Age regulates plant regenerative capacity.
Abstract
Plant cells are totipotent and competent to regenerate from differentiated organs. It has been shown that two phytohormones, auxin and cytokinin, play critical roles within this process. As in animals, the regenerative capacity declines with age in plants, but the molecular basis for this phenomenon remains elusive. Here, we demonstrate that an age-regulated microRNA, miR156, regulates shoot regenerative capacity. As a plant ages, the gradual increase in miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors leads to the progressive decline in shoot regenerative capacity. In old plants, SPL reduces shoot regenerative capacity by attenuating the cytokinin response through binding with the B-type ARABIDOPSIS RESPONSE REGULATORs, which encode the transcriptional activators in the cytokinin signaling pathway. Consistently, the increased amount of exogenous cytokinin complements the reduced shoot regenerative capacity in old plants. Therefore, the recruitment of age cues in response to cytokinin contributes to shoot regenerative competence.
INTRODUCTION
Regeneration of a multicellular organism from a piece of adult somatic tissue is a prevalent phenomenon that occurs in both plants and animals (Birnbaum and Sánchez Alvarado, 2008). In contrast with animal cells, plant cells have been thought to maintain totipotency, and most plant tissues from already differentiated organs are able to regenerate whole plants under proper in vitro culture conditions (Duclercq et al., 2011; Sugimoto et al., 2011). It is well known that the ratio of two phytohormones, auxin and cytokinin, determines the developmental fate of regenerating tissue. A high cytokinin:auxin ratio directs regeneration of the shoot, whereas a low cytokinin:auxin ratio induces root differentiation (Skoog and Miller, 1957). However, little is understood concerning the mechanisms by which the auxin/cytokinin balance exerts these opposite effects. In addition, how cytokinin promotes the specification of apical/shoot fate during shoot regeneration remains elusive.
Cytokinin signal transduction involves a multistep phosphorelay signaling cascade from ligand perception at the cell membrane to transcriptional activation in the nucleus (Hwang et al., 2012; Kieber and Schaller, 2014). In Arabidopsis thaliana, cytokinin is perceived by the cytokinin receptors ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and AHK4. Ligand binding triggers autophosphorylation at a conserved His residue in the receiver domain and subsequent transfer of the phosphoryl group to a conserved Asp residue in the attached transmitter domain. The phosphoryl group on the Asp residue is then passed on to one of five ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER proteins and then to a group of nucleus-localized B-type ARABIDOPSIS RESPONSE REGULATORs (ARRs). B-type ARRs activate the expression of cytokinin-responsive genes and A-type ARRs. A-type ARRs, in turn, interfere with the function of B-type ARR proteins through protein-protein interaction, which establishes a negative feedback loop to the signaling pathway (Werner and Schmülling, 2009; Hwang et al., 2012).
An interesting and common phenomenon in animals is the progressive reduction in regenerative capacity. For example, remyelination, a regenerative process that produces new myelin sheaths from adult stem cells in the central nervous system, declines with increasing age (Ruckh et al., 2012). Similarly, the mammalian heart appears to have the capacity to regenerate only within a brief period after birth (Porrello et al., 2011). To what extent and by which means age contributes to plant regenerative capacity is unknown.
miR156, which targets SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors, governs the age pathway in plants. The level of miR156, in response to endogenous sugar, gradually decreases with time (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009; Poethig, 2013; Yang et al., 2013; Yu et al., 2013). The onset of adult phase is defined by miR156 level: overexpression of miR156 prolongs the juvenile phase, whereas a reduction in miR156 activity leads to an accelerated expression of adult traits (Wu et al., 2009). It has been shown that miR156-targeted SPLs regulate diverse age-related developmental processes, such as embryonic pattern formation, juvenile-to-adult phase transition, flowering time, inflorescence trichome initiation, and anthocyanin biosynthesis (Wang et al., 2009; Wu et al., 2009; Nodine and Bartel, 2010; Yu et al., 2010; Gou et al., 2011; Bergonzi et al., 2013; Zhou et al., 2013; Rubio-Somoza et al., 2014).
Here, we show that old plants exhibit lower shoot regenerative capacity than young plants, largely due to the reduced cytokinin response. Our mutant characterizations, expression analyses, and protein-protein interaction assays further indicate that the increased level of miR156-targeted SPLs in old plants dampens shoot regeneration by interfering with the function of B-type ARRs, thus establishing a molecular link between developmental timing and cytokinin-mediated shoot regeneration.
RESULTS
The Progressive Decline in Shoot Regenerative Capacity with Age
To reveal whether the shoot regenerative capacity is changed as plants age, we performed in vitro regeneration assays using Arabidopsis and tobacco (Nicotiana tabacum) leaves. In Arabidopsis, shoot regeneration requires two steps: in the first step, callus, a pluripotent cell mass, is formed from explants on auxin-rich callus-inducing medium (CIM). Subsequently, culture of the callus on shoot-inducing medium (SIM), which contains a high cytokinin:auxin ratio, induces the differentiation of callus into shoot (Duclercq et al., 2011). By contrast, tobacco can be regenerated directly from leaf dics, which enables us to eliminate the effect of callus induction on shoot regeneration.
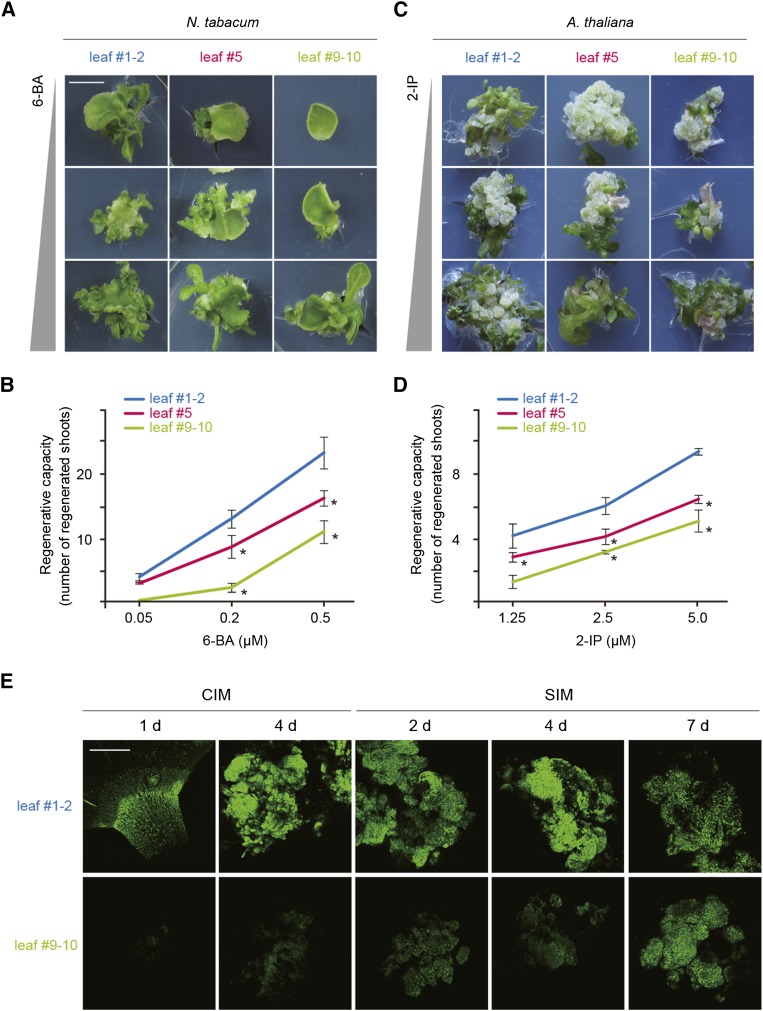
We compared shoot regenerative rates of the first/second (early), fifth (mid), and ninth/tenth (late) tobacco leaves. To avoid the impact of leaf age on regenerative capacity, leaves of the same developmental stage (1 cm in length) were used. In the absence of 6-benzylaminopurine (6-BA), a synthetic cytokinin, none of the leaf dics was competent to regenerate (Supplemental Figure 1A). Early tobacco leaves exhibited a higher regenerative rate than late leaves on Murashige and Skoog (MS) medium supplemented with 6-BA (Figures 1A and 1B). In Arabidopsis, the callus inductive rate was not changed as plants aged (Supplemental Figure 1B). However, the same difference in regenerative rate between early and late leaves was observed (Figures 1C and 1D). Thus, the shoot regenerative capacity was inversely correlated with plant age.
