Arabidopsis CEK4 may be the plasma membrane-localized isoform of the choline/ethanolamine kinase family involved in the rate-limiting step of phosphatidylcholine biosynthesis and embryo development.
Abstract
Phospholipids are highly conserved and essential components of biological membranes. The major phospholipids, phosphatidylethanolamine and phosphatidylcholine (PtdCho), are synthesized by the transfer of the phosphoethanolamine or phosphocholine polar head group, respectively, to the diacylglycerol backbone. The metabolism of the polar head group characterizing each phospholipid class is poorly understood; thus, the biosynthetic pathway of major phospholipids remains elusive in Arabidopsis thaliana. The choline/ethanolamine kinase (CEK) family catalyzes the initial steps of phospholipid biosynthesis. Here, we analyzed the function of the four CEK family members present in Arabidopsis. Knocking out of CEK4 resulted in defective embryo development, which was complemented by transformation of genomic CEK4. Reciprocal genetic crossing suggested that CEK4 knockout causes embryonic lethality, and microscopy analysis of the aborted embryos revealed developmental arrest after the heart stage, with no defect being found in the pollen. CEK4 is preferentially expressed in the vasculature, organ boundaries, and mature embryos, and CEK4 was mainly localized to the plasma membrane. Overexpression of CEK4 in wild-type Arabidopsis increased the levels of PtdCho in seedlings and mature siliques and of major membrane lipids in seedlings and triacylglycerol in mature siliques. CEK4 may be the plasma membrane-localized isoform of the CEK family involved in the rate-limiting step of PtdCho biosynthesis and appears to be required for embryo development in Arabidopsis.
INTRODUCTION
Phospholipids are essential cellular components of prokaryotes and eukaryotes and are a major component of biological membranes. Furthermore, phospholipids function as signaling molecules and as substrates for the biosynthesis of storage lipids. The phospholipids phosphatidylcholine (PtdCho) and phosphatidylethanolamine (PtdEtn) are abundant in plant tissues. In Arabidopsis thaliana and many other plant species, these phospholipids are produced by transferring the polar head group phosphocholine (P-Cho) or phosphoethanolamine (P-Etn), respectively, to sn-1,2-diacylglycerol (DAG). The DAG biosynthetic pathway has been studied extensively because DAG is the common substrate for the different classes of glycerolipids, namely, galactolipids, sulfolipids, phospholipids, and triacylglycerols. However, the proposed biosynthetic pathway underlying the biosynthesis of the polar head group awaits validation via isolation and knockout studies of the genes encoding the enzymes involved in this pathway.
A currently proposed biosynthetic pathway for the polar head group in Arabidopsis begins with the conversion of an amino acid Ser to ethanolamine (Etn) by Ser decarboxylase (SDC) (Rontein et al., 2001), followed by phosphorylation by choline/ethanolamine kinase (CEK) to produce P-Etn (Figure 1A). P-Etn is a common substrate for the biosynthesis of both PtdEtn and PtdCho: PtdEtn is produced via cytidine diphosphoethanolamine by CTP:phosphorylethanolamine cytidylyltransferase (PECT) (Mizoi et al., 2006) and amino alcohol:aminophosphotransferase (Dewey et al., 1994), whereas PtdCho synthesis requires trimethylation of P-Etn to P-Cho catalyzed by S-adenosyl-l-methionine:phosphoethanolamine N-methyltransferase (PMT) (Bolognese and McGraw, 2000). P-Cho is converted by CTP:phosphorylcholine cytidylyltransferase to cytidine diphospho-choline (Inatsugi et al., 2002), which is then incorporated into the DAG backbone by amino alcohol:aminophosphotransferase to produce PtdCho. In addition, phospholipid catabolism by phospholipases may provide a biosynthetic intermediate. For example, PtdCho is hydrolyzed by nonspecific phospholipase C (Nakamura et al., 2005) or phospholipase D combined with CEK to provide P-Cho (Figure 1A, broken arrows). Therefore, the initial pathway from Ser to P-Etn appears to be the gateway that channels amino acid metabolism into phospholipid biosynthesis and, hence, the rate-limiting step in PtdCho and PtdEtn biosynthesis. In Arabidopsis, knocking out of PECT1 causes embryonic lethality, and knocking down of PECT1 increases PtdCho levels at the expense of PtdEtn levels (Mizoi et al., 2006; Nakamura et al., 2014b). In addition, knocking out of PMT1 (or PEAMT1, XPL1) reduces PtdCho levels slightly (Cruz-Ramírez et al., 2004). Furthermore, a leaky mutant of SDC showed a severe growth defect, which suggests that SDC knockout is lethal (Kwon et al., 2012). Therefore, P-Etn synthesis may be crucial for the biosynthesis of PtdCho and PtdEtn. However, the lack of knowledge of the CEK gene family in Arabidopsis hampers our understanding of the critical step in phospholipid biosynthesis.
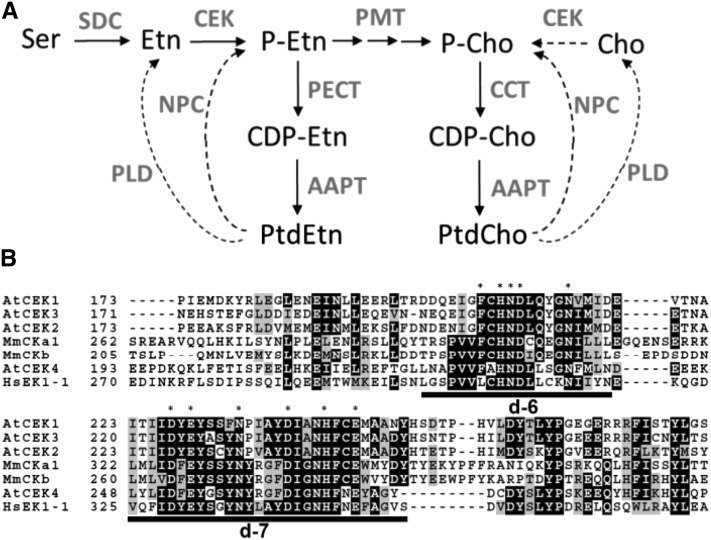
Figure 1.
The CEK Family in Arabidopsis.
(A) Proposed biosynthetic pathway of PtdCho and PtdEtn in Arabidopsis. Arrows indicate de novo biosynthesis, and dashed arrows indicate phospholipid hydrolysis pathways that provide phosphor-base compounds. Compounds are indicated in black letters and enzymes in gray. AAPT, aminoalcohol aminophosphotransferase; CCT, CTP:phosphorylcholine cytidylyltransferase; CDP-Cho, cytidine diphosphocholine; CDP-Etn, cytidine diphosphoethanolamine; Cho, choline; NPC, nonspecific phospholipase C; PECT, CTP:phosphorylethanolamine cytidylyltransferase; PLD, phospholipase D; SDC, serine decarboxylase.
(B) Amino acid sequence alignment of putative catalytic motifs in CEKs. Asterisks indicate highly conserved amino acid residues, and catalytic domains d-6 (Brenner’s phosphotransferase motif) and d-7 (putative CEK motif) defined by (Aoyama et al. 2004) are underlined. Hs, Homo sapiens; Mm, Mus musculus.
In this study, we characterized four putative CEK genes, CEK1, 2, 3, and 4, in Arabidopsis by isolating the T-DNA-tagged knockout mutants of these genes. We found that CEK4 knockout led to an embryo-lethal phenotype due to the developmental arrest of embryos beyond the heart stage. Overexpression of CEK4 increased the levels of PtdCho both in seedlings and in mature siliques, which subsequently increased the levels of major membrane lipid classes in seedlings and triacylglycerol (TAG) in mature siliques. We suggest that CEK4 is the plasma membrane-localized isoform of the CEK family involved in the rate-limiting step of the de novo biosynthesis of PtdCho, which is required for embryo development in Arabidopsis.
RESULTS
Arabidopsis Has Four CEK Homologs
On the basis of homology with known mammalian choline/ethanolamine kinases (Aoyama et al., 2004; Wu and Vance, 2010), we identified four genes encoding putative CEKs, which we designated CEK1, 2, 3, and 4 (Supplemental Table 1). An additional putative kinase (At1g34100) was previously suggested to be a CEK (Tasseva et al., 2004); however, At1g34100 is likely a pseudogene because it has no suitable start codon (The Arabidopsis Information Resource, www.arabidopsis.org). We therefore excluded this gene from our study. The CEKs encode proteins of ∼40 kD that lack a predicted transmembrane domain (Supplemental Table 1). The level of amino acid sequence identity among these CEKs ranges from 33% (CEK2 and CEK4) to 64% (CEK1 and CEK2). Multiple amino acid sequence alignment showed highly conserved regions, including d-6 and d-7 domains (Figure 1B) (Aoyama et al., 2004). In d-6, Brenner’s phosphotransferase motif (Brenner, 1987) is well conserved among CEKs (critical residues shown by asterisks), particularly CEK4, which shares extended sequence similarity with mammalian orthologs (Figure 1B). The d-7 domain contains a putative CEK motif (Figure 1B, underlined) (Aoyama et al., 2000), which is well conserved among CEKs. These sequence similarities suggest that CEKs encode functional enzymes in Arabidopsis.
