3K27me3 during meristem development.
Importantly, our data showing that thousands of genes gained or lost H3K27me3 during the SAM-to-IM transition and that downregulation of SDG711 or mutation of JMJ703 eliminated the gain of H3K27me3 from many genes and affected panicle development (Figures 1, 5, and 7) indicate that genome-wide reprogramming of H3K27me3 was critical for IM activity. The results are in line with previous data showing that PRC2-mediated regulation represents a robust system controlling plant vegetative-to-reproductive developmental transition by regulating transcriptional master regulators (Bouyer et al., 2011). The enrichment of transcription factor and protein kinase genes with different H3K27me3 levels in IM (Table 2) suggested an important role of this epigenetic modification in reprogramming gene expression and signaling during the transition. This assumption is further supported by the correlation between upregulation and loss of H3K27me3 of several key genes known to be required for the SAM-to-IM transition and for panicle development (Table 1). Previous data showed that members of the TCP, CONSTANS-like, and GRAS families are targeted by H3K27me3 specifically in the SAM in Arabidopsis, while other genes (such as homeobox genes STM, KNAT2, and KNAT6) that are specifically expressed in SAM are marked by H3K27me3 in leaves (Lafos et al., 2011). Our data indicated that several families of transcription factors were enriched for loss (e.g., TCP, MYB, bHLH, and AP2) or gain (e.g., ARF and Aux/IAA) of H3K27me3 in IM, while MADS family members showed both gain or lose of the mark in IM (Supplemental Figure 9), confirming that specific transcription factor gene families are preferentially and dynamically targeted by H3K27me3 (Lafos et al., 2011), which may be involved in gene expression reprogramming during meristem development. Consistent with previous data showing that auxin biosynthesis, transport, perception, and signaling genes are regulated by H3K27me3 in Arabidopsis during meristem cell differentiation (Lafos et al., 2011), our data revealed that both auxin and cytokinin-related genes are regulated by H3K27me3 during the SAM-to-IM transition. In particular, our data suggest that targeting of cytokinin oxidase genes by H3K27me3 is likely to play an important role in IM activity and panicle development.
However, there was no general correlation between differential H3K27me3 and gene expression change (>2-fold) in IM (Table 2). This is consistent with the data showing that although large numbers of gene loci lose H3K27me3 in Arabidopsis mutants of PRC genes, only a minority of the genes are derepressed (Lafos et al., 2011). Moreover, H3K27me3 is only partially removed upon gene activation in Drosophila (Schuettengruber et al., 2007). It is suggested that H3K27me3 is only critical for maintaining repression of a subset of the marked genes and that gain or loss of H3K27me3 at other loci does not lead to their repressed or increased expression due to the absence of specifically expressed transcription factors (Adrian et al., 2010; Eskeland et al., 2010; Farrona et al., 2011; Margueron and Reinberg, 2011).
Although only a relatively small portion of genes showed gain or loss of H3K4me3 in IM compared with SAM (Figure 5B), the amounts (TPM) of H3K4me3 were found to increase in many genes in IM (Figure 3). Data from the analysis of quantitative correlation between ChIP-seq and RNA-seq reads indicated that change of H3K27me3/H3K4me3 ratio was an important factor for differential expression of many genes after SAM-to-IM transition (Figure 3). This indicates that the two methylation marks are functionally connected and that the relative abundance of the two marks is related to gene activity. Although it is possible that chromatin regions modified by one mark are in a different cell population than the other mark, both marks may occur concomitantly in the same chromatin fragment. This type of bivalent domain was recently identified for many gene in Arabidopsis seedlings (Sequeira-Mendes et al., 2014). In animal embryonic stem cells, many promoters harbor the bivalent marks. These bivalent domains are considered to poise the expression of developmental genes, allowing timely activation while maintaining repression in the absence of differentiation signals (Young et al., 2011; Voigt et al., 2013). Considering that SAM and IM contain mostly undifferentiated cells, it can be suggested that many of the H3K27me3 and H3K4me3-marked genes identified in this study may contain the bivalent domains. As it is thought that bivalent domains have a function to finely tune gene expression during development (Young et al., 2011; Voigt et al., 2013), the dynamic change of the H3K27me3/H3K4me3 ratio may also alter bivalent marking and, thus, the fine-tuning of gene expression during the SAM-to-IM transition.