Figure 1.
The Developmental Decline in Shoot Regenerative Capacity.
(A) and (C) Shoot regeneration of tobacco (A) and Arabidopsis (C). The leaves from plants of different ages were used as explants for regeneration assays. For tobacco, shoots were induced on MS medium supplemented with 6-BA of different concentrations. For Arabidopsis, calli were induced from explants on CIM and shoots were then regenerated on MS medium supplemented with 0.9 μM indole-3-acetic acid (an auxin) and 2-isopentenyladenine (2-IP; a cytokinin) of different concentrations. Bar = 0.5 cm.
(B) and (D) Quantitative analyses of regenerative capacity in tobacco (B) and Arabidopsis (D). The regenerative rate was represented by the number of regenerated shoots. Nine (B) or eight (D) explants were examined. Asterisks indicate significant differences from leaf #1-2 (Student’s t test, P < 0.05). Data are means ± sd of three biological replicates.
(E) Visualization of the ProTCS:GFP reporter in explants cultured on CIM or SIM. Explants derived from early or late leaves were examined (n = 10). The number of days after transfer to CIM or SIM is indicated. Bar = 400 μm.
Shoot regeneration is directed by a high ratio of cytokinin to auxin. The regenerative rates were elevated as cytokinin level increased in both tobacco and Arabidopsis (Figures 1A to 1D). Notably, the regenerative rate of the late tobacco leaves on MS medium supplemented with 0.5 μM 6-BA was 5-fold higher than that on MS medium with 0.2 μM 6-BA (Figure 1C). Therefore, the increased amount of exogenous cytokinin complements the reduced regenerative capacity in old plants. In agreement with this observation, the expression of ProTCS:GFP (for green fluorescent protein), a synthetic cytokinin reporter (Müller and Sheen, 2008), was lower in late leaves than in early leaves on CIM and SIM (Figure 1E).
miR156 Contributes to Developmental Decline in Shoot Regenerative Capacity
miR156, which targets SPL transcription factors, regulates the juvenile-to-adult phase transition in many plant species, including Arabidopsis, Arabis alpina, Cardamine flexuosa, Populus × canadensis, and Zea mays (Chuck et al., 2007; Wu et al., 2009; Wang et al., 2011; Bergonzi et al., 2013; Zhou et al., 2013). In tobacco, miR156 level was correlated with age, being most abundant in seedlings (Supplemental Figure 2A). To determine whether miR156 plays a role in shoot regeneration in tobacco, we generated transgenic plants that overexpressed either miR156 (Pro35S:MIR156) or a target mimic of miR156 (Pro35S:MIM156), in which miR156 is inactivated (Supplemental Figures 2B to 2D) (Franco-Zorrilla et al., 2007).
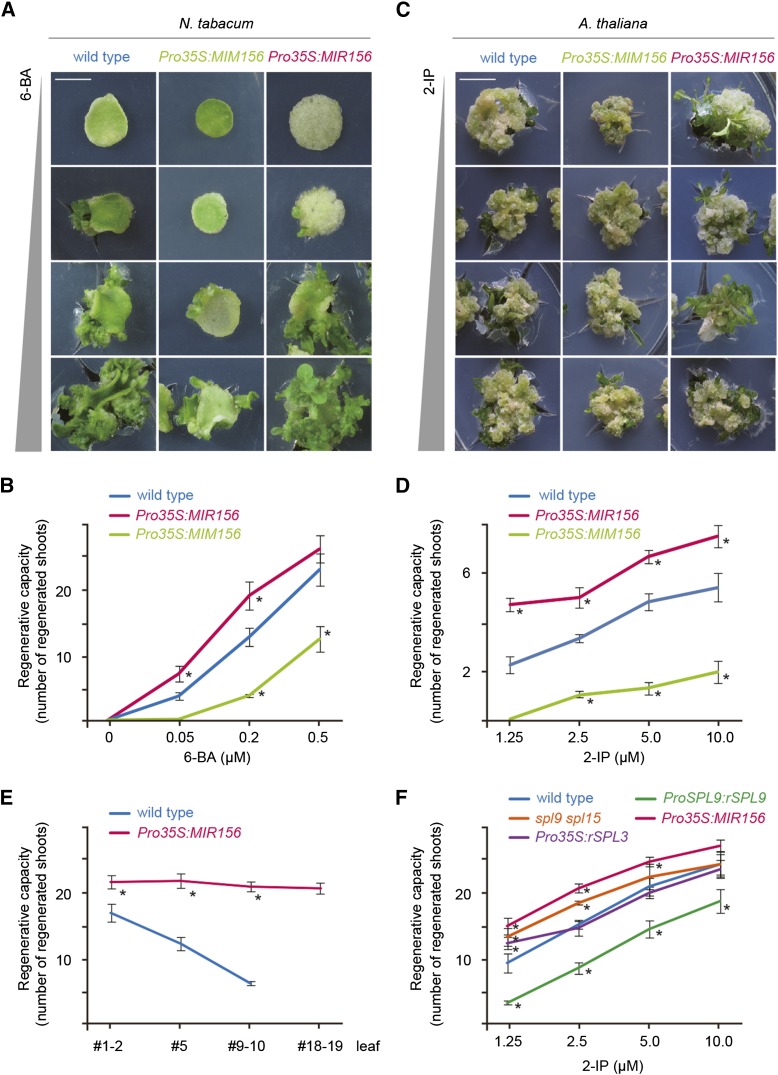
We performed regeneration assays using the fifth (mid) tobacco leaf. Compared with the wild type, Pro35S:MIR156 exhibited an increased regenerative capacity, whereas Pro35S:MIM156 exhibited a reduced regenerative capacity (Figures 2A and 2B). In addition, overexpression of miR156 suppressed the developmental decline in shoot regeneration that was evident in the wild type (Figure 2E; Supplemental Figure 3A). In the fourth (mid) Arabidopsis leaf, there was no difference in callus inductive rate between the wild type and Pro35S:MIR156 or Pro35S:MIM156. However, we observed a similar correlation between mR156 level and shoot regenerative capacity (Figures 2C and 2D). It has been shown that the manipulation of mR156 level in Arabidopsis results in a dramatic change in leaf morphology (Wang et al., 2008; Wu et al., 2009). We obtained comparable results using wild-type, Pro35S:MIR156, and Pro35S:MIM156 hypocotyls as explants, eliminating the effect of leaf morphology or leaf age on shoot regenerative capacity (Supplemental Figures 3B and 3C). Furthermore, elevation of cytokinin concentration in SIM rescued the low regenerative capacity of Pro35S:MIM156 plants (Figures 2B and 2D). Taken together, these results indicate that miR156 contributes to shoot regenerative competence.
