Abstract
To understand the mechanisms of male sterility in cotton (Gossypium spp.), combined histological, biochemical and transcription analysis using RNA-Seq was carried out in the anther of the single-gene recessive genic male sterility system of male sterile line 1355A and male fertile line 1355B, which are near-isogenic lines (NILs) differing only in the fertility trait. A total of 2,446 differentially expressed genes were identified between the anthers of 1355AB lines, at three different stages of development. Cluster analysis and functional assignment of differentially expressed genes revealed differences in transcription associated with pollen wall and anther development, including the metabolism of fatty acids, glucose, pectin and cellulose. Histological and biochemical analysis revealed that a major cellular defect in the 1355A was a thicker nexine, consistent with the RNA-seq data, and further gene expression studies implicated differences in fatty acids synthesis and metabolism. This study provides insight into the phenotypic characteristics and gene regulatory network of the genic male sterile line 1355A in upland cotton.
Cotton (Gossypium spp.) is one of the most important economic crops due to the value of its fibre. Heterosis is one strategy to improve cotton fibre productivity and quality. Male sterility, as an effective and economical pollination control system, is used worldwide for the exploitation of cotton heterosis. There have been identified 17 types of genic male sterile (GMS) lines in cotton1, among which 1355A belongs to the single-gene recessive GMS system. In the 1355A male sterile plant, the exine spine is not produced at the uninucleate stage, and the thickness of the pollen wall increases2. However, the pollen wall fine structure defect was still unknown. Physiological and biochemical analysis indicates that starch levels are lower in male sterile plants throughout the entire period of pollen development, and the soluble sugar level in young stamen of sterile plants are lower than in fertile plants, but the opposite is the case in mature stamens1. Recently RNA-seq was used to investigate the ‘Dong A’ male sterile mutant, showing that several key branch-point genes involved in hormone signaling, carbon and energy metabolism and pollen wall development were differentially expressed in the ‘Dong A’ male sterile mutant anther3. However, relatively little has been reported on the molecular genetic mechanism of 1355A male sterility.
In flowering plants, the haploid male sperm cells are protected in the pollen grains by the complex cell walls that are divided into two layers: the exine and the intine. The exine is a multilayered structure that is divided into sexine and nexine, and is primarily composed of sporopollenin, composed of fatty acids and phenolics4,5,6. The intine is structurally simpler and is composed of cellulose, hemicellulose, pectin polymers, hydrolytic enzymes and hydrophobic proteins7,8,9.
Pollen wall development begins in the newly formed microspores within the tetrad that are covered by the callose wall, which compartmentalises individual microspores when the primexine is laid down10. The primexine is the site of self-assembly of the glycocalyx and sporopollenin receptors11. The primexine has been described as a sporopollenin receptor because of the organised manner in which it accumulates sporopollenin at specific places12. During the tetrad stage, an undulating surface structure is formed in the plasma membrane of the microspore, which might be associated with normal primexine formation13. At the end of the tetrad stage, the microspores are released from the callose wall, and the exine exhibits a considerable increase in thickness because of the increased deposition and polymerisation of tapetally derived sporopollenin precursors9. In addition, the spines and other features of pollen ornamentation, which can be observed in some plants such as cotton, also appear during this stage. The mature exine structure is anatomically complete by the bicellular pollen stage. When the tapetum degrades, the remnants deposit as tryphine to fill the exine cavities, and the mature pollen wall is formed.
Recent molecular genetic studies have increased our understanding of pollen wall development. For example in Arabidopsis, CALLOSE SYNTHETASE5 (CALS5) is involved in callose synthesis14; NO EXINE FORMATION1 (NEF1), DEFECTIVE IN EXINE FORMATION1 (DEX1) and NO PRIMEXINE AND PLASMA MEMBRANE UNDULATION (NPU) play critical roles in primexine formation15,16,17; and MALE STERILITY1 (MS1), DIHYDROFLAVONOL 4-REDUCTASE-LIKE1/TETRAKETIDE–PYRONE REDUCTASE1 (DRL1/TKPR1), ECERIFERUM1 (CER1), MALE STERILITY2 (MS2), FACELESS POLLEN1 (FLP1), RUPTURED POLLEN GRAIN1 (RPG1), ACYL-COA SYNTHETASE5 (ACOS5), CYTOCHROME P450 703A2 (CYP703A2) and CYTOCHROME P450 703B1 (CYP704B1), TETRAKETIDE–PYRONE REDUCTASE2 (TKPR2), LAP6/POLYKETIDE SYNTHASE A (PKSA), and LAP5/POLYKETIDE SYNTHASE B (PKSB) are required for exine formation18,19,20,21,22,23,24,25,26,27,28.
Here, we report a transcriptome profiling analysis of anther development in the upland cotton male sterile line 1355A compared to that of the male fertile line 1355B at three different stages, using RNA-Seq. By identification of differentially expressed genes (DEGs), we identified pathways altered in 1355A male sterility. Anatomical studies show that the thicker nexine in 1355A contributes to male sterility and are consistent with pathways implicated by the RNA-seq data analysis, and suggest abnormal sporopollenin synthesis and deposition might lead to male sterility in 1355A.
Results
Phenotypic characteristics of the 1355A line
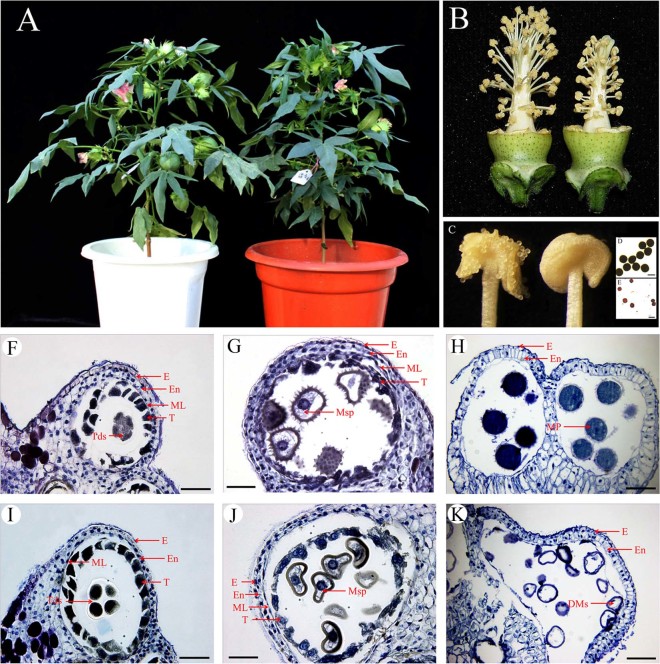
Morphological comparison of lines 1355A and 1355B revealed that the vegetative and floral development appears normal in 1355A plants (Fig. 1A). However, stamen filaments were shorter in 1355A (Fig. 1B) and anther dehiscence was abnormal (Fig. 1C) with pollen grains failing to show staining by I2-KI (Fig. 1D and E).
