Signal-transducing phospholipases A2 of two different plants are inhibited by alkaloids produced in the same but not in the other plant thus preventing continuous overproduction.
Abstract
The overproduction of specialized metabolites requires plants to manage the inherent burdens, including the risk of self-intoxication. We present a control mechanism that stops the expression of phytoalexin biosynthetic enzymes by blocking the antecedent signal transduction cascade. Cultured cells of Eschscholzia californica (Papaveraceae) and Catharanthus roseus (Apocynaceae) overproduce benzophenanthridine alkaloids and monoterpenoid indole alkaloids, respectively, in response to microbial elicitors. In both plants, an elicitor-responsive phospholipase A2 (PLA2) at the plasma membrane generates signal molecules that initiate the induction of biosynthetic enzymes. The final alkaloids produced in the respective plant inhibit the respective PLA, a negative feedback that prevents continuous overexpression. The selective inhibition by alkaloids from the class produced in the “self” plant could be transferred to leaves of Nicotiana benthamiana via recombinant expression of PLA2. The 3D homology model of each PLA2 displays a binding pocket that specifically accommodates alkaloids of the class produced by the same plant, but not of the other class; for example, C. roseus PLA2 only accommodates C. roseus alkaloids. The interaction energies of docked alkaloids correlate with their selective inhibition of PLA2 activity. The existence in two evolutionary distant plants of phospholipases A2 that discriminate “self-made” from “foreign” alkaloids reveals molecular fingerprints left in signal enzymes during the evolution of species-specific, cytotoxic phytoalexins.
INTRODUCTION
The evolutionary success of plant specialized metabolites reflects not only their potential usefulness in defense or communication but also an effective management of the burdens inherent to secondary biosynthesis, including competition for resources, metabolic derangement, and potential self-intoxication. Plants have developed several strategies to keep secondary biosynthesis compatible with the fitness of the producer. Much is known about the compartmentation and channeling of enzymes and metabolites that separate intermediates and products from basic metabolism, as exemplified by precursor pools, metabolons, and intracellular trafficking, e.g., in the biosynthesis of flavonoids or benzylisoquinolines (for a review, see Klein and Roos, 2009). Less information exists about the metabolic detoxification of end products, as exemplified by the recycling of benzophenanthridine alkaloids (Weiss et al., 2006; Müller et al., 2014). In a few cases, we know of mutated target structures that cause self-resistance to the produced toxin, e.g., in camptothecin-producing plants (Sirikantaramas et al., 2008).
These measures do not provide perfect solutions to the aforementioned burdens, as they require coordinate changes of a multitude of cellular activities and tend to increase the costs of biosynthesis. This is especially critical in cells that overexpress secondary biosynthetic enzymes as a defense against pathogens or herbivores. Self-regulatory mechanisms appear indispensable to keep the overproduction of these specialized metabolites (phytoalexins) within a manageable range.
The self-control of secondary metabolite production remains to be investigated in detail. The existence of undisclosed regulatory circuits has only accidentally been realized by the failure or unexpected outcome of naive attempts to overproduce valuable plant specialized compounds by overexpressing transcription factors or rate-limiting enzymes (Leonard et al., 2009). Here, we present a self-regulatory mechanism that stops the expression of biosynthetic enzymes by blocking the transfer of the inducing signals.
Much experimental work has been done with the pathogen-triggered biosynthesis of benzophenanthridines in the Papaveracea Eschscholzia californica (California poppy). Cultured cells respond to a yeast glycoprotein elicitor by overproducing these antimicrobial alkaloids (Schumacher et al., 1987; Gundlach et al., 1992; Roos et al., 1998, 2006; Cho et al., 2007), which intercalate in double-stranded DNA and inhibit a number of SH-dependent enzymes (Schmeller et al., 1997; Wink et al., 1998; Slaninová et al., 2001; Barták et al., 2003). The biosynthetic sequence of benzophenanthridines has long been known (Zenk, 1994), and over the last decades several biosynthetic enzymes have been characterized at the molecular level (for comprehensive and recent reviews, see Ziegler and Facchini, 2008; Hagel and Facchini, 2013). Several groups have confirmed that the elicitor-triggered alkaloid production occurs via transcriptional activation and expression of biosynthetic and related enzymes (Dittrich and Kutchan, 1991; Blechert et al., 1995; Viehweger et al., 2006; Cho et al., 2007; Angelova et al., 2010), making Eschscholzia cell cultures a model system for analyzing the expression of plant defense genes (Haider et al. 2000).