Figure 2.
miR156 Regulates Shoot Regenerative Capacity.
(A) and (C) Shoot regeneration of wild-type, Pro35S:MIR156, and Pro35S:MIM156 plants. The fifth tobacco leaf (A) and the fourth Arabidopsis leaf (C) were used as explants for regeneration assays. 2-IP, 2-isopentenyladenine. Bars = 0.5 cm.
(B) and (D) Quantitative analyses of regenerative capacity in tobacco (B) and Arabidopsis (D). Nine (B) or eight (D) explants were examined. Asterisks indicate significant differences from the wild type (Student’s t test, P < 0.05).
(E) miR156 overexpression suppressed the progressive decline in shoot regeneration in tobacco. Leaves from plants of different ages were used as the explants for regeneration assays. Shoots were induced on MS medium supplemented with 0.2 μM 6-BA. Nine explants were examined. Asterisks indicate significant differences from the wild type (Student’s t test, P < 0.05).
(F) Roles of SPL3 and SPL9 group genes in shoot regeneration. Eight explants were examined. Asterisks indicate significant differences from the wild type (Student’s t test, P < 0.05).
SPL9-Group Genes, but Not SPL3-Group Genes, Regulate Shoot Regeneration
miR156-targeted SPLs can be structurally divided into two groups, represented by SPL3 (including SPL3, SPL4, and SPL5) and SPL9 (including SPL2, SPL6, SPL9, SPL10, SPL11, SPL13, and SPL15) (Xing et al., 2010). SPL9 differs from SPL3 because it harbors a C-terminal domain responsible for protein-protein interaction (Yu et al., 2012). To determine whether both groups of genes play a role in shoot regeneration, we examined the shoot regenerative rate of the transgenic plants that overexpressed miR156-nontargetable SPL3 (rSPL3) and rSPL9 (Pro35S:rSPL3 and ProSPL9:rSPL9). Pro35S:rSPL3 exhibited the same regenerative rate as the wild type, whereas ProSPL9:rSPL9 markedly impaired shoot regeneration (Figure 2F), indicating that SPL9 group genes, but not SPL3, regulate shoot regenerative capacity. Earlier work revealed that SPL9 and SPL15 play major but redundant roles within the SPL9 group (Schwarz et al., 2008; Wang et al., 2008). The shoot regenerative capacity of the spl9-1 spl15-2 double mutant was higher than that of the wild type but lower than that of the miR156 overexpressor (Figure 2F), suggesting the functional redundancy of miR156-targeted SPL9 group genes in shoot regeneration.
miR156 Regulates Shoot Regenerative Capacity through Modulating the Cytokinin Response
The high regenerative capacity of Pro35S:MIR156 plants can result from a high concentration of cytokinin in vivo. Cytokinin measurement revealed that Pro35S:MIR156 accumulated the same amount of cytokinin as the wild type (Supplemental Table 1), suggesting that miR156 does not regulate shoot regenerative capacity through modulating endogenous cytokinin levels.
Cytokinin signaling transduction involves a multistep phosphorelay signaling cascade from ligand perception at the cell membrane to transcriptional activation in the nucleus. In Arabidopsis, four B-type ARR transcription factors, ARR1, ARR2, ARR10, and ARR12, play essential roles in cytokinin signaling transduction (Hwang and Sheen, 2001; Mason et al., 2005; Ishida et al., 2008). Compared with the wild type, the regenerative capacity was markedly reduced in arr2-4 arr12-1 and completely lost in the arr1-3 arr10-5 arr12-1 mutant. Pro35S:MIR156 did not restore the impaired shoot regenerative capacity of arr2-4 arr12-1 and arr1-3 arr10-5 arr12-1 (Supplemental Figure 4), suggesting that miR156 and its target SPLs modulate shoot regenerative capacity through B-type ARRs.
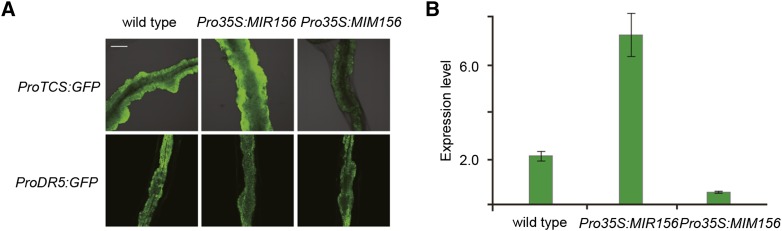
Compared with the wild type, ProTCS:GFP expression was elevated in Pro35S:MIR156 but reduced in Pro35S:MIM156 (Figures 3A and 3B). By contrast, the auxin response, as visualized by the ProDR5:GFP reporter (Sabatini et al., 1999), was not changed in either Pro35S:MIR156 or Pro35S:MIM156 (Figure 3A), indicating that miR156-SPL does not regulate shoot regeneration through modulating the auxin response.
Figure 3.
miR156 Regulates Shoot Regenerative Capacity through Modulating the Cytokinin Response.
(A) Visualization of ProTCS:GFP and ProDR5:GFP reporters 2 d after transfer to SIM. Explants derived from hypocotyls were examined (n = 10). Bar = 200 μm.