SDG711 Is Required for Gain of H3K27me3 in IM and Gene Repression during Panicle Development
Our data showing the effects of downregulation of SDG711 on panicle size, genome-wide H3K27me3, and gene expression in IM indicate that SDG711 plays an important role in IM activity and panicle development. This is further supported by the observation that SDG711 RNAi caused H3K27me3 loss and activation of many key panicle developmental and hormone genes (Table 1). However, it cannot be excluded that some of the differential gene expression and H3K27me3 between the RNAi lines and the wild type may be due to slight differences in development stages of the harvested IM samples. Considering that in the RNAi lines, there was only partial downregulation of SDG711 transcripts (Figure 1A), the RNAi effect on H3K27me3 may be underestimated. Our data suggest that SDG711-mediated H3K27me3 and gene repression may control IM activity and panicle development in several aspects. First, SDG711 may function to attenuate gene activation in IM, as many genes are highly activated in the IM, especially those involved in plant metabolism (such as photosynthesis and glycolysis) were found to lose H3K27me3 and to be further activated in SDG711 RNAi IM (Table 1; Supplemental Data Set 1). Second, SDG711 may regulate cytokinin accumulation and signaling in IM. This is supported by the upregulation of several CKX genes (including CKX2/Gn1a) in SDG711 RNAi and downregulation in SDG711 overexpressing IM and the correlating changes of active cytokinin levels (Table 1, Figure 7; Supplemental Figure 3). Cytokinins are involved in rice inflorescence meristem activity and reduced expression of CKX2/Gn1a leads to higher cytokinin accumulation in rice IM and larger panicle sizes (Ashikari et al., 2005). Third, SDG711 may have a function to maintain the repression of many H3K27me3-marked genes in IM until floral organogenesis, as many of those genes lost H3K27me3 and were derepressed in the RNAi IM (Figure 6; Supplemental Data Set 3). This is consistent with the decreased expression of SDG711 during panicle development (Figure 1), which may be a prerequisite for the timely expression of the genes during floral organogenesis. Collectively, the analysis revealed the importance of SDG711-mediated H3K27me3 in developmental switch of gene expression during panicle development.
SDG711 and JMJ703 Have Synergistic Functions in H3K27me3 and in Panicle Development
Our data showing the similar effects of SDG711 RNAi and jmj703 mutation on genome-wide H3K27me3 and panicle development (Figures 5, 7, and 8) indicate that the two genes may have synergistic functions in histone methylation and gene expression in the IM. The effects of double knockdown/knockout of the genes on panicle size and H3K27me3 and H3K4me3 modifications and expression of CKX2/Gn1a support this hypothesis. Possibly, JMJ703-dependent removal of H3K4me3 may be required for SDG711 to mediate H3K27me3 on a subset of genes in the IM. This is consistent with the observations that increased H3K27me3 levels were accompanied by decreased H3K4me3 in a subset of genes during the SAM-to-IM transition (Figure 3A). Likely, a functional interplay between the two enzymes may act in the negative crosstalk between the two histone methylation marks on a subset of genes. The observation that overexpression of JMJ703 or SDG711 led to decreased H3K4me3 and increased H3K27me3 in CKX2/Gn1a, while RNAi or mutation of the genes produced opposite effects on the modifications, corroborate this hypothesis. In mouse embryonic stem cells, the H3K4 demethylase JARID1a/RBBP2 is associated with the PRC2 complex to mediate PcG complex-dependent transcriptional repression (Pasini et al., 2008). In addition, the PRC2 complex recruits JARID1a/RBBP2 to its target genes to mediate repressive activity during embryonic stem cell differentiation (Pasini et al., 2008). The observation that JMJ703 and SDG711 proteins were both associated with the CKX2/Gn1a locus raises the possibility of a physical interaction between JMJ703 and a PRC2 complex on target genes in rice. Whether the two proteins are implicated in the generation of bivalent domains in rice meristem cells, as JARID1a/RBBP2 and PRC2 proteins do in animal stem cells (Young et al., 2011; Voigt et al., 2013), awaits further analysis.