Abstract
Background
Addictions to alcohol and tobacco, known risk factors for cancer, are complex heritable disorders. Addictive behaviors have a bidirectional relationship with pain. We hypothesize that the associations between alcohol, smoking, and opioid addiction observed in cancer patients have a genetic basis. Therefore, using bioinformatics tools, we explored the underlying genetic basis and identified new candidate genes and common biological pathways for smoking, alcohol, and opioid addiction.
Results
Literature search showed 56 genes associated with alcohol, smoking and opioid addiction. Using Core Analysis function in Ingenuity Pathway Analysis software, we found that ERK1/2 was strongly interconnected across all three addiction networks. Genes involved in immune signaling pathways were shown across all three networks. Connect function from IPA My Pathway toolbox showed that DRD2 is the gene common to both the list of genetic variations associated with all three addiction phenotypes and the components of the brain neuronal signaling network involved in substance addiction. The top canonical pathways associated with the 56 genes were: 1) calcium signaling, 2) GPCR signaling, 3) cAMP-mediated signaling, 4) GABA receptor signaling, and 5) G-alpha i signaling.
Conlusions
Cancer patients are often prescribed opioids for cancer pain thus increasing their risk for opioid abuse and addiction. Our findings provide candidate genes and biological pathways underlying addiction phenotypes, which may be future targets for treatment of addiction. Further study of the variations of the candidate genes could allow physicians to make more informed decisions when treating cancer pain with opioid analgesics.
Electronic supplementary material
The online version of this article (doi:10.1186/s12918-015-0167-x) contains supplementary material, which is available to authorized users.
Keywords: Pain, Opioid, Smoking, Alcohol, Addiction, Genes, Inflammation, Cancer
Background
Pain is a debilitating problem that cancer patients face, impairing their quality of life. Pain may be related to multiple factors, including radiotherapy, chemotherapy, surgery, and cancer progression. In order to mitigate therapy-related pain or cancer-related pain, physicians often prescribe opioid analgesics to cancer patients [1, 2]. The prescription of opioids for pain carries risk for opioid abuse and addiction. Because of the increased survival rate in cancer patients, their exposure to prescriptions of opioids are also prolonged, further increasing their risk for opioid abuse and addiction [3–5].
Studies showed that opioid abuse was associated with past histories of drug and alcohol abuse in patients treated for cancer-related pain with opioid analgesics [6, 7]. Several clinical trials also found that patients with a history of cigarette smoking and illicit drug abuse had a significantly higher risk for opioid addiction than those without the history [8–11]. Taken together, these studies suggest that past addictive behaviors to various substances may predict opioid addiction in cancer patients with opioid prescriptions for pain. However, very few studies have explored whether there exists a genetic basis and common pathways to the relationship between smoking, alcohol, and opioid addiction.
Bioinformatics uses methods and software tools to organize and analyze biological data [12]. Specifically, gene network analyses have been used frequently to identify genes associated with drug abuse and addiction [13–15]. However, there has been limited application of bioinformatics in understanding multiple addiction phenotypes, specifically, smoking, alcohol and opioid addiction. We hypothesize that the associations between alcohol, smoking, and opioid addiction observed in the clinical setting have a genetic basis.
The goal of the current study is to use bioinformatics tools to determine whether there exists a genetic basis and common pathways to the relationship between smoking, alcohol, and opioid addiction and identify new candidate target genes. Understanding the genetic bases of addiction will underscore the importance of integrating genetic studies into the process of drug administration, as well as allow clinicians to more accurately tailor a patient’s drugs and dosage based on medical history and genetic risk factors [16].
Methods
With the goal of identifying commonly shared genes for alcohol, smoking and opioid addiction we conducted a literature search as described below. Subsequently, using genes pooled from literature as a starting point, we performed gene network analyses: a) specific to each phenotype (Phenotype Specific Biological Network) and b) commonly shared between alcohol, smoking and opioid addiction (Common Biological Network). Finally, we used the Connect function from IPA My Pathway toolbox to connect the commonly shared genes of the three phenotypes to the signaling network involved in neuronal adaptation/plasticity in substance addiction [17, 18].
Literature search
Each substance of abuse was searched on the PubMed database using the keywords “addiction” and “SNPs” in July 2014. Specifically, we used the term “alcohol addiction SNPs” for alcohol addiction, “smoking/nicotine/tobacco addiction SNPs” for smoking addiction, and “opioid addiction SNPs” for opioid addiction (Fig. 1). No limitations were placed on the year of publication. Non-human trials, literature reviews, and meta-analyses were excluded. Articles about treatment of drug addiction and drug addiction in patients with mental illnesses were also excluded. The genes reported in the literature to be statistically significantly associated with one of the addiction phenotypes were included in the pathway analysis and are called focus genes. The genes that were not replicated in an independent study were excluded. Figure 1 shows the criteria of the literature search.
Fig. 1.
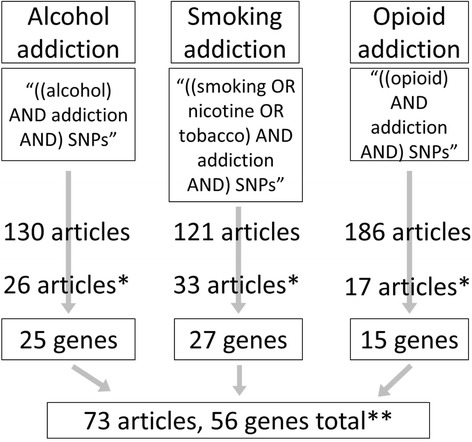
Literature search flowchart. *Subset after using the following Exclusion criteria: Literature review/meta-analysis, non-human experiments, other mental disorders, recovery/withdrawal, unrelated to phenotype, genes that were not replicated in or confirmed by at least one independent study. **Some overlaps between phenotypes for articles and genes
Ingenuity pathway analysis
Ingenuity Pathway Analysis (IPA) was used to produce a comprehensive analysis of the genes commonly shared in these addiction pathways. IPA is a software used to connect molecules based on the Ingenuity Knowledge Base, its database of information on biomolecules and their relationships [19]. The Core Analysis function was used to compare genes pooled from literature for each phenotype of addiction with the genes and other molecules in IPA’s database and generates gene networks based on their interactions.
We first designated a set of criteria for the molecules included in the Core Analysis. The following criteria were used: genes and endogenous chemicals, maximum molecules per network (140) and networks per analysis (25), humans, tissues and primary cells. Figure 2 illustrates the steps of the network generation process [20]. The resulting networks are then scored based on the negative base 10 logarithm of the p-value obtained using the Fisher’s exact test (i.e., -log10(p-value)), with the null hypothesis being that the molecules within the networks were connected based on chance.
Fig. 2.
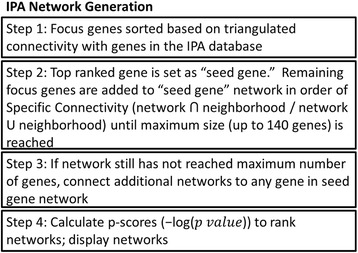
IPA network generation process
Phenotype specific biological network
Gene networks were created for each addiction phenotype. Only the networks with a p-score of 5 or higher were considered significant (i.e., p-value ≤ 10−5), a nominal significance used in previous studies [21]. The genes in each network were ranked based on number of edges, or interactions with other genes in the network.
Common biological network
In order to identify the shared genes, the opioid addiction network was compared with alcohol and smoking addiction networks. In addition, another network was generated by combining all 56 focus genes for all three addiction phenotypes (Fig. 3). In these analyses, only the network with a score ≥ 5 was considered significant [21]. Supplementary to the gene network, IPA also provides a list of top canonical pathways associated with the focus genes, along with a Fisher’s exact test p-value and the ratio between the number of focus genes in the canonical pathway and the total number of molecules in the canonical pathway. In this study, we also reported the top canonical pathways associated with all 56 focus genes for all three addiction phenotypes.
Fig. 3.

Network generated by pooling all 56 focus genes for alcohol, nicotine and opioid addiction (p-score = 45)
Finally, in order to understand the biological context of the gene network (association of genetic variations with addiction to opioids, alcohol and nicotine), we used the Connect function from IPA My Pathway toolbox to connect the commonly shared genes of three phenotypes to the signaling network involved in neuronal adaptation/plasticity in substance addiction [17, 18]. The Connect function adds specific interactions between molecules. While performing this analysis, we limited the interactions from only human studies. All results were generated through the use of Ingenuity® iReport [19].