(B) Expression of ProTCS:GFP. Expression of GFP, as examined by quantitative real-time PCR, was normalized to that of TUB. The expression level in the wild type was set to 1.0. Data are means ± sd of three biological replicates.
miR156-Targeted SPL Binds to B-Type ARRs
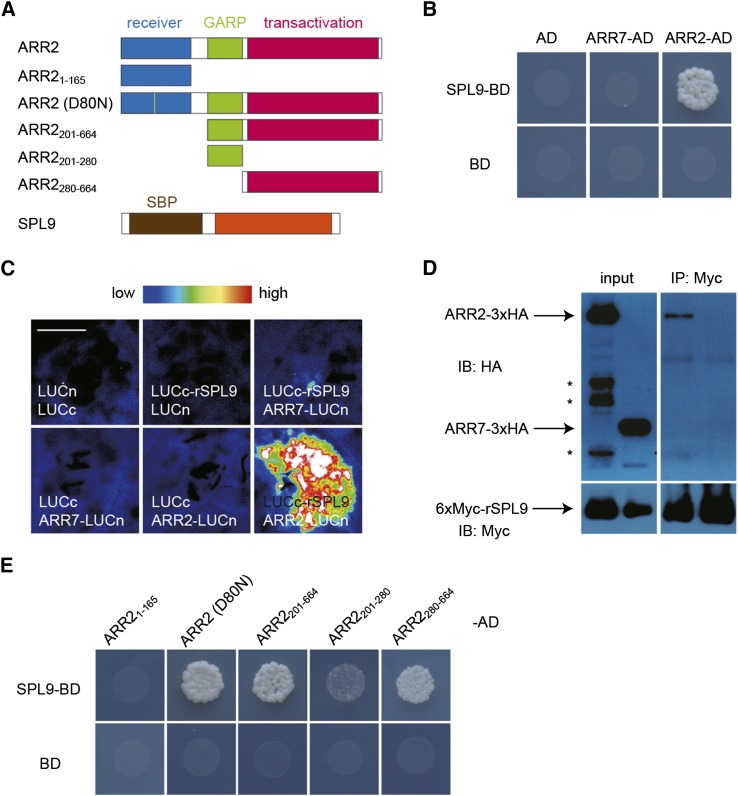
The above results suggest that miR156 regulates shoot regenerative capacity through modulating the cytokinin response rather than the cytokinin level. Therefore, we proceeded to determine whether SPLs affect the cytokinin response through binding to B-type ARRs. Yeast two-hybrid analyses showed that SPL9, as well as two other SPLs belonging to the SPL9 group, SPL2 and SPL10, bound to B-type ARRs including ARR1, ARR2, ARR10, and ARR12 but not to an A-type ARR such as ARR7 (Figure 4B; Supplemental Figure 5). Consistent with the negligible role of SPL3 in shoot regeneration, we did not observe a direct interaction between SPL3 and B-type ARRs in yeast (Supplemental Figure 5A).
Figure 4.
miR156-Targeted SPL Binds to B-Type ARR.
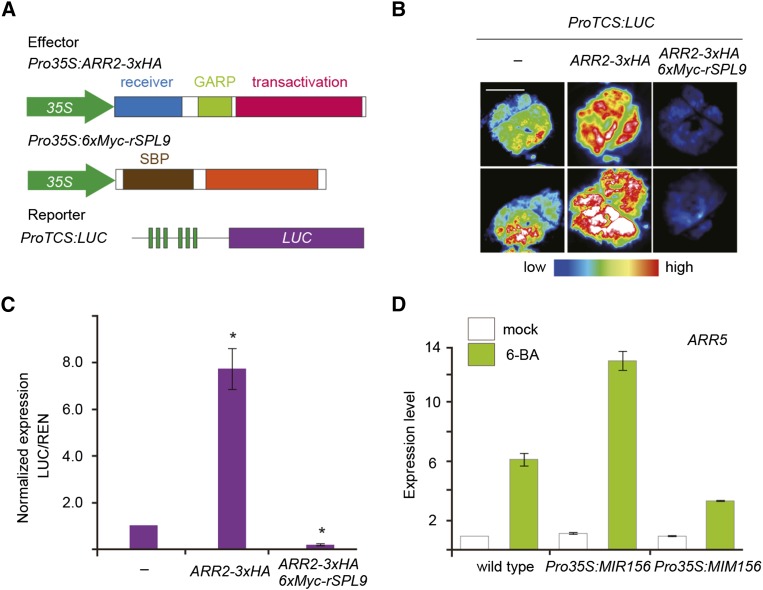
(A) Diagrams of ARR2 and SPL9 constructs for interaction studies. The receiver, GARP, and transactivation domains of ARR2 are shown. The brown and orange boxes indicate the SBP domain and the C-terminal protein-protein interaction domain of SPL9, respectively.
(B) Yeast two-hybrid assay. ARR2 and ARR7 were fused to the GAL4 activation domain (AD) and SPL9 was fused to the GAL4 DNA binding domain (BD). Interaction assays were performed on an SD-Leu-Trp-His plate with 25 mM 3-amino-1,2,4-triazole.
(C) BiLC assay in N. benthamiana leaves. ARR2 or ARR7 was fused to the N-terminal domain of LUC (LUCn) and rSPL9 was fused to the C-terminal domain of LUC (LUCc). Bar = 0.5 cm.
(D) CoIP assay. ARR2-3xHA and 6xMyc-rSPL9 or ARR7-3xHA and 6xMyc-rSPL9 fusion proteins were transiently expressed in N. benthamiana leaves. Protein extract was immunoprecipitated (IP) with anti-Myc agarose beads and immunoblotted (IB) against anti-HA or anti-Myc antibody. Asterisks indicate degraded products of ARR2-3xHA.
(E) Yeast two-hybrid assay. The mutated or truncated form of ARR2 was fused to the GAL4-AD domain. Interaction assays were performed on an SD-Leu-Trp-His plate with 25 mM 3-amino-1,2,4-triazole.
To confirm the interaction between ARR2 and SPL9, we performed bimolecular luminescence complementation (BiLC) assays in Nicotiana benthamiana. To this end, ARR2, ARR7, and rSPL9 were fused to the N- or C-terminal domain of LUCIFERASE (LUC). The reconstituted luminescence was observed in leaves that coexpressed Pro35S:ARR2-LUCn and Pro35S:LUCc-rSPL9. By contrast, leaves infiltrated with combinations of Pro35S:LUCc and Pro35S:ARR2-LUCn, Pro35S:LUCc-rSPL9 and Pro35S:LUCn, or Pro35S:LUCc-rSPL9 and Pro35S:ARR7-LUCn barely produced LUC signals (Figure 4C). Using the same BiLC assays, we observed direct interaction between SPL9 and ARR1, ARR10, or ARR12 (Supplemental Figure 6). Coimmunoprecipitation (CoIP) experiments showed that 3× hemagglutinin (HA)-tagged ARR1 (ARR1-3xHA) or ARR2 (ARR2-3xHA) protein, but not 3× HA-tagged ARR7 (ARR7-3xHA), strongly interacted with 6xMyc-tagged rSPL9 (6xMyc-rSPL9) in vivo (Figure 4D; Supplemental Figure 7).