Differential Gene Expression Pattern of Four CEKs
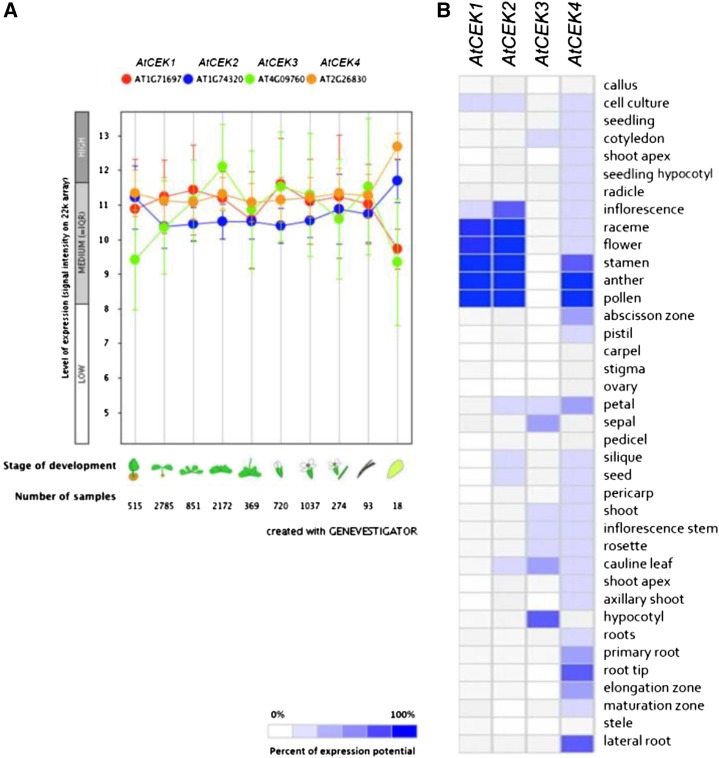
To investigate the differential gene expression of the four CEKs, we used the publicly available gene expression database (Genevestigator; www.genevestigator.com). First, we identified differential expression patterns at different stages of development among the four isoforms (Figure 2A); CEK3 expression was increased during vegetative growth but was reduced on entry into the reproductive phase. Although expression of the other three isoforms was ubiquitous, CEK2 and 4 showed increased expression in senescent tissues. Next, except for CEK3, most CEKs were expressed in flowers (Figure 2B); CEK4 was highly expressed in male reproductive organs, and CEK1 and 2 were highly expressed in whole flowers. The expression of CEK3 was low overall but relatively high in hypocotyls and cauline leaves. However, some discrepancy was found between the data presented in Figures 2A and 2B; for example, Figure 2A shows that the expression level of CEK3 is similar to that of CEK1 or CEK4 in flowers, whereas CEK3 was not found to be expressed in flowers in the data shown in Figure 2B. Thus, we performed a tissues-specific expression study for CEK4 using a β-glucuronidase (GUS) reporter system for the main isoform discussed in this study (Figure 6; Supplemental Figure 2). Because the GUS staining experiment confirmed that CEK4 has the highest expression in male reproductive organs, the gene expression data in Figure 2B may be useful for outlining the tissue specificity of the other CEK isoforms whose expression patterns have not been studied by GUS staining.
Figure 2.
Gene Expression Pattern of CEK1, 2, 3, and 4.
(A) Developmental stage-specific expression patterns. Left to right, germinating seed, seedling, young rosette, developed rosette, bolting rosette, young flower, developed flower, flowers and siliques, mature siliques, and senescence. “HIGH,” “MEDIUM,” and “LOW” expression were calculated by microarray assay. The number of samples indicates microarray gene expression data collected by Genevestigator.
(B) Heat map of tissue-specific expression pattern of CEK1, 2, 3, and 4. Data were analyzed with Genevestigator.
Figure 6.
Tissue-Specific Expression of CEK4 by Histochemical GUS Staining of ProCEK4:CEK4-GUS cek4-1/-.
(A) to (F) Time-course GUS staining profile of germinating seeds. Seeds were stratified in sterile water for 1 d and then placed on Murashige and Skoog agar plates. Seeds upon imbibition ([A] and [B]), 1 d after imbibition (upon planting on a Murashige and Skoog plate) (C), 1 d after plating (D), 2 d after plating (E), and 3 d after plating (F).
(G) Five-day-old seedling.
(H) Ten-day-old seedling.
(I) Trichomes on the cotyledon of a 10-d-old seedling.
(J) Joint between roots and shoots.
(K) Branch of lateral roots.
(L) Root tip.
(M) A whole rosette of a bolting plant.
(N) An inflorescence stem.
(O) An inflorescence.
(P) Flowers at different developmental stages.
(Q) Developing siliques.
Bars = 500 μm in (A) to (L), 5 mm in (M) to (O), and 1 mm in (P) and (Q).
Isolation of cek Mutants in Arabidopsis
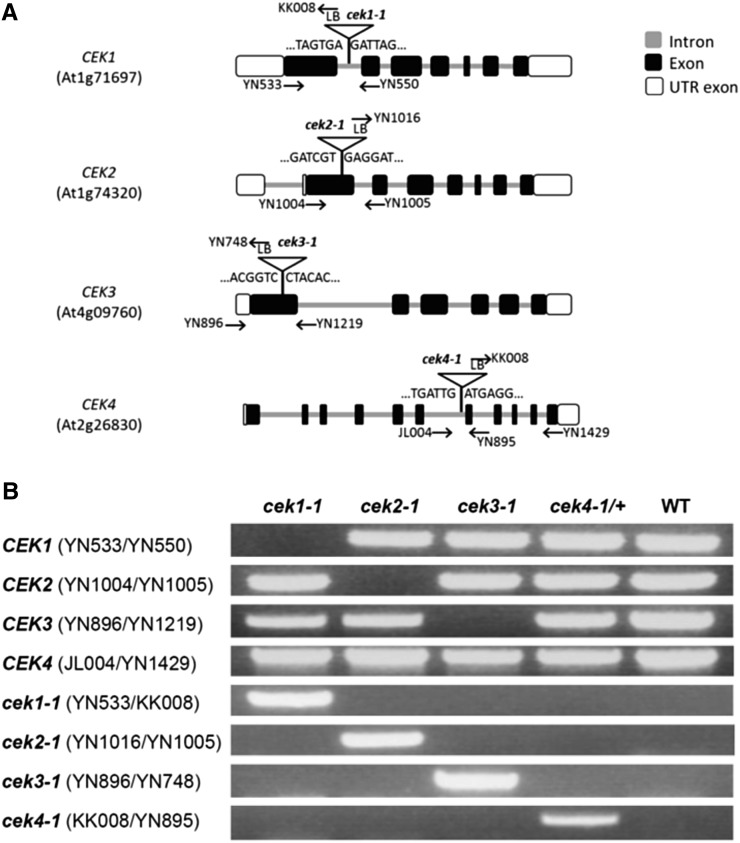
To study the in vivo function of the four CEKs, we isolated T-DNA knockout mutants of the genes: cek1-1 (SALK_069908), cek2-1 (GK-124F01), cek3-1 (SALK_026906), and cek4-1 (SALK_057727). We confirmed the position of the T-DNA insertion by sequencing (Figure 3A). PCR-based genotyping isolated homozygous mutants for CEK1, 2, and 3 but not CEK4 (Figure 3B), whose homozygotes were not isolated after screening more than 100 seedlings containing T-DNA, suggesting that the homozygous cek4-1 mutant may be lethal.
Figure 3.
Isolation of cek Mutants.
(A) Positions of T-DNA and primers used for PCR-based genotyping.
(B) Identification of homozygous (cek1-1, cek2-1, and cek3-1) and heterozygous (cek4-1/+) mutants by PCR-based genotyping. See Supplemental Table 10 for the sequences of primers used for genotyping.
