Abstract
Determining the distribution of a drug and its metabolites within tissue is a key facet of evaluating drug candidates. Drug distribution can have a significant implication in appraising drug efficacy and potential toxicity. The specificity and sensitivity of mass spectrometry imaging (MSI) make it a perfect complement to the analysis of drug distributions in tissue. The detection of lapatinib as well as several of its metabolites in liver tissue was determined by MSI using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) coupled to high resolving power Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometers. IR-MALDESI required minimal sample preparation while maintaining high sensitivity. The effect of the electrospray solvent composition on IR-MALDESI MSI signal from tissue analysis was investigated and an empirical comparison of IR-MALDESI and UV-MALDI for MSI analysis is also presented.
Keywords: MALDI, IR-MALDESI, mass spectrometry imaging, drug distribution
1 INTRODUCTION
In drug discovery and development, it is essential to determine and understand the pharmacokinetic (PK) properties of drug candidates including their absorption, distribution, metabolism, and excretion (ADME) [1]. Drug efficacy is at least partially dependent on its localization toward its intended target; however, untargeted accumulation of the drug or its metabolites in tissue can result in adverse toxicological effects. While drug levels in plasma do play an important role in pharmacokinetic and pharmacodynamic (PK/PD) studies, it has been suggested that plasma concentrations alone do not accurately reflect the localized drug concentration in tissue [2].
In 1997 Caprioli et al. demonstrated the molecular imaging of peptides and proteins from biological tissue sections using matrix-assisted laser desorption ionization (MALDI) MSI[3]. There are several key attributes that make MALDI MSI a good candidate for the mapping of pharmaceuticals in tissue. MALDI is known to be a highly sensitive technique that is capable of detecting attomole quantities of analyte [4]. Also, detection based on MS allows for simultaneous image acquisition of multiple species as well as discrimination between parent drug and metabolite distributions. An added benefit is that MS does not require the use of radiolabeled standards and therefore distributions studies can be performed as a more cost effective methodology earlier in the drug discovery process to refine the list of potential drug candidates to those that exhibit desirable PK/PD properties. Troendle et al. demonstrated the direct detection of pharmaceuticals from tissue sections by MALDI MS [5] and Caprioli's group presented the first MALDI MSI analysis of dosed drugs in tissue [6,7]. Because whole body autoradiography (WBA) and LC-MS are more commonly used for drug distribution analysis, several groups have validated the observed distributions and relative intensities obtained from MSI by performing autoradiography or LC-MS on serial sections [7-28].
The multiplexing capabilities of MSI can be realized if a large m/z range is acquired from each pixel enabling simultaneous acquisition of spatial information for hundreds of endogenous as well as exogenous molecules (xenobiotics). The additional information obtained from these endogenous molecular distributions could prove to be useful when correlated with drug accumulation [19,25,29-32]. However, because most MALDI MSI analyses are performed with lower resolution mass analyzers, isobaric interferences in the low m/z range from endogenous molecules or matrix-related peaks necessitate the use of MS/MS imaging to follow a specific transition which precludes multiplex image acquisition. Several sample preparation methods have been described to circumvent isobaric matrix interferences [21,33]. Despite being less common, the examples of high spectral resolution MSI for determining drug distribution in tissue demonstrate the benefits of using FTICR or Orbitrap MS for this type of study [23,28,30,31,34-39]. Most of these groups report on the resolution of several unique species with distinct spatial distributions all resolved at the same nominal mass. This higher resolving power (RP) decreases the risk of an unresolved interfering species, therefore allowing for full mass range MSI to be performed and the benefits of multiplex image acquisition to be realized. In addition to high RP, these instruments also provide high mass measurement accuracy (MMA) which, for small molecules, can provide a unique elemental composition to aid in ion identification [40].
A majority of the challenges with MALDI MSI are directly related to the matrix application process. This process commonly involves spraying or spotting a saturated solution of organic matrix to extract analyte from within the tissue for co-crystallization with the matrix on the tissue surface [41]. Therefore, variation in extraction efficiency relating either to tissue type or analyte solubility in the matrix solvent can impact the observed relative ion intensities. There is also some debate on the degree of molecular diffusion that can take place during this process. In addition, the homogeneity of matrix application and the size of the resulting matrix crystals directly impact the achievable spatial resolution making sample preparation the most important step in the MALDI MSI process. Because of the large number of variables associated with matrix application (i.e. choice of matrix, choice of matrix solvent composition, application method), there are no standard procedures that work best for all samples. Thus, the decision of which method to use is typically determined empirically for a specific analyte or tissue type. Also, given that the matrix molecules ionize very well and are present in such large excess, the low mass range is typically dominated by matrix-related peaks making the analysis of small molecules more challenging. Despite these analytical challenges, MALDI has been the most widely implemented method of ionization for imaging of drugs and metabolites in tissues.
Other ionization methods that don't require such extensive sample preparation show a great deal of promise in MSI. Desorption electrospray ionization (DESI) has been used on several occasions for drug distribution studies [14,16,28] and liquid extraction surface analysis (LESA) is capable of providing a quick profiling of drug distribution [42-46]. Another ambient ionization method that has demonstrated potential in MSI is infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) [47]. MALDESI was the first ambient ionization technique to combine atmospheric pressure matrix-assisted laser desorption with electrospray post-ionization [48]. While first described for UV desorption, the MALDESI process is independent of laser wavelength given that any wavelength laser could be used, provided an appropriate matrix (endogenous or exogenous) is chosen that strongly absorbs the corresponding laser wavelength. The use of an infrared laser is particularly appealing because it allows for the use of ice as the laser energy absorbing matrix which does not produce mass spectral interferences and can be easily applied to the sample if necessary.