ARR2 harbors three structural domains: a receiver domain at the N terminus, a GRAP DNA binding domain in the middle, and a transactivation domain at the C terminus (Figure 4A) (Heyl and Schmülling, 2003). The transactivation domain of ARR2 was able to bind to SPL9, whereas the deletion of the receiver domain or DNA binding domain had no effect on the SPL9 interaction (Figure 4E). B-type ARRs are phosphorylated at Asp in the receiver domain in response to cytokinin treatment (Hwang et al., 2012). To test whether the binding of ARR2 with SPL9 requires its phosphorylation, we mutated Asp-80 in ARR2 to Asn (D80N; Figure 4A). Both wild-type and mutated ARR2 (D80N) bound to SPL9 in yeast (Figure 4E), suggesting that the phosphorylation status does not play a role in the ARR2-SPL interaction.
SPL9 Impairs the Cytokinin Response and Shoot Regeneration
To investigate whether the binding of SPL9 to ARR2 affects the cytokinin response, we performed a tobacco transient assay with ProTCS:LUC (Figure 5A). Overexpression of ARR2-3xHA (Pro35S:ARR2-3xHA) resulted in a 7.5-fold enhancement in LUC activity. The LUC signal was substantially suppressed when 6xMyc-rSPL9 was overexpressed (Pro35S:6xMyc-rSPL9) (Figures 5B and 5C; Supplemental Figure 8A). Notably, LUC expression in Pro35S:ARR2-3xHA Pro35S:6xMyc-rSPL9 was much lower than that in the mock-treated sample, suggesting that SPL9 overexpression was sufficient to inhibit the endogenous cytokinin response in tobacco leaves. Similar results were obtained using the combinations Pro35S:ARR1-3xHA/Pro35S:6xMyc-rSPL9, Pro35S:ARR10-3xHA/Pro35S:6xMyc-rSPL9, and Pro35S:ARR12-3xHA/Pro35S:6xMyc-rSPL9 (Supplemental Figure 9). In Arabidopsis, consistent with these observations, the induction of A-type ARRs, including ARR5, ARR6, and ARR15, by 6-BA was elevated in Pro35S:MIR156 but reduced in Pro35S:MIM156 (Figure 5D; Supplemental Figures 8B and 8C).
Figure 5.
miR156-Targeted SPL Regulates the Cytokinin Response.
(A) Diagrams of effector and reporter constructs for transactivation studies. The green boxes in the ProTCS:LUC construct indicate B-type ARR binding sites.
(B) Transactivation assays. N. benthamiana leaves were infiltrated with different combinations of effector and reporter. Bar = 0.5 cm.
(C) Quantitative analyses of LUC activity in N. benthamiana leaves. Pro35S:REN was used as an internal control. Quantification was performed by normalizing LUC activity to that of REN. The LUC activity in ProTCS:LUC without effector was set to 1.0. Data are means ± sd of three biological replicates. Asterisks indicate significant differences from mock sample (Student’s t test, P < 0.05).
(D) Expression of ARR5. Seven-day-old Arabidopsis seedlings were treated with DMSO (mock) or 5 μM 6-BA for 40 min. The expression of ARR5 in mock wild-type sample was set to 1.0. Data are means ± sd of three biological replicates.
We then performed the same experiment using ProDR5:LUC as reporter. In agreement with the above findings that the auxin response was not changed in either Pro35S:MIR156 or Pro35S:MIM156 explants (Figure 3A), overexpression of ARR2 and SPL9 did not affect DR5 expression (Supplemental Figure 10).
To further investigate the repression of ARR2 transcriptional activity by SPL9, we generated an effector in which the transactivation domain of ARR2 (ΔARR2) was fused to the DNA binding domain of yeast GAL4 (Pro35S:BD-ΔARR2) and a Pro6xUAS:LUC reporter in which LUC was expressed from six repeats of the GAL4 target sequence (Supplemental Figure 11A). The LUC signal was markedly elevated by Pro35S:BD-ΔARR2 but not by Pro35S:ΔARR2, which did not harbor GAL4-BD. The activation of LUC by Pro35S:BD-ΔARR2 was suppressed when 6xMyc-rSPL9 was coexpressed (Supplemental Figures 11B and 11C).
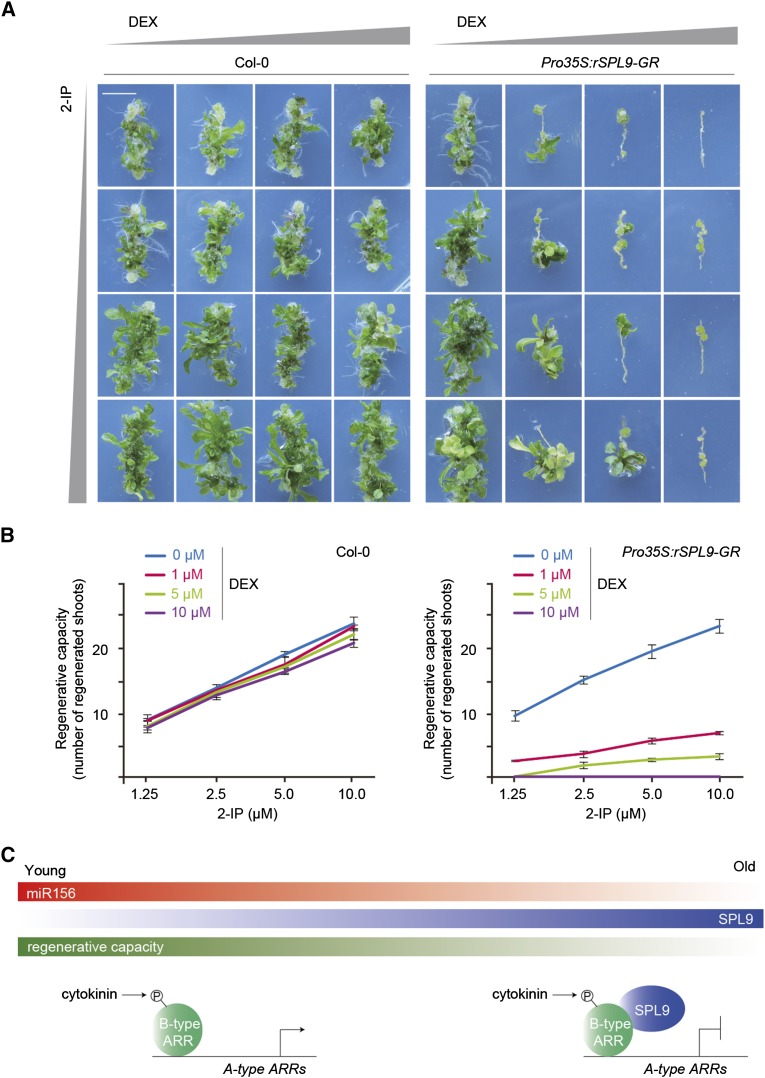
To explore the role of SPL9 in shoot regeneration, we compared the regenerative capacity among the wild type, the spl9-1 spl15-2 double mutant, and Pro35S:rSPL9-GR transgenic plants, in which rSPL9 was fused to the hormone binding domain of rat glucocorticoid receptor (GR) and expressed from the 35S promoter (Wu et al., 2009). Treatment with the steroid hormone ligand dexamethasone (DEX), which leads to a translocation of the rSPL9-GR fusion protein from the cytoplasm to the nucleus, resulted in the same phenotype as the transgenic plants expressing rSPL9 (ProSPL9:rSPL9) (Wu et al., 2009). The spl9-1 spl15-2 double mutant exhibited higher regenerative capacity than the wild type. Notably, shoot regeneration of the spl9-1 spl15-2 double mutant was not affected by DEX treatment (Figures 6A and 6B; Supplemental Figure 12). We observed an inverse correlation between the concentration of DEX (i.e., the level of rSPL9-GR fusion protein in the nucleus) and shoot regenerative capacity (Figures 6A and 6B). The addition of 10 μM DEX in SIM resulted in a complete loss of shoot regenerative capacity at all the hormone concentrations tested. At lower concentrations of DEX, the increased level of 6-BA complemented the reduced shoot regenerative capacity caused by rSPL9-GR.