More recently, transcriptome analyses in cell cultures of the related species Papaver somniferum (Opium poppy) have identified a few genes related to alkaloid biosynthesis (e.g., the one encoding the rate-limiting enzyme) among the multitude of elicitor-induced genes (Zulak et al., 2007, 2009; Desgagné-Penix et al., 2010). In our lab, cellular and molecular events of the signal chain preceding gene activation have been elucidated in some detail (Viehweger et al., 2002, 2006; Färber et al., 2003; Roos et al., 2006; Heinze et al., 2013; Müller et al., 2014). Figure 1 provides an overview of the main biosynthetic and signaling steps relevant to this study. The following signaling elements are crucial for this work: (1) Elicitor contact activates a plasma membrane bound phospholipase A2 (PLA2) via G-protein-controlled conformational transfer. In the isolated plasma membrane, PLA2 activity is stimulated by yeast elicitor in presence of GTP (Roos et al., 1999; Viehweger et al., 2006; Heinze et al., 2007). PLA2 is part of a membrane-associated protein complex that contains variable amounts of Gα and a cyclophilin, which likely catalyzes conformational transfer between Gα and its target. The inhibitory effect of Gα is abolished by GTP+elicitor or by Gα dimerization (Heinze et al., 2007, 2013). Mutant cell strains that express low amounts of Gα show little or no elicitor-triggered activation of PLA2 or alkaloid production (Viehweger et al., 2006; Heinze et al., 2013). (2) Lysophosphatidylcholine (LPC) produced by PLA2-catalyzed lipid hydrolysis acts as a second messenger by activating vacuolar H+/Na+ antiporters. The LPC level in the cytoplasm peaks after elicitor contact (Viehweger et al., 2002) and is controlled by its PLA2-catalyzed generation and rapid enzymic reacylation (Schwartze and Roos, 2008). If added to permeabilized cells in low micromolar concentrations, LPC causes a Na+-dependent efflux of vacuolar protons, which is blocked by amiloride, an inhibitor of vacuolar sodium/proton antiporters (Viehweger et al., 2002). (3) The resulting peak of cytoplasmic H+ is necessary and sufficient to activate biosynthetic genes. Artificially triggered pH peaks cause a similar expression of biosynthetic genes (e.g., the berberine bridge enzyme) as the yeast elicitor (Viehweger et al., 2006). The highest increase of transcript level was found for the gene encoding the rate-limiting enzyme, 3′-hydroxy N-methylcoclaurine 4'-O-methyltransferase both after elicitor treatment or artificial pH shifts (Angelova et al., 2010). Bracketing of vacuolar and cytoplasmic pH reversibly blocks the elicitation of alkaloid biosynthesis (Roos et al., 1998).
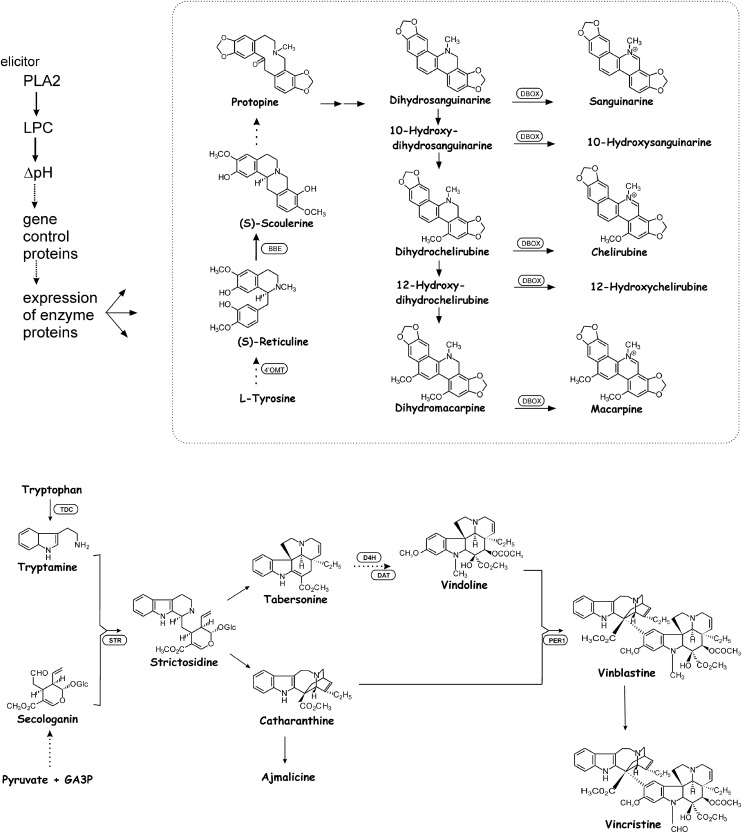
Figure 1.
Signal and Biosynthetic Steps Relevant for This Work.
Top: Benzophenanthridine alkaloids, investigated in E. californica cultures, and main steps of their biosynthesis (according to Zenk, 1994) and the preceding signal chain (Roos et al., 2006). Bottom: Monoterpenoid indole alkaloids, investigated in C. roseus cultures, and selected steps of their biosynthesis (according to Facchini and De Luca, 2008). DHBOx, dihydrobenzophenanthridine oxidase; 4'OMT, 3′-hydroxy N-methylcoclaurine 4'-O-methyltransferase; BBE, berberine bridge enzyme; D4H, desacetoxyvindoline 4-hydroxylase; DAT, deacetylvindoline 4-O-acetyltransferase; PER, peroxidase; STR, strictosidine synthase.
In summary, the enzyme PLA2 at the plasma membrane initiates a signal transduction pathway that is required for the overexpression of alkaloid biosynthesis. In the actual study, we show that specific properties of this enzyme allow a far-reaching feedback mechanism that controls this process at the stage of early signaling.
RESULTS
Cloning and Silencing of PLA2 Confirms Its Essential Role in Elicitor-Triggered Signaling
The molecular identification of the elicitor-responsive PLA2 was essential for this study. Starting with tryptic peptides from purified plasma membrane and facilitated by the detection of their coding sequences in an EST database, cDNA fragments containing sequences typical of plant PLA2 were identified. These sequences finally allowed the rapid amplification of cDNA ends (RACE)-assisted amplification of a complete open reading frame (ORF). The translated peptide sequence of PLA2 of E. californica is shown in Figure 2 and aligned with selected proteins of plant homologs. PLA2 contains consensus regions that code, e.g., for the DGGX(=V)RG motif (24 to 29) and the boxes around the catalytic dyad, Ser-65 plus Asp-217. The complete gene and protein sequences of PLA2 from E. californica are deposited in GenBank (see the Accession Numbers section). According to a recently proposed nomenclature (Scherer et al., 2010), the enzyme is homologous to the group of pPLA-II phospholipases.