Results
Literature search
A total of 73 unique articles were extracted based on the PubMed search for a thorough review. Figure 1 illustrates how the PubMed search produced this final list of articles for literature review. The articles associated with the corresponding type of addiction were summarized in tabular format (Tables 1, 2, 3). This resulted in a list of 56 focus genes total (Fig. 1), and each of these genes was used in the IPA Core Analysis. Opioid receptor genes [22] were frequently studied for alcohol and opioid addiction [22–28]. Nicotinic acetylcholine receptor genes were widely explored for alcohol and nicotine addiction [29–46]. Dopamine receptor genes were frequently explored in all three phenotypes [5, 27, 47–53]. Several overlapping focus genes across the three addiction phenotypes were observed, including DRD2 and CRHR1 for all three phenotypes, OPRM1 for alcohol and opioid addiction network, and BDNF and CNR1 for nicotine and opioid addiction network (Table 4). The 56 focus genes were subsequently used as seed genes in Ingenuity Pathway Analysis.
Table 1.
Summary of literature search - alcohol addiction
| Author | Ethnicity | Sample size | Phenotype | Salient gene(s) | Salient SNP(s) | Statistical analysis |
|---|---|---|---|---|---|---|
| Batel et al. [47] | EA | 134 | Alcohol dependence | DRD1 | rs686 | P = 0.0008 |
| Bierut et al. [77] | EA, AA | 5632 | Increased aversion from alcohol | ADH1B | rs1229984 | OR = 0.34 P = 6.6E-10 |
| Cao et al. [78] | Han Chinese | 603 | Alcohol addiction | 5-HTR | rs6313 | OR = 0.71 P = 0.001 |
| Chen et al. [79] | EA, AA | 3627 | Alcohol addiction | PKNOX2 | rs1426153 rs11220015 rs11602925 rs750338 rs12273605 rs10893365 rs10893366 rs12284594 | P = 5.75E-5, 6.86E-5, 4.24E-5, 4.26E-5, 3.0E-4, 1.72E-5, 1.37E-5, 1.97E-6 |
| Deb et al. [25] | South Asian | 144 | Alcohol addiction | OPRM1 | rs1799971 | P = 0.02 |
| Desrivieres et al. [80] | E | 145 | Drinking behavior | P13K | rs2302975 rs1043526 | P = 0.0019, 0.0379 |
| Enoch et al. [81] | AA | 360 | Alcohol addiction | HTR3B | rs1176744 | P = 0.002 |
| Ehringer et al. [35] | EA, Hisp, AA | 108 | Alcohol response | CHRNB2 | rs2072658 | |
| Haller et al. [37] | EA, AA | 1315 | Alcohol addiction | CHRNB3 | rs149775276 | P = 2.6E-4 for EA, P = 0.006 for AA |
| Hill et al. [82] | EA | 1000 | Alcohol dependence | KIAA0040 | rs2269650 rs2861158 rs1008459 rs2272785 rs10912899 rs3753555 | P = 0.033, 0.037, 0.014, 0.062, 0.035, 0.020 |
| Kalsi et al. [83] | EA, AA | 847 | Alcohol addiction | DKK2 | rs427983 rs419558 rs399087 | P = 0.007 |
| Kumar et al. [26] | Bengali/Hindu | 310 | Alcohol addiction | OPRM1 | rs16918875 rs702764 rs963549 | P = 0.0364 |
| Kuo et al. [84] | E | 1238 | Initial sensitivity to alcohol | GAD | P = 0.002 | |
| London et al. [85] | EA | Risk for alcohol addiction | ANKK1 | rs1800497 | P = 0.001 | |
| Mignini et al. [51] | E | 560 | Dopaminergic system; alcohol dependence | DRD2/ANKK1 | rs1800497 | P = 0.023 |
| Munoz et al. [86] | E | 1533 | Number of drinks per day | ADH1B, ADH6 | rs1229984 in ADH1B rs3857224 in ADH6 | rs1229984: OR = 0.19, P = 4.77E-10 for men, OR = 0.48, P = 0.0067 for women; rs38572: OR = 1.61, P = 1.01E-3 for women, NS for men |
| Novo-Veleiro et al. [87] | E | 457 | Risk for alcohol addiction | miR-146a | rs2910164 | OR = 1.615 P = 0.023 |
| Preuss et al. [88] | E (German & Polish) | 3091 | Alcoholism | ADH4 | rs1800759 rs1042364 | rs1800759: OR = 0.88 rs1042364: OR = 0.87 |
| Ray et al. [89] | CA, As, Latino, NA, AA | 124 | Level of response to alcohol/drinking problems | GABRG1 | rs1497571 | P < 0.01 |
| Samochowiec et al. [90] | EA | 275 | Alcohol dependence | MMP-9 | rs3918242 | P < 0.01 |
| Schumann et al. [91] | E | 1544 | Alcohol dependence | NR2A, MGLUR | OR = 2.35, 1.69 | |
| Treutlein et al. [92] | E | 296 | Potential alcohol dependence | CRHR1 | ||
| Wang et al. [42] | EA, AA | 2309 | Alcohol dependence | CHRNA5 | rs680244 | P = 0.003 |
| Wang et al. [93] | EA | 2010 | Alcohol dependence | C15orf53 | rs12903120 rs12912251 | rs12903120: P = 5.45E − 8 |
| Xuei et al. [94] | EA | 1923 | Risk for alcohol addiction | GABRR1, GABRR2 | rs17504587 rs282129 rs13211104 rs9451191 rs2821211 rs6942204 | P = 0.04. 0.03, 0.03, 0.021, 0.025, 0.04 |
| Yang et al. [95] | EA, AA | 3564 | Alcohol dependence | HTR3B | rs3891484 rs375898 | D’ > 7 |
Table 2.
Summary of literature search - smoking addiction
| Author | Ethnicity | Sample size | Phenotype | Salient gene(s) | Salient SNP(s) | Statistical analysis |
|---|---|---|---|---|---|---|
| Agrawal et al. [96] | EA | 1929 | Nicotine dependence | GABRA4, GABRA2, GABRE | P = 0.030 | |
| Agrawal et al. [97] | EA | 1921 | Nicotine dependence | GABRA4, GABRA2 | P = 0.002 | |
| Anney et al. [98] | E | 815 | Cigarette dose | CHRM5 | rs7162140 | P = 0.01 |
| Baker et al. [31] | EA | 886 | Nicotine dependence | CHRNA5-A3-B4 | P = 0.04 | |
| Berrettini et al. [99] | EA | 1276 | Nicotine addiction | CYP2A6 | rs410514431 | P = 1.0E-12 |
| Beuten et al. [100] | EA, AA | 2037 | Nicotine dependence | BDNF | rs6484320 rs988748 rs2030324 rs7934165 | P = 0.002 |
| Beuten et al. [101] | EA, AA | Nicotine dependence | GABAB2 | rs2491397 rs2184026 rs3750344 rs1435252 rs378042 rs2779562 rs3750344 | P = 0.003 | |
| Beuten et al. [102] | EA, AA | Nicotine dependence | COMT | rs933271 rs4680 rs174699 | P = 0.0005 | |
| Broms et al. [32] | E | 1428 | Nicotine dependence | CHRNA5, CHRNA3, CHRNB4 | rs2036527 rs578776 rs11636753 rs11634351 rs1948 rs2036527 | P = 0.000009, 0.0001, 0.0059, 0.0069, 0.0071, 0.0003 |
| Chen et al. [103] | 688 | Nicotine dependence | CNR1 | rs2023239 rs12720071 rs806368 | P < 0.001 | |
| Chen et al. [79] | EA, AA | 3627 | Nicotine addiction | PKNOX2 | rs1426153 rs11220015 rs11602925 rs750338 rs12273605 rs10893365 rs10893366 rs12284594 | P = 0.0159, 0.0163, 0.0136, 0.0491, 0.0921, 0.0411, 0.0621, 0.0239 |
| Conlon et al. [33] | EA | 1122 | Nicotine dependence | CHRNA5, CHRNA3, AGPHD1 | rs16969968 rs578776 rs8034191 | OR = 3.2, 2.8, 0.3 |
| Culverhouse et al. [34] | AA, EA | 18500 | Nicotine dependence | CHRNB3, CHRNA7 | rs13273442 | P = 0.00058 for EA, 0.05 for AA |
| Docampo et al. [104] | E | 752 | Lower risk for smoking behavior | NRXN3 | rs1424850 rs221497 rs221473 | rs1424850: OR = 0.55, P = 0.0002 |
| rs221497: OR = 0.47, P = 0.0020 | ||||||
| rs221473: OR = 0.54, P = 0.0009 | ||||||
| Ehringer et al. [35] | EA, Hisp, AA | 108 | Nicotine response | CHRNB2 | rs2072658 | |