Figure 6.
miR156-Targeted SPL Regulates Regenerative Capacity.
(A) Shoot regeneration assay using the wild type (Columbia-0 [Col-0]) and Pro35S:rSPL9-GR. The hypocotyls were cultured on CIM and then transferred to MS medium supplemented with different concentrations of 2-isopentenyladenine (2-IP) and DEX. Bar = 0.5 cm.
(B) Quantitative analyses of regenerative capacity. The regenerative rate was represented by the number of regenerated shoots on six explants. Data are means ± sd of three biological replicates.
(C) Model for the regulation of shoot regenerative capacity by a microRNA timer. In old plants, the amount of SPL9 is increased due to the developmental decline of miR156. SPL9 inhibits the transcriptional activity of B-type ARRs, thereby reducing the shoot regenerative capacity (see text for details).
To confirm the role of the ARR2-SPL9 interaction in shoot regeneration, we performed regeneration assays using ProSPL9:SPL9, Pro35S:ARR2, and Pro35S:ARR2 ProSPL9:SPL9 plants. Consistent with previous findings, Pro35S:ARR2 showed elevated regenerative capacity in comparison with the wild type (Hwang and Sheen, 2001). Pro35S:ARR2 rescued the reduced shoot regenerative capacity of ProSPL9:SPL9 (Supplemental Figure 13).
DISCUSSION
Our results reveal an important role of miR156, the master regulator of juvenility, in shoot regeneration (Figure 6C). Young plants exhibit a high cytokinin response and regenerative capacity. As a plant ages, miR156 levels decline, alleviating the repression of its SPL targets. SPL directly inhibits the transcriptional activity of B-type ARR and thereby impairs shoot regenerative capacity. In Caenorhabditis elegans, the transitions between stages of larval development are mediated by increases in the expression of two sequentially expressed microRNAs, lin-4 and let-7 (Ambros, 2011). A recent report demonstrated that overexpression of let-7 in the juvenile state impairs tissue repair, a type of regeneration in mouse (Shyh-Chang et al., 2013). Thus, these observations highlight that plants and animals, although they evolved independently, adopt a similar molecular mechanism by which the regenerative capacity is governed by the microRNA timer.
Growing lines of evidence showed that SPL9 exerts dual molecular roles. It has been shown that SPL9 regulates the vegetative phase transition and trichome production on floral organs through activating MIR172B and TRICHOMELESS1 (Wu et al., 2009; Yu et al., 2010), whereas it acts as a transcriptional repressor on DIHYDROFLAVONOL REDUCTASE through binding with the MYB transcription factor PRODUCTION OF ANTHOCYANIN PIGMENTS1 (Gou et al., 2011). Yeast two-hybrid assays demonstrated that SPL9 binds to the transactivation domain of ARR2 (Figures 4A and 4E). In this scenario, we speculate that the binding of SPL9 to ARR2 changes the conformation of ARR2, thereby impairing its transcriptional activation toward downstream targets.
It is well known that old plants have lower capacities for both shoot and root regeneration than young plants. However, this phenomenon could not be explained concurrently by the altered cytokinin:auxin ratio in old plants, because a low cytokinin:auxin ratio inhibits shoot production but induces root regeneration. Our results revealed that old plants exhibit a lower cytokinin response, which is responsible for the decreased shoot regeneration. The molecular mechanism causing the reduced root regenerative capacity in old plants and whether this process is mediated by an altered auxin response await further investigations.
In vitro regeneration is influenced by the type of explants used and by environmental factors such as culture medium, plant hormones, and gelling agent strength. Although the protocol for in vitro regeneration varies greatly between plant species, most of the explants, such as cotyledon, hypocotyl, petiole, and early leaves, are collected from juvenile plants. It has been shown that, during vegetative regeneration of the heather Calluna vulgaris, the number of newly formed sprouts decreased significantly in plants that were more than 6 years old (Berdowski and Siepel, 1998). Similarly, the shoot regenerative capacity declines as Quercus euboica ages (Kartsonas and Papafotiou, 2007). These observations are consistent with our findings that the level of miR156 is correlated with shoot regenerative competence and that overexpression of miR156 increases the regenerative rate in both Arabidopsis and tobacco. Because miR156 is highly conserved in land plants from moss to flowering plants (Axtell and Bowman, 2008), these results further suggest that age cues serve as a common element behind plant cell totipotency. Manipulating miR156 levels during regeneration will thus be of great value for the in vitro propagation of all plant species, especially for some rare and endangered trees.
METHODS
Plant Materials
Arabidopsis thaliana (ecotype Columbia-0), tobacco (Nicotiana tabacum cv SR1), and Nicotiana benthamiana were grown at 21°C (day)/19°C (night) in long-day conditions (16 h of light/8 h of dark). Pro35S:MIR156, Pro35S:MIM156, Pro35S:rSPL3, ProSPL9:rSPL9, spl9-1 spl15-2, arr2-4, arr1-3 arr10-5 arr12-1, and ProTCS:GFP were described (Mason et al., 2005; Ishida et al., 2008; Müller and Sheen, 2008; Wang et al., 2008). The arr2-4 arr12-1 double mutant was identified by PCR genotyping.
For transgenic Arabidopsis plants, the binary constructs were delivered into Agrobacterium tumefaciens GV3101 (pMP90) by the freeze-thaw method. Transgenic plants were generated by the floral dipping method (Clough and Bent, 1998) and screened with 0.05% glufosinate (Basta) on soil, 40 μg/mL hygromycin, or 50 μg/mL kanamycin on half-strength MS plates.