Figure 2.
Alignment of PLA2 Amino Acid Sequences of E. californica and Selected Plant Species.
Line 1: The PLA2 protein coded by the E. californica gene JQ886492 identified and used in this work. Line 2: The PLA2 protein coded by the C. roseus gene Caros024612.1 (ORCAE library, University of Ghent) identified and used in this work. Line 3: Protein sequence of the patatin-like PLA2 from Solanum cardiophyllum that was used as the template for homology modeling. The following lines contain a selection of plant PLA2 sequences found in the NCBI BLASTp database as annotated items. Red colored amino acids mark noncompatible exchanges.
To test whether this PLA2 gene encodes the enzyme required for the elicitor-triggered alkaloid response, we used RNA interference (RNAi)-based silencing to reduce PLA2 transcript levels (see Methods and Supplemental Figures 2 and 3). From several transformed clones, we characterized two cell strains that displayed a strong depression of PLA2 mRNA levels (RT-PCR in Supplemental Figure 3). These RNAi strains almost completely lacked elicitor-triggered alkaloid production, and their plasma membrane was devoid of elicitor-stimulatable PLA2 activity (Figure 3). It is therefore justified to assume that the cloned PLA2 gene encodes the elicitor-responsive enzyme that is essential for the signal transfer to induce the overexpression of alkaloid biosynthesis. The silencing of PLA2 as performed in this study did not substantially reduce the PLA2 activity measurable in the absence of elicitor (Figure 3, columns 1 and 3). This activity might represent a different PLA2 species that was not targeted by the sequence used for gene silencing. The coexistence of elicitor-responsive and nonresponsive PLA2 enzymes in the plasma membrane is consistent with earlier activity measurements by Heinze et al. (2007).
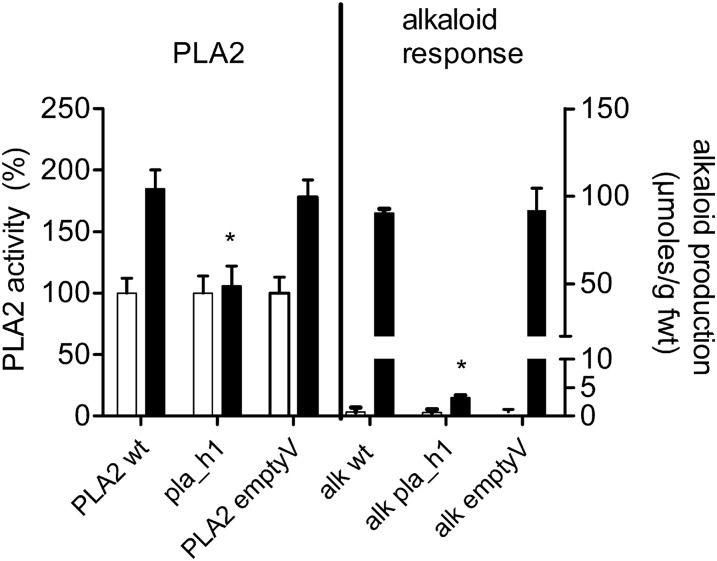
Figure 3.
Silencing of PLA2 Prevents Elicitor-Triggered Responses.
PLA2 activity and elicitor stimulation of alkaloid production are compared between the wild-type culture (PLA2 wt), the RNAi line (pla_h1), and the control culture transformed with the empty vector (emptyV). Each pair of columns collates data measured in the absence of elicitor (open column) and after elicitor contact (black column). Left: PLA2 activity measured in the isolated plasma membrane. Data are means ± sd; n = 3 different membrane preparations. Right: Total alkaloid production (sum of dihydrobenzophenanthridines plus benzophenanthridines) during 24 h. Data are means ± sd; n = 3 cultures of the same batch. Asterisks indicate a significant difference (P < 0.05) of elicited mutant cultures from the elicited wild type. The experiment was repeated with another mutant cell line (pla_h2), which yielded similar results. The elicitor-triggered events “increase in PLA2 activity” and “alkaloid overproduction,” expressed in percentage of the wild type, can be compared as follows: pla_h1, 6% and 2.7%; pla_h2, 3% and 4.3%. In cultures transformed by the empty vector, these stimulations did not significantly differ from the wild type.
E. californica Alkaloids Suppress Elicitor-Triggered PLA2 Activity and Alkaloid Overproduction
Knowledge about the function of PLA2 in the elicitor-triggered responses was instrumental to our investigation of the regulation of gene expression in alkaloid metabolism. Figures 4 and 5A (columns 1 and 2) show the typical burst of alkaloid production in an E. californica cell suspension culture, triggered by a fungal elicitor. The response to elicitor can be completely suppressed by sanguinarine, the first benzophenanthridine in the biosynthetic sequence, if this alkaloid is present at the time of elicitor contact (Figure 4, columns 3 to 5). The suppression is not observed if the alkaloid is added 30 min or more after the elicitor (columns 6 to 8). Elicitor-induced synthesis of new biosynthetic enzymes requires at least 3 h (Viehweger et al., 2006; Angelova et al., 2010); therefore, this result indicates that benzophenanthridines interfere with an event prior to gene expression, likely a step in the elicitor-initiated signal chain. Based on the known function of PLA2 in signal transduction (as explained in the Introduction), we tested the in vitro activity of this enzyme in the presence of individual benzophenanthridines, i.e., the final products of alkaloid biosynthesis in this cell culture. The detailed analysis shown in Figure 5B revealed a remarkable selectivity: All tested benzophenanthridines inhibit PLA2 activity by more than 40%, but they display individual differences, with 10-hydroxychelerythrine being by far the strongest inhibitor. The degree of inhibition of PLA2 activity comports well with the depression of the elicitor-triggered biosynthesis exerted by each alkaloid (Figures 5A and 5B). Alkaloids of a different family, e.g., monoterpenoid indoles, were also tested and caused neither substantial nor specific inhibition (see below).