| Ella et al. [105] | Japanese | 2521 | Nicotine addiction | DBH | rs5320 | P = 0.030 |
| Gabrielsen et al. [36] | Norwegian | 155941 | Smoking status(cigarettes per day, duration, packs per year) | CHRNA5/A3/B4 | rs16969968 | P = 3.15E-25, 1.11E-6, 3.01E-23 (respectively for phenotypes) |
| Huang et al. [106] | EA, AA | 3403 | Nicotine dependence | ANKK1 | rs2734849 | P = 0.0026 |
| Lang et al. [107] | E | 320 | Smoking behavior | BDNF | P = 0.045 | |
| Li et al. [38] | EA, AA | 2037 | Nicotine dependence | CHRNA4 | rs2273504 rs1044396 rs3787137 rs2236196 | |
| Liu et al. [108] | EA, AA | 2091 | Smoking behavior | IL15 | rs4956302 | P = 8.8E-8 |
| Ma et al. [109] | EA, AA | 2037 | Nicotine dependence | DDC | rs3735273 rs1451371 rs3757472 rs3735273 rs1451371 rs2060762 | P = 0.005, 0.006 |
| Mobascher et al. [110] | German | 5500 | smoking behavior/nicotine addiction | CHRM2 | rs324650 | OR = 1.17 |
| Nees et al. [39] | E, EA | 965 | Nicotine dependence | CHRNA5/A3/B4 | rs578776 | P < 0.05 |
| Sherva et al. [40] | EA, AA | 435 | Smoking | CHRNA5 | rs16969968 | P = 0.0001 |
| Rice et al. [29] | EA, AA | 3365 | Nicotine dependence | CHRNB3 | rs1451240 | P = 2.4E-8 |
| Sarginson et al. [30] | EA, Asian, AA, Hispanic | 577 | Smoking behavior | CHRNA5/A3/B4 | rs16969968 rs1051730 | P < 0.0001 |
| Sorice et al. [41] | E | 2272 | Smoking behavior | CHRNA5-A3-B4 | rs1051730 | P = 0.0151, 0.022, 0.22 for three populations |
| Voisey et al. [52] | EA | 378 | Nicotine dependence | DRD2 | rs1800497 | P = 0.0003 |
| Wang et al. [43] | EA, AA | 3622 | ND (smoking quantity and FTND) | CHRNA2, CHRNA6 | EA: rs3735757 rs2472553 | EA: P = 0.0068 for FTND, AA: P = 0.0043 for SQ and 0.00086 for FTND |
| Wassenaar et al. [44] | E | 860 | Nicotine dependence | CYP2A6 and CHRNA5-A3-B4 | rs1051730 | P =0.036 |
| Weiss et al. [45] | E | 2827 | Nicotine dependence | CHRNA5-A3-B4 | rs17486278 | P = 0.0005 |
| Zeiger et al. [46] | EA, Hisp | 1056 | Response to smoking | CHRNA6, CHRNB3 | rs4950 rs13280604 rs2304297 | P = 0.043, 0.011, 0.053 |
Table 3.
Summary of literature search - opioid addiction
| Author | Ethnicity | Sample size | Phenotype | Salient gene(s) | Salient SNP(s) | Statistical analysis |
|---|---|---|---|---|---|---|
| Beer et al. [22] | E | 284 | Opioid dependence | GAL, OPRD1 | rs948854 rs2236861 | P = 0.001 |
| Bunten et al. [23] | 184 | Opioid addiction | OPRM1 | rs1799971 | P = 0.0046 | |
| Compton et al. [24] | EA | 109 | Opioid addiction | OPRM1 | rs1799971 | |
| Clarke et al. [111] | Han Chinese | 858 | Opioid dependence | PDYN | rs1997794 rs1022563 | P = 0.019, 0.006 |
| Clarke et al. [48] | EA, AA | 992 | Opioid addiction | DRD2 | rs1076560 | OR = 1.29, P = 0.0038 |
| Crist et al. [112] | EA, AA | 671 | Opioid addiction | WLS | rs3748705 (AA) rs983034 rs1036066 (EA) | AA: P = 0.025EA: P = 0.043, 0.045 |
| de Cid et al. [113] | E | 91 | Opioid Addiction | BDNF | ||
| Doehring et al. [49] | CA | 184 | Opioid addiction | DRD2 | rs1076560 rs1799978 rs6277 rs12364283 rs1799732 rs6468317 rs6275 rs1800498 rs1800497 | P = 0.022, 0.048 |
| Gelernter et al. [114] | EA, AA | 8246 | Opioid dependence | KCNG2 | rs62103177 | P = 3.60E-10 |
| Herman et al. [115] | EA, AA | 1367 | Opioid dependence | CNR1 | rs6928499 rs806379 rs1535255 rs2023239 | |
| Ho et al. [50] | Chinese | 252 | Opioid dependence | DRD4 | P = 0.041 | |
| Kumar et al. [116] | South Asian | 260 | Opioid dependence | CREBBP | rs3025684 | P < 0.0001 |
| Kumar et al. [26] | Bengali/Hindu | 330 | Opioid addiction | OPRM1 | rs16918875 rs702764 rs963549 | P = 0.0264 |
| Levran et al. [117] | 74 | Opioid addiction | CYP2B6 | |||
| Liu et al. [118] | African | 3627 | Opioid addiction | NCK2 | rs2377339 | P = 1.33E-11 |
| Nagaya et al. [28] | Asian | 160 | Opioid addiction | OPRM1 | rs1799972 | OR = 1.77, P < 0.0001 |
| Zhu et al. [53] | Chinese | 939 | Opioid dependence/addiction | DRD1 | rs686 | P = 0.0003 |
Table 4.
Overlapping genes for networks of nicotine, alcohol and opioid addiction; focus genes from literature are bolded
| A: Opioids ∩ Alcohol | B: Opioids ∩ Nicotine | C: Opioids ∩ Alcohol ∩ Nicotine | |||
|---|---|---|---|---|---|
| Molecule | Edges in opioid network/edges in alcohol network | Molecule | Edges in opioid network/edges in nicotine network | Molecule | Edges in opioid network/edges in alcohol network/edges in nicotine network |
| NFkB (complex) | 112/86 | ERK1/2 | 74/76 | ERK1/2 | 74/62/76 |
| ERK1/2 | 74/62 | ARRB2 | 8/3 | DRD2 | 6/3/4 |
| IL1R1 | 7/4 | DRD2 | 6/4 | TAP1 | 5/5/3 |
| IL1 | 6/8 | HSPD1 | 5/4 | SAA | 4/3/4 |
| DEFB4A/DEFB4B | 6/4 | TAP1 | 5/3 | PSMB9 | 4/3/3 |
| DRD2 | 6/3 | SAA | 4/4 | TAPBP | 4/3/3 |
| ELANE | 5/6 | PSMB9 | 4/3 | ELF3 | 4/3/2 |
| F2RL1 | 5/6 | TAPBP | 4/3 | TAC1 | 4/3/2 |
| TAP1 | 5/5 | ELF3 | 4/2 | CLEC11A | 3/4/2 |
| F2R | 5/3 | TAC1 | 4/2 | SMPD2 | 3/3/3 |
| ADRBK1 | 5/2 | PSMB10 | 3/3 | CXCL3 | 3/3/2 |
| Ikb | 4/4 | SMPD2 | 3/3 | P2RY6 | 3/3/2 |
| CXCL2 | 4/3 | AKAP13 | 3/2 | PSMB10 | 3/2/3 |
| ELF3 | 4/3 | CLEC11A | 3/2 | AKAP13 | 3/2/2 |
| FPR2 | 4/3 | CXCL3 | 3/2 | TLR6 | 3/2/2 |
| PSMB9 | 4/3 | P2RY6 | 3/2 | CRHR1 | 2/4/3 |
| SAA | 4/3 | TLR6 | 3/2 | CD244 | 2/3/3 |
| TAC1 | 4/3 | CD244 | 2/3 | CXCL5 | 2/3/2 |
| TAPBP | 4/3 | CRHR1 | 2/3 | CCL21 | 2/2/2 |
| DEFB103A/DEFB103B | 4/2 | CCL21 | 2/2 | GMFG | 2/2/2 |
| LTF | 3/5 | CNR1 | 2/2 | ||
| TNFSF11 | 3/5 | CXCL5 | 2/2 | ||
| TNFSF15 | 3/5 | GMFG | 2/2 | ||
| CLEC11A | 3/4 | GPRASP1 | 2/2 | ||
| TLR1 | 3/4 | BDNF | 2/1 | ||
| CXCL3 | 3/3 | ||||
| KLF6 | 3/3 | ||||
| P2RY6 | 3/3 | ||||
| SMPD2 | 3/3 | ||||
| AKAP13 | 3/2 | ||||
| ARF6 | 3/2 | ||||
| IER3 | 3/2 | ||||
| PSMB10 | 3/2 | ||||
| TLR6 | 3/2 | ||||
| TRPC6 | 3/2 | ||||
| CRHR1 | 2/4 | ||||
| CCL22 | 2/3 | ||||
| CD244 | 2/3 | ||||
| CXCL5 | 2/3 | ||||
| CC2D1A | 2/2 | ||||
| CCL21 | 2/2 | ||||
| GMFG | 2/2 | ||||
| SH3GLB2 | 2/2 | ||||
| STAB2 | 2/2 | ||||
| TSC22D3 | 2/2 | ||||
| OPRM1 | 1/2 | ||||
IPA – Phenotype-specific biological network
Individual gene networks were generated through IPA’s Core Analysis for each addiction phenotype (Additional file 1: Figures S1-S3). TNF, NF-κB, and ERK1/2 were present as highly interconnected genes for alcohol addiction (103, 86, and 62 edges, respectively). For nicotine addiction, TNF, ERK1/2 and Akt had the most edges (85, 76, and 53, respectively). NF-κB, RELA, and ERK1/2 were most interconnected for opioid addiction (112, 92, and 74 edges respectively).