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa spp japonica) material used in this study for ChIP-seq and RNA-seq analysis were from the ‘DongJin’ (DJ) background, including wild-type, SDG711 T-DNA insertion (3A-60654.R), overexpression, and RNAi lines (Liu et al., 2014) as well as JMJ703 T-DNA insertion line (3A-00550) (Chen et al., 2013). To construct the JMJ703 and SDG711 double knockdown plants, we transformed jmj703 T-DNA mutant plants with the SDG711 RNAi construct (Liu et al., 2014). The double transformants were selected by PCR using the primers muJ3-F, muJ3-R, a T-DNA-specific primer 2715L1, pMCG-F, and pMCG-R (Supplemental Data Set 4). The Ubi-JMJ703-FLAG transgenic plants were produced in the Zhonghua11 (ZH11) background. For that, the JMJ703 full-length cDNA was amplified using the primer set OXJMJ703-F and OXJMJ703-R (Supplemental Data Set 4), then inserted into the overexpression vector pU1301-3xFLAG under the control of the maize (Zea mays) ubiquitin gene promoter (Sun and Zhou, 2008). The construct was transformed into ZH11.
The germinated rice seedlings of all genotypes were transplanted in field at the beginning of May in Wuhan area and grown till the beginning of June, which corresponded to the 6- to 7-leaf stage, to collect SAM (<0.5 mm, observed with a microscope, as a criteria for sampling). Rice plants were grown until mid June (∼10- to 12-leaf stage) to start IM collection (1 to 2 mm, observed with a microscope, as criteria). During the period from May to early June, the daylength at Wuhan area is <13.5 h (which is short day for rice flowering) but afterward becomes >13.5 h (which is long day for rice flowering). Considering the short period of the long-day condition between early to mid June, which may accelerate flowering of SDG711 RNAi plants (Liu et al., 2014), we harvested IM of the RNAi plants earlier (June 17th to 19th) than that of wild-type and jmj703 plants (June 21st to 24th). About 400 plants were used to collect SAM and IM, and apices were hand-dissected from apices and frozen in liquid nitrogen for RNA extraction or fixed in formaldehyde for chromatin cross-linking.
RNA-seq and Data Analysis
Rice SAM and IM total RNA samples were isolated using TRIzol reagent (Invitrogen). RNA libraries were prepared according to the protocol at http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseqstrandedmrna/truseq-stranded-mrna-sample-prep-guide-15031047-e.pdf. Briefly, 1 to 4 μg of total RNA was used for mRNA purification and cDNA synthesis. After a single “A” nucleotide was added to the 3′ ends of the blunt fragments of the double-stranded cDNAs, indexing adapters are ligated to the ends of the cDNA. PCR was performed to amplify the DNA fragments. The amplified DNA fragments were purified and sequenced with the Illumina HiSeq-2000 system.
Sequence reads were filtered using Trimmomatic (version 0.32) (Bolger et al., 2014). Clean tags were mapped to the reference genome of rice (RGAP version 7.0) using TopHat (version 2.0.13). These data were then analyzed for differentially expressed genes using Cufflinks (version 2.2.1) (Trapnell et al., 2012). Genes with an expression change fold >2 with P value<0.05 were defined as a differentially expressed. For pairwise scatterplots, RPKM was used. For gene pathway analysis, enrichment was calculated by comparing the percentage of differentially regulated pathway genes among all differentially regulated genes with the percentage of pathway genes in the rice genome. Then P values were calculated by Fisher’s test using the different gene numbers to determine the significance of difference of the comparisons. FDRs were used to adjust the P value.
ChIP
About 2 g of SAM or IM (∼1 to 2 mm) was cross-linked in 1% formaldehyde under vacuum. Chromatin was extracted and fragmented to 200 to 750 bp by sonication, and ChIP was performed using the following antibodies: H3K27me3 (a2363; ABclonal) and H3K4me3 (07-473; Millipore). The specificity of the antibodies was checked by immunoblots and dot blots using peptides containing H3K27me1-3 or H3K4me1-3 modifications. The precipitated and input DNA samples were analyzed either by high-throughput sequencing or by real-time PCR with gene-specific primers listed in Supplemental Data Set 4. All assays were performed at least three times from two biological replicates.