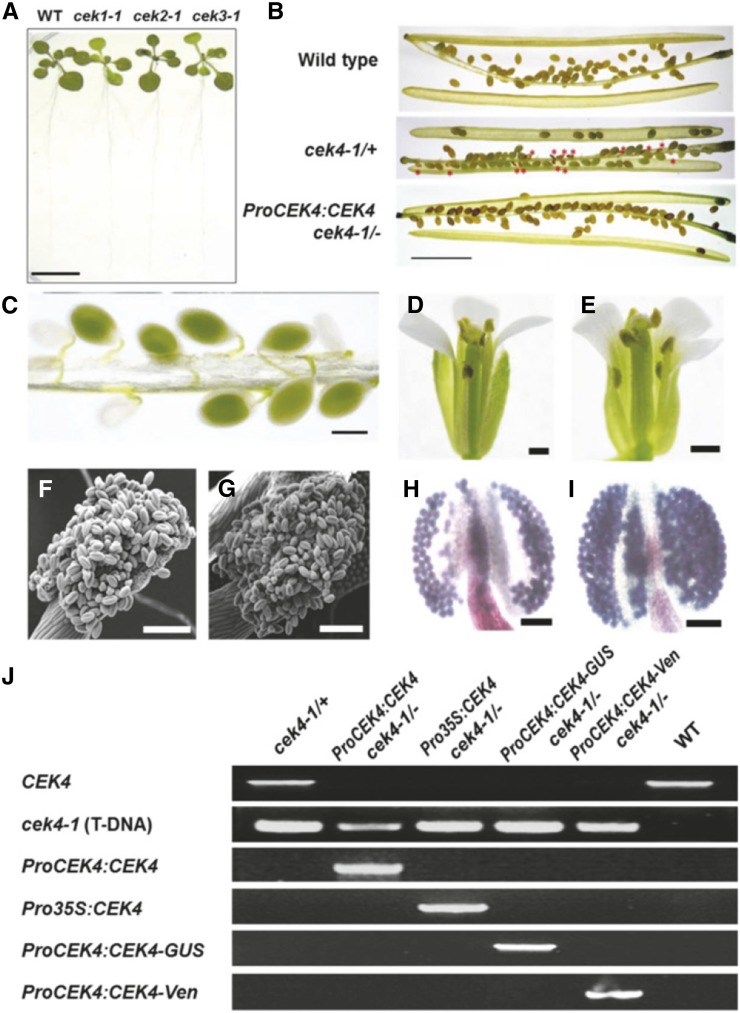
Homozygous seedlings of cek1-1, cek2-1, and cek3-1 showed no visible phenotypes under normal growth conditions (Figure 4A). To investigate whether CEK1, CEK2, or CEK3 is involved in glycerolipid metabolism, we analyzed the polar glycerolipid composition of the seedlings of cek1-1, cek2-1, and cek3-1 (Supplemental Tables 2 and 3). The result showed no significant difference in the glycerolipid contents (Supplemental Table 2) or their fatty acid composition (Supplemental Table 3) among these cek mutants and two wild-type lines, Col-0 and Col-8. In addition, we analyzed TAG and DAG contents in mature siliques of these mutants; however, no significant difference was found in their amount (Supplemental Table 4) or fatty acid composition (Supplemental Table 5). These results suggest that these CEK isoforms may have redundant functions in glycerolipid metabolism and plant development.
Figure 4.
Phenotype of cek Mutants.
(A) The phenotype of 14-d-old homozygous cek1-1, cek2-1, and cek3-1 mutants and the wild type. Bar = 10 mm.
(B) Siliques of the wild type, cek4-1/+ (producing wrinkled seeds; red asterisks), and ProCEK4:CEK4 cek4-1/- (producing normal seeds). Bar = 5 mm.
(C) Developing seeds (green or white) in a silique of cek4-1/+. Bar = 500 μm.
(D) and (E) Flower of the wild type (D) and cek4-1/+ (E). One petal was removed for observation. Bars = 1 mm.
(F) and (G) Scanning electron microscopy image of wild-type (F) and cek4-1/+ (G) pollen. Bars = 100 μm.
(H) and (I) Alexander’s staining of wild-type (H) and cek4-1/+ (I) pollen. Bars = 100 μm.
(J) PCR-based genotyping of complemented cek4-1/- plants by ProCEK4:CEK4, ProCEK4:CEK4-GUS, or ProCEK4:CEK4-Ven. See Supplemental Figure 3 for annealing positions and Supplemental Table 10 for sequences of the primers used.
Knocking Out of CEK4 Caused an Embryo-Lethal Phenotype
The siliques of cek4-1/+ frequently produced wrinkled seeds (Figure 4B, red asterisks), indicative of the embryonic lethality of cek4-1/-. Siliques of cek4-1/+ but not the wild type showed a number of underdeveloped embryos (Figure 4C); the occurrence was 16 of 65 embryos, nearly 25% of the total number of embryos. We observed phenotype of cek4-1/+ during vegetative growth; however, overall growth (Supplemental Figures 1A and 1B), leaf shape (Supplemental Figures 1C and 1D), or flowering time (Supplemental Figure 1E) showed no significant differences between cek4-1/+ and wild-type Col-0. We therefore observed reproductive organs of cek4-1/+. Overall structure of mature flower was indistinguishable between the wild type (Figure 4D) and cek4-1/+ (Figure 4E). To examine whether any defect is observed in the anther and pollen, which were shown to have the highest expression level of CEK4 in the publicly available gene expression database (Figure 2B), we obtained scanning electron microscopy images of wild-type (Figure 4F) and cek4-1/+ (Figure 4G) pollen. However, no abnormally shaped pollen was found. We further examined the viability of pollen using Alexander’s staining (Alexander, 1969), which stains viable pollens purple and nonviable pollens green; however, the pollen of the wild type (Figure 4H) and cek4-1/+ (Figure 4I) all stained purple, indicating viability. We counted the number of pollen from cek4-1/+ and the wild type (Col-0) that stained green, and the occurrence of green pollen was <0.01% after scoring more than 1000 pollen grains, respectively. These observations suggest that pollen of cek4-1/+ are functional, while wrinkled seeds are found in the siliques.
To further investigate the embryonic lethal phenotype, we crossed cek4-1/+ pollen with the stigmas of cek4-1/+ (Table 1), which produced aborted embryos at a frequency of ∼25%; the genotype of F1 seedlings was WT:cek4-1/+ = 1:2 (wild type [WT]). Pollen of cek4-1/+ crossed with wild-type stigmas produced no aborted embryos; the segregation ratio of F1 seedlings was cek4-1/+:WT = 1:1. Next, we found that wild-type pollen crossed with cek4-1/+ stigmas produced F1 seedlings with a segregation ratio of cek4-1/+:WT = 1:1, without aborted embryos. Furthermore, manually self-crossing ProCEK4:CEK4 cek4-1/- produced no aborted embryos, yet produced an F1 population of 100% cek4-1/- genetic background. These results suggest that cek4-1 is not defective in the male or female gametophyte but is deficient in embryo development.
Table 1. Reciprocal Crossing of the cek4-1/+ Mutant.
| Reciprocal Crosses |
F1 Embryo Phenotypes |
F1 Seedling Genotypes |
|||||
|---|---|---|---|---|---|---|---|
| Female | Male | Normal | Aborted | Ratio (Normal:Aborted) | WT | Heterozygote | Homozygote |
| cek4-1/+ | cek4-1/+ | 49 | 16 | 3:1 | 16 | 33 | 0 |
| cek4-1/+ | WT | 59 | 0 | 1:0 | 30 | 29 | 0 |
| WT | cek4-1/+ | 59 | 0 | 1:0 | 29 | 30 | 0 |
| cek4-1/- ProCEK4:CEK4 | cek4-1/- ProCEK4:CEK4 | 54 | 0 | 1:0 | 0 | 0 | 54 |
WT, wild type.
To examine whether this embryonic defect was due to CEK4 knockout, we transformed the genomic sequence of CEK4 driven by its native promoter (ProCEK4:CEK4) into cek4-1/+ and searched for a viable cek4-1/- harboring ProCEK4:CEK4 in the next generation (Figures 4B and 4J). ProCEK4:CEK4 cek4-1/- produced siliques without aborted embryos (Figure 4B, lowest panel). Moreover, the complementation of cek4-1/- (Figure 4J) was observed with different transgenes used later in this study, including ProCEK4:CEK4-GUS, ProCEK4:CEK4-Ven, and Pro35S:CEK4. Thus, knocking out CEK4 caused an embryo-lethal phenotype.