IPA – Common biological network
Table 4 lists overlapping genes for alcohol and opioids (A), smoking and opioids (B), and all three addiction phenotypes (C). Genes were ranked by the number of edges within the opioid network. The network for opioid addiction was found to have the most number of genes that overlap with the network for alcohol addiction relative to the smoking addiction genes. ERK1/2 was found to be very strongly interconnected across all three addiction networks with 74 edges in opioid network, 62 edges in alcohol network and 76 edges in nicotine network (Table 4, panel C). ERK1/2 also shows with highest number of edges in opioid and nicotine network (Table 4, panel B) and second highest edges in opioid and alcohol network (Table 4, panel A). We also noticed that some commonly shared genes are involved in the immune response. Specifically, the immune response genes that were common in the three networks (panel C) were: corticotropin-releasing hormone receptor 1 (CRHR1), chemokine ligand 21 (CCL21), chemokine ligand 3 (CXCL3), chemokine ligand 5 (CXCL5) and toll-like receptor 6 (TLR6). In addition to the above genes, the following immune response genes were also found in opioid and alcohol genes networks (panel A): beta-defensin 103 (DEFB103A/DEFB103B), beta-defensin 2 (DEFB4A/DEFB4B), elastase neutrophil expressed (ELANE), protease activated receptor 2 (F2RL1), lactoferrin (LTF), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappa B), toll-like receptor 1 (TLR1), TSC22 domain family protein 3 (TSC22D3), chemokine ligand 22 (CCL22), chemokine ligand 2 (CXCL2), interleukin 1 receptor type 1 (IL1R1), tumor necrosis factor ligand superfamily member 11 and 15 (TNFSF11 and TNFSF15).
By pooling all 56 focus genes from three addiction phenotypes, a total of 8 networks were generated by using IPA Core Analysis. Figure 3 shows the network with the highest statistical significance (p-value = 10−45). Figure 4 shows the top canonical pathways for the combined focus genes, including calcium signaling, GPCR signaling, cAMP-mediated signaling, GABA receptor signaling, and Gαi signaling (p-values = 1.26E-12, 4.45E-12, 1.71E-11, 6.3E-10, 4.29E-8).
Fig. 4.

Top canonical pathways obtained by pooling all 56 focus genes for alcohol, nicotine and opioid addiction. Blue bars: p-score for each of the canonical pathways. Yellow lines: ratio for each of the canonical pathways, calculated as the number of focus genes included in the canonical pathway divided by the total number of genes that constitute the canonical pathway
Biological context
Finally, we used the “Connect” function from IPA My Pathway toolbox to connect the commonly shared genes (i.e., overlapping genes) related to addiction to opioids, alcohol and nicotine (Table 4, panel C) to the signaling network involved in neuronal adaptation/plasticity in substance addiction (Fig. 5) [17, 18]. Particularly, DRD2 is the gene common to both the list of genetic variations associated with substance addiction and the components of the brain neuronal signaling network involved in substance addiction. IPA identified multiple links between components of these 2 lists of genes. ERK1/2 was directly connected to DRD1 and indirectly connected to RAC1, FOS, ERK, Creb, PI3K, BDNF and Pka in the signaling network in neuronal adaptation/plasticity in substance addiction (i.e., reward circuit). All the commonly shared immune response genes for the three addiction phenotypes, including TLR6, CXCL5, CXCL3, CRHR1 and CCL21, were indirectly linked to NFkB in the reward circuit. Gene CCL21 was also indirectly linked to Akt and ERK in the reward circuit.
Fig. 5.
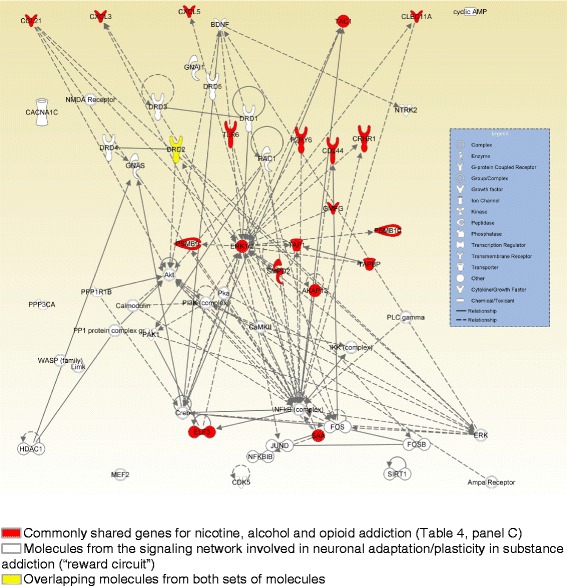
The links of genes associated with addiction to opioids, tobacco and alcohol to components of the brain “reward circuit”
Discussion
One of the most challenging areas of oncologic medicine is the management and treatment of severe, chronic pain that arises from cancer therapies, including surgery, chemotherapy, and radiation, as well as cancer itself. Opioids remain the drugs of choice for cancer pain management [54], however, the use of opioids for treatment of chronic pain in cancer patients remains debatable. An increasing concern is the potential rise in aberrant drug-taking behaviors of cancer patients undergoing treatment for chronic pain [3, 55]. Given that addictions to alcohol and tobacco are known risk factors for cancer, exploring genetic markers of risk for these addiction phenotypes in cancer patients may help in risk stratification. Indeed, studies have begun to show that genetic vulnerability to different substances of addiction may partly overlap [56]. The primary aims of this study were to determine whether there exists a genetic basis to the relationship between smoking, alcohol, and opioid addiction, and to identify candidate genes associated with the three phenotypes for further study.
We used IPA, a bioinformatics tool, to identify commonly shared genes for alcohol, smoking, and opioid addiction. Of the 20 genes commonly shared across the alcohol, smoking and opioid addiction phenotypes, extracellular-signal-regulated kinases 1 and 2 (ERK1/2) was found to have the most interconnections across all three addiction networks as indicated by the number of edges (biological interactions; Table 4). Recent studies suggest the relevance of ERK pathway in drug addiction. Several studies have cited the role of ERK in brain’s response to drugs of abuse [57–59]. Specifically, Valjent et al. [59] demonstrated that multiple drugs of abuse increased activation of ERK1/2. Molecular mechanisms underlying ERK1/2 activation by drugs of abuse and the role of ERK1/2 signaling in long-term neuronal plasticity in the striatum may provide novel targets for therapeutic intervention in addiction [60]. Moreover, studies exploiting ERK activation for cancer therapy have been promising, including the use of MEK inhibitors to block ERK activation in acute lymphoblastic leukemia for instance [61]. Future studies are needed to assess the potential clinical relevance of ERK1/2 for addiction, e.g., to genotype ERK1/2 and stratify patients for prompt intervention, or to determine appropriate dosage of opioid analgesics to patients with specific genotypes.
Of note, the identified shared genes for the three addiction phenotypes are involved in immune response. This is consistent with recent research that implicates immune signaling in drug addiction. Dafney et al. demonstrated that certain immunosuppressive treatments controlled morphine withdrawal in rats [62, 63]. More recent studies demonstrated that blocking pro-inflammatory glial activation could block the elevation of dopamine induced by opioid receptor activity [64, 65]. Hutchinson et al. have also found evidence that toll-like receptors (TLRs), a class of innate immune receptors, interact with opioids and glial cells, contributing to opioid reward behaviors [65]. Our recent studies also showed that cytokine genes are implicated in pain, depressed mood, and fatigue in cancer patients [66–68], and these cytokines may serve as biomarkers of risk for persistent pain in cancer patients.