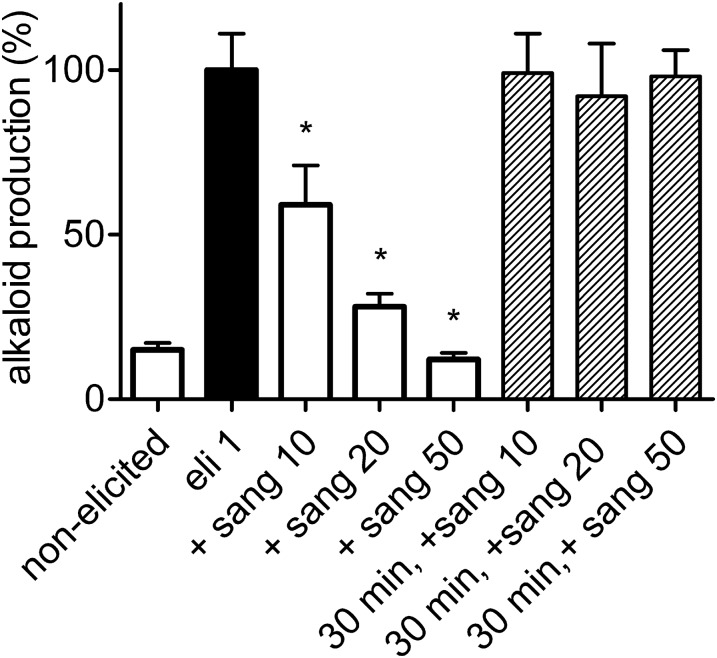
Figure 4.
Effect of Sanguinarine on Elicitor-Triggered Alkaloid Biosynthesis.
The increase of total alkaloid content in 24 h (sum of dihydrobenzophenanthridines plus benzophenanthridines) is plotted for untreated and elicitor-treated cultures. Sanguinarine was added together with the elicitor (columns 3 to 5) or 30 min after elicitor (columns 6 to 8) at a final concentration printed next to the alkaloid (in µM). Data are means ± se; n = 3 cultures of the same batch. Values are normalized to the content of elicitor treated cells (usually between 30 and 50 µg/g fresh weight) that was set to 100%. Significant differences (P < 0.05) from this level are indicated by asterisks.
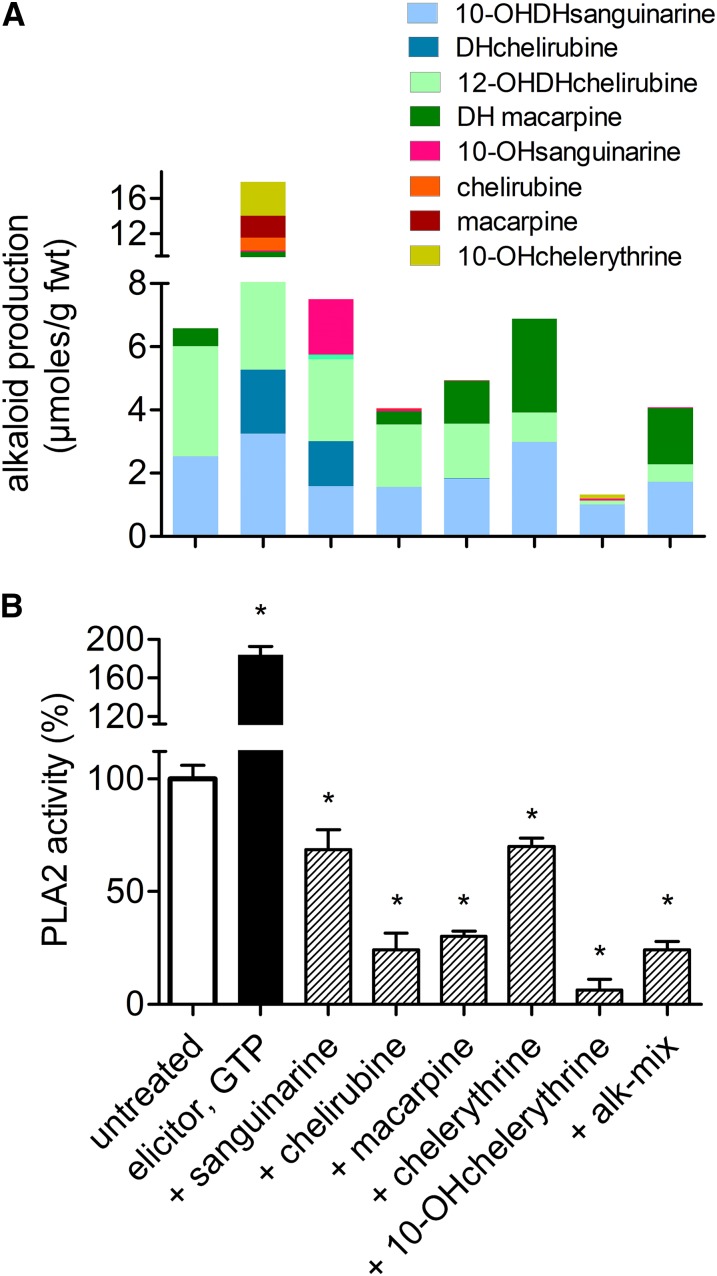
Figure 5.