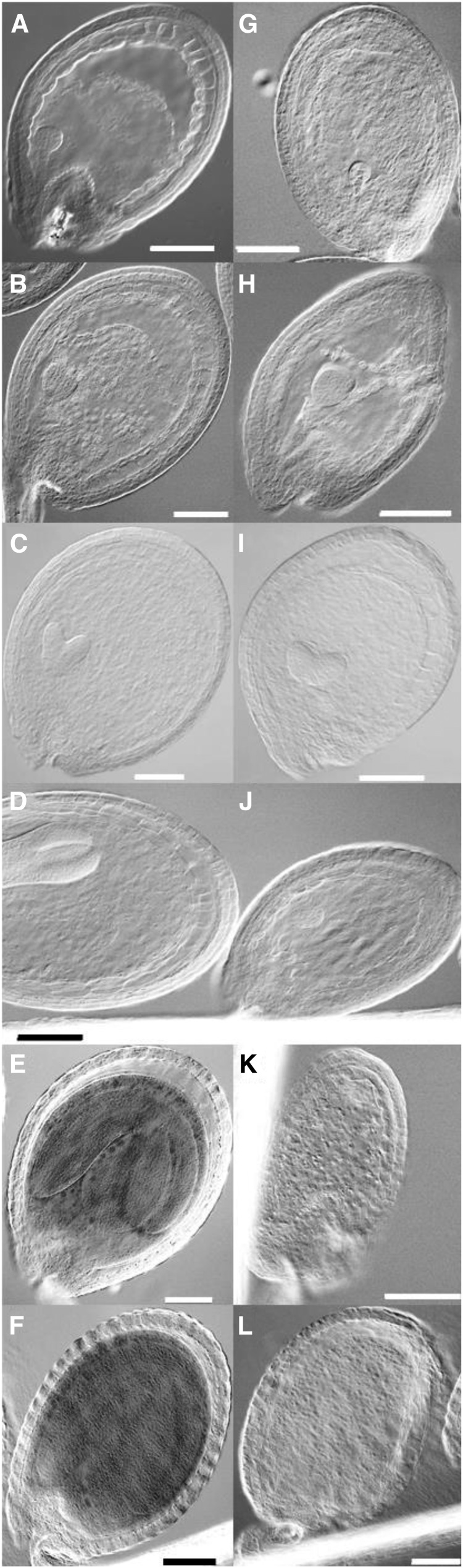
Defective Embryo Maturation in cek4
To further investigate the defective development of embryos in cek4-1, we observed the pale embryos (Figure 4C) produced at different developmental stages of cek4-1 (Figure 5). Octant and globular stages showed no significant structural alteration from wild-type embryos (Figures 5A, 5B, 5G, and 5H). However, when normal embryos in the silique are in the heart stage (Figures 5C and 5I), defective embryos remained round-shaped (Figure 5I) compared with the wild type (Figure 5C), with no further significant development at subsequent developmental stages (Figures 5D to 5F and 5J to 5L). Thus, disruption of CEK4 arrested embryo development after the heart stage.
Figure 5.
Microscopy Analysis of Embryo Development.
A normal wild-type (WT) embryo ([A] to [F]) and aborted embryo in cek4-1/+ ([G] to [L]) at the octant ([A] and [G]), globular ([B] and [H]), heart ([C] and [I]), torpedo ([D] and [J]), bent cotyledon ([E] and [K]), and mature embryo ([F] and [L]) stages. Bars = 100 μm.
Tissue-Specific Localization of CEK4
We investigated the tissue-specific expression pattern of CEK4 using a histochemical GUS reporter assay (Figure 6). We transformed cek4-1/+ plants with a plasmid vector harboring the genomic sequence of CEK4 including the promoter and open reading frame followed by a GUS reporter (ProCEK4:CEK4-GUS). In the T2 population, we recovered fully viable ProCEK4:CEK4-GUS cek4-1/- plants, indicating that ProCEK4:CEK4-GUS was functional in vivo (Figure 4J, lane 4).
GUS staining of ProCEK4:CEK4-GUS cek4-1/- plants during germination was first observed in entire embryos (Figures 6A to 6C), and then restricted to the hypocotyls (Figures 6D to 6G), vasculature (Figures 6G and 6H), and trichomes (Figure 6I) of cotyledons. In addition, strong GUS staining was observed at the shoot apical meristem (Figures 6G and 6H) and at the point of connection between the root and shoot (Figure 6J). In roots, GUS staining was seen at the lateral branch (Figure 6K) but not at the root apical meristem (Figure 6L). In rosette leaves, GUS staining was observed in the vasculature (Figure 6M). Weaker but significant staining was observed at the node of inflorescence stems (Figures 6N and 6O). During flower development, GUS staining was first observed in the whole stamen but was later limited to the vasculature in petals, the joint between the pedicel and flower, the stigma and the filament of the stamens (Figure 6P). In developing siliques but not embryos, GUS staining was limited to organ boundaries (Figure 6Q). The GUS staining patterns shown here are in agreement with the gene expression data presented in Figure 2, in that the male floral organs are the primary tissue of CEK4 expression. However, GUS staining in the root tips and lateral roots was not as strong as in the male floral organs, whereas the data in Figure 2B show that CEK4 expression is strong in roots. While the gene expression data in public databases have limited usefulness, our stable CEK4-GUS transformant, which complemented the cek4-1/+ phenotype and therefore is functional in vivo, accurately reveals the expression pattern. Thus, our GUS reporter assay indicated that CEK4 was expressed in different tissues, particularly in the vasculature and at organ boundaries.
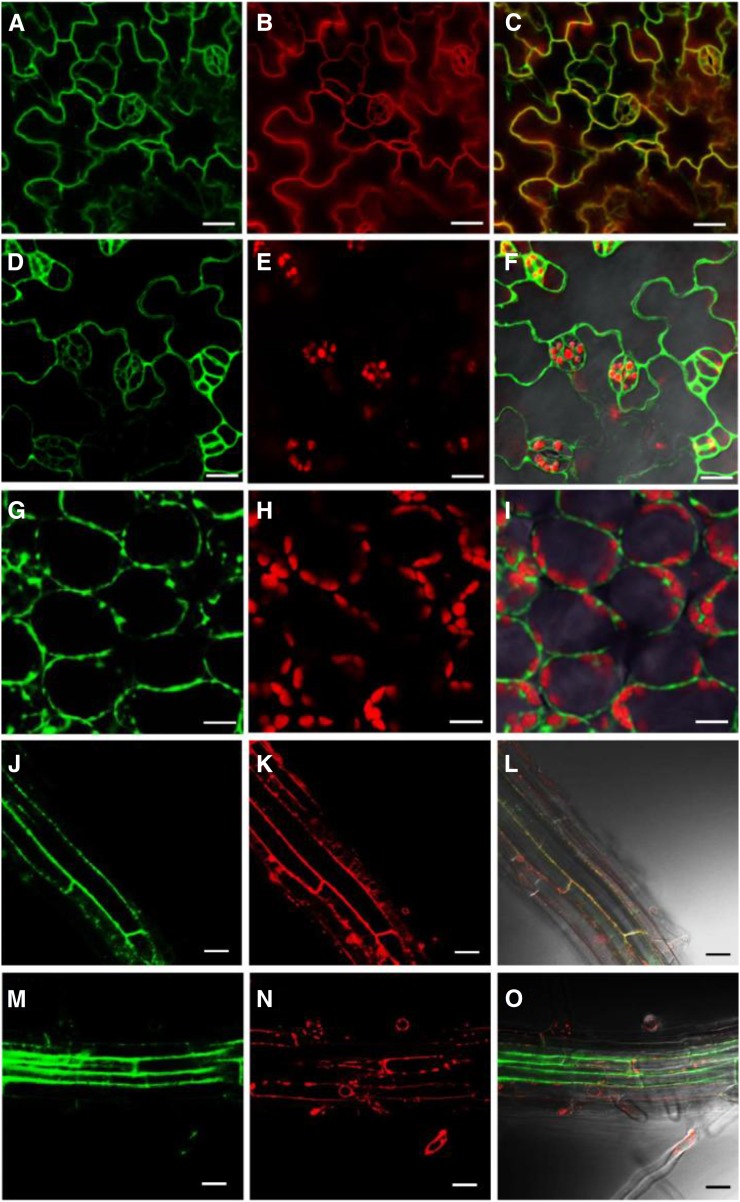
Subcellular Localization of CEK4
To investigate the subcellular localization of CEK4, we constructed a plasmid vector containing the genomic sequence of CEK4 fused C-terminally to the triple repeat of Venus (Ven) fluorescent protein (ProCEK4:CEK4-Ven) and transformed it into cek4-1/+ heterozygotes. This construct was functional in vivo because we isolated viable ProCEK4:CEK4-Ven cek4-1/- plants (Figure 4J, lane 5). Confocal laser scanning microscopy showed that Ven fluorescence colocalized with the plasma membrane marker FM4-64 (Figures 7A to 7C), but not with chloroplast autofluorescence in the leaf epidermis (Figures 7D to 7F). Although the circumference of some of the chloroplasts showed overlapping yellow color (Figure 7F), we did not observe any overlap in Ven fluorescence and chlorophyll autofluorescence in the leaf mesophyll cells, which contained many chloroplasts (Figures 7G to 7I). We next examined the possibility that CEK4 is localized to the endoplasmic reticulum (ER), which is the primary site of phospholipid biosynthesis. Whereas Ven fluorescence clearly overlapped with the plasma membrane marker FM4-64 in roots (Figures 7J to 7L), it did not overlap with the ER marker ER-Tracker Red (Figures 7M to 7O). These observations suggest that CEK4 is mainly localized to the plasma membrane, although they cannot rule out the possibility that CEK4 is also localized at the ER.