ChIP-seq and Data Analysis
DNA libraries (ChIP) were prepared by strictly following the protocol at http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseqchip/truseq-chip-sample-prep-guide-15023092-b.pdf. Briefly, 10 ng precipitated DNA was end-repaired, single “A” nucleotide was added to the 3′ end of the blunt ends of the fragments followed by adapter ligation, library fragments of 250 to 300 bp (insert plus adapter sequences) were isolated from an agarose gel, and PCR was performed with a PCR primer cocktail to enrich DNA fragments. After purification, the DNA fragments were sequenced with the Illumina HiSeq-2000.
Sequence reads from all libraries were filtered by Trimmomatic (version 0.32) (Bolger et al., 2014), and clean tags were mapped to the reference genome of rice (RGAP version 7.0) using Bowtie2 (version 2.1.0) (Trapnell et al., 2012). Reads that could be mapped equally well to multiple locations without mismatch or with identical mismatches were assigned to one position at random and were retained for further analyses as described previously (Wang et al., 2009b). Samtools (version 0.1.17) was used to remove potential PCR duplicates, and MACS software (Zhang et al., 2008) was used to call histone modification peaks with the default parameters (bandwidth, 300 bp; model fold, 10, 30; P value, 1.00e-5) (Wang et al., 2009b). After the positions of the peaks on the chromosomes were found, genes (including the 2-kb upstream and 2-kb downstream regions) overlapping with the peaks were considered to have the epigenetic marks. The wig files of the analysis pipeline were used for viewing the data in the GBrowse 2.0 software.
For pairwise contour plots and scatterplots, read counts for a gene from each library were normalized to TPM, which divides the read number of each gene by the total read number aligned to genome successfully and multiplied by 106 (He et al., 2010).
Tissue Specificity and Heat Map of Gene Expression
We estimated tissue specificity by calculating Shannon entropy (Schug et al., 2005; Zhang et al., 2007; Makarevitch et al., 2013) using the Rice Expression Profile Database (RiceXPro) (http://ricexpro.dna.affrc.go.jp/). Expression data of 12 tissues including panicle (0.6 to 10 mm), anther (0.3 to 2 mm), pistil (5- to 18-cm inflorescence), palea (1.5- to 7-mm floret), lemma (1.5- to 7-mm floret), ovary (1 to 7 d after flowering [DAF]), embryo (7 to 10 DAF), endosperm (7 to 10 DAF), root (27 d after tillering [DAT]), stem (83 DAT), leaf blade (27 DAT), and leaf sheath (27 DAT) are used to calculate the gene’s tissue specificity. Tissue specificity score = 1 − H (p)/2.5, and entropy was calculated as:
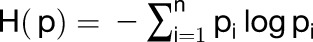 |
where Pi is a relative abundance of the gene’s transcript in tissue i.
Genes with expression (IM) <400 and expression (other tissue)/expression (IM) >2 are defined as repressed in IM but expressed in other tissue. A heat map was generated by the R package heatmap2 in the R programming environment.
RT-PCR
Total RNAs from rice stems, leaves, seedlings, and panicles of different sizes were isolated using TRIzol reagent (Invitrogen). Three micrograms of total RNA was reverse-transcribed in a reaction of 20 μL using DNase I and SuperScript III (Invitrogen) according to the manufacturer’s instructions to obtain cDNA. Real-time PCR was performed in an optical 96-well plate that included SYBR Premix EX Taq and 0.5 μL of Rox Reference Dye II (Takara), 1 μL of the reverse transcription reaction, and 0.25 μM of each gene-specific primer in a final volume of 25 μL on a PRISM 7500 PCR instrument (Applied Biosystems). The reactions were performed at 95°C for 10 s, 45 cycles of 95°C for 5 s, and 60°C for 40 s. Disassociation curve analysis was performed as follows: 95°C for 15 s, 60°C for 20 s, and 95°C for 15 s. Data were collected using the ABI PRISM 7500 sequence detection system following the instruction manual. The relative expression levels were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The rice ACTIN1 gene was used as the internal control. The primers for real-time PCR are listed in Supplemental Data Set 4.