Figure 1. Phenotypic comparison between the 1355B and the 1355A lines.
(A) A 1355B plant (left) and a 1355A plant (right) at full-bloom stage. (B) A 1355B flower (left) and a 1355A flower (right), with petals removed. (C) In a 1355B anther (left), dehiscence is normal; while in a 1355A anther (right), dehiscence is abnormal. (D) 1355B pollen grains stained with 1% I2-KI solution at stage 12 showing mature pollen grains that are dyed black. (E) 1355A pollen grains stained with 1% I2-KI solution at stage 12 showing mature pollen grains that are not dyed black. (F–K) Locules from the anther section of the 1355B (F–H) and 1355A (I–K) plants during stage 7, stage 8 and stage 12. (F) and (I) stage 7. (G) and (J) stage8. (H) and (K) stage 12. There are no differences between the 1355B (F) and 1355A (I) plants at stage 7. Compared to those of the 1355B plant (G), the spines could not be identified on the surface of pollen in the 1355A plant (J). The microspore cytoplasm was stained deeply in the 1355B plants (H), but the microspores aborted in 1355A plants (K). E, epidermis; En, endothecium; ML, middle layer; T, tapetal layer; Mp, mature pollen; DMs, degenerated microspores; Msp, microspores and Tds, tetrads. Bars, 100 μm in (D) and (E); Bars, 50 μm in (F) to (K). Acknowledge the authors Yuanlong Wu and Li Yang for photographing (A), Yuanlong Wu for photographing (B), (D) and (E), and Zancheng Wu for photographing (C) and (F) to (K).
To identify the relationship between bud length (from the nectary to the top of the bud) and anther development stages, cross sections were performed on the 1355B anther. Fourteen stages of anther development were identified based on the distinctive cellular events of Arabidopsis thaliana and G. hirsutum (Coker 315) anther development stages29,30 (Supplementary Table S1 and Supplementary Fig. S1).
To investigate the cellular defects of the 1355A male sterile plants during pollen development, cross sections were compared to those of the 1355B anther at stage 7, stage 8 and stage 12, referred to our previous observation that the first detectable sign of male sterility occurs at stage 8 in the 1355A. And in order to know what changed in mature pollen, we selected stage 12 when the pollen is mature. No detectable differences were observed between the 1355B and the 1355A anthers prior to stage 7 (Fig. 1F and I), while significant differences in anther development were observed after the tetrad stage (stage 8). When the microspores of 1355B plants were released from the callose walls, the microspores were normal, and the exine with spines was formed (Fig. 1G). In contrast, the 1355A microspores were shrunken in appearance, and the exine lacked spines (Fig. 1J). On the day of anthesis (stage 12), the cytoplasm of mature pollen of 1355B anthers stained strongly with toluidine blue (Fig. 1H), while 1355A pollen cytoplasm was empty, and only residual defective pollen walls were present in the anther locules (Fig. 1K), which means the cytoplasm is degraded.
Identification of the genes that were differentially expressed between lines 1355A and 1355B
Previous studies and our phenotypic observations indicated that the first detectable sign of male sterility occurs during the early uninucleate microspore stage (stage 8) in the 1355A plants. Therefore, anthers were collected at the tetrad stage (stage 7), the uninucleate microspore stage (stage 8) and the mature pollen stage (stage 12) with a view to identifying differentially expressed genes (DEGs) between 1355B and 1355A, using RNA-Seq technology.
The results of the classification of raw reads and randomness assessment indicate that high-quality libraries were generated (Supplementary Fig. S2A and B). The six libraries include a total of 44,391,107 clean reads (Table 1), and each read was approximately 50 bp in length. A sequencing saturation analysis indicated that up to 7.3 million clean reads for each sample were created, which is sufficient for the quantitative analysis of gene expression (Supplementary Fig. S2C). By mapping the read sequences to a reference database (cotton unigenes from NCBI) containing 20,671 unigene sequences, we obtained a total of 16,949,140 (38.18%) mapped reads, but there were only 15,473,443 (34.86%) unique match reads and the number of unique match reads ranged from 2,399,000 to 2,885,941 in six libraries. A total of 27,441,967 (61.82%) clean reads were not mapped to the cotton unigenes assembly (Table 1).
Table 1. Summary of tag numbers.