Furthermore, it is also speculated that synaptic plasticity induced by substances of abuse in the neuronal circuits of reward may underlie behavioral changes that characterize addiction. Importantly, NF-kappa B may be the link between inflammation and neuronal/synaptic plasticity involved in behavioral changes in addiction, as we have shown that all the commonly shared immune response genes of three addiction phenotypes were linked to NF-kappa B in the reward circuit (Fig. 5). NF-kappa B is one of several transcription factors present at the synapse, and it is activated by brain-specific activators such as glutamate (via AMPA/KA and NMDA receptors) and neurotrophins [69]. To date, there are currently no pharmacotherapies for drug addiction targeting immune signaling.
Our results also showed the top canonical pathways associated with all the 56 focus genes of three addiction phenotypes were: 1) calcium signaling, 2) GPCR signaling, 3) cAMP-mediated signaling, 4) GABA receptor signaling, and 5) Gαi signaling. These pathways have been confirmed to be associated with substance addiction in the literature [70–74]. They are the post-receptor signaling pathways for the glutaminergic, dopaiminergic and GABAergic neurons involved in the “reward circuitry” in mammalian brains [75]. Whether these pathways can be used as targets for drug addiction therapy needs to be explored. Our approach of identifying genetic variations associated with addiction to multiple substances and linking to known the neural signaling network involved in substance addiction in the brain has clarified the functional significance of many of the genetic associations to substance addiction. This bioinformatics approach has also identified signaling pathways that may be targeted by drugs. Promising research has shown that allosteric modulators of GPCRs can be used to treat addiction by altering the affinity of the GPCR to its ligand or impacting its downstream signaling responses [72]. Other studies have also suggested positive allosteric modulation of GABAB as a therapeutic strategy for treatment of addiction [71, 76].
Among the limitations of this study is that edges are simplified in the IPA designates only a single edge between each pair of molecules in a network regardless of the number of interactions the two molecules share. Furthermore, this bioinformatics analysis is hypothesis-generating, and the findings must be further investigated and validated experimentally.
Conclusions
Studying smoking, alcohol, and opioid addiction phenotypes in conjunction allowed us to identify molecules and pathways involved in multiple types of drug addiction. IPA is able to use large-scale information to produce comprehensive networks of genes and underlying biological pathways implicated in a phenotype [19]. Most of the current literature on addiction genes focuses on genes specific to each type of addiction, while in this study we studied genes relating to multiple addiction phenotypes. Our findings show immune signaling and ERK1/2 as novel genetic markers for multiple addiction phenotypes including alcohol, smoking and opioid addiction. Future studies are needed to validate our findings in large cohorts of patients.
Additional files
Network generated using 25 focus genes for alcohol addiction (p-score = 16). Figure S2. Network generated using 27 focus genes for nicotine addiction (p-score = 31). Figure S3. Network generated using 15 focus genes for opioid addiction (p-score = 10).
Footnotes
Christine Yuan and Jian Wang contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CRG, CY, JW, SY, SS made substantial contributions to conception and design, analysis and interpretation of data; CRG, CY, SY, JW and SS final approval of the version to be published; and all agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Contributor Information
Cielito C. Reyes-Gibby, Email: creyes@mdanderson.org
Christine Yuan, Email: cyuan95@yahoo.com.
Jian Wang, Email: jianwang@mdanderson.org.
Sai-Ching J. Yeung, Email: syeung@mdanderson.org
Sanjay Shete, Email: sshete@mdanderson.org.
References
- 1.Arai YC, Matsubara T, Shimo K, Suetomi K, Nishihara M, Ushida T, et al. Low-dose gabapentin as useful adjuvant to opioids for neuropathic cancer pain when combined with low-dose imipramine. J Anesth. 2010;24(3):407–410. doi: 10.1007/s00540-010-0913-6. [DOI] [PubMed] [Google Scholar]
- 2.Smith EM, Pang H, Cirrincione C, Fleishman S, Paskett ED, Ahles T, et al. Effect of duloxetine on pain, function, and quality of life among patients with chemotherapy-induced painful peripheral neuropathy: a randomized clinical trial. JAMA. 2013;309(13):1359–1367. doi: 10.1001/jama.2013.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koyyalagunta D, Burton AW, Toro MP, Driver L, Novy DM. Opioid abuse in cancer pain: report of two cases and presentation of an algorithm of multidisciplinary care. Pain Physician. 2011;14(4):E361–371. [PubMed] [Google Scholar]
- 4.Rauenzahn S, Del Fabbro E. Opioid management of pain: the impact of the prescription opioid abuse epidemic. Curr Opin Support Palliat Care. 2014;8(3):273–278. doi: 10.1097/SPC.0000000000000065. [DOI] [PubMed] [Google Scholar]
- 5.Sweitzer MM, Donny EC, Hariri AR. Imaging genetics and the neurobiological basis of individual differences in vulnerability to addiction. Drug Alcohol Depend. 2012;123(Suppl 1):S59–71. doi: 10.1016/j.drugalcdep.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granata R, Bossi P, Bertulli R, Saita L. Rapid-onset opioids for the treatment of breakthrough cancer pain: two cases of drug abuse. Pain Med. 2014;15(5):758–761. doi: 10.1111/pme.12382. [DOI] [PubMed] [Google Scholar]
- 7.Koyyalagunta D, Bruera E, Aigner C, Nusrat H, Driver L, Novy D. Risk stratification of opioid misuse among patients with cancer pain using the SOAPP-SF. Pain Med. 2013;14(5):667–675. doi: 10.1111/pme.12100. [DOI] [PubMed] [Google Scholar]
- 8.Erlich PM, Hoffman SN, Rukstalis M, Han JJ, Chu X, Linda Kao WH, et al. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum Genet. 2010;128(5):491–499. doi: 10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- 9.Hojsted J, Ekholm O, Kurita GP, Juel K, Sjogren P. Addictive behaviors related to opioid use for chronic pain: a population-based study. Pain. 2013;154(12):2677–2683. doi: 10.1016/j.pain.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 10.Log T, Hartz I, Handal M, Tverdal A, Furu K, Skurtveit S. The association between smoking and subsequent repeated use of prescribed opioids among adolescents and young adults–a population-based cohort study. Pharmacoepidemiol Drug Saf. 2011;20(1):90–98. doi: 10.1002/pds.2066. [DOI] [PubMed] [Google Scholar]
- 11.Skurtveit S, Furu K, Selmer R, Handal M, Tverdal A. Nicotine dependence predicts repeated use of prescribed opioids. Prospective population-based cohort study. Ann Epidemiol. 2010;20(12):890–897. doi: 10.1016/j.annepidem.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Readhead B, Dudley J. Translational Bioinformatics Approaches to Drug Development. Adv Wound Care. 2013;2(9):470–489. doi: 10.1089/wound.2012.0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atreya RV, Sun J, Zhao Z. Exploring drug-target interaction networks of illicit drugs. BMC Genomics. 2013;14(Suppl 4):S1. doi: 10.1186/1471-2164-14-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han S, Yang BZ, Kranzler HR, Liu X, Zhao H, Farrer LA, et al. Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. Am J Hum Genet. 2013;93(6):1027–1034. doi: 10.1016/j.ajhg.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li CY, Mao X, Wei L. Genes and (common) pathways underlying drug addiction. PLoS Comput Biol. 2008;4(1):e2. doi: 10.1371/journal.pcbi.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman AD, Cohen MJ, Manley GT, Butte AJ. Altering physiological networks using drugs: steps towards personalized physiology. BMC Med Genomics. 2013;6(Suppl 2):S7. doi: 10.1186/1755-8794-6-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010;33(6):267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.“FOSB”. Wikipedia: The free encyclopedia [http://en.wikipedia.org/wiki/FOSB#Delta_FosB]
- 19.Ingenuity Pathway Analysis software [http://www.ingenuity.com/]