Figure 7.
Subcellular Localization of CEK4 in the Leaf and Root Tissues of ProCEK4:CEK4-Ven cek4-1/- Using Confocal Laser Scanning Microscopy.
(A) to (C) Venus fluorescence (A), FM4-64 staining (B), and merged (C) images of ProCEK4:CEK4-Ven cek4-1/- leaf epidermal cells.
(D) to (F) Venus fluorescence (D), chlorophyll autofluorescence (E), and merged (F) images of leaf epidermal cells.
(G) to (I) Venus fluorescence (G), chlorophyll autofluorescence (H), and merged (I) images of leaf mesophyll cells.
(J) to (L) Venus fluorescence (J), FM4-64 staining (K), and merged (L) images at root cortex cells.
(M) to (O) Venus fluorescence (M), ER-Tracker Red (Invitrogen) staining (N), and merged (O) images at the root cortex cells. Bars = 20 μm.
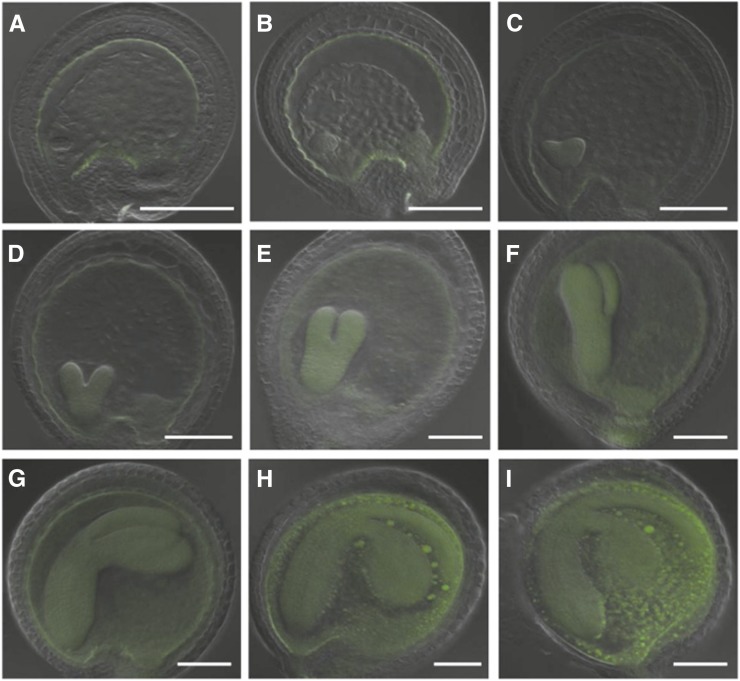
Localization of CEK4 during Embryo Development
Because the embryogenesis of the cek4-1/- mutant was arrested beyond the heart stage (Figure 5), we observed the localization of CEK4 in the mature seeds of ProCEK4:CEK4-GUS cek4-1/- by histochemical GUS staining. In intact mature seeds, we observed GUS staining in the embryo and chalazal region (Supplemental Figure 2A), as well as in the aleurone layer (Supplemental Figure 2B). In the embryo separated from the testa, we observed a strong GUS signal not only in the embryo but also in the endosperm region (Supplemental Figure 2C). Moreover, we observed a clear GUS signal surrounding cells in the aleurone layer that had been separated from the seed coat (Supplemental Figure 2D). To investigate the localization of CEK4 during embryo development more precisely, we observed fluorescence in ProCEK4:CEK4-Ven cek4-1/- embryos at different stages of development (Figure 8). During the globular stage and the transition from globular to heart stage, Ven fluorescence was initially observed at the periphery of the endothelium (Figure 8A) and then gradually expanded to the embryos, chalazal endosperm, and nucleated cytoplasm (Figures 8B and 8C). Between the heart stage and torpedo stage, the embryo sac undergoes a sequential process of endosperm cellularization (Mansfield and Briarty, 1990; Bewley et al., 2012). At this stage, Ven fluorescence was weak at the endothelium but enhanced at the embryos, cellular endosperm, peripheral endosperm, and the chalazal region (Figures 8C to 8F). Eventually, from the bent cotyledon to mature embryo stage, an intense fluorescence signal for CEK4-Ven was observed in the whole embryo sac (Figures 8G to 8I). No fluorescence signal was observed in embryos not containing ProCEK4:CEK4-Ven. This localization of fluorescence signal is in agreement with the result of our GUS staining analysis (Supplemental Figure 2). Thus, CEK4 showed dynamic changes in sub-embryo localization during embryo development and enhanced expression during the mature stages.
Figure 8.
Sub-Embryo Localization of CEK4-Ven in ProCEK4:CEK4-Ven cek4-1/- Using Confocal Laser Scanning Microscopy.
Embryos at the globular stage ([A] and [B]), heart stage ([C] and [D]), torpedo stage ([E] and [F]), bent cotyledon stage ([G] and [H]), and mature stage (I). Green, Ven fluorescence. Bars = 100 μm.
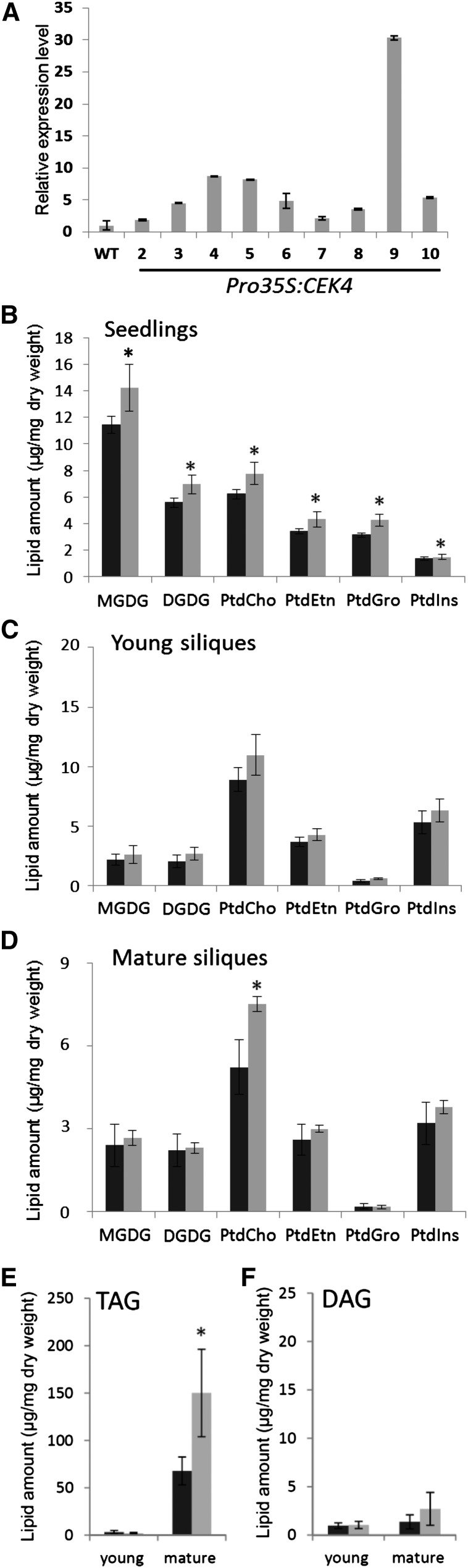
Involvement of CEK4 in Phospholipid Biosynthesis in Vivo
To study the involvement of CEK4 in phospholipid biosynthesis, we first analyzed the polar glycerolipid contents in young and mature siliques of cek4-1/+. However, no significant changes were observed (Supplemental Table 6). Therefore, we overexpressed CEK4 using a CaMV 35S promoter (Pro35S) in wild-type plants and measured the levels of different glycerolipid classes in seedlings and young and mature siliques (Figure 9). We first transformed Pro35S:CEK4 into cek4-1/+ and isolated viable Pro35S:CEK4 cek4-1/- plants (Figure 4J, lane 3), which validated the functionality of the transgene Pro35S:CEK4 in vivo. Next, we isolated multiple lines of the wild type harboring Pro35S:CEK4 and examined the expression level of CEK4. The expression of CEK4 was upregulated in these lines (Figure 9A). In particular, line 9 showed about a 30-fold increase in expression of CEK4 as compared with the wild type (Figure 9A). Line 9 Pro35S:CEK4 seedlings showed an increase in the levels of PtdCho and PtdEtn by 25 and 27%, respectively, compared with the wild type (Figure 9B). Moreover, other lipid classes such as monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and phosphatidylglycerol showed an increase (Figure 9B), maybe because an increase in the biosynthesis of these phospholipid classes funnels them into lipids that are in high demand during vegetative growth. We further analyzed the fatty acid composition of these glycerolipid classes but found no difference in composition between Pro35S:CEK4 cek4-1/- line 9 and the wild type (Supplemental Table 7).