| Stage 7 | Stage 8 | Stage 12 | Total | ||||
|---|---|---|---|---|---|---|---|
| 1355B | 1355A | 1355B | 1355A | 1355B | 1355A | ||
| Clean reads | 7 317 252 | 7 438 268 | 7 351 770 | 7 467 140 | 7 492 466 | 7 324 211 | 44 391 107 |
| Total Mapped Reads | 2 851 673 | 2 873 607 | 2 755 481 | 2 704 315 | 2 635 598 | 3 128 466 | 16 949 140 |
| (%) of Total Mapped Reads | 38.97% | 38.63% | 37.48% | 36.22% | 35.18% | 42.71% | 38.18% |
| Unique Match | 2 606 158 | 2 617 802 | 2 486 204 | 2 478 338 | 2 399 000 | 2 885 941 | 15 473 443 |
| (%) of Unique Match | 35.62% | 35.19% | 33.82% | 33.19% | 32.02% | 39.40% | 34.86% |
| Total Unmapped Reads | 4 465 579 | 4 564 661 | 4 596 289 | 4 762 825 | 4 856 868 | 4 195 745 | 27 441 967 |
| (%) of Total Unmapped Reads | 61.03% | 61.37% | 62.52% | 63.78% | 64.82% | 57.29% | 61.82% |
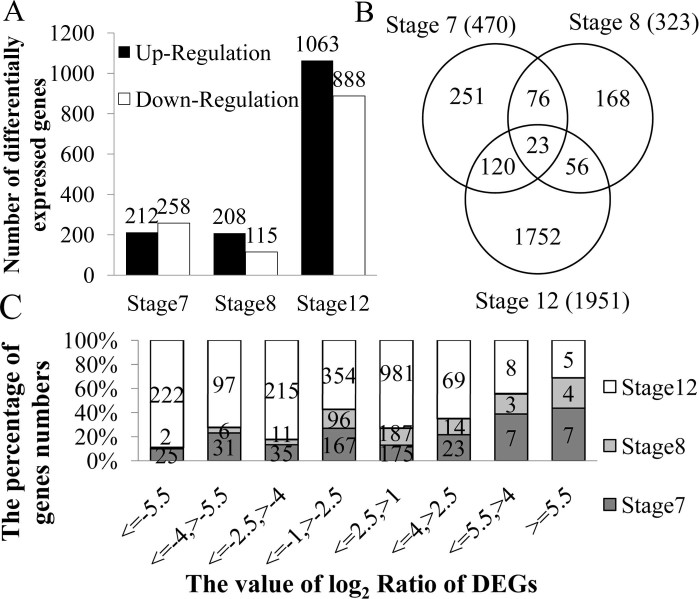
The expression levels of genes were determined by calculating the number of unique match reads for each gene and then normalising this number to Reads Per Kb per Million reads (RPKM), which associates the read numbers with the gene expression levels. A total of 20,113 genes were detected during the three stages of anther development in the 1355AB lines (Supplementary Table S2-1). Among these genes, 2,446 differentially expressed genes were filtered with FDR ≤ 0.001 and absolute value of log2 Ratio ≥ 1 (Supplementary Table S2-2). The number of DEGs that were up- or down- regulated during different pollen development stages demonstrates that the least number of the DEGs occurs at stage 8 rather than at stage 7, and at stage 7, more down-regulated DEGs than up-regulated DEGs were identified. At stage 8 and stage 12, more up-regulated DEGs than down-regulated DEGs were identified (Fig. 2A). This may indicate that the reversed up- and down- regulated DEG ratios at stage 7 and stage 8 contains critical information linked to male sterility.
Figure 2. The statistical analysis of differentially expressed genes in the 1355A plants compared to those of the 1355B plants.
(A) Number of differentially expressed genes that were up- or down-regulated during analyzed stages. (B) Showing the relationship of differentially expressed genes in three pollen development stages using a Venn diagram. The overlapping regions indicate the number of DEGs that are present in more than one stage, and the central region corresponds to the expressed genes that are present in all three of the stages. (C) The distribution of the value log2 Ratio of the DEGs in the 1355A plants compared to the 1355B plants.
The number of DEGs during analysed stages was 470, 323 and 1,951 (Fig. 2B). Among these genes, only 23 were present in all three of the stages. However, more than half of the DEGs exhibited specific expression in the three stages of anther development (Fig. 2B). By analysing the distribution of the DEGs log2 Ratio, we found that many DEGs have an absolute value of log2 Ratio range from 1 to 2.5 during stages 7 and 8. During the stage 12, the expression of the down-regulated DEGs was distributed widely (Fig. 2C). This result indicates that the male sterility gene affects little transcriptional changes in 1355A anther.
Using BLASTx against the non-redundant protein sequence (nr) database in GenBank, 2,446 DEGs were annotated. Among these, 1,793 genes (73.30% of the differentially expressed sequences) had an above-the-cut-off BLAST result; 436 genes (17.83%) belonged to the functional categories ‘unknown proteins’ or ‘predicted proteins’; 217 genes (8.87%) had ‘no hits’ results (Supplementary Table S2-2). However, 2,138 genes (87.41%) had an orthologue in Arabidopsis (Supplementary Table S3).
To evaluate the potential functions of the DEGs, we added gene ontology (GO) terms to DEGs at stages 7 and 8 (Supplementary Fig. S3A and B). In terms of annotated biological processes, the difference between these genes was ‘cellular process’ (16% at stage 7 and 13% at stage 8) at level 2. This biological process was further studied at level 3. The results indicate that the ‘cellular metabolic process’ is significantly different to ‘cellular process’ (Supplementary Fig. S4A and B). Interestingly, ‘cellular homeostasis’ is particularly highlighted during stage 8 (Supplementary Fig. S4B), suggesting a role for the male sterility defect in anther metabolic homeostasis at stage 8.
Genes those are associated with male sterility in 1355A plant anthers
The bioinformatics analysis indicates that several subsets of genes, including those associated with metabolic events and cell wall-related genes, are differentially expressed during 1355B and 1355A anther development associated with male sterility.
Metabolic events
Basic metabolic events play an essential role in anther development31. Many metabolic genes were identified amongst the DEGs, particularly fatty acid biosynthesis-related genes, fatty acid metabolism-related genes and glucose- and phenylpropanoid pathway- related genes.
Fatty acids
Using the KEGG results and other annotations, several genes participating in fatty acid biosynthesis and fatty acid metabolism were identified by RNA-Seq analysis. Seven fatty acid biosynthesis genes, namely fatty acyl-ACP thioesterases B (ES849077), NAD(P)-binding Rossmann-fold superfamily protein (ES818872), acetyl-CoA carboxylase 1 (ES840243 and ES842786) and acetyl-CoA carboxylase (ES805503, ES796810 and DW482723), were down-regulated at stage 7 in the 1355A anthers (Supplementary Table S4-1). One gene encoding an NAD(P)-binding Rossmann-fold superfamily protein (ES806303) was up-regulated at stage 7 and stage 8 but down-regulated at stage 12. Interestingly, the expression patterns of fatty acid metabolism genes were different from those of fatty acid biosynthesis. Five fatty acid metabolism genes were up-regulated in the 1355A at stage 7, these encoded acyl-CoA oxidase 3 (EX164385), long-chain acyl-CoA synthetase 6 (ES813531), aldehyde dehydrogenase 3I1 (DW480953), GroES-like zinc-binding alcohol dehydrogenase family protein (ES797791) and acetoacetyl-CoA thiolase 2 (ES834402) (Supplementary Table S4-1).