Herein we report on the use of both IR-MALDESI and UV-MALDI coupled to FT-ICR for the MSI analysis of lapatinib dosed liver tissue. Both techniques provide complementary information on the detection of the parent drug and several of its metabolites. An investigation of the role of the electrospray solvent composition on IR-MALDESI signal is also presented.
2 EXPERIMENTAL
Materials
Formic acid was purchased from Sigma Aldrich (St. Louis, MO). HPLC grade acetonitrile, methanol, trifluoroacetic acid, and water were purchased from Burdick & Jackson (Muskegon, MI, USA) or Fisher Scientific (Pittsburgh, PA) and were used as received without further purification. 2,5-dihydroxybenzoic acid, (DHB, purity 98%) was purchased from Sigma Aldrich (St. Louis, MO) and purified by recrystallized prior to use. ITO coated glass microscope slides were purchased from Bruker Daltonics (Billerica, MA). Shandon colorfrost plus positively charged glass microscope slides were purchased from Fisher Scientific (Pittsburgh, PA).
2.1 Samples
All studies were conducted in accordance with the GSK Policy on the Care, Welfare and Treatment of Laboratory Animals and were reviewed the Institutional Animal Care and Use Committee either at GSK or by the ethical review process at the institution where the work was performed.
Following repeat oral administration of lapatinib, dog liver tissue was flash frozen in liquid nitrogen at necropsy and stored at −80 °C until sectioning. Frozen tissues were sectioned at 12 μm for UV-MALDI with serial 50 μm sections collected for IR-MALDESI using a Leica 350S cryostat (Wetzlar, Germany). Sections for IR-MALDESI were thaw mounted onto positively charged glass microscope slides and sections for UV-MALDI were thaw mounted onto ITO coated glass microscope slides.
IR-MALDESI Imaging
For a detailed description of the IR-MALDESI imaging source and source parameters please refer to the following references [47,49]. In short, the sample is placed on the liquid cooled Peltier stage which is cooled to −10 °C while open to the ambient in order to deposit a thin layer of ice over the surface of the tissue section. The energy from a mid IR laser (IR Opolette, Opotek, Carlsbad, CA, USA) pulse (2.94 μm) is strongly absorbed by the ice matrix to facilitate desorption of the sample. The ablated neutral sample molecules partition into the charged solvent droplets of an electrospray plume generated from an emitter capillary coupled to a syringe pump. Ion production occurs through an ESI-like process and ions are then sampled by the atmospheric pressure interface of the mass spectrometer. The IR-MALDESI source is coupled to a Thermo LTQ-FT Ultra (ThermoFisher Scientific, San Jose, CA, USA). The resolving power of the LTQ-FT was set at 100,000 at m/z 400. The automatic gain control (AGC), which controls the number of ions sent to the ICR cell to maintain high MMA with external mass calibration, was turned off for these experiments due to its incompatibility with the pulsed nature of the IR-MALDESI source. Internal calibration can be performed using the average frequency shift recalibration technique (described in detail elsewhere [50]) to maintain MMA within ± 1 ppm. For these experiments a large m/z window (m/z 150-1500) was collected in order to obtain spatial information on endogenous species as well as the drug-related peaks. Stage movement, laser ablation, and mass spectrometer acquisition is synchronized as described previously [47]. Three IR pulses are performed at each pixel which ablates all the way through the 50 μm thick tissue section with the ion injection time (length of time that ions are accumulated in the LTQ) set to 200 ms in order to accumulate ions from all three laser pulses (3 pulses at 20 Hz). In this manner, a single acquisition mass spectrum is obtained at each pixel location. Thus, the signal at each pixel in the ion image is representative of an average of ions from the entire voxel of material that is ablated. It should also be noted that liver morphology can vary within these 50 μm thick voxels. A rectangular region of interest (ROI) that defines the boundaries of the MSI experiment was drawn to encompass the entire tissue area The MSI experiments were performed at a spatial resolution of 150 μm.
2.2 UV -MA LDI Imag ing
Immediately following thaw-mounting of the tissue section on the ITO coated glass slide, a matrix solution consisting of DHB (30 mg/mL) in water/methanol/TFA (1:1:.05 v/v/v) was applied using an ImagePrep (Bruker Daltonics, Billerica, MA) automated matrix application device for all UV-MALDI tissue slides. All UV-MALDI (355 nm) experiments were performed using a Solarix 7T Fourier transform-ion cyclotron resonance mass spectrometer (Bruker Daltonics, Billerica, MA). Data were acquired at a spatial resolution of 150 μm and spectra at each position were generated using 100 laser shots at a frequency of 1 kHz. Dog liver tissue sections consisted of 2962 pixels. Ion images for tissue sections were acquired in 1 hour at a peak resolution of ~125,000 (FWHM, m/z 400).
2.3 Data A na lys is
The .raw files from the Thermo instrument were converted into mzXML files using the freely available MSConvert tool from Proteowizard [51]. The Bruker files were exported as .ascii files using CompassXport (free data export tool from Bruker). Analysis of both imaging files was performed using the freely available standalone version of the MSiReader software [52]. All images shown have not been normalized or interpolated in order to demonstrate the true quality of the raw data. The observed accurate masses were used to assign the identification of lapatinib and its metabolites. The nomenclature for the observed metabolites is based on a previously published metabolite descriptions of lapatinib [53]. For the pixel by pixel calculation of the percentage of drug-related material (% DRM), the pixel intensities for lapatinib and all of its metabolites were exported from MSiReader by exporting the intensity data for all pixels containing drug related material. These intensity values for all drug-related m/z's were summed on a pixel by pixel basis to obtain a heat map of the total intensity for all DRM. This new heat map was then loaded back into MSiReader using the load custom data function. By normalizing lapatinib or one of its metabolites with this custom heat map, the percentage of total DRM at each pixel can be determined for each drug component. These heat maps show the distribution of the composition of DRM in each pixel where the intensity scale is % DRM for that particular compound (lapatinib or metabolites). While this is not an exact measure of the % DRM due to potential differences in ionization efficiency, it can provide valuable information about the relative distribution of the parent and its metabolites.