Figure 9.
Construction and Analysis of Pro35S:CEK4.
(A) Expression of CEK4 among 9 different transgenic plant lines harboring Pro35S:CEK4.
(B) to (D) Polar glycerolipid levels in 10-d-old seedlings (B), young siliques (C), and mature siliques (D) of Pro35S:CEK4 line 9 (gray bars) and the wild type (black bars).
(E) and (F) Amount of TAG (E) and DAG (F) detected in the young and mature siliques of Pro35S:CEK4 line 9 (gray bars) and the wild type (black bars). The wild type in (A) and (B) is Col-0 and in (C) to (F) is Col-8. Data are mean ± sd from three biological replicates. Asterisks indicate significance by Student’s t test (P value < 0.02).
MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; PtdGro, phosphatidylglycerol; PtdIns, phosphatidylinositol.
Prompted by the arrested embryo development beyond heart stage (Figure 5) and the intense fluorescence observed for CEK4-Ven plants during the mature stage of embryo development (Figure 8), we next analyzed the polar glycerolipid levels in young (Figure 9C) and mature (Figure 9D) siliques. While young siliques of Pro35S:CEK4 line 9 showed no significant change from the wild type in polar glycerolipid levels (Figure 9C), a marked increase in PtdCho levels was found in the mature siliques of Pro35S:CEK4 line 9 (Figure 9D). No significant changes were observed in the fatty acid composition of these glycerolipid classes in young (Supplemental Table 8) or mature (Supplemental Table 9) siliques. This increase in PtdCho in mature siliques prompted us to quantify TAG levels in Pro35S:CEK4 line 9 because the major glycerolipid metabolic flux in mature seeds drives the accumulation of TAG as the primary seed storage reserve. Indeed, PtdCho is a substrate of TAG biosynthesis; it is a substrate of PtdCho:diacylglycerol acyltransferase (PDAT) (Dahlqvist et al., 2000) or is first converted to DAG by phospholipases and then serves as a substrate of acyl-CoA:diacylglycerol acyltransferase or PDAT (Zhang et al., 2009). In young siliques, no difference was found in the levels of TAG (Figure 9E) or DAG (Figure 9F) between Pro35S:CEK4 line 9 and the wild type. However, the mature siliques of Pro35S:CEK4 line 9 had nearly 2-fold higher levels of TAG than the wild type (Figure 9E), while DAG levels were indistinguishable from those of the wild type (Figure 9F). This result indicates that overexpression of CEK4 increases PtdCho and further increases TAG contents in the mature siliques.
Therefore, overexpression of CEK4 increased PtdCho levels both in seedlings and mature siliques, boosting the metabolism of major membrane lipid classes in seedlings and TAG in mature siliques. Thus, CEK4 may be involved in the rate-limiting step of phospholipid biosynthesis in Arabidopsis in vivo.
DISCUSSION
We investigated an unexplored CEK gene family in Arabidopsis. Comprehensive isolation of gene knockout mutants of CEK1, 2, 3, and 4 suggested that CEK4 is a major CEK because the cek4-1 mutant showed the embryo-lethal phenotype. The arrested development of embryos at the heart stage, along with functional complementation of the phenotype with transgenic CEK4 as well as the reporter assay during embryo development, indicated that CEK4 is required for the later stages of embryo development. Overexpression of CEK4 in Arabidopsis wild-type plants increased the levels of PtdCho both in seedlings and mature siliques and further increased the abundance of major membrane lipid classes in seedlings and of TAG in mature siliques. This suggests that CEK4 may be involved in the de novo biosynthesis of phospholipids. The plasma membrane localization of a CEK4-Ven fusion protein suggests that the initial step of the phospholipid biosynthetic pathway may be localized to the plasma membrane, although phospholipids are finally assembled in the endoplasmic reticulum. Thus, CEK4 may be involved in the rate-limiting step of phospholipid biosynthesis and indispensable for embryo development in Arabidopsis.
Role of CEK4 in Phospholipid Biosynthesis
The overexpression of CEK4 in a wild-type background increased levels of both PtdCho and PtdEtn in seedlings and PtdCho in mature siliques. A possible reason why increased PtdEtn was not observed in mature siliques is that PtdCho is the predominant phospholipid class in mature seeds and siliques (Devaiah et al., 2006). Interestingly, this resulted in an increase in primary membrane glycerolipid classes in seedlings and TAG in mature siliques. Vegetative growth requires a source of cellular membranes; therefore, membrane lipid biosynthesis is more active than storage lipid accumulation. In mature siliques, membrane lipids are converted to TAG, which serves as a storage compound in mature seeds. No significant change was observed for the fatty acid composition of these lipid classes, suggesting that this change was due to stimulation of polar head group metabolism, a pathway that does not involve modification of acyl groups. Therefore, these lipid profiles suggest that CEK4 is involved in the rate-limiting step in the de novo biosynthesis of PtdCho, which influences the biosynthesis of the predominant glycerolipid classes in seedlings and mature siliques. Possible substrates of CEK4 are Cho and Etn; therefore, CEK4 may function at least as an Etn kinase, which is rate-limiting for de novo phospholipid biosynthesis. Because Etn is produced from Ser by SDC (Figure 1A), this Ser-Etn-P-Etn metabolic flow may be a gateway to phospholipid biosynthesis, which channels the flux of amino acid metabolism into the biosynthesis of phospholipids. The reaction product of CEK4, P-Etn, can be used for PtdEtn biosynthesis and for PtdCho biosynthesis via P-Cho through three steps of methylation by PMTs. Indeed, this was found to be a predominant PtdCho biosynthetic pathway in the malaria parasite Plasmodium falciparum (Pessi et al., 2004). An analogous pathway may exist for PtdCho biosynthesis in Arabidopsis.
Role of CEK4 in Embryo Development
The knockout mutant cek4-1 showed defective embryonic development. This phenotype was complemented by transformation with ProCEK4:CEK4, ProCEK4:CEK4-GUS, ProCEK4:CEK4-Ven, or 35S:CEK4 (Figure 4), which confirmed that the embryo-lethal phenotype was due to the CEK4 knockout. Reciprocal crossing revealed that neither male nor female gametophytes were defective, and in developing embryos, growth was arrested after the heart stage (Figure 5). A sub-embryo localization study using CEK4-Ven showed that CEK4-Ven expression was induced at the mature stage of embryo development (Figure 8). No defect was found in the male floral parts in cek4-1/+ (Figures 4D to 4I). These observations suggest that CEK4 function may be critical during embryo development. The embryo lethality of cek4-1/+ may be due to a deficiency in phospholipid biosynthesis because mutation in PECT, which catalyzes the reaction following CEK, affects PtdEtn biosynthesis and results in an embryo-lethal phenotype (Mizoi et al., 2006). Moreover, a null mutation in SDC may be lethal (Kwon et al., 2012). Furthermore, mutation in the gene encoding lysophosphatidic acid acyltransferase1 (LPAAT1/ATS2), a key enzyme in de novo glycerolipid biosynthesis (Kennedy pathway), results in an embryo-lethal phenotype (Kim and Huang, 2004; Yu et al., 2004). Thus, phospholipid biosynthesis may be essential during embryo maturation. Moreover, developmental arrest in the later stages of embryo maturation could be associated with accumulation of TAG, the primary storage compounds in seeds. PtdCho is the substrate of one of two major TAG-biosynthetic pathways catalyzed by PDAT (Dahlqvist et al., 2000). Indeed, double knockout of PDAT and diacylglycerol acyltransferase, which mediates the other major TAG biosynthesis pathway, causes defects in embryo maturation (Zhang et al., 2009), which supports the notion that TAG accumulation is required for maturation of embryos. Indeed, overexpression of CEK4 increased not only PtdCho but also TAG in mature siliques (Figure 9). The defective embryo development of cek4-1/- suggests that CEK4 is involved in phospholipid biosynthesis and possibly also in TAG accumulation during embryo maturation. Future studies are required to further dissect the spatiotemporal role of CEK4 during embryo maturation.
METHODS
Plant Growth Conditions
Arabidopsis thaliana plants (Columbia-0 ecotype) were grown under 16 h light/8 h dark at 22°C. Murashige and Skoog medium was used at half-strength concentration for plant culture (Murashige and Skoog, 1962).
Plant Materials
The following mutant seeds were obtained from the ABRC: cek1-1 (SALK_069908), cek2-1 (GK-124F01), cek3-1 (SALK_026906), and cek4-1 (SALK_057727). Homozygous plants were isolated by PCR-based genotyping with gene-specific primers and T-DNA-specific primers as illustrated in Figure 3. The primers used were for cek1-1 (YN533/YN550 and YN533/KK008), cek2-1 (YN1004/YN1005 and YN1016/YN1005), cek3-1 (YN896/YN1219 and YN748/YN896), and cek4-1 (JL004/YN1429 and KK008/YN895). See Supplemental Table 10 for the oligonucleotide sequences used in this study. Two wild-type Columbia lines, Col-0 and Col-8, were used for lipid analysis as controls.