Glucose
Subsets of genes participating in multiple branches of the energy metabolism pathway, including starch and sucrose metabolism, glycolysis/gluconeogenesis and pyruvate metabolism, were identified. The expression patterns of the starch and sucrose metabolism genes were up-regulated during analyzed stages in the 1355A line, although four genes were down-regulated, including the gene of UDP-D-glucuronate 4-epimerase 3 (GQ292790; Supplementary Table S4-2). In contrast, the expression patterns of glycolysis/gluconeogenesis genes were mostly down-regulated at stage 7 (Supplementary Table S4-2). Genes are associated with pyruvate metabolism were up-regulated during analyzed stages in the 1355A, similar to the genes involved in starch and sucrose metabolism (Supplementary Table S4-2).
Cell wall component: pectin and cellulose
Genes related to cell wall components were identified. Down-regulated in the 1355A were three cellulose synthase genes (ES811740, ES815864 and ES805098), seven pectin lyase-like superfamily protein genes (EX164190, DW484863, EX166022, U09717, EX165880, ES831677 and ES804623), two plant invertase/pectin methylesterase inhibitor superfamily genes (EX164972 and EX165781) and one pectin methylesterase gene (DT560826; Supplementary Table S5-1). These genes might contribute to the change of intine formation in 1355A.
Comparison of DEGs data with qRT-PCR
To confirm the expression profiles in the 1355B and 1355A anthers obtained by RNA-seq, qRT-PCR was performed on 17 genes, including fatty acid biosynthesis genes (Supplementary Fig. S5A), fatty acid metabolism genes (Supplementary Fig. S5B), pectin-related genes (Supplementary Fig. S5C) and other metabolic-related genes (Supplementary Fig. S5D), during the three anther development stages.
Pearson's correlation coefficient was calculated by SPSS to assess the correlation between different platforms. Overall, the 29 DEGs of the qPCR measurements were moderately correlated with the RNA-Seq results (Supplementary Fig. S6. A, R = 0.854; correlation is significant at the 0.01 level). The correlations during analyzed stages were 0.578, 0.909 and 0.914 (Supplementary Fig. S6B to D). These results indicate that the RNA-seq method is an accurate and reliable way to identify genes that are differentially expressed during cotton anther development.
The defective pollen wall in the 1355A male sterile line
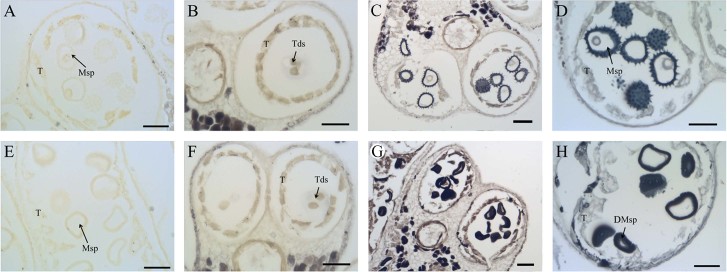
The RNA-seq results suggest that lipid metabolism in the 1355A anther might be abnormal. To confirm this, the lipid stain Sudan black B was used to stain the anther cross sections of the 1355AB plants with non-stained anther cross sections at stage 9 as negative controls (Fig. 3A and E). At stage 7, weak positive staining was detected in the tetrads of both the 1355B and 1355A anthers (Fig. 3B and F). Compared with 1355B (Fig. 3C), the staining sharply accumulated in the pollen wall of 1355A microspores at stage 8 (Fig. 3G). At stage 9, the staining was strong in both the 1355B and 1355A microspores (Fig. 3D and H). This result suggests that abnormal lipid metabolism be associated with abnormal pollen wall development at stage 8 in the 1355A anther. SEM analysis also demonstrates that 1355A anthers exhibit defective pollen walls, whereby the mature pollen wall lacks spines, consistent with the observations from the cross sections (Supplementary Fig. S7).
Figure 3. Sudan Black B staining of anthers of the 1355B and1355A plants.
The black staining signals indicate the lipid accumulation. (A) and (E) The non-stained anthers of the 1355B (A) and 1355A (E) plants at stage 8 are the negative signals. (B) and (F) The stained anthers of the 1355B (B) and 1355A (F) plants at stage 7. (C) and (G) The stained anthers of the 1355B (C) and 1355A (G) plants at stage 8. (D) and (H) The stained anthers of the 1355B (D) and 1355A (H) plants at stage 9. T, tapetal layer; Tds, tetrads; Msp, microspores; DMsp, degenerated microspores; Bars = 50 μm.
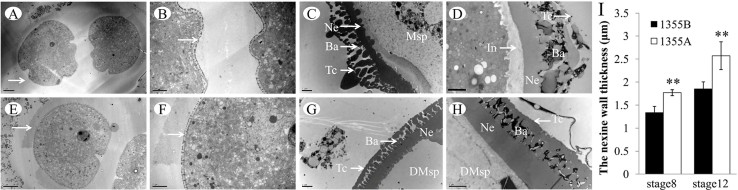
To further study the defects in the 1355A pollen wall formation, the fine structure of the pollen wall in the 1355AB plants was observed by TEM. At tetrad stage, characteristic callose walls (Fig. 4A and E) and the formation of primexine (Fig. 4B and F) were detected in both 1355B and 1355A microspores. As development proceeded, the basic exine structure, with an undulating sexine formed at the young uninucleate pollen stage, was seen in 1355B microspores (Fig. 4C). However, a thickened nexine and smooth sexine were observed in the 1355A microspores at this stage (Fig. 4G). There was obvious intine in the mature 1355B pollen (Fig. 4D); however, the 1355A pollen intine was not formed (Fig. 4H). Direct measurements showed that the pollen nexine of 1355A is thicker than that of 1355B (Fig. 4I).
Figure 4. TEMS of pollen walls from the 1355B and 1355A plants.