2.4 IR-MAL DESI Imaging Solvent Composition Analysis
To investigate the effect of the electrospray solvent composition on the observed signal from an IR-MALDESI MSI experiment, the syringe pump was replaced with a HPLC pump (Chorus 220, Leap Technologies, Carrboro, NC). Both methanol and acetonitrile were investigated as the organic solvent. The solvent compositions were as follows; solvent A: 95% water 5% organic (methanol or acetonitrile) 0.2% formic acid and solvent B: 5% water 95% organic (methanol or acetonitrile) 0.2% formic acid. The following solvent gradient was performed during an imaging analysis within the tissue section at a flow rate of 3 μL/min.; 0-5 min 95% A, 5-20 min 95% A to 95% B, 20-25 min 95%B. A rectangular ROI was selected such that every pixel in the imaging experiment was on tissue (Fig. 2a-b). Three separate experiments were performed for each solvent system on serial sections of lapatinib dosed liver tissue. The extracted ion chromatograms for several lipid species (369.3516, cholesterol, [M+H-H2O]+; 758.5694, PC(34:2)/PE(37:2), [M+H]+; 786.6007, PC(36:2)/PE(39:2), [M+H]+; 806.5694, PC(38:6)/PE(41:6), [M+H]+; 810.6007, PC(38:4)/PE(41:4), [M+H]+), lapatinib and its metabolites, and several ambient ions (371.1012, pentadimethylcyclocyloxane, [M+H]+; 519.1388, heptadimethylcyclocyloxane, [M+H]+; 536.1654, heptadimethylcyclocyloxane, [M+NH4]+; 593.1576, octadimethylcyclocyloxane, [M+H]+; 610.1841, octadimethylcyclocyloxane, [M+NH2]+) were obtained for each experiment using Xcalibur (ThermoFisher Scientific) and exported into Excel. The intensities for each group (lipids, DRM, and ambient ions) were summed for each experiment and the plots in Fig 2c) represent a 20 point moving average of all three imaging experiments for both solvent systems.
Fig. 2.
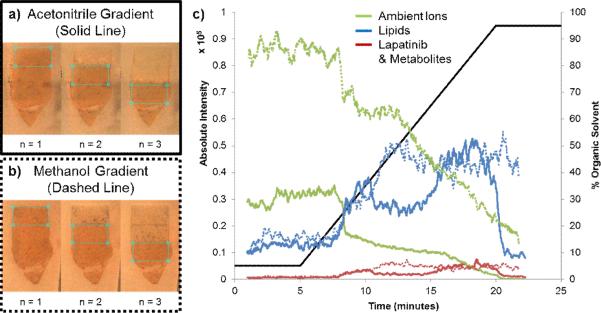
Influence of solvent composition on IR-MALDESI MSI signal of tissue related material. a) Optical images showing the region of interest for the three imaging experiments performed with an acetonitrile gradient. b) Optical images showing the region of interest for the three imaging experiments performed with a methanol gradient. c) The summed extracted ion chromatograms for several lipids (blue lines), ambient ions (green lines), and lapatinib and its metabolites (red lines). The solid lines represent data from the acetonitrile gradient and the dashed lines represent data from the methanol gradient
3 RESULTS and DISCUSSION
3.1 Preliminary IR-MALDESI Imaging
IR-MALDESI MSI was performed on a 50 μm thick section of dog liver tissue from an animal dosed with the tyrosine kinase inhibitor lapatinib. Because of the increased penetration depth of an IR laser compared to UV, a significantly larger amount of material is ablated with a single IR laser pulse [54]. While drug related signal was observed with 12 μm thick tissue sections, a previous investigation on the effect of tissue thickness on overall IR-MALDESI signal demonstrated that the use of thicker tissue with multiple pulses per pixel provided more consistent results (data not shown). This, however, does come at the cost of loss of spatial resolution in the z-direction where tissue morphology can vary within the thicker tissue sections. The tissue section was placed on the liquid cooled Peltier stage of the IR-MALDESI source and a thin layer of ice was deposited over the top of the tissue section just prior to analysis. This ice layer has been shown to improve sensitivity and pixel to pixel reproducibility in IR-MALDESI imaging experiments [47,49] as it resonantly absorbs the laser energy and facilitates efficient desorption of the tissue-related material.