Specificity of Inhibition Exerted by Benzophenanthridines on Elicitor-Triggered Events in E. californica Cell Suspensions.
(A) Top: Effect of individual benzophenanthridines at the elicitor-triggered alkaloid production. Columns show total benzophenanthridine contents, produced in 24 h after elicitor contact, and consist of the color-coded individual compounds. Alkaloids indicated at the bottom abscissa were added to the cultures 20 min prior to the elicitor at 10 µM concentrations (no GTP added). Note that the main increase after elicitor contact occurs with the dihydro(DH)benzophenanthridines. The figure plots a typical, single experiment that was repeated with three different culture batches and yielded similar relations between the total alkaloids of each sample, although the alkaloid contents differed substantially between these batches. Standard deviations of the HPLC-based determination of the individual alkaloids typically range from 7 to 12%, as shown in test experiments with the alkaloid mix.
(B) Bottom: Elicitor-stimulated PLA2 activity in the presence of the same alkaloids. The indicated alkaloids were added 10 min prior to the substrate (together with elicitor plus GTP) to give a final concentration of 10 µM. Data are means ± se; n = 5. Asterisks indicate significant (P < 0.05) differences from the untreated culture. Alk mix: 10 µM alkaloids extracted from an E. californica culture 48 h after elicitor contact, mainly 20.8% 10-hydroxychelerythrine, 41.9% chelirubine, and 17.7% macarpine.
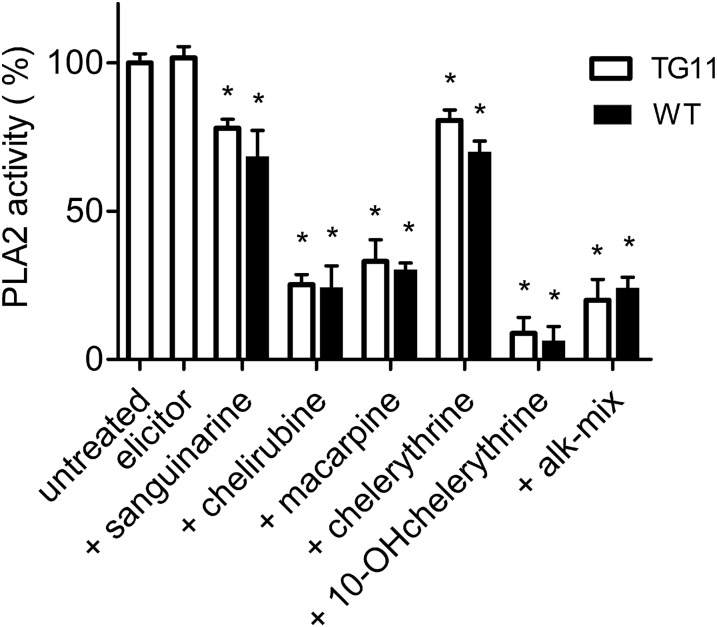
As mentioned in the Introduction, stimulation of PLA2 by elicitor is mediated by an adherent complex of Gα proteins and cyclophilin (Heinze et al., 2007, 2013). To reveal whether the inhibitory benzophenanthridines targeted the enzyme PLA2 directly or influenced its Gα-dependent activation, we measured the degree of inhibition of PLA2 activity in the nonelicited state of the wild type and in a mutant cell line (TG11) with reduced Gα content. No significant differences were detectable between these unstimulated conditions (Figure 6) and the elicitor-stimulated enzyme in wild-type cells (Figure 5B). These data suggest that the presence or activation of Gα is not required for the inhibition of PLA2 caused by the alkaloids.
Figure 6.
Effect of Benzophenanthridines on PLA2 in Nonelicited and Low-Gα Cultures.
PLA2 activities of plasma membranes isolated from the low Gα mutant TG11 (open columns) or from the wild type (black columns) are compared. The plasma membrane of the TG11 cell strain contains ∼14% of the Gα of the wild type and has an unchanged basal PLA2 activity (Viehweger et al., 2006; Heinze et al., 2013) The indicated alkaloids were added 10 min prior to the substrate to give a final concentration of 10 µM. Data are means ± se; n = 5. Asterisks indicate that all alkaloid treatments significantly (P < 0.05) inhibited enzyme activity.
To this point, our results show that E. californica cells harbor a unique plasma membrane-bound PLA2 that is essential for the elicitor-triggered alkaloid response and prone to inhibition by E. californica benzophenanthridine alkaloids.
The alkaloid concentrations required for a substantial inhibition were ∼10 µM, which is well in the range found in the spent medium of elicited cell cultures (Weiss et al., 2006). Moreover, the local concentrations in the cell wall region, i.e., close to the plasma membrane, tend to be higher than in the surrounding medium, since the quarternary benzophenanthridines accumulate at (cellulosic) cell wall components, as visualized in elicited cells (Färber et al., 2003; Weiss et al., 2006; Viehweger et al., 2006; Müller et al., 2014).
Therefore, the inhibition of the plasma membrane-associated PLA2 by the “own” overproduced alkaloids is likely to occur in elicited cells or tissue, and the alkaloid sensitivity of this enzyme argues for a regulatory circuit that stops the induction of biosynthetic enzymes once a critical concentration of benzophenanthridines has been reached. This mechanism has the potential to protect elicited cells from self-intoxication.