Plasmid Vector Construction and Transgenic Plant Production
For ProCEK4:CEK4 cek4-1/-, 4922 bp of the genomic sequence for CEK4 (ProCEK4:CEK4) was amplified by PCR with the primers YN1164 and YN1295. The fragment was cloned into the pENTR_D_TOPO plasmid vector (Invitrogen) to obtain pYL11, which was then recombined into the pBGW destination vector with the use of LR Clonase (Invitrogen) (Karimi et al., 2005) to obtain pYL13. The pYL13 plasmid was transformed into cek4-1/+ by Agrobacterium tumefaciens-mediated transformation. Twenty-four transformants were selected on soil by spraying with 0.1% Basta solution. Resistant plants were genotyped for cek4-1/+, and those carrying T-DNA (i.e., cek4-1/+ or cek4-1/-) were selected for harvesting of T2 seeds individually. The obtained T2 seeds underwent double selection using Basta and Km, and resistant plants homozygous for cek4-1 and harboring ProCEK4:CEK4 were selected by genotyping. To distinguish transgenic CEK4 (ProCEK4:CEK4) from endogenous CEK4, specific primers were designed to amplify transgenic CEK4 (JL004 and KK097) or endogenous CEK4 (Figure 4J; Supplemental Figure 3).
For Pro35S:CEK4 cek4-1/- and Pro35S:CEK4 wild type, 1125 bp of the open reading frame (ORF) for CEK4 was amplified with the primers YN1442 and YN1443. The fragment was cloned into the XhoI and BamHI sites of pYN2047 (pENTR plasmid vector containing 2xPro35S) to obtain pJL8, which was then recombined into the pBGW destination vector using LR Clonase (Karimi et al., 2005) to obtain pJL10. The pJL10 plasmid was transformed into the wild type or cek4-1/+ via Agrobacterium-mediated transformation. In all, 24 T1 plants were selected by repeatedly spraying soil-grown seedlings with 0.1% Basta solution. For Pro35S:CEK4 cek4-1/-, seedlings with a cek4-1/+ background were selected for harvesting of T2 seeds individually. The obtained T2 seeds underwent double-selection using Basta and Km, and resistant plants homozygous for cek4-1 and harboring Pro35S:CEK4 were selected by genotyping. To distinguish transgenic Pro35S:CEK4 from endogenous CEK4, the specific primers YN894 and KK096 were designed (Figure 4J; Supplemental Figure 3).
For ProCEK4:CEK4-GUS cek4-1/-, to create the GUS reporter construct translationally fused to the C terminus of the ORF of CEK4, the stop codon of pYL11 was removed by PCR-based site-directed mutagenesis (Sawano and Miyawaki, 2000) with the primer YN1427. The obtained pYL28 was recombined into the pGWB533 destination vector (Nakagawa et al., 2007) using LR Clonase. The resulting plasmid pJL1 was transformed into cek4-1/+ via Agrobacterium-mediated transformation. In all, 48 T1 plants were selected by spraying 0.1% Basta solution onto seedlings growing in soil, and those with the cek4-1/+ background were selected for harvesting of seeds individually. The obtained T2 seeds were double-screened using Basta and Km, and resistant plants homozygous for cek4-1 and harboring ProCEK4:CEK4-GUS were selected by genotyping. To distinguish transgenic ProCEK4:CEK4-GUS from endogenous CEK4, specific primers (JL004 and KK098) were designed (Figure 4J; Supplemental Figure 3). Line No. 54 was used for the observations in Figure 6.
For ProCEK4:CEK4-Ven cek4-1/-, to create the triple (3X) repeat of a Venus fluorescent reporter construct fused translationally to the C terminus of the ORF of CEK4, the 3xVen cassette was inserted into the SfoI site of pYL18. The obtained entry vector pCC102 was recombined into a pBGW destination vector using LR Clonase (Karimi et al., 2005), and the resulting plasmid pCC106 was transformed into cek4-1/+ via Agrobacterium-mediated transformation. In total, 48 T1 plants were selected by spraying soil-grown seedlings with 0.1% Basta solution, and then those with a cek4-1/+ background were selected for harvesting of seeds individually. The obtained T2 seeds were double-screened using Basta and Km, and resistant plants homozygous for cek4-1 and harboring ProCEK4:CEK4-Ven were selected by genotyping. To distinguish transgenic ProCEK4:CEK4-Ven from endogenous CEK4, specific primers (JL004 and KK104) were designed (Figure 4J; Supplemental Figure 3). Line 46 was used for the observations in Figures 7 and 8.
Lipid Analysis
Polar glycerolipids were analyzed as described (Nakamura et al., 2003). Briefly, total lipids were extracted from 10-d-old or 14-d-old seedlings as described (Bligh and Dyer, 1959; Nakamura et al., 2014a), and each glycerolipid class was separated on a silica-gel thin-layer chromatography plate with a solvent system of chloroform:methanol:aqueous ammonia = 120:80:8 (by volume) for the first dimension and chloroform:methanol:acetic acid:water = 170:20:15:3 (by volume) for the second dimension. Each lipid spot was visualized by spraying with purimuline solution, which was scraped off, and acyl moieties were hydrolyzed and methylesterified with HCl-methanol containing pentadecanoic acid (C15:0) as an internal standard. Fatty acid methyl esters were quantified using a gas chromatograph (GC-2010; Shimadzu) equipped with a ULBON HR-SS-10 column (Shinwa Chemical Industries). Data are the mean ± sd of three biologically independent samples.
Regarding sample harvesting of siliques for lipid analysis, young siliques were defined as being shorter than 12 mm and narrower than 1 mm, with embryos that were in the globular stage, as determined using a stereomicroscope. Mature siliques were defined as being longer than 12 mm and wider than 2 mm, with embryos beyond the heart stage. Prior to lipid extraction, samples were incubated in hot isopropanol containing 0.05% (v/v) butylated hydroxytoluene at 75°C for 15 min to inactivate phospholipase activity. Polar glycerolipids were analyzed as described above. TAG and DAG were separated on silica gel thin-layer chromatography plates by one-dimensional development with the solvent system of hexane:diethylether:acetic acid = 160:40:4 (by volume).
GUS Staining
Harvested fresh tissues were immediately immersed in ice-cold 90% (v/v) acetone for 15 min and then with GUS staining solution (10 mM EDTA, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.1% [w/v] Triton X-100, and 0.5 mg/mL X-Gluc [5-bromo-4-chloro-3-indolyl-β-d-glucuronide] in 100 mM phosphate buffer]. After incubation in the dark at 37°C overnight, staining was stopped by replacing the solution with 70% ethanol. For colored tissues, pigments were removed by immersing the tissue in ethanol:acetic acid = 6:1 (volume).
Cryo-Scanning Electron Microscopy Analysis
Fresh flower samples were frozen in liquid nitrogen slush and transferred to a preparation chamber at −160°C for 5 min. Sublimation was performed at −85°C for 15 min. Subsequently, the samples were coated with platinum (Pt) at −130°C, transferred to a scanning electron microscopy chamber, and observed at −160°C using Cryo-SEM (FEI Quanta 200 SEM/Quorum Cryo System PP2000TR FEI) at 20 kV.
Alexander’s Staining of Pollen
For pollen viability observations, stamens were placed on microscope slides and mounted in a drop of Alexander’s staining buffer (10 mL 95% ethanol, 1 mL malachite green [1% in 95% ethanol], 5 mL fuchsine acid [1% in water], 0.5 mL orange G [1% in water], 5 g phenol, 5 g chloral hydrate, 2 mL glacial acetic acid, 25 mL glycerol, and 50 mL deionized water) (Schoft et al., 2011) under a cover slip for observation. After incubation, viable pollen grains stain purple while aborted grains stain green. Images were taken using an upright microscope (Zeiss Axio Imager Z1).
Microscopy Analysis of Embryogenesis
Embryos were first cleared using clearance solution (chloral hydrate:glycerol:water = 8:2:1 by volume) and then Nomarski (differential interference contrast) images were acquired using an upright microscope (Zeiss Axio Imager Z1).