In Situ Hybridization
All the tested materials were fixed, dehydrated, embedded, sliced, and attached to slides as previously reported (Zhao et al., 2009). For preparation of the digoxigenin-labeled RNA probes, the coding regions of SDG711 and CKX2 were amplified via PCR using primers (Insitu-SDG711-F and Insitu-SDG711-R, or Insitu-OsCKX2-F and Insitu-OsCKX2-R, respectively) (Supplemental Data Set 4). The PCR products were cloned into pGEM-T vectors, linearized, and used as templates for amplifying digoxigenin-labeled sense and antisense RNA probes. Tissue sections were cleared, dehydrated, dried, hybridized, and washed. The labeled probes were detected and images were photographed with a microscope (Leica DM2500B).
Cytokinin Dosage
The rice IM (<0.5 cm) of DJ, SDG711 overexpression, RNAi, and T-DNA insertion (3A-60654.R) was used for measuring cytokinin content. The samples were homogenized in liquid nitrogen and extracted in cold extraction buffer at –20°C overnight. The material was analyzed for three technical repeats. One hundred milligrams (fresh weight) of material and 10 ng of each internal standard were used for each technical repeat. The extraction buffer was a mixture of methanol‐water‐formic acid (15:4:1, v/v/v). The extraction was performed according to those reported by Dobrev and Kamínek (2002).
An ultrafast liquid chromatograph with an autosampler (Shimadzu) was used for liquid chromatography. The Atlantis T3 column (2.1 × 150 mm, 5 μm; Waters) was used at ambient temperature. The hybrid triple quadrupole/linear ion trap mass spectrometer of ABI 4000Q‐Trap (Applied Biosystems) outfitted with an electrospray ion source was used for measurement of cytokinin content according to the method by Liu et al. (2012), except that a positive ionization mode was used for measuring the contents of cytokinin and the internal standards.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: SDG711, Os06g16390; JMJ703, Os05g10770; CKX1, Os01g09260; CKX2, Os01g10110; CKX3, Os10g34230; CKX4, Os01g71310; CKX5, Os01g56810; CKX6, Os02g12770; CKX7, Os02g12780; CKX8, Os04g44230; CKX9, Os05g31040; CKX10, Os06g37500; and CKX11, Os08g35860. The ChIP-seq and RNA-seq data described in this article have been deposited into the Gene Expression Omnibus database (accession number GSE68299).
Supplemental Data
Supplemental Figure 1. Rice SAM and IM RNA-seq data.
Supplemental Figure 2. Overlap of differentially expressed genes in IM versus SAM and those in SDG711 RNAi IM compared with the wild type.
Supplemental Figure 3. Expression of rice cytokinin oxidase (CKX) gene family members and accumulation of active cytokinins in IM of SDG711 transgenic and mutant plants.
Supplemental Figure 4. Experimental validation of RNA-seq and ChIP-seq data.
Supplemental Figure 5. Genome browser screenshots of randomly selected six regions (100 kb each) with H3K27me3 modification in different samples.
Supplemental Figure 6. Relative proportions of H3K27me3 and H3K4me3 ChIP-seq peaks in genic (TE and non-TE) and intergenic regions and in promoter and gene body regions.
Supplemental Figure 7. Pairwise scatterplots between amounts (TMP) of H3K4me3.
Supplemental Figure 8. Tissue specificity scores of genes marked by H3K27me3 in SAM or IM (one tissue) or in both (two tissues) compared with unmarked genes.
Supplemental Figure 9. Relative fold changes of transcription factor family genes that gained or lost H3K27me3 in IM compared with SAM.
Supplemental Table 1. Pathway analysis of up- and downregulated genes in wild-type IM/SAM and SDG711 RNAi IM/wild type.