- 20.IPA Network Generation Algorithm [http://www.ingenuity.com/wp-content/themes/ingenuitytheme/pdf/ipa/IPA-netgen-algorithm-whitepaper.pdf]
- 21.Muurling T, Stankovic KM. Metabolomic and network analysis of pharmacotherapies for sensorineural hearing loss. Otol Neurotol. 2014;35(1):1–6. doi: 10.1097/MAO.0000000000000254. [DOI] [PubMed] [Google Scholar]
- 22.Beer B, Erb R, Pavlic M, Ulmer H, Giacomuzzi S, Riemer Y, et al. Association of polymorphisms in pharmacogenetic candidate genes (OPRD1, GAL, ABCB1, OPRM1) with opioid dependence in European population: a case-control study. PLoS One. 2013;8(9):e75359. doi: 10.1371/journal.pone.0075359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunten H, Liang WJ, Pounder DJ, Seneviratne C, Osselton D. Interindividual variability in the prevalence of OPRM1 and CYP2B6 gene variations may identify drug-susceptible populations. J Anal Toxicol. 2011;35(7):431–437. doi: 10.1093/anatox/35.7.431. [DOI] [PubMed] [Google Scholar]
- 24.Compton P, Geschwind DH, Alarcon M. Association between human mu-opioid receptor gene polymorphism, pain tolerance, and opioid addiction. Am J Med Genet Part B Neuropsychiatr Genet. 2003;121B(1):76–82. doi: 10.1002/ajmg.b.20057. [DOI] [PubMed] [Google Scholar]
- 25.Deb I, Chakraborty J, Gangopadhyay PK, Choudhury SR, Das S. Single-nucleotide polymorphism (A118G) in exon 1 of OPRM1 gene causes alteration in downstream signaling by mu-opioid receptor and may contribute to the genetic risk for addiction. J Neurochem. 2010;112(2):486–496. doi: 10.1111/j.1471-4159.2009.06472.x. [DOI] [PubMed] [Google Scholar]
- 26.Kumar D, Chakraborty J, Das S. Epistatic effects between variants of kappa-opioid receptor gene and A118G of mu-opioid receptor gene increase susceptibility to addiction in Indian population. Prog Neuropsychopharmacol Biol Psychiatry. 2012;36(2):225–230. doi: 10.1016/j.pnpbp.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Levran O, Peles E, Randesi M, Li Y, Rotrosen J, Ott J, et al. Stress-related genes and heroin addiction: a role for a functional FKBP5 haplotype. Psychoneuroendocrinology. 2014;45:67–76. doi: 10.1016/j.psyneuen.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaya D, Ramanathan S, Ravichandran M, Navaratnam V. A118G mu opioid receptor polymorphism among drug addicts in Malaysia. J Integr Neurosci. 2012;11(1):117–122. doi: 10.1142/S0219635212500082. [DOI] [PubMed] [Google Scholar]
- 29.Rice JP, Hartz SM, Agrawal A, Almasy L, Bennett S, Breslau N, et al. CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarginson JE, Killen JD, Lazzeroni LC, Fortmann SP, Ryan HS, Schatzberg AF, et al. Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am J Med Genet Part B, Neuropsychiatr Genet. 2011;156B(3):275–284. doi: 10.1002/ajmg.b.31155. [DOI] [PubMed] [Google Scholar]
- 31.Baker TB, Weiss RB, Bolt D, von Niederhausern A, Fiore MC, Dunn DM, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine Tobacco Res. 2009;11(7):785–796. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broms U, Wedenoja J, Largeau MR, Korhonen T, Pitkaniemi J, Keskitalo-Vuokko K, et al. Analysis of detailed phenotype profiles reveals CHRNA5-CHRNA3-CHRNB4 gene cluster association with several nicotine dependence traits. Nicotine Tobacco Res. 2012;14(6):720–733. doi: 10.1093/ntr/ntr283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conlon MS, Bewick MA. Single nucleotide polymorphisms in CHRNA5 rs16969968, CHRNA3 rs578776, and LOC123688 rs8034191 are associated with heaviness of smoking in women in Northeastern Ontario, Canada. Nicotine Tobacco Res. 2011;13(11):1076–1083. doi: 10.1093/ntr/ntr140. [DOI] [PubMed] [Google Scholar]
- 34.Culverhouse RC, Johnson EO, Breslau N, Hatsukami DK, Sadler B, Brooks AI, et al. Multiple distinct CHRNB3-CHRNA6 variants are genetic risk factors for nicotine dependence in African Americans and European Americans. Addiction. 2014;109(5):814–822. doi: 10.1111/add.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet Part B Neuropsychiatr Genet. 2007;144B(5):596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- 36.Gabrielsen ME, Romundstad P, Langhammer A, Krokan HE, Skorpen F. Association between a 15q25 gene variant, nicotine-related habits, lung cancer and COPD among 56,307 individuals from the HUNT study in Norway. Eur J Hum Genet. 2013;21(11):1293–1299. doi: 10.1038/ejhg.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haller G, Kapoor M, Budde J, Xuei X, Edenberg H, Nurnberger J, et al. Rare missense variants in CHRNB3 and CHRNA3 are associated with risk of alcohol and cocaine dependence. Hum Mol Genet. 2014;23(3):810–819. doi: 10.1093/hmg/ddt463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14(9):1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 39.Nees F, Witt SH, Lourdusamy A, Vollstadt-Klein S, Steiner S, Poustka L, et al. Genetic risk for nicotine dependence in the cholinergic system and activation of the brain reward system in healthy adolescents. Neuropsychopharmacology. 2013;38(11):2081–2089. doi: 10.1038/npp.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherva R, Wilhelmsen K, Pomerleau CS, Chasse SA, Rice JP, Snedecor SM, et al. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103(9):1544–1552. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorice R, Bione S, Sansanelli S, Ulivi S, Athanasakis E, Lanzara C, et al. Association of a variant in the CHRNA5-A3-B4 gene cluster region to heavy smoking in the Italian population. Eur J Hum Genet. 2011;19(5):593–596. doi: 10.1038/ejhg.2010.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14(5):501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, DvdV A, Xu Q, Seneviratne C, Pomerleau OF, Pomerleau CS, et al. Significant associations of CHRNA2 and CHRNA6 with nicotine dependence in European American and African American populations. Hum Genet. 2014;133(5):575–586. doi: 10.1007/s00439-013-1398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103(17):1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17(5):724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]
- 47.Batel P, Houchi H, Daoust M, Ramoz N, Naassila M, Gorwood P. A haplotype of the DRD1 gene is associated with alcohol dependence. Alcohol Clin Exp Res. 2008;32(4):567–572. doi: 10.1111/j.1530-0277.2008.00618.x. [DOI] [PubMed] [Google Scholar]
- 48.Clarke TK, Weiss AR, Ferarro TN, Kampman KM, Dackis CA, Pettinati HM, et al. The dopamine receptor D2 (DRD2) SNP rs1076560 is associated with opioid addiction. Ann Hum Genet. 2014;78(1):33–39. doi: 10.1111/ahg.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doehring A, Hentig N, Graff J, Salamat S, Schmidt M, Geisslinger G, et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenet Genomics. 2009;19(6):407–414. doi: 10.1097/FPC.0b013e328320a3fd. [DOI] [PubMed] [Google Scholar]
- 50.Ho AM, Tang NL, Cheung BK, Stadlin A. Dopamine receptor D4 gene -521C/T polymorphism is associated with opioid dependence through cold-pain responses. Ann N Y Acad Sci. 2008;1139:20–26. doi: 10.1196/annals.1432.054. [DOI] [PubMed] [Google Scholar]
- 51.Mignini F, Napolioni V, Codazzo C, Carpi FM, Vitali M, Romeo M, et al. DRD2/ANKK1 TaqIA and SLC6A3 VNTR polymorphisms in alcohol dependence: association and gene-gene interaction study in a population of Central Italy. Neurosci Lett. 2012;522(2):103–107. doi: 10.1016/j.neulet.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Voisey J, Swagell CD, Hughes IP, van Daal A, Noble EP, Lawford BR, et al. A DRD2 and ANKK1 haplotype is associated with nicotine dependence. Psychiatry Res. 2012;196(2-3):285–289. doi: 10.1016/j.psychres.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 53.Zhu F, Yan CX, Wen YC, Wang J, Bi J, Zhao YL, et al. Dopamine D1 receptor gene variation modulates opioid dependence risk by affecting transition to addiction. PLoS One. 2013;8(8):e70805. doi: 10.1371/journal.pone.0070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.World Health Organization Cancer Pain Relief . With a Guide to Opioid Availability. 2. Geneva, Switzerland: World Health Organization; 1996. [Google Scholar]