(A) and (E) Anthers of the 1355B (A) and 1355A (E) plants at stage 7 showing tetrads. The white arrows indicate the callose wall of the tetrads. (B) and (F) The higher magnification of (A) and (E) respectively. The white arrows indicate primexine and probacula formation. (C) and (G) Pollen wall of the 1355B (C) and 1355A (G) plants at stage 8 exhibiting early uninuclear microspores. (D) and (H) Pollen wall of the 1355B (D) and 1355A (H) plants at stage 12 exhibiting mature microspores. (I) The nexine wall thickness at stages 8 and 12. Msp, microspores; DMsp, degenerated microspores; In, intine; Ne, nexine; Ba, bacula; Te, tectum. The error bars represent SD (student's t test, ** p < 0.01, p = 5.34E-07 at stage 8, p = 1.39E-05 at stage 12). Bars, 5 μm in (A) and (E); Bars, 2 μm in (B) to (D), and (F) to (H).
Homologous genes of pollen wall development
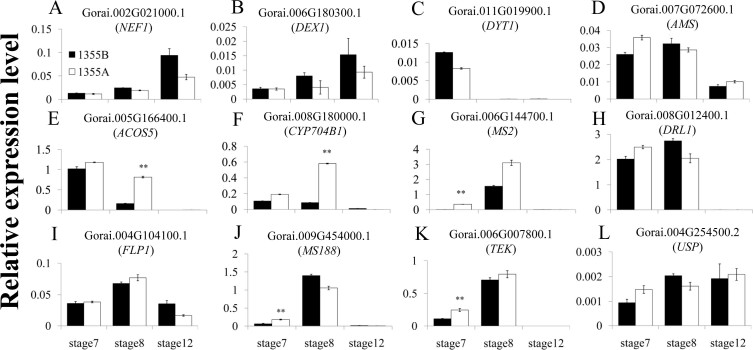
To further explore the molecular mechanism of male sterility in the 1355A mutant, expression of homologous genes from Arabidopsis thaliana, associated with pollen wall development, was analysed by qRT-PCR in both cotton lines. Expression of the primexine genes NEF1 and DEX1 was not significantly differentially expressed between the 1355A and 1355B anthers (Fig. 5A and B). The tapetum development genes DYT1 and AMS also were not significantly differentially expressed between the 1355A and 1355B anthers (Fig. 5C and D). However, the sporopollenin synthesis-related genes ACOS5 and CYP704B1 were differently expressed at stage 8 between 1355B and 1355A (Fig. 5E and F), and the MS2 gene was differently expressed at stage 7 between 1355B and 1355A (Fig. 5G). Interestingly, the substrates of CYP704B1, ACOS5 and MS2, fatty acids and the fatty acids derivatives are also constituents of the pollen wall. The sporopollenin synthesis-related genes DRL1 and FLP1 did not exhibit significant differential expression between 1355B and 1355A (Fig. 5H and I). The MS188 and TEK for sexine and nexine formation, respectively, were differently expressed at stage 7 between 1355B and 1355A (Fig. 5J and K). However, the UDP-sugar pyrophosphorylase (USP) gene for intine synthesis did not exhibit significant differential expression between 1355B and 1355A (Fig. 5L).
Figure 5. The expression pattern of homologous genes in pollen development.
(A) and (B) The primexine formation genes. (C) and (D) The tapetum development genes. (E) to (I) The sporopollenin synthesis-related genes. (J) The sexine formation genes. (K) The nexine formation gene. (L) The intine synthesis gene. The white and black columns refer to the 1355A male sterile line and 1355B male fertile line, respectively. The error bars represent SD (student's t test, ** p < 0.01 and absolute value of log2 Ratio ≥ 1, p = 8.06E-07 in (E), p = 8.16E-09 in (F), p = 1.73E-05 in (G), p = 3.34E-04 in (J) and p = 5.64E-04 in (K)). Three biological replicates were performed.
Discussion
Through comparing the definition of anther development stages in Arabidopsis thaliana and G. hirsutum of Coker 315 line and 1355B line (Supplementary Table S1), we conclude that cotton anther development is similar to that of Arabidopsis thaliana. One important difference between anther development in cotton and Arabidopsis thaliana is that the tapetal cell degradation commences earlier in cotton than in Arabidopsis thaliana (Supplementary Table S1), the other is the structure of the pollen wall. Arabidopsis thaliana pollen has a reticulate structure9, while the pollen wall of cotton is more complex. The mature pollen is generally spiny in the exine32, similar to the family Compositae33. These results, which are similar by identifying the anther development stages in G. hirsutum (Coker 315 and 1355B) in different indicative sizes, provide a lot of information for the cotton anther development research.
In this study, an RNA-seq bioinformatics-guided histological analysis reveals that the lack of the spines, as pollen wall ornamentation, is a hallmark of the pollen wall of the 1355A male sterile line. Further studies suggest that defective metabolic events, particularly fatty acid, affect pollen wall formation in the mutant.
The pollen wall functions as a barrier against unfavourable environment conditions and maintains pollen viability34, and the exine, which is divided into sexine and nexine, and the intine are the two principal layers of the pollen wall35. Previous studies indicate that the AMS gene directly regulated the genes of MS188 and TEK in Arabidopsis thaliana, which are the key regulators for the sexine and nexine formation respectively36,37. In our study, we did not detect significantly different in the cotton homologues of AMS expression, while the expression patterns of MS188 and TEK homologues were significantly up-regulated at stage 7 (Fig. 5J and K), in agreement with the histological analysis results that show a lack of ornamentation (spines) in the sexine (Fig. 4G) and thicker nexine (Fig. 4I) in the 1355A pollen wall. For the MS188 and TEK homologues, two transcriptional regulation factors, they may lead to the phenotype delay upon the gene expression change happened. This is the reason why the MS188 and TEK homologues showed significantly different in expression at stage 7, while the phenotypes were present at stage 8. We propose that the up-regulated expression of MS188 and TEK homologues at stage 7 might affect the sexine lacking spines and the thicker nexine formation at stage 8 in 1355A, respectively. These results may indicate that the male sterile gene may act as a repressor for the MS188 and TEK, which is independent of AMS.