Supplemental Table 2. H3K27me3 and H3K4me3 ChIP-seq reads and analysis data of rice SAM and IM of wild-type, SDG711 RNAi, and jmj703 plants.
Supplemental Table 3. Pathway analysis of genes that gained or lost H3K27me3 in IM/SAM and SDG711 RNAi/wild type IM.
Supplemental Table 4. Enrichment of genes that lost H3K27me3 in SDG711 RNAi and jmj703 mutant versus wild-type IM and in wild-type IM versus SAM for gain or loss of H3K4me3.
Supplemental Data Set 1. Relative transcript levels of kinase, transcription factor, and metabolic pathway genes that lost or gained H3K27me3 in IM/SAM and SDG711 RNAi/wild type IM.
Supplemental Data Set 2. List of differentially expressed in IM compared SAM which lost H3K27me3 and further induced in SDG711 RNAi IM.
Supplemental Data Set 3. List of IM silent and floral organ active genes that lost H3K27me3 and were activated in SDG711 RNAi IM.
Supplemental Data Set 4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Qinglu Zhang and Xianghua Li for help in field experiments and management. This work was supported by grants from National Science Foundation of China (31371241), the 863 Key Project “Rice Functional Genomics” of the Chinese Ministry of Science and Technology (2012AA10A303), the special transgenic program of the Chinese Ministry of Agriculture (2014ZX08009-002B), and grants from the Bill and Melinda Gates Foundation.
AUTHOR CONTRIBUTIONS
X.L., S.Z., W.W., Y.Y., Y.Z., Q.X., C.Z., F.T., and S.C. performed research. S.Z., X.L., and D.-X.Z. analyzed data. D.-X.Z., X.L., and S.Z. wrote the article.
Glossary
- IM
inflorescence meristem
- SAM
shoot apical meristem
- ChIP
chromatin immunoprecipitation
- H3K27me3
H3 lysine 27 trimethylation
- H3K4me3
H3 lysine 4 trimethylation
- FDR
false discovery rate
- RNAi
RNA interference
- TE
transposable element
- TPM
tags per million
- RPKM
reads per kilobase per million reads
- DAF
days after flowering
- DAT
days after tillering
References
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Avramova Z. (2009). Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int. J. Dev. Biol. 53: 371–381. [DOI] [PubMed] [Google Scholar]
- Berr A., Shafiq S., Shen W.H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 1809: 567–576. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yuan Z., Chen M., Yin C., Luo Z., Zhao X., Liang W., Hu J., Zhang D. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5: 3476. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276. [DOI] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Chen X., Wang Q., Zhang F., Lou Z., Zhang Q., Zhou D.X. (2013). Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet. 9: e1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hu Y., Zhou D.X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809: 421–426. [DOI] [PubMed] [Google Scholar]
- Cui R., Han J., Zhao S., Su K., Wu F., Du X., Xu Q., Chong K., Theissen G., Meng Z. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61: 767–781. [DOI] [PubMed] [Google Scholar]
- Cui X., Jin P., Cui X., Gu L., Lu Z., Xue Y., Wei L., Qi J., Song X., Luo M., An G., Cao X. (2013). Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. USA 110: 1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev P.I., Kamínek M. (2002). Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 950: 21–29. [DOI] [PubMed] [Google Scholar]
- Eskeland R., Leeb M., Grimes G.R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A.I., Wutz A., Bickmore W.A. (2010). Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Thorpe F.L., Engelhorn J., Adrian J., Dong X., Sarid-Krebs L., Goodrich J., Turck F. (2011). Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Parenicova L., Falasca G., Pelucchi N., Masiero S., Ciannamea S., Lopez-Dee Z., Altamura M.M., Colombo L., Kater M.M. (2004). Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 135: 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Avramova Z. (2014). ATX1/AtCOMPASS and the H3K4me3 marks: how do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 21: 75–82. [DOI] [PubMed] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., Guo C., Kong H., Xue H., Zhang D. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153: 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Yang H., Jiao Y. (2014). Regulation of inflorescence architecture by cytokinins. Front. Plant Sci. 5: 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Derkacheva M. (2009). Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 25: 414–423. [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu D., Zhong X., Zhang C., Zhang Q., Zhou D.X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA 109: 5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K., Maekawa M., Izawa T., Itoh J., Nagato Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69: 168–180. [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Harada J.J., Goldberg R.B., Fischer R.L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 98: 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., Yoshida H., Nagamura Y., Kyozuka J. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Villar C.B. (2008). Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 18: 236–243. [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- Lafos M., Kroll P., Hohenstatt M.L., Thorpe F.L., Clarenz O., Schubert D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.Y., An G. (2012). Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69: 445–461. [DOI] [PubMed] [Google Scholar]