- 55.Dev R, Parsons HA, Palla S, Palmer JL, Del Fabbro E, Bruera E. Undocumented alcoholism and its correlation with tobacco and illegal drug use in advanced cancer patients. Cancer. 2011;117(19):4551–4556. doi: 10.1002/cncr.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10(4):225–231. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cahill E, Salery M, Vanhoutte P, Caboche J. Convergence of dopamine and glutamate signaling onto striatal ERK activation in response to drugs of abuse. Front Pharmacol. 2014;4:172. doi: 10.3389/fphar.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brami-Cherrier K, Roze E, Girault JA, Betuing S, Caboche J. Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse. J Neurochem. 2009;108(6):1323–1335. doi: 10.1111/j.1471-4159.2009.05879.x. [DOI] [PubMed] [Google Scholar]
- 59.Valjent E, Pages C, Herve D, Girault JA, Caboche J. Addictive and non-addictive drugs induce distinct and specific patterns of ERK activation in mouse brain. Eur J Neurosci. 2004;19(7):1826–1836. doi: 10.1111/j.1460-9568.2004.03278.x. [DOI] [PubMed] [Google Scholar]
- 60.Pascoli V, Cahill E, Bellivier F, Caboche J, Vanhoutte P. Extracellular signal-regulated protein kinases 1 and 2 activation by addictive drugs: a signal toward pathological adaptation. Biological psychiatry 2014, 76(12):917–926. [DOI] [PubMed]
- 61.Knight T, Irving JA. Ras/Raf/MEK/ERK pathway activation in childhood acute lymphoblastic leukemia and its therapeutic targeting. Front Oncol. 2014;4:160. doi: 10.3389/fonc.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dafny N, Dougherty PM, Drath D. Immunosuppressive agent modulates the severity of opiate withdrawal. NIDA Res Monogr. 1990;105:553–555. [PubMed] [Google Scholar]
- 63.Hutchinson MR, Watkins LR. Why is neuroimmunopharmacology crucial for the future of addiction research? Neuropharmacology. 2014;76 Pt B:218–227. doi: 10.1016/j.neuropharm.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bland ST, Hutchinson MR, Maier SF, Watkins LR, Johnson KW. The glial activation inhibitor AV411 reduces morphine-induced nucleus accumbens dopamine release. Brain Behav Immun. 2009;23(4):492–497. doi: 10.1016/j.bbi.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hutchinson MR, Northcutt AL, Hiranita T, Wang X, Lewis SS, Thomas J, et al. Opioid activation of toll-like receptor 4 contributes to drug reinforcement. J Neurosci. 2012;32(33):11187–11200. doi: 10.1523/JNEUROSCI.0684-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reyes-Gibby CC, Spitz M, Wu X, Merriman K, Etzel C, Bruera E, et al. Cytokine genes and pain severity in lung cancer: exploring the influence of TNF-alpha-308 G/A IL6-174G/C and IL8-251T/A. Canc Epidemiol Biomarkers Prev. 2007;16(12):2745–2751. doi: 10.1158/1055-9965.EPI-07-0651. [DOI] [PubMed] [Google Scholar]
- 67.Reyes-Gibby CC, Wang J, Spitz M, Wu X, Yennurajalingam S, Shete S. Genetic variations in interleukin-8 and interleukin-10 are associated with pain, depressed mood, and fatigue in lung cancer patients. J Pain Symptom Manage. 2013;46(2):161–172. doi: 10.1016/j.jpainsymman.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reyes-Gibby CC, Swartz MD, Yu X, Wu X, Yennurajalingam S, Anderson KO, et al. Symptom clusters of pain, depressed mood, and fatigue in lung cancer: assessing the role of cytokine genes. Support Care Canc. 2013;21(11):3117–3125. doi: 10.1007/s00520-013-1885-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salles A, Romano A, Freudenthal R. Synaptic NF-kappa B pathway in neuronal plasticity and memory. Journal of physiology, Paris 2014, 108(4-6):256–262. [DOI] [PubMed]
- 70.Blendy JA, Maldonado R. Genetic analysis of drug addiction: the role of cAMP response element binding protein. J Mol Med (Berl) 1998;76(2):104–110. doi: 10.1007/s001090050197. [DOI] [PubMed] [Google Scholar]
- 71.Kumar K, Sharma S, Kumar P, Deshmukh R. Therapeutic potential of GABA(B) receptor ligands in drug addiction, anxiety, depression and other CNS disorders. Pharmacol Biochem Behav. 2013;110:174–184. doi: 10.1016/j.pbb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 72.Nickols HH, Conn PJ. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol Dis. 2014;61:55–71. doi: 10.1016/j.nbd.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rivero G, Gabilondo AM, Garcia-Fuster MJ, La Harpe R, Garcia-Sevilla JA, Meana JJ. Differential regulation of RGS proteins in the prefrontal cortex of short- and long-term human opiate abusers. Neuropharmacology. 2012;62(2):1044–1051. doi: 10.1016/j.neuropharm.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 74.Robison AJ. Emerging role of CaMKII in neuropsychiatric disease. Trends in neurosciences 2014, 37(11):653–662. [DOI] [PubMed]
- 75.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14(9):609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filip M, Frankowska M, Sadakierska-Chudy A, Suder A, Szumiec L, Mierzejewski P, et al. GABAB receptors as a therapeutic strategy in substance use disorders: focus on positive allosteric modulators. Neuropharmacology 2015, 88:36-47. [DOI] [PubMed]
- 77.Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17(4):445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao J, Liu X, Han S, Zhang CK, Liu Z, Li D. Association of the HTR2A gene with alcohol and heroin abuse. Hum Genet. 2014;133(3):357–365. doi: 10.1007/s00439-013-1388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Cho K, Singer BH, Zhang H. The nuclear transcription factor PKNOX2 is a candidate gene for substance dependence in European-origin women. PLoS One. 2011;6(1):e16002. doi: 10.1371/journal.pone.0016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desrivieres S, Krause K, Dyer A, Frank J, Blomeyer D, Lathrop M, et al. Nucleotide sequence variation within the PI3K p85 alpha gene associates with alcohol risk drinking behaviour in adolescents. PLoS One. 2008;3(3):e1769. doi: 10.1371/journal.pone.0001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Enoch MA, Gorodetsky E, Hodgkinson C, Roy A, Goldman D. Functional genetic variants that increase synaptic serotonin and 5-HT3 receptor sensitivity predict alcohol and drug dependence. Mol Psychiatry. 2011;16(11):1139–1146. doi: 10.1038/mp.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hill SY, Jones BL, Zezza N, Stiffler S. Family-based association analysis of alcohol dependence implicates KIAA0040 on Chromosome 1q in multiplex alcohol dependence families. Open J Genet. 2013;3(4):243–252. doi: 10.4236/ojgen.2013.34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalsi G, Kuo PH, Aliev F, Alexander J, McMichael O, Patterson DG, et al. A systematic gene-based screen of chr4q22-q32 identifies association of a novel susceptibility gene, DKK2, with the quantitative trait of alcohol dependence symptom counts. Hum Mol Genet. 2010;19(12):2497–2506. doi: 10.1093/hmg/ddq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, Alexander J, et al. Associations of glutamate decarboxylase genes with initial sensitivity and age-at-onset of alcohol dependence in the Irish Affected Sib Pair Study of Alcohol Dependence. Drug Alcohol Depend. 2009;101(1-2):80–87. doi: 10.1016/j.drugalcdep.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.London ED, Berman SM, Mohammadian P, Ritchie T, Mandelkern MA, Susselman MK, et al. Effect of the TaqIA polymorphism on ethanol response in the brain. Psychiatry Res. 2009;174(3):163–170. doi: 10.1016/j.pscychresns.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Munoz X, Amiano P, Celorrio D, Dorronsoro M, Sanchez MJ, Huerta JM, et al. Association of alcohol dehydrogenase polymorphisms and life-style factors with excessive alcohol intake within the Spanish population (EPIC-Spain) Addiction. 2012;107(12):2117–2127. doi: 10.1111/j.1360-0443.2012.03970.x. [DOI] [PubMed] [Google Scholar]