The Overexpression of Benzophenanthridine and Monoterpenoid Indole Alkaloids Share Common Regulatory Mechanisms
The results obtained with E. californica cell cultures raised the question as to whether other alkaloid producing plant species use similar mechanisms to control alkaloid production. In a first attempt, we tested cultured cells of Catharanthus roseus (Madagascar periwinkle) as they overproduce toxic alkaloids in response to external stimuli that activate genes encoding biosynthetic enzymes (Pasquali et al., 1992; Moreno et al.,1996) via jasmonate-dependent signalling (van der Fits and Memelink, 2001; Memelink et al., 2001; Mujib et al., 2012). Similar to what we know from E. californica (Roos et al., 1998; Färber et al., 2003; Viehweger et al., 2006), the expression of biosynthetic enzymes in C. roseus cultures occurs independently of the oxidative burst (Pauw et al., 2004). The main phytoalexins of C. roseus are monoterpenoid indole alkaloids (MIAs), whose biosynthesis is also well investigated at the chemical and enzymatic level (Facchini and De Luca, 2008). The last part of the biosynthetic pathway is shown in Figure 1, at the bottom. Our experiments readily revealed regulatory features of C. roseus cells that imply similarities with essential signal steps known from E. californica.
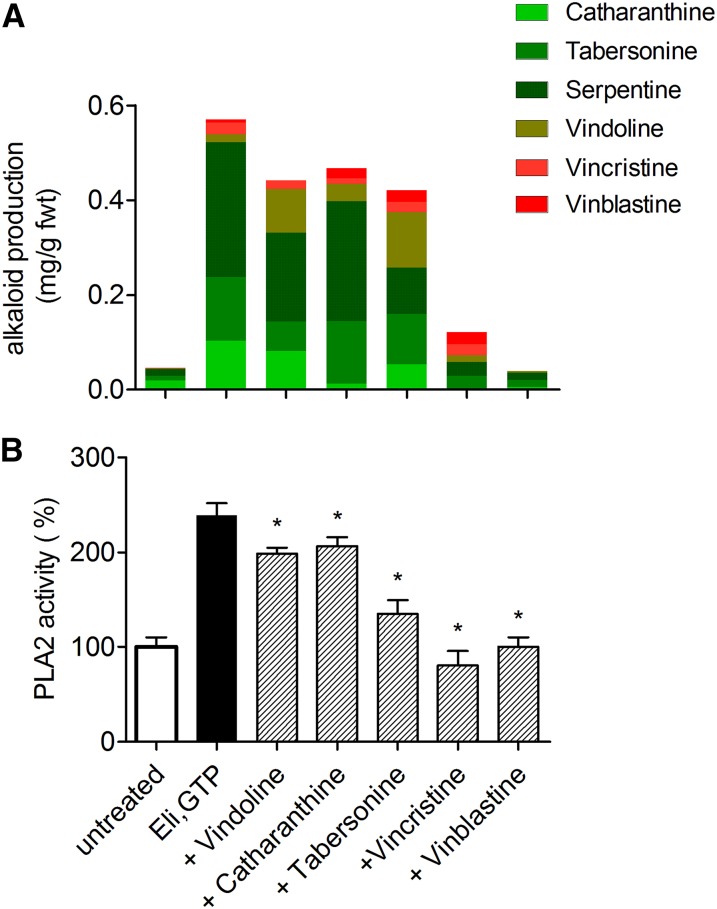
First, cell cultures of C. roseus respond to the same yeast elicitor preparation that is active in E. californica, and this elicitor induces enhanced alkaloid production also in C. roseus (Figure 7A, columns 1 and 2). In the strain used here, this response includes the formation of the late, dimeric alkaloids vinblastine and vincristine, which are not generally found in cultured cells (Vázquez-Flota et al., 2002). Analogous to E. californica, elicitor-triggered overproduction is depressed if monoterpenoid indole alkaloids are present together with the elicitor (Figure 7A, columns 3 to 7), and the ultimate products of biosynthesis, i.e., the dimeric alkaloids vincristine and vinblastine, exert the strongest effects.
Figure 7.
Effects of C. roseus Alkaloids on Elicitation of C. roseus Cells.
(A) Top: Elicitor-triggered alkaloid formation, influenced by selected MIAs. The “effector” alkaloids, added 10 min prior to the elicitor (10 µM each), are named at the bottom. Data are total alkaloid contents produced in 24 h after elicitor contact and consist of the color-coded individual compounds. The figure represents a typical, single experiment that was repeated with two different batches and yielded similar relations between the total alkaloids of each sample. Standard deviation of the HPLC-based determination of the extracted individual alkaloids ranges from 8 to 13%, as shown in test experiments with added vincristine and catharanthine.
(B) Bottom: Effects of the same MIAs at the elicitor-stimulated PLA2 in the plasma membrane. If indicated, an alkaloid was added 10 min prior to the substrate, together with elicitor plus GTP, to give a final concentration of 10 µM. Data are means ± se; n = 5. Asterisks indicate significant (P < 0.05) inhibition by alkaloids.