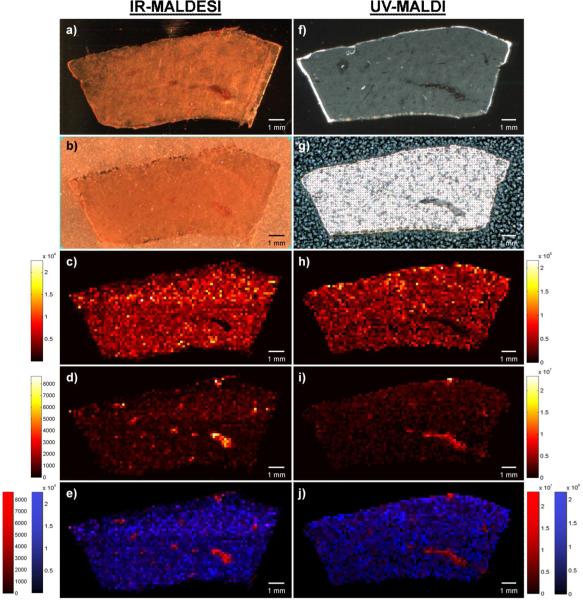
The results of the IR-MALDESI MSI experiment are shown in Fig. 1a-e. In addition to detecting lapatinib, IR-MALDESI MSI was capable of determining the spatial distribution for eleven of its metabolites (see supplemental Fig. S1 for ion maps of all drug-related material). Optical images of the tissue section before (Fig. 1a) and after the ice matrix deposition (Fig. 1b) demonstrate the uniformity of matrix application. At 150 μm spatial resolution, the distribution of the parent drug as [M+H]+ appears to be homogenously distributed throughout the liver with exception to the area around the blood vessels where little to no lapatinib was detected (Fig. 1c). To highlight the location of these blood vessels, the distribution for heme b as [M]+ is presented in Fig. 1d and an overlay of the lapatinib distribution (blue) with the heme distribution (red) is shown in Fig. 1e. Additionally, there were over 700 peaks relating to endogenous species that were also detected. Based on an accurate mass search of this peak list in the Scripps METLIN database [55], a majority of these endogenous species were likely to be lipids (primarily phosphocholines). The lower mass limit for the subsequent UVMALDI analysis was set to m/z 340 to avoid low mass interferences from matrix related peaks. Given that the matrix for the IR-MALDESI imaging experiments is ice, there are no low mass matrix interferences allowing for acquisition of a lower mass range (m/z 150 for these experiments). Of the endogenous species that were detected, over 40 of them were below the lower mass range for the UV-MALDI acquisition (340 m/z). These low m/z peaks include endogenous species such as phenylalanine, spermine, and adenosine just to name a few. The ability to acquire imaging data at such low m/z without matrix interference becomes even more significant when investigating the distribution of drugs or endogenous species that fall within this range.
Fig. 1.
IR-MALDESI (a-e) and UV-MALDI (f-j) MSI of lapatinib dosed liver tissue. a) Optical image of liver tissue section analyzed by IR-MALDESI. b) Optical image of liver tissue section after deposition of ice matrix. c) Ion map of lapatinib [M+H]+. d) Ion map of heme b [M]+ showing the distribution of the blood vessels. e) Overlay of c) and d) where blue is the lapatinib and red is the heme b distribution. f) Optical image of liver tissue section analyzed by UV-MALDI. g) Optical image of liver tissue section after matrix application. h) Ion map of lapatinib [M+H]+. i) Ion map of SM(34:0) or PE-Cer(37:1) [M+H]+ showing the distribution of the blood vessels. j) Overlay of h) and i) where blue is the lapatinib and red is the blood vessel distribution
3.2 UV-MALDI Imaging
UV-MALDI MSI was performed on a 12 μm thick section of dog liver that was a serial cut to the one analyzed by IR-MALDESI to allow for empirical comparison between the two techniques. The UV-MALDI results are shown in Fig. 1f-j. Images depicting the section before and after the matrix application process are shown in Fig. 1f and 1g respectively. The spatial resolution of the MALDI experiment was set to 150 μm to match the resolution currently attainable by IR-MALDESI. The conditions used for the matrix application step in MALDI IMS can be adjusted to match the desired spatial resolution. Higher spatial resolution experiments (e.g. 10 μm) require higher quality application (e.g. sublimation) of the matrix to achieve a homogenous coating and avoid introducing artificial variation of the analyte signal. In these experiments, given the relatively low spatial resolution, the matrix application step was adjusted to enhance for sensitivity by increasing the volume of matrix solution applied during each spray cycle to increases the analyte extraction from the tissue. As a result, it is apparent from Fig. 1g that the aggressive matrix application method used in this instance resulted in heterogeneous crystal formation on the tissue surface. This heterogeneity can produce variation in analyte signal that correlates with the local matrix concentration. This technique, however, led to the observation of lapatinib as [M+H]+ (Fig. 1h) as well as twenty-four metabolites of lapatinib directly from the dog liver tissue. The distributions for these metabolites are similar to the one shown for lapatinib (Fig. 1h) but can be found in supplemental Fig. S2. Lapatinib and its metabolites were detected homogeneously throughout the liver at this spatial resolution. As was observed in the IR-MALDESI MSI analysis, lapatinib was not detected in the regions around the blood vessels. The ion image for m/z 725.5575 (Fig. 1i), tentatively identified as SM(34:0) or PE-Cer(37:1), relates to the location of these blood vessels and an overlay with the lapatinib distribution is given in Fig. 1j. Comparison of Fig. 1c (IR-MALDESI lapatinib distribution) and Fig. 1h (UV-MALDI lapatinib distribution) highlights that both techniques provide similar results. The UV-MALDI MSI technique was able to detect more of the lesser abundant metabolites, however, because these MALDI and MALDESI experiments were performed on two different mass spectrometers, it is difficult to discern whether this improved sensitivity is directly related to differences in the ionization sources.
3.3 Influence of Electros pray Solvent Composition
Comparison of the preliminary IR-MALDESI MSI results with those obtained from UV-MALDI revealed that the latter may be more sensitive as more of the lesser abundant metabolites were observed. In order to improve sensitivity, an optimization of the IR-MALDESI source geometry for tissue imaging experiments was performed [49]. In addition to the source geometry optimization, the influence of the electrospray solvent composition on IR-MALDESI signal was also investigated. It was hypothesized that the composition of the electrospray solvent would impact analyte partitioning into the charged solvent droplets and prior results from other groups using similar ionization sources support this hypothesis [56,57].