The main composition of the exine is sporopollenin9. Sporopollenin synthesis requires many genes that are precisely regulated. Several studies have demonstrated that the ACOS5 gene is most highly expressed during anther development at stage 8 in Arabidopsis thaliana21; the CYP704B1 gene is mainly expressed in flower buds at stage 9 and 10 (corresponding to the anther development from stage 5 to 9) in Arabidopsis thaliana24, and the expression of MS2 is initiated at stage 738. These results are consistent with the expression pattern of cotton homologues in 1355B (Fig. 5E to G). The mutations in Arabidopsis thaliana ACOS5 gene cause an apparent loss of exine pollen wall21; Mutations in the Arabidopsis thaliana CYP704B1 gene cause a lack of a normal exine layer and a striped surface24, and the mutations in the Arabidopsis thaliana MS2 gene caused a poorly constructed pollen wall38. These results indicate that the down-regulation of the ACOS5, CYP704B1 and MS2 genes leads to decreased sporopollenin biosynthesis. Interestingly, in the 1355A anthers, the expression patterns of ACOS5 and CYP704B1 homologues were dramatically up-regulated at stage 8, compared to the 1355B anthers (Fig. 5E and F), and the expression patterns of MS2 homologue was also significantly up-regulated at stage 7 (Fig. 5G). The expression change of ACOS5, CYP704B1 homologues, which is in agree with the stage of phenotype emerged, was more dramatically than MS2 homologue, which indicated the ACOS5, CYP704B1 homologues were more important than MS2 homologue for the phenotype of 1355A. We propose that some sporopollenin synthesis genes that are up-regulated at stage 8 might lead to excessively accumulating sporopollenin and finally to male sterility.
While the DRL1 gene is involved in pollen wall development23 and the FLP1 gene is involved in sporopollenin biosynthesis during the latter stages of anther development17, there were no significant differences (Fig. 5H and I) between these 1355A and 1355B anthers; it seems that the transcriptional regulation of these genes is not relevant to the 1355A male sterility phenotype.
We also found that the intine was lacking in the 1355A pollen (Fig. 4D and H). The intine is composed of hydrolytic enzymes, hydrophobic proteins, cellulose, hemicellulose, and pectic polymers39. In addition, the microspore cytoplasm was degraded, and the 1355A pollen only maintained the pollen wall after the later uninucleate pollen stage, which might be partly explained by most protease genes being up-regulated during the late development stage in the 1355A anthers (Supplementary Table S5-2), and cell wall-related genes, such as pectin and cellulose genes, were down-regulated (Supplementary Table S5-1). However, the homologue of UDP-sugar pyrophosphorylase (USP), which is important in intine synthesis40, was no significant differences (Fig. 5L) between 1355A and 1355B anthers. Previous studies indicated that the nexine acts as a template for the intine formation37, in agreement with our results. We propose that laking of intine possibly is not the direct reason for the 1355A male sterility, but the thicker nexine.
The production of a functional pollen wall requires precisely coordinated metabolic events33, particularly fatty acid, pectin and glucose metabolism. Due to anther development requiring large amounts of sugar41, it has been proposed that sporopollenin, which is the main exine wall component, may contain derivatives of aliphatics, such as fatty acids derivatives and phenolics5,42,43. During pollen development, the pollen wall is rich in lipids5,42,44. Several studies have demonstrated that fatty acids are subjected to hydroxylation by CYP703B124. Hydroxylauric acid serves as a substrate for ACOS5, which is available for conversion to hydroxylauryl-CoA21. The MS2 gene product converts hydroxylauryl-CoA to fatty alcohols, which are the sporopollenin monomeric constituents38. Our RNA-seq results indicate that, in 1355A anthers the FATB gene (ES849077) and the ACC1 gene (ES840243) were down-regulated compared to 1355B anthers at stage 7; one ENR1 (ES806303) gene was up-regulated at stage 7, while a second ENR1 gene (ES818872) was down-regulated at stages 7 and 8 but up-regulated at stage 12. These genes are related to fatty acid biosynthesis. Other studies have also demonstrated that the FATB fatty acid biosynthesis gene (ES849077) mutant reduces the cytosolic supply of palmitate45 and the levels of straight-chain C20-24 components46 in Arabidopsis thaliana. The FATB gene also produces free fatty acids for sporopollenin biosynthesis47. The ACC1 fatty acid biosynthesis gene (ES840243) catalyses the rate-limiting step of fatty acid de novo biosynthesis48. The ENR1 fatty acid biosynthesis gene is a subunit of the fatty acid synthase complex49. In contrast, in 1355A anthers, the ACX3 fatty acid metabolism gene (EX164385) was up-regulated at stage 7 compared to 1355B anthers. The ACX3 gene has medium-chain-length (C8:0 to C14:0) substrate specificity50. These results indicate that the proportion of fatty acid and fatty acid derivatives may change at stage 7 and that de novo fatty acid biosynthesis may be blocked at stage 7 in 1355A anthers, although the fatty acid synthase complex may not be affected at that stage. Lipid stain also validated the results by showing more lipids being deposited in the exine wall at stage 8 (Fig. 3C and G).
Therefore after the tetrad stage, a key period for pollen wall formation33,51,52, abundant sporopollenin synthesis, which is related to the abnormal fatty acid pathway, leads to the 1355A male sterility as shown by failing to form spins on the pollen wall surface, a thicker nexine and lack of intine. Ultimately, defective pollen walls lead to male sterility in 1355A plants.
Methods
Plant materials
The recessive genic male sterile (RGMS) two-type line (1355AB) was used as the plant material. The 1355AB line was maintained by full sib-mating (1355A × 1355B); therefore the male sterile line 1355A and the male fertile line 1355B are near-isogenic lines (NILs) differing only in the fertility trait. 1355AB plants were cultivated in the field during the normal cotton planting season and in the greenhouse during the winter in Wuhan, China, using standard farming practices. During anthesis, male sterile plants were identified by staining the pollen grains with I2-KI solution and were photographed under a microscope (Leica DM2500, Germany). Bud lengths ranging from 6–7, 7–8, and >24 mm corresponding to stage 7 (tetrad pollen period, TTP), stage 8 (uninucleate pollen, UNP) and stage 12 (mature pollen period, MTP) respectively were collected, and the anthers were sampled for RNA-seq. All of the harvested samples were immediately deep-frozen in liquid nitrogen and stored at −70°C before use.