Second, the plasma membrane of C. roseus cells harbors PLA2 activity, which is strongly stimulated by elicitor in the presence of GTP (Figure 7B, columns 1 and 2). To learn whether this enzyme activity is involved in the expression of alkaloid biosynthesis, we tested for an elicitor-like effect of its product, LPC, which functions as a second messenger in the elicitor-triggered expression of alkaloid biosynthesis in E. californica (Viehweger et al., 2002, 2006; see the Introduction). We found that LPC stimulated the production of MIAs in C. roseus cultures to a similar degree as the yeast elicitor: Addition of 10 or 20 µM LPC caused 60% ± 9% or 109% ± 12%, respectively, of the maximum alkaloid response seen after elicitor treatment (cytotoxic effects of LPC were observed only above 30 µM). This might suggest that a product of PLA2 activity also affects the expression of alkaloid biosynthesis in C. roseus.
Third, the elicitor-stimulated PLA2 activity in the plasma membrane of C. roseus cells was severely inhibited by the dimeric end products vincristine or vinblastine, but less by their monomeric precursor alkaloids (Figure 7B, columns 3 to 7). This selectivity is consistent with the depression of elicitor-triggered alkaloid formation shown before (compare Figures 7A and 7B). The inhibitory alkaloids affected the elicitor-stimulated enzyme to a similar degree as the unstimulated PLA2 (compare Figures 7B and 8B), which argues again for direct interference with this enzyme rather than with its elicitor-dependent activation mechanism.
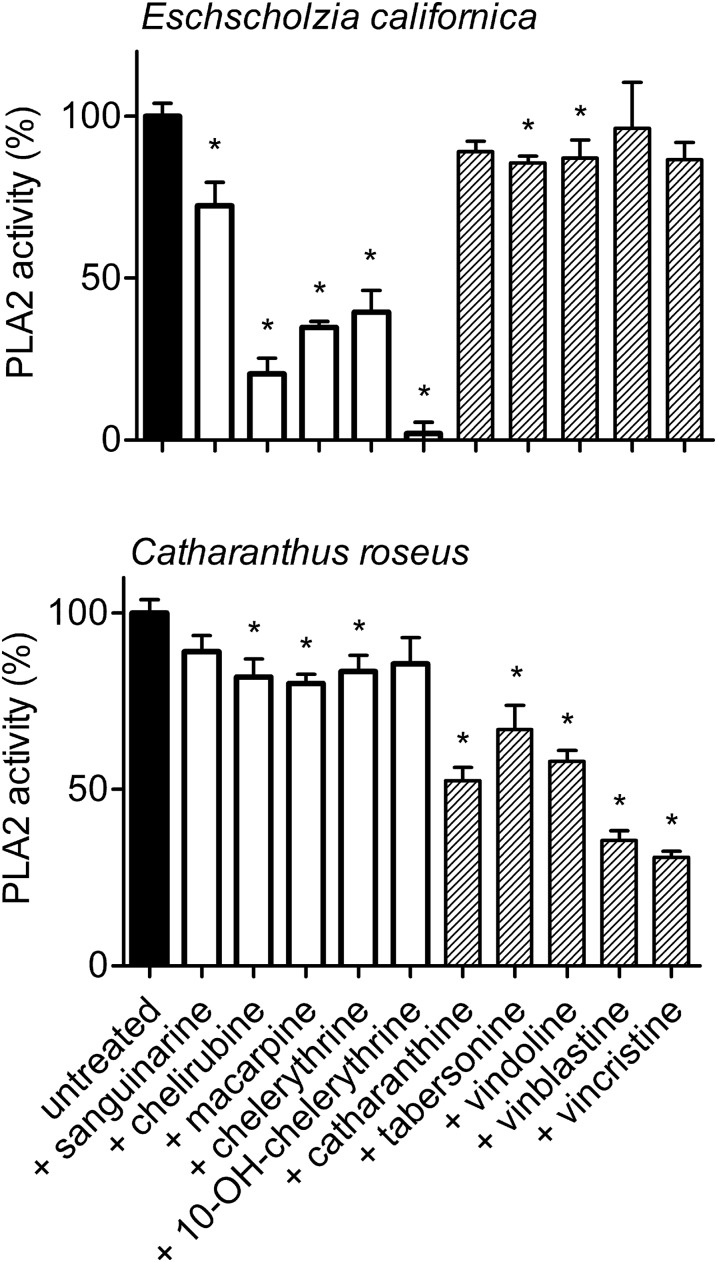
Figure 8.
Inhibition of PLA2 by Benzophenanthridines and MIAs in Plasma Membranes of E. californica and C. roseus.
Enzyme activities are compared in the nonstimulated state between plasma membranes isolated from either plant as described in Methods. Each alkaloid, named at the bottom abscissa, was added 10 min prior to the substrate to give a final concentration of 10 µM. Data are means ± se; n = 4 cell samples. They are normalized to the PLA2 activity of parallel samples in the absence of alkaloids, which is set to 100% (∼5 pkat/mg membrane protein). Asterisks indicate significant (P < 0.05) inhibition of this activity by alkaloids. This experiment was done with one culture batch of the respective plant. Two repetitions with other batches yielded a similar outcome.
Seen together, the data from C. roseus cultures indicate similarities with the elicitor-triggered alkaloid response of E. californica cells. Therefore, it appears that the inhibition by self-made alkaloids of the PLA2 enzyme required for their own overexpression is a regulatory feature of phytoalexin production shared by these two plants.
PLA2 Enzymes of E. californica and C. roseus Discriminate between “Self-Made” and “Foreign” Alkaloids
The feedback inhibition caused by the tested alkaloids shows remarkable species specificity. In Figure 8, the effects of E. californica and C. roseus alkaloids are compared for the plasma membrane-associated PLA2 of the plant producing these compounds. Benzophenanthridines strongly reduce PLA2 enzyme activity in E. californica cells but show little or no inhibitory effect on the PLA2 from C. roseus; the opposite is true for the tested MIAs. Thus, the enzyme of either plant is inhibited by alkaloids produced in the same, but little or not by those produced in the other plant. This implies that each PLA2 can discriminate between self-made and foreign alkaloids.