For transgenic tobacco plants, the overnight culture of Agrobacterium was resuspended with infection buffer (30 g/L glucose) to OD600 = 0.8. Tobacco seeds were sterilized with 20% NaClO for 15 min and germinated on an MS plate (4.4 g of MS basal medium with vitamin powder [PhytoTechnology Laboratories], 0.5 g/L methylester sulfonate, 20 g/L sucrose, and 8 g/L agar, pH 5.7). Leaf discs (1 cm in diameter) were infected with Agrobacterium suspension for 30 min. The explants were briefly dried, transferred to sterile filter paper, and kept in darkness at 25°C for 2 d. The explants were then transferred to selection shooting medium (4.4 g of MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, 2 μg/mL 6-BA, 100 μg/mL kanamycin, 250 μg/mL timentin, and 8 g/L agar, pH 5.7) and incubated at 25°C for 2 weeks under long-day conditions. The explants were subcultured at 3-week intervals. The whole explants together with the shoots were transferred to selection rooting medium (4.4 g of MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, 100 μg/mL kanamycin, 250 μg/mL timentin, and 8 g/L agar, pH 5.7). Rooted plants were transferred to soil and grown in a growth chamber. Independent T2 lines were used for regeneration assays.
Constructs
The oligonucleotide primers for all constructs are given in Supplemental Table 2. For yeast two-hybrid constructs, the pGBKT7 (Clontech) series of SPL2, SPL3, SPL9, and SPL10 has been described (Yu et al., 2012). ARR1, ARR2, ARR7, ARR10, and ARR12 were cloned into pGADT7 (Clontech). ARR2 (D80N) was generated by PCR-mediated mutagenesis.
BiLC constructs were generated as described (Gou et al., 2011). The cDNA of rSPL9 was amplified and cloned into the JW772 vector behind LUCc under the control of the 35S promoter. The coding region of ARR1, ARR2, ARR7, ARR10, or ARR12 was cloned into the JW771 vector in front of LUCn under the control of the 35S promoter.
For CoIP constructs, ARR1, ARR2, and ARR7 were cloned into the JW819 vector with a 3xHA C-terminal fusion. rSPL9 was cloned into the JW1016 vector with a 6xMyc N-terminal fusion tag.
For Pro35S:rSPL9-GR, the rSPL9-GR fragment was PCR amplified using JW66 as template and cloned into JW807 behind the 35S promoter.
For tobacco transient assays, the TCS or DR5 promoter was cloned into the TQ108 vector in front of LUC. Pro35S:ARR2 was generated by cloning ARR2 into JW807 behind the 35S promoter. Pro35S:rSPL9 has been described (Gou et al., 2011). For Pro6xUAS:LUC, 6xUAS (6xCGGGTGACAGCCCTCCG) was fused with the minimal 35S promoter and cloned into TQ108 in front of LUC. For Pro35S:ΔARR2 and Pro35S:BD-ΔARR2, the fragment of ΔARR2 or BD-ΔARR2 (ΔARR2 fused with the GAL4-BD domain) was cloned into JW807 behind the 35S promoter.
Regeneration Experiments
Tobacco regeneration assays were performed on MS medium with different concentrations of 6-BA as indicated.
For Arabidopsis regeneration assays, Arabidopsis seeds were sterilized with 15% bleach and germinated on half-strength MS plates (2.21 g of MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, and 8 g/L agar, pH 7.5). Explants (leaves or hypocotyls) were excised and transferred to CIM (4.4 g of MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, 2.2 μM 2,4-D, 0.2 μM kinetin, and 8 g/L agar, pH 5.7) for 7 d. The calli were then transferred to SIM (4.4 g of MS basal medium with vitamin powder, 0.5 g/L methylester sulfonate, 20 g/L sucrose, 0.9 μM indole-3-acetic acid, and 8 g/L agar, pH 5.7) with different concentrations of 2-isopentenyladenine and incubated at 25°C under long-day conditions.
The numbers of explants and regenerated shoots were scored. The regenerative capacity was represented by the number of regenerated shoots in a given number of explants. Three independent experiments (biological triplicates) were performed.
Yeast Two-Hybrid Assay
Plasmids were transformed into yeast strain AH109 (Clontech) by the LiCl-polyethylene glycol method. The transformants were selected on SD-Leu-Trp plates. The interactions were tested on SD-Leu-Trp-His or SD-Ade-Leu-Trp-His plates with 3-amino-1,2,4-triazole. At least 10 individual clones were analyzed.
CoIP and Immunoblot Analyses
Agrobacteria-infiltrated N. benthamiana leaves were used for CoIP analyses. The soluble protein was extracted in extraction buffer (50 mM HEPES, 10 mM EDTA, 50 mM NaCl, 10% glycerol, 1% polyvinylpolypyrrolidone, 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10 μM MG-132, and 1× protease inhibitor cocktail, pH 7.5). Immunoprecipitation was performed with anti-Myc beads (Sigma-Aldrich; E6654) for 3 h (for ARR1 or ARR2 with SPL9) at 4°C. The beads were washed three times with wash buffer (50 mM HEPES, 150 mM NaCl, 10 mM EDTA, 0.1% Triton X-100, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride, pH 7.5). 3xHA or 6xMyc fusion proteins were detected by immunoblot with anti-HA-peroxidase (Roche; 12013819001) or anti-Myc (Millipore; 05-724) antibody.
BiLC Analysis and Tobacco Transient Assay
Agrobacterium was resuspended in infiltration buffer (10 mM methylester sulfonate, 10 mM MgCl2, and 150 µM acetosyringone, pH 5.7) at OD600 = 0.8. Pro35S:P19-HA (Papp et al., 2003) was coinfiltrated to inhibit gene silencing. Luciferin (1 mM) was infiltrated before LUC activity was monitored after 3 d. The LUC signal was photographed with a cool CCD camera. To quantify LUC activity, we used a dual-LUC reporter system in which Pro35S:RENILLA (REN) was used as an internal control (Hellens et al., 2005). The firefly luciferase activity was quenched before REN activity was measured with a luminometer (Promega 20/20). The LUC activity was calculated by normalizing the values to those of REN. The expression of 6xMyc-rSPL9, ARR2-3xHA, and P19-HA was examined by immunoblot (Supplemental Figure 8A). Three independent experiments (biological triplicates) were performed.
The BiLC assay was performed as described (Gou et al., 2011). Agrobacterium was resuspended in infiltration buffer at OD600 = 0.8.
Expression Analyses
For cytokinin treatment, 7-d-old wild-type Arabidopsis seedlings grown in half-strength MS liquid medium under long-day conditions were treated with DMSO (mock) or 5 μM 6-BA for 40 min. Total RNA was extracted with Trizol reagent (Invitrogen). Total RNA (1 µg) was treated with 1 μL of DNase I (1 unit/μL; Fermentas) and used for cDNA synthesis with oligo(dT) primer (Fermentas). The average expression levels and se values were calculated from 2-ΔΔCt values. Biological triplicates with technical duplicates were performed. The quantitative RT-PCR primers for TUB have been described (Wang et al., 2009). The oligonucleotide primers for all genes are given in Supplemental Table 2. Quantitative RT-PCR on mature miR156 was performed as described (Yu et al., 2013). The expression of miR156 in tobacco was normalized to that of the ribosomal protein gene Nt-L25.