Histological analyses
To identify the anther development stage, flower buds were harvested from plants in the field and fixed in FAA [10% formalin, 5% acetic acid, and 50% ethanol (v/v)]. Detailed cross-sections were performed as previously described53.
Lipid staining
Lipid staining was conducted as described by Dun et al. (2011)54. The anther section of the 1355AB lines during stage 7, stage 8 and stage 9 were deparaffinised by incubating in dimethylbenzene twice for 30 min each and were rehydrated in an ethanol gradient for 5 min. The sections were stained in 1% Sudan Black B in 70% ethanol at 58°C for 45 min. Sections were washed with 70% ethanol to remove excess colour and were observed by light microscopy.
Illumina sequencing and data processing
Total RNA was isolated from collected anthers using a modified guanidine thiocyanate method55. RNA-seq was performed by the Beijing Genomics Institute (Shenzhen, China). mRNA was enriched and broken into short fragments (approximately 200 bp). Using the mRNA fragments as templates, double stranded cDNA was synthesised. After attaching the sequencing adaptors, the library fragments were purified by agarose gel electrophoresis and enriched by PCR amplification, after which the fragments were sequenced (Illumina HiSeq™ 2000). Clean reads were acquired after certain steps of raw sequences data processing.
Clean reads were mapped to the cotton contigs assembly using SOAPaligner/soap2 mismatches; no more than 2 bases were allowed in the alignment. The number of clean reads for each gene was calculated and then normalised to Reads Per Kb per Million reads (RPKM), which associates the read number with the gene expression levels.
Screening of differentially expressed genes
A rigorous algorithm was used to screen the differentially expressed genes in cotton anther development. A false discovery rate (FDR) ≤ 0.001 and an absolute value of log2 Ratio ≥ 1 were used as thresholds to determine the significance of the gene expression difference. A bioinformatics analysis of the gene expression patterns was performed according to Xu et al. (2011)56.
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) was used to validate RNA-seq data in identifying genes that were related to 1355A male sterility, according to Xu et al. (2011)56. Gene-specific primers (Supplementary Table S6) were designed according to the sequences (Supplementary Table S7) with Primer Premier 5 (http://www.premierbiosoft.com/crm/jsp/com/pbi/crm/clientside/ProductList.jsp).
Electron microscopy
To compare the pollen wall differences between the 1355A and 1355B plants, mature pollen was collected and fixed in 2.5% (v/v) glutaraldehyde. Scanning electron microscopy (SEM) was performed as previously described by Min et al. (2013)57.
For transmission electron microscopy (TEM), different stages of anthers were dissected, and the middle parts of the anther tissues were immediately prefixed in 2.5% glutaraldehyde (v/v)/0.1 M phosphate buffer (pH 7.2) at 4°C and then vacuum-infiltrated until the samples sank to the bottom of container as described by Zhou et al. (2009)58. The nexine wall thickness of the microspores was measured using Nano measurer 1.2 software (http://emuch.net/html/201402/7022970.html). The measurements were repeated 10 times for each sample.
Supplementary Material
supplementary information
supplementary table s2
supplementary table s3
supplementary table s4
supplementary table s5
supplementary table s6
supplementary table s7
Acknowledgments
The authors would like to thank Jianbo Cao, Yuan Tan and Fangmei Zhang (Huazhong Agricultural University, China) for their helps in scanning electron microscopy and transmission electron microscopy. This work was supported by funding from the National High Technology Research and Development Program of China (grant number: 2011AA10A102; 2013AA100601-4).
Footnotes
Author Contributions Y.W. carried out the experiment with L.M., Z.W. and L.Y., and wrote the main manuscript text. X.Z. designed and supervised the research with L.M., L.Z. and X.Y., and revised the manuscript. Y.W. and D.Y. performed the statistical analysis of the RNA-seq data. X.G. provided the cotton seed. All authors reviewed the manuscript.
References
- Tang W. Study on morphological, cytological and biochemical characteristics in anthers of recessive genetic sterile line of upland cotton (Gossypium hirsutum L.). Master degree thesis, Sichuan Agricultural University, Ya an, (2008).
- Wang G., Guo X. & Zhang X. Sterile stability of four genetic male sterile lines and genetic male sterile gene localization in 1355A. Journal of Huazhong Agricultural University 25, 351–354 (2006). [Google Scholar]
- Wei M., Song M., Fan S. & Yu S. Transcriptomic analysis of differentially expressed genes during anther development in genetic male sterile and wild type cotton by digital gene-expression profiling. BMC Genomics 14, 97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermann R., Ahlers F. & Schmitz-Thom I. [Sporopollenin]. Biopolymers Vol. 1 [Stenbuchel A., & Hofrichter M., eds. (eds.)] [209–227] (Wiley-VCH Verlag, Weinheim, 2001). [Google Scholar]
- Ahlers F., Thom I., Lambert J., Kuckuk R. & Rolf W. 1H NMR analysis of sporopollenin from Typha Angustifolia. Phytochemistry 50, 1095–1098 (1999). [Google Scholar]
- Guilford W. J., Schneider D. M., Labovitz J. & Opella S. J. High resolution solid state 13C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol. 86, 134–136 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen H. & Makaroff C. A. Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185, 7–21 (1995). [Google Scholar]
- Huang L. et al. The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. J. Exp. Bot. 60, 301–313 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T. & Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62, 437–460 (2011). [DOI] [PubMed] [Google Scholar]
- Honys D., Renák D. & Twell D. [Male gametophyte development and function]. Floriculture, ornamental and plant biotechnology: advances and topical issues Vol.1 [Silva J. T., ed. (ed.)] [76–87] (Global Science Books, London, 2006). [Google Scholar]
- Gabarayeva N. & Hemsley A. R. Merging concepts: The role of self-assembly in the development of pollen wall structure. Rev. Palaeobot. Palynol. 138, 121–139 (2006). [Google Scholar]
- Skvarla J. J. & Larson D. A. Fine structural studies of Zea mays pollen I: cell membranes and exine ontogeny. Am. J. Bot. 53, 1112–1125 (1966). [Google Scholar]
- Suzuki T., Masaoka K., Nishi M., Nakamura K. & Ishiguro S. Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol. 49, 1465–1477 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M. & Verma D. P. S. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42, 315–328 (2005). [DOI] [PubMed] [Google Scholar]
- Ariizumi T. et al. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39, 170–181 (2004). [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders D. M., Dodrill C. H., Owen H. A. & Makaroff C. A. DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127, 1739–1749 (2001). [PMC free article] [PubMed] [Google Scholar]