- 87.Novo-Veleiro I, Gonzalez-Sarmiento R, Cieza-Borrella C, Pastor I, Laso FJ, Marcos M. A genetic variant in the microRNA-146a gene is associated with susceptibility to alcohol use disorders. Eur Psychiatr. 2014;29(5):288–292. doi: 10.1016/j.eurpsy.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 88.Preuss UW, Ridinger M, Rujescu D, Samochowiec J, Fehr C, Wurst FM, et al. Association of ADH4 genetic variants with alcohol dependence risk and related phenotypes: results from a larger multicenter association study. Addict Biol. 2011;16(2):323–333. doi: 10.1111/j.1369-1600.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 89.Ray LA, Hutchison KE. Associations among GABRG1, level of response to alcohol, and drinking behaviors. Alcohol Clin Exp Res. 2009;33(8):1382–1390. doi: 10.1111/j.1530-0277.2009.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Samochowiec A, Grzywacz A, Kaczmarek L, Bienkowski P, Samochowiec J, Mierzejewski P, et al. Functional polymorphism of matrix metalloproteinase-9 (MMP-9) gene in alcohol dependence: family and case control study. Brain Res. 2010;1327:103–106. doi: 10.1016/j.brainres.2010.02.072. [DOI] [PubMed] [Google Scholar]
- 91.Schumann G, Johann M, Frank J, Preuss U, Dahmen N, Laucht M, et al. Systematic analysis of glutamatergic neurotransmission genes in alcohol dependence and adolescent risky drinking behavior. Arch Gen Psychiatry. 2008;65(7):826–838. doi: 10.1001/archpsyc.65.7.826. [DOI] [PubMed] [Google Scholar]
- 92.Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, et al. Genetic association of the human corticotropin releasing hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Mol Psychiatry. 2006;11(6):594–602. doi: 10.1038/sj.mp.4001813. [DOI] [PubMed] [Google Scholar]
- 93.Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. 2013;18(11):1218–1224. doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xuei X, Flury-Wetherill L, Dick D, Goate A, Tischfield J, Nurnberger J, Jr, et al. GABRR1 and GABRR2, encoding the GABA-A receptor subunits rho1 and rho2, are associated with alcohol dependence. Am J Med Genet Part B Neuropsychiatr Genet. 2010;153B(2):418–427. doi: 10.1002/ajmg.b.30995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang J, Li MD. Association and interaction analyses of 5-HT3 receptor and serotonin transporter genes with alcohol, cocaine, and nicotine dependence using the SAGE data. Hum Genet. 2014;133(7):905–918. doi: 10.1007/s00439-014-1431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agrawal A, Pergadia ML, Saccone SF, Hinrichs AL, Lessov-Schlaggar CN, Saccone NL, et al. Gamma-aminobutyric acid receptor genes and nicotine dependence: evidence for association from a case-control study. Addiction. 2008;103(6):1027–1038. doi: 10.1111/j.1360-0443.2008.02236.x. [DOI] [PubMed] [Google Scholar]
- 97.Agrawal A, Pergadia ML, Balasubramanian S, Saccone SF, Hinrichs AL, Saccone NL, et al. Further evidence for an association between the gamma-aminobutyric acid receptor A, subunit 4 genes on chromosome 4 and Fagerstrom Test for Nicotine Dependence. Addiction. 2009;104(3):471–477. doi: 10.1111/j.1360-0443.2008.02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anney RJ, Lotfi-Miri M, Olsson CA, Reid SC, Hemphill SA, Patton GC. Variation in the gene coding for the M5 muscarinic receptor (CHRM5) influences cigarette dose but is not associated with dependence to drugs of addiction: evidence from a prospective population based cohort study of young adults. BMC Genet. 2007;8:46. doi: 10.1186/1471-2156-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berrettini WH, Doyle GA. The CHRNA5-A3-B4 gene cluster in nicotine addiction. Mol Psychiatry. 2012;17(9):856–866. doi: 10.1038/mp.2011.122. [DOI] [PubMed] [Google Scholar]
- 100.Beuten J, Ma JZ, Payne TJ, Dupont RT, Quezada P, Huang W, et al. Significant association of BDNF haplotypes in European-American male smokers but not in European-American female or African-American smokers. Am Jo Med Genet Part B Neuropsychiatr Genet. 2005;139B(1):73–80. doi: 10.1002/ajmg.b.30231. [DOI] [PubMed] [Google Scholar]
- 101.Beuten J, Ma JZ, Payne TJ, Dupont RT, Crews KM, Somes G, et al. Single- and multilocus allelic variants within the GABA(B) receptor subunit 2 (GABAB2) gene are significantly associated with nicotine dependence. Am J Hum Genet. 2005;76(5):859–864. doi: 10.1086/429839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beuten J, Payne TJ, Ma JZ, Li MD. Significant association of catechol-O-methyltransferase (COMT) haplotypes with nicotine dependence in male and female smokers of two ethnic populations. Neuropsychopharmacology. 2006;31(3):675–684. doi: 10.1038/sj.npp.1300997. [DOI] [PubMed] [Google Scholar]
- 103.Chen X, Williamson VS, An SS, Hettema JM, Aggen SH, Neale MC, et al. Cannabinoid receptor 1 gene association with nicotine dependence. Arch Gen Psychiatry. 2008;65(7):816–824. doi: 10.1001/archpsyc.65.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Docampo E, Ribases M, Gratacos M, Bruguera E, Cabezas C, Sanchez-Mora C, et al. Association of neurexin 3 polymorphisms with smoking behavior. Genes Brain Behav. 2012;11(6):704–711. doi: 10.1111/j.1601-183X.2012.00815.x. [DOI] [PubMed] [Google Scholar]
- 105.Ella E, Sato N, Nishizawa D, Kageyama S, Yamada H, Kurabe N, et al. Association between dopamine beta hydroxylase rs5320 polymorphism and smoking behaviour in elderly Japanese. J Hum Genet. 2012;57(6):385–390. doi: 10.1038/jhg.2012.40. [DOI] [PubMed] [Google Scholar]
- 106.Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, Inohara N, et al. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African-American sample. Neuropsychopharmacology. 2009;34(2):319–330. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- 107.Lang UE, Sander T, Lohoff FW, Hellweg R, Bajbouj M, Winterer G, et al. Association of the met66 allele of brain-derived neurotrophic factor (BDNF) with smoking. Psychopharmacology (Berl) 2007;190(4):433–439. doi: 10.1007/s00213-006-0647-1. [DOI] [PubMed] [Google Scholar]
- 108.Liu YZ, Pei YF, Guo YF, Wang L, Liu XG, Yan H, et al. Genome-wide association analyses suggested a novel mechanism for smoking behavior regulated by IL15. Mol Psychiatry. 2009;14(7):668–680. doi: 10.1038/mp.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma JZ, Beuten J, Payne TJ, Dupont RT, Elston RC, Li MD. Haplotype analysis indicates an association between the DOPA decarboxylase (DDC) gene and nicotine dependence. Hum Mol Genet. 2005;14(12):1691–1698. doi: 10.1093/hmg/ddi177. [DOI] [PubMed] [Google Scholar]
- 110.Mobascher A, Rujescu D, Mittelstrass K, Giegling I, Lamina C, Nitz B, et al. Association of a variant in the muscarinic acetylcholine receptor 2 gene (CHRM2) with nicotine addiction. Am J Med Genet Part B Neuropsychiatr Genet. 2010;153B(2):684–690. doi: 10.1002/ajmg.b.31011. [DOI] [PubMed] [Google Scholar]
- 111.Clarke TK, Krause K, Li T, Schumann G. An association of prodynorphin polymorphisms and opioid dependence in females in a Chinese population. Addict Biol. 2009;14(3):366–370. doi: 10.1111/j.1369-1600.2009.00151.x. [DOI] [PubMed] [Google Scholar]
- 112.Crist RC, Ambrose-Lanci LM, Zeng A, Yuan C, Kampman KM, Pettinati HM, et al. Case-control association study of WLS variants in opioid and cocaine addicted populations. Psychiatry Res. 2013;208(1):62–66. doi: 10.1016/j.psychres.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Cid R, Fonseca F, Gratacos M, Gutierrez F, Martin-Santos R, Estivill X, et al. BDNF variability in opioid addicts and response to methadone treatment: preliminary findings. Genes Brain Behav. 2008;7(5):515–522. doi: 10.1111/j.1601-183X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 114.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Herman AI, Kranzler HR, Cubells JF, Gelernter J, Covault J. Association study of the CNR1 gene exon 3 alternative promoter region polymorphisms and substance dependence. Am J Med Genet Part B Neuropsychiatr Genet. 2006;141B(5):499–503. doi: 10.1002/ajmg.b.30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kumar D, Deb I, Chakraborty J, Mukhopadhyay S, Das S. A polymorphism of the CREB binding protein (CREBBP) gene is a risk factor for addiction. Brain Res. 2011;1406:59–64. doi: 10.1016/j.brainres.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 117.Levran O, Peles E, Hamon S, Randesi M, Adelson M, Kreek MJ. CYP2B6 SNPs are associated with methadone dose required for effective treatment of opioid addiction. Addict Biol. 2013;18(4):709–716. doi: 10.1111/j.1369-1600.2011.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu Z, Guo X, Jiang Y, Zhang H. NCK2 is significantly associated with opiates addiction in African-origin men. Sci World J. 2013;2013:748979. doi: 10.1155/2013/748979. [DOI] [PMC free article] [PubMed] [Google Scholar]