Cytokinin Measurement
Wild-type and Pro35S:MIR156 seedlings were used for cytokinin measurements. Samples were purified, derivatized by propionylation, and quantified by liquid chromatography-mass spectrometry analysis according to the published protocol (Nordström et al., 2004). Chromatographic separation was performed on a reverse-phase analytical column (150 × 1 mm BetaMax Neutral, 5-μm particle size; Thermo Hypersil).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ARR1 (At3g16857), ARR2 (At4g16110), ARR10 (At4g31920), ARR12 (At2g25180), ARR5 (At3g48100), ARR6 (At5g62920), ARR7 (At1g19050), ARR15 (At1g74890), SPL2 (At5g43270), SPL3 (At2g33810), SPL9 (At2g42200), SPL10 (At1g27370), SPL15 (At3g57920), TUB (At5g62690), and Nt-L25 (L18908).
Supplemental Data
Supplemental Figure 1. Shoot Regeneration of N. tabacum and A. thaliana.
Supplemental Figure 2. Transgenic N. tabacum Plants.
Supplemental Figure 3. miR156 Regulates Shoot Regeneration.
Supplemental Figure 4. Genetic Interaction between miR156 and B-Type ARRs.
Supplemental Figure 5. Interactions between B-Type ARRs and miR156-Targeted SPLs.
Supplemental Figure 6. Validation of the Interactions between B-Type ARRs and SPLs by BiLC Assays.
Supplemental Figure 7. Validation of ARR1-SPL9 Interaction by CoIP Assays.
Supplemental Figure 8. SPL Negatively Regulates Cytokinin Response.
Supplemental Figure 9. ProTCS:LUC Transactivation Assays.
Supplemental Figure 10. ProDR5:LUC Transactivation Assays.
Supplemental Figure 11. SPL9 Suppresses ARR2 Transcriptional Activation.
Supplemental Figure 12. spl9 spl15 Shoot Regeneration Assays.
Supplemental Figure 13. Genetic Interaction between SPL9 and B-Type ARRs.
Supplemental Table 1. Cytokinin Measurement.
Supplemental Table 2. Oligonucleotide Primer Sequences.
Supplementary Material
Acknowledgments
We thank the ABRC for seeds; Bruno Müller for the ProTCS:GFP reporter construct; Hongxia Zhang for tobacco seeds; Gun Löfdahl and Xiao-Shu Gao for skillful technical assistance; and members of the J.-W. Wang laboratory, Ignacio Rubio-Somoza, Yingbo Mao, Daiyin Chao, and Ling-Jian Wang, for discussion and comments on the article. This work was supported by the National Natural Science Foundation of China (Grants 31430013, 31222029, and 912173023), the State Key Basic Research Program of China (Grant 2013CB127000), the Shanghai Pujiang Program (Grant 12PJ1409900), the Recruitment Program of Global Expects (China), the National Key Laboratory of Plant Molecular Genetics Key Research Program, the Ministry of Education, Youth, and Sports, Czech Republic (Grant LO1204 from the National Program of Sustainability I), the Swedish Governmental Agency for Innovation Systems, and the Swedish Research Council.
AUTHOR CONTRIBUTIONS
T.-Q.Z. and J.-W.W. designed the research. T.-Q.Z., H. Lian, H.T., C.-M.Z., S.Y., K.D., J.-H.C., and Q.C. performed research. All authors analyzed data. J.-W.W. wrote the article.
Glossary
- CIM
callus-inducing medium
- SIM
shoot-inducing medium
- 6-BA
6-benzylaminopurine
- MS
Murashige and Skoog
- BiLC
bimolecular luminescence complementation
- CoIP
coimmunoprecipitation
- DEX
dexamethasone
References
- Ambros V. (2011). MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349. [DOI] [PubMed] [Google Scholar]
- Berdowski J.J.M., Siepel H. (1998). Vegetative regeneration of Calluna vulgaris at different ages and fertilizer levels. Biol. Conserv. 46: 85–93. [Google Scholar]
- Bergonzi S., Albani M.C., Ver Loren van Themaat E., Nordström K.J., Wang R., Schneeberger K., Moerland P.D., Coupland G. (2013). Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097. [DOI] [PubMed] [Google Scholar]
- Birnbaum K.D., Sánchez Alvarado A. (2008). Slicing across kingdoms: Regeneration in plants and animals. Cell 132: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Cigan A.M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Duclercq J., Sangwan-Norreel B., Catterou M., Sangwan R.S. (2011). De novo shoot organogenesis: From art to science. Trends Plant Sci. 16: 597–606. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A., Schmülling T. (2003). Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 6: 480–488. [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389. [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J., Müller B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63: 353–380. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49: 47–57. [DOI] [PubMed] [Google Scholar]
- Kartsonas E., Papafotiou M. (2007). Mother plant age and seasonal influence on in vitro propagation of Quercus euboica Pap, an endemic, rare and endangered oak species of Greece. Plant Cell Tissue Organ Cult. 90: 111–116. [Google Scholar]
- Kieber J.J., Schaller G.E. (2014). Cytokinins. The Arabidopsis Book 12: e0168, doi/10.1199/tab.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M.D., Bartel D.P. (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 24: 2678–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A., Tarkowski P., Tarkowska D., Norbaek R., Astot C., Dolezal K., Sandberg G. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101: 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I., Mette M.F., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J. (2003). Evidence for nuclear processing of plant microRNA and short interfering RNA precursors. Plant Physiol. 132: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (2013). Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 105: 125–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Zhou C.M., Confraria A., Martinho C., von Born P., Baena-Gonzalez E., Wang J.W., Weigel D. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24: 2714–2719. [DOI] [PubMed] [Google Scholar]
- Ruckh J.M., Zhao J.W., Shadrach J.L., van Wijngaarden P., Rao T.N., Wagers A.J., Franklin R.J. (2012). Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell 10: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M.T., Tsanov K.M., Nguyen L., Asara J.M., Cantley L.C., Daley G.Q. (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155: 778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11: 118–130. [PubMed] [Google Scholar]
- Sugimoto K., Gordon S.P., Meyerowitz E.M. (2011). Regeneration in plants and animals: Dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 21: 212–218. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Park M.Y., Wang L.J., Koo Y., Chen X.Y., Weigel D., Poethig R.S. (2011). miRNA control of vegetative phase change in trees. PLoS Genet. 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538. [DOI] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Salinas M., Höhmann S., Berndtgen R., Huijser P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22: 3935–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xu M., Koo Y., He J., Poethig R.S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Cai W.J., Wang S., Shan C.M., Wang L.J., Chen X.Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Cao L., Zhou C.M., Zhang T.Q., Lian H., Sun Y., Wu J., Huang J., Wang G., Wang J.W. (2013). Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Galvão V.C., Zhang Y.C., Horrer D., Zhang T.Q., Hao Y.H., Feng Y.Q., Wang S., Schmid M., Wang J.W. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.M., Zhang T.Q., Wang X., Yu S., Lian H., Tang H., Feng Z.Y., Zozomova-Lihová J., Wang J.W. (2013). Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340: 1097–1100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.