To systematically explore how solvent composition influences the observed IR-MALDESI signal, the syringe pump that normally supplies the isocratic flow of electrospray solvent was replaced with an LC pump to provide a solvent gradient. In short, an imaging experiment was performed over the lapatinib dosed liver tissue while an LC gradient from 5% organic to 95% organic was performed. Both methanol and acetonitrile were investigated as the organic solvent. The results of these experiments are provided in Fig. 2. Optical images of the ROI for each imaging experiment are provided in Fig. 2a for the acetonitrile gradient and Fig. 2b for the methanol gradient. The summed extracted ion chromatograms of several lipids are represented in blue, lapatinib and its metabolites are shown in red, and several ambient ions are shown in green. Also provided is an overlay of the gradient plotted on the secondary y-axis. For both solvent systems the abundance of the tissue-related ions (lipids and DRM) was low with the high aqueous composition but started to increase by around 20% B. The acetonitrile gradient (solid lines) shows an initial spike around 35% B then the most abundant signal for both groups of tissue-related ions was observed at around 75% B. For the methanol gradient (dashed lines) the tissue-related ions reach their maximum abundance around 50% B then the abundance pretty much plateaus. At the optimal solvent composition for both solvent systems, the maximum abundance for both tissue-related ion groups (lipids and DRM) was essentially the same. However, the major difference between the two was the abundance of the ambient ions. As can be observed in Fig. 2c, at 50% B in the methanol gradient, where the tissue-related signal is maximized, the ambient ions are more abundant than at 75% B in the acetonitrile gradient. Because these ions are being used as internal calibrants for mass recalibration, it is important that they are relatively abundant. Therefore, 50% aqueous methanol with 0.2% formic acid was chosen as the optimal solvent composition for IR-MALDESI MSI of these lapatinib dosed liver tissue sections. Even though this experiment did not exhibit much selectivity between the two tissue-related groups (lipids vs. DRM) it did lead to an overall improvement in tissue-related signal.
3.4 IR-MALDESI Imaging with Optimized Geometry and Solvent Composition
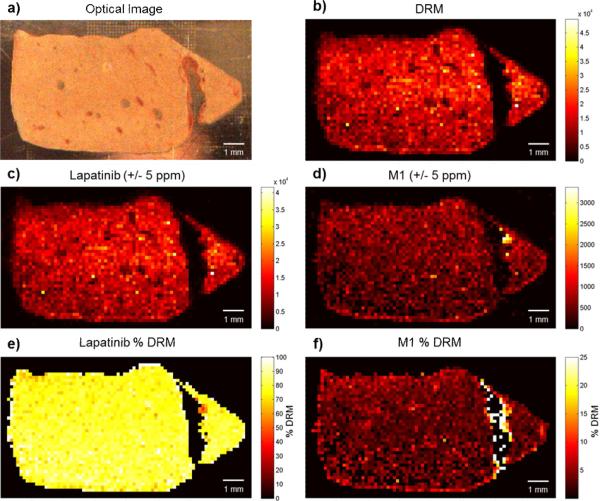
Sections of the lapatinib dosed liver tissue were examined by IR-MALDESI using the optimal solvent composition and recently optimized source geometry [49]. This analysis resulted in the observation of several more metabolites that were not previously detected by IR-MALDESI (supplemental Fig. S3) as well as an improvement signal reproducibility (pixel-to-pixel variability). A summary of the results from the optimized IR-MALDESI MSI analysis is shown in Fig. 3. The optical image of the tissue as well as the distributions for lapatinib and the metabolite M1 are presented in Fig. 3a, c, and d respectively. Again, the distribution is homogeneous throughout the tissue as was observed in the previous experiments. A new method for evaluating the distribution of drug-related ions is also presented. By summing the intensities for all drug-related ions on a pixel-by-pixel basis, the image in Fig. 3b can be obtained. Normalizing lapatinib or one of its metabolites to this total DRM image provides the composition of DRM at each pixel. For lapatinib, the % DRM image (Fig. 3e) shows that it makes up roughly 70% of all drug-related intensity homogeneously over the tissue while not being detected in the blood vessels. In contrast, M1 (Fig. 3f) was roughly 3% of the total DRM over most of the tissue but around the blood vessels the M1 makes up a larger portion of the total DRM. This implies that it is one of the only metabolites observed in this region. This information is not as obvious when looking at the M1 ion distribution (Fig. 3d) alone which supports the utility of determining the % DRM distribution.
Fig. 3.
IR-MALDESI MSI of lapatinib dosed liver tissue section after optimization of solvent composition and geometrical parameters. a) Optical image of the liver tissue section prior to ice matrix deposition. b) Ion map depicting the pixel-by-pixel sum of all drug related material. c) Ion map of lapatinib [M+H]+. d) Ion map of metabolite M1 [M+H]+. e) Distribution of the lapatinib composition of the total drug related material. f) Distribution of the M1 composition of the total drug related material
4 CONCLUSIONS
These experiments serve as proof of principle that IR-MALDESI MSI coupled to FT-ICR can be used to determine the distribution of a dosed drug and its metabolites directly from tissue. The influence of the electrospray solvent composition on IR-MALDESI signal for tissue related ions was also investigated and it was determined that a 50% aqueous methanol solution provided the best results. Furthermore, through analysis of a serial tissue section with the more commonly used UV-MALDI MSI, it was demonstrated that both techniques offer similar information. There are inherent benefits and drawbacks to both of these techniques. While analysis at atmospheric pressure reduces sample restrictions compared to analysis under vacuum, the number of ions that are sampled into the mass spectrometer is often times a fraction of the total number of ions produced. In addition, the process of matrix application continues to be a key issue in sample preparation for MALDI analyses. Due to the heterogeneity of the matrix crystallization that was observed for the UV-MALDI experiments, attention must be paid to assure an even coating and reduction in analytical variability. In addition, the IR-MALDESI source is currently being coupled to the Q Exactive which has demonstrated improved ion transmission compared to the LTQ-FT and could result in observation of the lesser abundant metabolites.