- Li H., Freeling M., Lisch D. (2010). Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 107: 22184–22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chen X., Zhong X., Zhao Y., Liu X., Zhou S., Cheng S., Zhou D.X. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25: 4725–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhou C., Zhao Y., Zhou S., Wang W., Zhou D.X. (2014). The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 5: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Cai B.-D., Feng Y.-Q. (2012). Rapid determination of endogenous cytokinins in plant samples by combination of magnetic solid phase extraction with hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 891-892: 27–35. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luo M., Platten D., Chaudhury A., Peacock W.J., Dennis E.S. (2009). Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2: 711–723. [DOI] [PubMed] [Google Scholar]
- Makarevitch I., Eichten S.R., Briskine R., Waters A.J., Danilevskaya O.N., Meeley R.B., Myers C.L., Vaughn M.W., Springer N.M. (2013). Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez G., Slotkin R.K. (2012). Developmental relaxation of transposable element silencing in plants: functional or byproduct? Curr. Opin. Plant Biol. 15: 496–502. [DOI] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Ohtsu K., et al. (2007). Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J. 52: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Hansen K.H., Christensen J., Agger K., Cloos P.A., Helin K. (2008). Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 22: 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S., Grossniklaus U. (2007). Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta 1769: 375–382. [DOI] [PubMed] [Google Scholar]
- Qi W., Sun F., Wang Q., Chen M., Huang Y., Feng Y.Q., Luo X., Yang J. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157: 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N.N., Prasad K., Kumar P.R., Vijayraghavan U. (2008). Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl. Acad. Sci. USA 105: 3646–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C.H., Lee S., Cho L.H., Kim S.L., Lee Y.S., Choi S.C., Jeong H.J., Yi J., Park S.J., Han C.D., An G. (2009). OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 32: 1412–1427. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Matsuoka M. (2008). Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11: 209–214. [DOI] [PubMed] [Google Scholar]
- Schubert D., Clarenz O., Goodrich J. (2005). Epigenetic control of plant development by Polycomb-group proteins. Curr. Opin. Plant Biol. 8: 553–561. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745. [DOI] [PubMed] [Google Scholar]
- Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J. Jr. (2005). Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6: R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. (2008). Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20: 266–273. [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J., Aragüez I., Peiró R., Mendez-Giraldez R., Zhang X., Jacobsen S.E., Bastolla U., Gutierrez C. (2014). The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., et al. (2012). Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct. Integr. Genomics 12: 229–248. [DOI] [PubMed] [Google Scholar]
- Steffen P.A., Ringrose L. (2014). What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 15: 340–356. [DOI] [PubMed] [Google Scholar]
- Sun Q., Zhou D.X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. USA 105: 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Tsuji H., Matsumoto A., Fujita A., Shimatani Z., Terada R., Sakamoto T., Kurata T., Shimamoto K. (2015). FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc. Natl. Acad. Sci. USA 112: E901–E910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. (2013). A double take on bivalent promoters. Genes Dev. 27: 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yin H., Qian Q., Yang J., Huang C., Hu X., Luo D. (2009a). NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res. 19: 598–611. [DOI] [PubMed] [Google Scholar]
- Wang X., Elling A.A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X.S., Deng X.W. (2009b). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61: 421–442. [DOI] [PubMed] [Google Scholar]
- Yaish M.W., El-Kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.M., Rothstein S.J. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Willson T.A., Wakefield M.J., Trounson E., Hilton D.J., Blewitt M.E., Oshlack A., Majewski I.J. (2011). ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 39: 7415–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Chen X. (2011). Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14: 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.