- Chang H. S. et al. NO PRIMEXINE AND PLASMA MEMBRANE UNDULATION is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 158, 264–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Z. A., Morroll S. M., Dawson J. & Tighe P. J. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28, 27–39 (2001). [DOI] [PubMed] [Google Scholar]
- Tang L. K., Chu H., Yip W. K., Yeung E. C. & Lo C. An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 181, 576–587 (2009). [DOI] [PubMed] [Google Scholar]
- Aarts M. G., Keijzer C. J., Stiekema W. J. & Pereira A. Molecular characterization of the CER1 gene of arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7, 2115–2127 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarts M. G. M. et al. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12, 615–623 (1997). [DOI] [PubMed] [Google Scholar]
- Ariizumi T. et al. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. Rep. 53, 107–116 (2003). [DOI] [PubMed] [Google Scholar]
- Guan Y. F. et al. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147, 852–863 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C. et al. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21, 507–525 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M. et al. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19, 1473–1487 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. A. et al. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151, 574–589 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. S. et al. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22, 4045–4066 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. A. et al. LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153, 937–955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P. M. et al. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322 (1999). [Google Scholar]
- Xu Y., Iacuone S., Li S. F. & Parish R. W. MYB80 homologues in Arabidopsis, cotton and Brassica: regulation and functional conservation in tapetal and pollen development. BMC plant biol. 14, 1471–2229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Zhang Z. & Cao J. Pollen wall development: the associated enzymes and metabolic pathways. Plant Biology 15, 249–263 (2013). [DOI] [PubMed] [Google Scholar]
- Takahashi M. & Kouchi J. Ontogenetic development of spinous exine in Hibiscus syriacus (Malvaceae). Am. J. Bot. 75, 1549–1558 (1988). [Google Scholar]
- Blackmore S., Wortley A. H., Skvarla J. J. & Rowley J. R. Pollen wall development in flowering plants. New Phytol. 174, 483–498 (2007). [DOI] [PubMed] [Google Scholar]
- Scott R. J., Spielman M. & Dickinson H. G. Stamen structure and function. Plant Cell 16, S46–S60 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl G. M., Zwiebel B. I., Grier D. G. & Preuss D. Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126, 5431–5440 (1999). [DOI] [PubMed] [Google Scholar]
- Zhang Z. B. et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52, 528–538 (2007). [DOI] [PubMed] [Google Scholar]
- Lou Y. et al. The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 5, 3855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. et al. Male sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157, 842–853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox R. B. & Heslop-Harrison J. Pollen-Wall Proteins: The fate of intine-held antigens on the stigma in compatible and incompatible pollinations of Phalaris Tuberosa L. J. Cell Sci. 9, 239–251 (1971). [DOI] [PubMed] [Google Scholar]
- Schnurr J., Storey K., Jung H. J., Somers D. & Gronwald J. UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta 224, 520–532 (2006). [DOI] [PubMed] [Google Scholar]
- Oliver S. N., Dennis E. S. & Dolferus R. ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in Rice. Plant Cell Physiol. 48, 1319–1330 (2007). [DOI] [PubMed] [Google Scholar]
- Meuter-Gerhards A., Riegert S. & Wiermann R. Studies on sporopollenin biosynthesis in Cucurbita maxima (DUCH.) -II. the involvement of aliphatic metabolism. J Plant Physiol. 154, 431–436 (1999). [Google Scholar]
- Schulze Osthoff K. & Wiermann R. Phenols as integrated compounds of sporopollenin from Pinus Pollen. J Plant Physiol. 131, 5–15 (1987). [Google Scholar]
- Jung K. H. et al. Wax-deficient anther1 is involved in cuticle and wax production in Rice anther walls and is required for pollen development. Plant Cell 18, 3015–3032 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15, 1020–1033 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I., Ohlrogge J. B. & Pollard M. Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J. 53, 437–449 (2008). [DOI] [PubMed] [Google Scholar]
- Quilichini T. D., Grienenberger E. & Douglas C. J. The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry (2014) Available at: http://dx.doi.org/10.1016/j.phytochem.2014.05.002. (Accessed: 3rd June 2014) [DOI] [PubMed] [Google Scholar]
- Wakil S. J., Stoops J. K. & Joshi V. C. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 52, 537–579 (1983). [DOI] [PubMed] [Google Scholar]
- Mou Z., He Y., Dai Y., Liu X. & Li J. Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–417 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J. et al. Promoter trapping of a novel medium-chain Acyl-CoA Oxidase, which is induced transcriptionally during Arabidopsis seed germination. J. Biol. Chem. 275, 34375–34381 (2000). [DOI] [PubMed] [Google Scholar]
- Hemsley A. R. & Gabarayeva N. I. Exine development: the importance of looking through a colloid chemistry “window”. Plant Syst. Evol. 263, 25–49 (2007). [Google Scholar]
- Ariizumi T. & Toriyama K. Pollen exine pattern formation is dependent on three major developmental processes in Arabidopsis thaliana. Int J Plant Dev Biol 1, 106–115 (2007). [Google Scholar]
- Min L. et al. Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 164, 1293–1308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun X. et al. BnaC.Tic40, a plastid inner membrane translocon originating from Brassica oleracea, is essential for tapetal function and microspore development in Brassica napus. Plant J. 68, 532–545 (2011). [DOI] [PubMed] [Google Scholar]
- Tu L. L. et al. Genes expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development. Plant Cell Rep. 26, 1309–1320 (2007). [DOI] [PubMed] [Google Scholar]
- Xu L. et al. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L. et al. Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J. 75, 823–835 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou Z., Wang L., Li J., Song X. & Yang C. Study on programmed cell death and dynamic changes of starch accumulation in pericarp cells of Triticum aestivum L. Protoplasma 236, 49–58 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information
supplementary table s2
supplementary table s3
supplementary table s4
supplementary table s5
supplementary table s6
supplementary table s7