Supplementary Material
Highlights.
FT-ICR Mass spectrometry imaging of Lapatinib dosed liver tissue.
Comparison of IR-MALDESI and UV-MALDI ionization techniques for MSI.
Optimization of ESI solvent composition for IR-MALDESI MSI of dosed liver tissue.
Concurrent detection of Lapatinib and several metabolites in liver tissue by MSI.
ACKNOWLEDGEMENTS
The author gratefully acknowledge the financial support received from the National Institutes of Health (R01GM087964), GlaxoSmithKline, the William R. Kenan, Jr. Fund for Engineering, Technology & Science, the W. M. Keck Foundation, and North Carolina State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lanao JM, Fraile MA. Drug Tissue Distribution: Study Methods and Therapeutic Implications. Curr Pharm Des. 2005;11(29):3829–3845. doi: 10.2174/138161205774580679. [DOI] [PubMed] [Google Scholar]
- 2.Eichler H-G, Müller M. Drug Distribution. Clin Pharmacokinet. 1998;34(2):95–99. doi: 10.2165/00003088-199834020-00001. [DOI] [PubMed] [Google Scholar]
- 3.Caprioli RM, Farmer TB, Gile J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 4.Jespersen S, Niessen WMA, Tjaden UR, van der Greef J, Litborn E, Lindberg U, Roeraade J, Hillenkamp F. Attomole detection of proteins by matrix-assisted laser desorption/ionization mass spectrometry with the use of picolitre vials. Rapid Commun Mass Spectrom. 1994;8(8):581–584. [Google Scholar]
- 5.Troendle FJ, Reddick CD, Yost RA. Detection of pharmaceutical compounds in tissue by matrix-assisted laser desorption/ionization and laser desorption/chemical ionization tandem mass spectrometry with a quadrupole ion trap. J Am Soc Mass Spectrom. 1999;10(12):1315–1321. [Google Scholar]
- 6.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36(4):355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 7.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38(10):1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh Y, Casale R, Fukuda E, Chen J, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun Mass Spectrom. 2006;20(6):965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 9.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct Molecular Analysis of Whole-Body Animal Tissue Sections by Imaging MALDI Mass Spectrometry. Anal Chem. 2006;78(18):6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 10.Drexler DM, Garrett TJ, Cantone JL, Diters RW, Mitroka JG, Prieto Conaway MC, Adams SP, Yost RA, Sanders M. Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues. J Pharmacol Toxicol Methods. 2007;55(3):279–288. doi: 10.1016/j.vascn.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh Y, Chen J, Korfmacher WA. Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. J Pharmacol Toxicol Methods. 2007;55(2):193–200. doi: 10.1016/j.vascn.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Signor L, Varesio E, Staack RF, Starke V, Richter WF, Hopfgartner G. Analysis of erlotinib and its metabolites in rat tissue sections by MALDI quadrupole time-of-flight mass spectrometry. J Mass Spectrom. 2007;42(7):900–909. doi: 10.1002/jms.1225. [DOI] [PubMed] [Google Scholar]
- 13.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int J Mass Spectrom. 2007;260(2–3):195–202. [Google Scholar]
- 14.Kertesz V, Van Berkel GJ, Vavrek M, Koeplinger KA, Schneider BB, Covey TR. Comparison of Drug Distribution Images from Whole-Body Thin Tissue Sections Obtained Using Desorption Electrospray Ionization Tandem Mass Spectrometry and Autoradiography. Anal Chem. 2008;80(13):5168–5177. doi: 10.1021/ac800546a. [DOI] [PubMed] [Google Scholar]
- 15.Trim PJ, Atkinson SJ, Princivalle AP, Marshall PS, West A, Clench MR. Matrix-assisted laser desorption/ionisation mass spectrometry imaging of lipids in rat brain tissue with integrated unsupervised and supervised multivariant statistical analysis. Rapid Commun Mass Spectrom. 2008;22(10):1503–1509. doi: 10.1002/rcm.3498. [DOI] [PubMed] [Google Scholar]
- 16.Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc Natl Acad Sci USA. 2008;105(47):18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acquadro E, Cabella C, Ghiani S, Miragoli L, Bucci EM, Corpillo D. Matrix-Assisted Laser Desorption Ionization Imaging Mass Spectrometry Detection of a Magnetic Resonance Imaging Contrast Agent in Mouse Liver. Anal Chem. 2009;81(7):2779–2784. doi: 10.1021/ac900038y. [DOI] [PubMed] [Google Scholar]
- 18.Goodwin RJA, Scullion P, MacIntyre L, Watson DG, Pitt AR. Use of a Solvent-Free Dry Matrix Coating for Quantitative Matrix-Assisted Laser Desorption Ionization Imaging of 4-Bromophenyl-1,4-diazabicyclo(3.2.2)nonane-4-carboxylate in Rat Brain and Quantitative Analysis of the Drug from Laser Microdissected Tissue Regions. Anal Chem. 2010;82(9):3868–3873. doi: 10.1021/ac100398y. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andrén PE. Fine Mapping the Spatial Distribution and Concentration of Unlabeled Drugs within Tissue Micro-Compartments Using Imaging Mass Spectrometry. PLoS One. 2010;5(7):e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koeniger SL, Talaty N, Luo Y, Ready D, Voorbach M, Seifert T, Cepa S, Fagerland JA, Bouska J, Buck W, Johnson RW, Spanton S. A quantitation method for mass spectrometry imaging. Rapid Commun Mass Spectrom. 2011;25(4):503–510. doi: 10.1002/rcm.4891. [DOI] [PubMed] [Google Scholar]