Confocal Laser Scanning Microscopy
Fluorescence of 3xVen in 14-d-old seedlings of the ProCEK4:CEK4-Ven cek4-1/- line 46 was observed under a microscope (LSM 510 Meta; Carl Zeiss) equipped with LCI Plan-Neofluar 63×/1.3-numerical aperture (NA) immersion, Plan-Apochromat 20×/0.8-NA, and Plan-Apochromat 10×/0.45-NA objectives. For plasma membrane or ER staining, samples were immersed in 5 μg/mL FM 4-64 (Molecular Probes, Invitrogen) or in the ER-Tracker Red dye (2 μM; Invitrogen), respectively, for 5 min. Images were captured using an LSM 510 v3.2 confocal microscope (Carl Zeiss) with filters for Venus (514-nm laser, band-pass 520 to 555 nm) and FM 4-64 or ER-Tracker Red dye (514-nm laser, band-pass 650 to 710 nm). Ven fluorescence in ProCEK4:CEK4-Ven cek4-1/- embryos of different stages was observed by confocal microscopy as above after the samples were cleared with clearance solution as described above.
RNA Extraction and Quantitative RT-PCR
Total RNA was isolated from 7-d-old seedlings use TRI reagent (Ambion) including DNase treatment, and cDNA was synthesized using the SuperScript III first-strand synthesis kit (Invitrogen). Quantitative RT-PCR was performed using the 7500 Real-Time PCR System (Applied Biosystems). The comparative threshold cycle method was used to determine relative gene expression, with the expression of ACT serving as an internal control (Liu et al., 2007). Data are mean ± sd from three biological replicates, with three technical replicates. The primer sets for quantitative RT-PCR were reported previously (Nakamura et al., 2014a).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers:CEK1 (At1g71697), CEK2 (At1g74320), CEK3 (At4g09760), and CEK4 (At2g26830).
Supplemental Data
Supplemental Figure 1. Vegetative growth phenotype of cek4-1/+ mutant at 28 d old.
Supplemental Figure 2. Expression of CEK4 in mature seeds by histochemical GUS staining of ProCEK4:CEK4-GUS cek4-1/-.
Supplemental Figure 3. Annealing position of primers used for genotyping in Figure 4J.
Supplemental Table 1. The Arabidopsis CEK family.
Supplemental Table 2. Polar glycerolipid composition (mol %) of the 14-d-old seedlings of homozygous cek1-1, cek2-1, and cek3-1 mutants and the wild types Col-0 and Col-8 as controls.
Supplemental Table 3. Fatty acid composition (mol %) of the polar glycerolipid classes in 14-d-old seedlings of cek1-1, cek2-1, cek3-1, and the wild types Col-0 and Col-8.
Supplemental Table 4. Levels of TAG and DAG (μg/mg dry weight) in mature siliques of homozygous cek1-1, cek2-1, and cek3-1 mutants and the wild type Col-8.
Supplemental Table 5. Fatty acid composition (mol %) of TAG in mature siliques of cek1-1, cek2-1, cek3-1, and the wild type Col-8.
Supplemental Table 6. Amounts of polar glycerolipid classes (μg/mg dry weight) in young siliques and mature siliques of cek4-1/+ and wild type Col-8.
Supplemental Table 7. Fatty acid composition (mol %) of the polar glycerolipid classes in seedlings of Pro35S:CEK4 line 9 and the wild type Col-0.
Supplemental Table 8. Fatty acid composition (mol %) of the polar glycerolipid classes in young siliques of Pro35S:CEK4 line 9 and the wild type Col-8.
Supplemental Table 9. Fatty acid composition (mol %) of the polar glycerolipid classes in mature siliques of Pro35S:CEK4 line 9 and the wild type Col-8.
Supplemental Table 10. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank Tsuyoshi Nakagawa (Shimane University, Japan) for kindly providing the pGWB533 plasmid vector. We also thank staff at the Institute of Plant and Microbial Biology, Academia Sinica, including Wann-Neng Jane, Mei-Jane Fang, Yi-Chia Chou, and Ji-Ying Huang, for technical assistance in microscopy observation, Chia-En Chen for constructing pCC102 and pCC106 plasmid vectors, and Kazue Kanehara for discussion and critical reading of the article. This research was supported by the core research budget provided by the Institute of Plant and Microbial Biology, Academia Sinica (Y.N.). Y.N. is supported by the EMBO Young Investigator Program.
AUTHOR CONTRIBUTIONS
Y.-C.L. performed overall experiments. Y.-c.L. performed a part of transgenic plant construction and provided technical support to Y.-C.L. Y.N. conceived the research, performed transgenic plant management, genetic crossing, a part of phenotype observation, and data analysis, and wrote the article. All authors commented on the article and approved the contents.
Glossary
- PtdCho
phosphatidylcholine
- PtdEtn
phosphatidylethanolamine
- P-Etn
phosphoethanolamine
- P-Cho
phosphocholine
- DAG
sn-1,2-diacylglycerol
- Etn
ethanolamine
- CEK
choline/ethanolamine kinase
- PMT
S-adenosyl-l-methionine:phosphoethanolamine N-methyltransferase
- TAG
triacylglycerol
- ER
endoplasmic reticulum
- PDAT
PtdCho:diacylglycerol acyltransferase
- ORF
open reading frame
- NA
numerical aperture
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Aoyama C., Liao H., Ishidate K. (2004). Structure and function of choline kinase isoforms in mammalian cells. Prog. Lipid Res. 43: 266–281. [DOI] [PubMed] [Google Scholar]
- Aoyama C., Yamazaki N., Terada H., Ishidate K. (2000). Structure and characterization of the genes for murine choline/ethanolamine kinase isozymes alpha and beta. J. Lipid Res. 41: 452–464. [PubMed] [Google Scholar]
- Bewley, J.D., Bradford, K., Hilhorst, H., Nonogaki, H. (2012). Seeds: Physiology of Development, Germination and Dormancy. (New York: Springer). [Google Scholar]
- Bligh E.G., Dyer W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Bolognese C.P., McGraw P. (2000). The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol. 124: 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1987). Phosphotransferase sequence homology. Nature 329: 21. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A., López-Bucio J., Ramírez-Pimentel G., Zurita-Silva A., Sánchez-Calderon L., Ramírez-Chávez E., González-Ortega E., Herrera-Estrella L. (2004). The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 16: 2020–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist A., Stahl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97: 6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry 67: 1907–1924. [DOI] [PubMed] [Google Scholar]
- Dewey R.E., Wilson R.F., Novitzky W.P., Goode J.H. (1994). The AAPT1 gene of soybean complements a cholinephosphotransferase-deficient mutant of yeast. Plant Cell 6: 1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatsugi R., Nakamura M., Nishida I. (2002). Phosphatidylcholine biosynthesis at low temperature: differential expression of CTP:phosphorylcholine cytidylyltransferase isogenes in Arabidopsis thaliana. Plant Cell Physiol. 43: 1342–1350. [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Kim H.U., Huang A.H. (2004). Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol. 134: 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Yu S.I., Lee H., Yim J.H., Zhu J.K., Lee B.H. (2012). Arabidopsis serine decarboxylase mutants implicate the roles of ethanolamine in plant growth and development. Int. J. Mol. Sci. 13: 3176–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. (1990). Endosperm cellularization in Arabidopsis thaliana L. Arab. Inf. Serv. 27: 65–72. [Google Scholar]
- Mizoi J., Nakamura M., Nishida I. (2006). Defects in CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18: 3370–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Awai K., Masuda T., Yoshioka Y., Takamiya K., Ohta H. (2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280: 7469–7476. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Arimitsu H., Yamaryo Y., Awai K., Masuda T., Shimada H., Takamiya K., Ohta H. (2003). Digalactosyldiacylglycerol is a major glycolipid in floral organs of Petunia hybrida. Lipids 38: 1107–1112. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Teo N.Z., Shui G., Chua C.H., Cheong W.F., Parameswaran S., Koizumi R., Ohta H., Wenk M.R., Ito T. (2014a). Transcriptomic and lipidomic profiles of glycerolipids during Arabidopsis flower development. New Phytol. 203: 310–322. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Andrés F., Kanehara K., Liu Y.C., Dörmann P., Coupland G. (2014b). Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 5: 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Kociubinski G., Mamoun C.B. (2004). A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc. Natl. Acad. Sci. USA 101: 6206–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W.I., Voelker D.R., Basset G., Hanson A.D. (2001). Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 276: 35523–35529. [DOI] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. (2000). Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft V.K., Chumak N., Choi Y., Hannon M., Garcia-Aguilar M., Machlicova A., Slusarz L., Mosiolek M., Park J.S., Park G.T., Fischer R.L., Tamaru H. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 108: 8042–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasseva G., Richard L., Zachowski A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566: 115–120. [DOI] [PubMed] [Google Scholar]
- Wu G., Vance D.E. (2010). Choline kinase and its function. Biochem. Cell Biol. 88: 559–564. [DOI] [PubMed] [Google Scholar]
- Yu B., Wakao S., Fan J., Benning C. (2004). Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 45: 503–510. [DOI] [PubMed] [Google Scholar]
- Zhang M., Fan J., Taylor D.C., Ohlrogge J.B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.