- 21.Manier ML, Reyzer M, Goh A, Dartois V, Via L, Barry C, III, Caprioli R. Reagent Precoated Targets for Rapid In-Tissue Derivatization of the Anti-Tuberculosis Drug Isoniazid Followed by MALDI Imaging Mass Spectrometry. J Am Soc Mass Spectrom. 2011;22(8):1409–1419. doi: 10.1007/s13361-011-0150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prideaux B, Dartois Vr, Staab D, Weiner DM, Goh A, Via LE, Barry Iii CE, Stoeckli M. High-Sensitivity MALDIMRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal Chem. 2011;83(6):2112–2118. doi: 10.1021/ac1029049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellino S, Groseclose MR, Sigafoos J, Wagner D, de Serres M, Polli JW, Romach E, Myer J, Hamilton B. Central Nervous System Disposition and Metabolism of Fosdevirine (GSK2248761), a Non-Nucleoside Reverse Transcriptase Inhibitor: An LC-MS and Matrix-Assisted Laser Desorption/Ionization Imaging MS Investigation into Central Nervous System Toxicity. Chem Res Toxicol. 2012;26(2):241–251. doi: 10.1021/tx3004196. [DOI] [PubMed] [Google Scholar]
- 24.Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos J-M, Bouzom F, Stauber J. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J Proteomics. 2012;75(16):4952–4961. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson A, Forngren B, Bjurström S, Goodwin RJA, Basmaci E, Gustafsson I, Annas A, Hellgren D, Svanhagen A, Andrén PE, Lindberg J. In Situ Mass Spectrometry Imaging and Ex Vivo Characterization of Renal Crystalline Deposits Induced in Multiple Preclinical Drug Toxicology Studies. PLoS One. 2012;7(10):1–10. doi: 10.1371/journal.pone.0047353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA. Quantitative MALDI Tandem Mass Spectrometric Imaging of Cocaine from Brain Tissue with a Deuterated Internal Standard. Anal Chem. 2012;85(2):1081–1089. doi: 10.1021/ac302960j. [DOI] [PubMed] [Google Scholar]
- 27.Takai N, Tanaka Y, Inazawa K, Saji H. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun Mass Spectrom. 2012;26(13):1549–1556. doi: 10.1002/rcm.6256. [DOI] [PubMed] [Google Scholar]
- 28.Vismeh R, Waldon DJ, Teffera Y, Zhao Z. Localization and Quantification of Drugs in Animal Tissues by Use of Desorption Electrospray Ionization Mass Spectrometry Imaging. Anal Chem. 2012;84(12):5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- 29.Atkinson SJ, Loadman PM, Sutton C, Patterson LH, Clench MR. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(7):1271–1276. doi: 10.1002/rcm.2952. [DOI] [PubMed] [Google Scholar]
- 30.Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3(21):2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- 31.Römpp A, Guenther S, Takats Z, Spengler B. Mass spectrometry imaging with high resolution in mass and space (HR2 MSI) for reliable investigation of drug compound distributions on the cellular level. Anal Bioanal Chem. 2011;401(1):65–73. doi: 10.1007/s00216-011-4990-7. [DOI] [PubMed] [Google Scholar]
- 32.Goodwin RJA, Iverson SL, Andren PE. The significance of ambient-temperature on pharmaceutical and endogenous compound abundance and distribution in tissues sections when analyzed by matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun Mass Spectrom. 2012;26(5):494–498. doi: 10.1002/rcm.6125. [DOI] [PubMed] [Google Scholar]
- 33.Shariatgorji M, Nilsson A, Goodwin RJA, Svenningsson P, Schintu N, Banka Z, Kladni L, Hasko T, Szabo A, Andren PE. Deuterated Matrix-Assisted Laser Desorption Ionization Matrix Uncovers Masked Mass Spectrometry Imaging Signals of Small Molecules. Anal Chem. 2012;84(16):7152–7157. doi: 10.1021/ac301498m. [DOI] [PubMed] [Google Scholar]
- 34.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR Imaging Mass Spectrometry of Drugs and Metabolites in Tissue. Anal Chem. 2008;80(14):5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dekker LJM, van Kampen JJA, Reedijk ML, Burgers PC, Gruters RA, Osterhaus ADME, Luider TM. A mass spectrometry based imaging method developed for the intracellular detection of HIV protease inhibitors. Rapid Commun Mass Spectrom. 2009;23(8):1183–1188. doi: 10.1002/rcm.3981. [DOI] [PubMed] [Google Scholar]
- 36.Végvári Á , Fehniger TE, Gustavsson L, Nilsson A, Andrén PE, Kenne K, Nilsson J, Laurell T, Marko-Varga G. Essential tactics of tissue preparation and matrix nano-spotting for successful compound imaging mass spectrometry. J Proteomics. 2010;73(6):1270–1278. doi: 10.1016/j.jprot.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Marko-Varga G, Fehniger TE, Rezeli M, Döme B, Laurell T, Végvári Á . Drug localization in different lung cancer phenotypes by MALDI mass spectrometry imaging. J Proteomics. 2011;74(7):982–992. doi: 10.1016/j.jprot.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 38.Fehniger TE, Végvári Á , Rezeli M, Prikk K, Ross P, Dahlbäck M, Edula G, Sepper R, Marko-Varga G. Direct Demonstration of Tissue Uptake of an Inhaled Drug: Proof-of-Principle Study Using Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging. Anal Chem. 2011;83(21):8329–8336. doi: 10.1021/ac2014349. [DOI] [PubMed] [Google Scholar]
- 39.Shahidi-Latham SK, Dutta SM, Prieto Conaway MC, Rudewicz PJ. Evaluation of an Accurate Mass Approach for the Simultaneous Detection of Drug and Metabolite Distributions via Whole-Body Mass Spectrometric Imaging. Anal Chem. 2012;84(16):7158–7165. doi: 10.1021/ac3015142. [DOI] [PubMed] [Google Scholar]
- 40.Herniman J, Langley G, Bristow T, O'Connor G. The validation of exact mass measurements for small molecules using FT-ICRMS for improved confidence in the selection of elemental formulas. J Am Soc Mass Spectrom. 2005;16(7):1100–1108. [Google Scholar]
- 41.Crossman L, McHugh NA, Hsieh Y, Korfmacher WA, Chen J. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun Mass Spectrom. 2006;20(2):284–290. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 42.Marshall P, Toteu-Djomte V, Bareille P, Perry H, Brown G, Baumert M, Biggadike K. Correlation of Skin Blanching and Percutaneous Absorption for Glucocorticoid Receptor Agonists by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging and Liquid Extraction Surface Analysis with Nanoelectrospray Ionization Mass Spectrometry. Anal Chem. 2010;82(18):7787–7794. doi: 10.1021/ac1017524. [DOI] [PubMed] [Google Scholar]
- 43.Blatherwick EQ, Berkel GJV, Pickup K, Johansson MK, Beaudoin M-E, Cole RO, Day JM, Iverson S, Wilson ID, Scrivens JH, Weston DJ. Utility of spatially-resolved atmospheric pressure surface sampling and ionization techniques as alternatives to mass spectrometric imaging (MSI) in drug metabolism. Xenobiotica. 2011;41(8):720–734. doi: 10.3109/00498254.2011.587550. [DOI] [PubMed] [Google Scholar]
- 44.Eikel D, Vavrek M, Smith S, Bason C, Yeh S, Korfmacher WA, Henion JD. Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Commun Mass Spectrom. 2011;25(23):3587–3596. doi: 10.1002/rcm.5274. [DOI] [PubMed] [Google Scholar]
- 45.Parson WB, Koeniger SL, Johnson RW, Erickson J, Tian Y, Stedman C, Schwartz A, Tarcsa E, Cole R, Van Berkel GJ. Analysis of chloroquine and metabolites directly from whole-body animal tissue sections by liquid extraction surface analysis (LESA) and tandem mass spectrometry. J Mass Spectrom. 2012;47(11):1420–1428. doi: 10.1002/jms.3068. [DOI] [PubMed] [Google Scholar]
- 46.Schadt S, Kallbach S, Almeida R, Sandel J. Investigation of Figopitant and Its Metabolites in Rat Tissue by Combining Whole-Body Autoradiography with Liquid Extraction Surface Analysis Mass Spectrometry. Drug Metab Dispos. 2012;40(3):419–425. doi: 10.1124/dmd.111.043562. [DOI] [PubMed] [Google Scholar]
- 47.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer. J Am Soc Mass Spectrom. 2013;24(1):92–100. doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sampson J, Hawkridge A, Muddiman D. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2006;17(12):1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Robichaud G, Barry JA, Muddiman DC. IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections using Ice as a Matrix. J Am Soc Mass Spectrom. 2014;25(3):319–328. doi: 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barry JA, Robichaud G, Muddiman DC. Mass Recalibration of FT-ICR Mass Spectrometry Imaging Data Using the Average Frequency Shift of Ambient Ions. J Am Soc Mass Spectrom. 2013;24(7):1137–1145. doi: 10.1007/s13361-013-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. J Am Soc Mass Spectrom. 2013;24(5):718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castellino S, O'Mara M, Koch K, Borts DJ, Bowers GD, MacLauchlin C. Human Metabolism of Lapatinib, a Dual Kinase Inhibitor: Implications for Hepatotoxicity. Drug Metab Dispos. 2012;40(1):139–150. doi: 10.1124/dmd.111.040949. [DOI] [PubMed] [Google Scholar]
- 54.Dreisewerd K, Berkenkamp S, Leisner A, Rohlfing A, Menzel C. Fundamentals of matrix-assisted laser desorption/ionization mass spectrometry with pulsed infrared lasers. Int J Mass Spectrom. 2003;226(1):189–209. [Google Scholar]
- 55.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 56.Peng IX, Ogorzalek Loo RR, Shiea J, Loo JA. Reactive-Electrospray-Assisted Laser Desorption/Ionization for Characterization of Peptides and Proteins. Anal Chem. 2008;80(18):6995–7003. doi: 10.1021/ac800870c. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Zhang C, Sun J, Ren X, Luo H. Laser desorption dual spray post-ionization mass spectrometry for direct analysis of samples via two informative channels. J Mass Spectrom. 2013;48(2):250–254. doi: 10.1002/jms.3145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.