In an attempt to substantiate this assumption, we transiently expressed the E. californica and C. roseus PLA2 genes in leaves of Nicotiana benthamiana, a plant not known to produce alkaloids of the benzophenanthridine or monoterpenoid indole type, and reassessed the alkaloid inhibition with the recombinant PLA2s. The plant expression system was chosen over yeast cells to minimize the risk of improper expression and/or processing of the membrane-associated PLA2 protein and because of its high expression efficiency (see Engler et al., 2009). Supplemental Figure 5 confirms the delivery of the PLA2 genes to the host plant cells and their expression at the mRNA level. The PLA2 activity of plasma membranes isolated from infected leaves did not significantly increase over those infected by the empty vector, if related to the membrane protein content. This points to a controlled expression of genes that encode plasma membrane-associated PLA2s, which is not unexpected as, at least in E. californica, this PLA2 is part of a protein complex with distinct stoichiometry (Heinze et al., 2013).
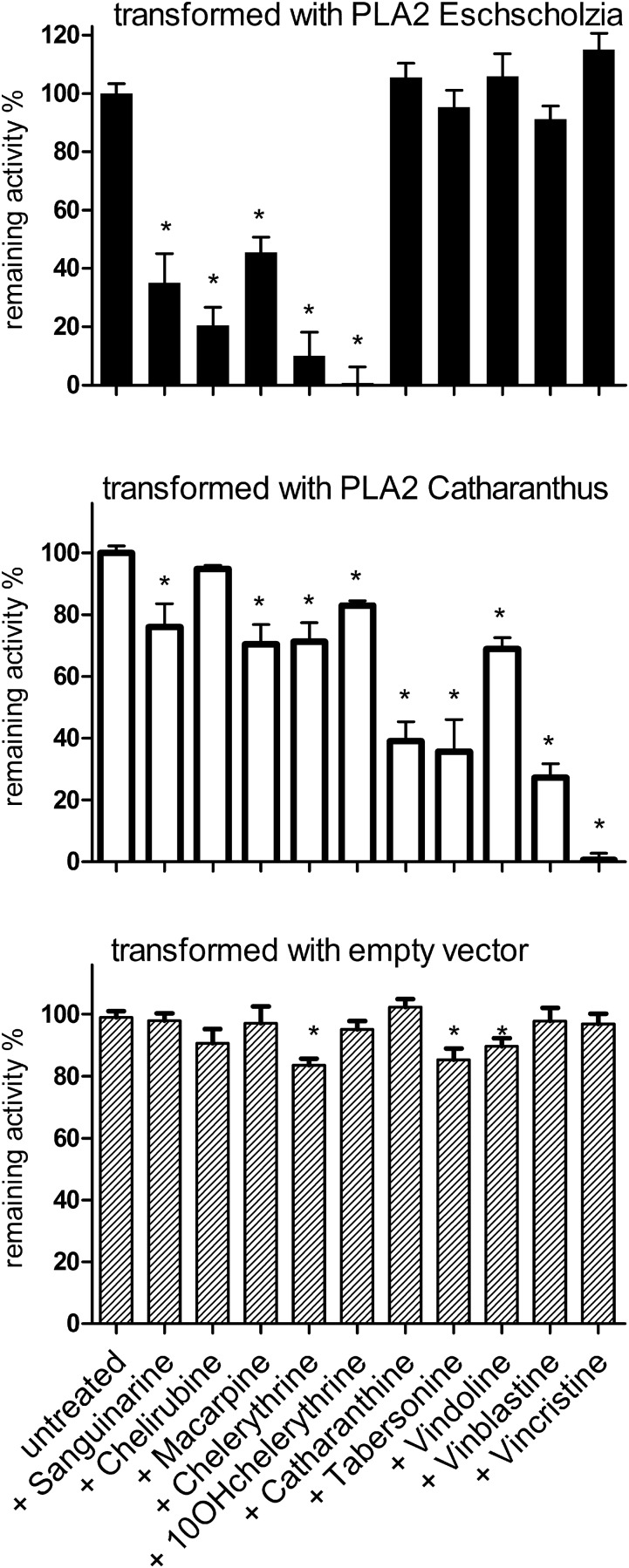
While coexistence of host PLA2 and recombinant PLA2 in the plasma membranes of infected leaves cannot be derived from the measurable enzyme activities, the inhibition patterns clearly indicate that the enzyme(s) encoded by the transgene and controlled by the Tobacco mosaic virus (TMV) promoter dominate the available activity. Specifically, the PLA2 of N. benthamiana is only weakly inhibited by benzophenanthridines or MIAs (e.g., chelerythrine and tabersonine) without a preference for either group of alkaloids (Figure 9, bottom). This PLA2 has low abundance in the infected leaves, as its nonselective inhibition pattern is rarely detectable; for example, tabersonine inhibits N. benthamiana PLA2 by ∼15% but does not significantly inhibit the PLA2 in leaves transformed with E. californica PLA2 (Figure 10, bottom and top). The background activity contributed by the host PLA2 can thus be assumed to stay within the limits of assay variability.
Figure 9.
Inhibition by Benzophenanthridines and MIAs of PLA2 Activities in Plasma Membranes of Transformed N. benthamiana Leaves.
Leaves of N. benthamiana were transiently infected via Agrobacterium tumefaciens with plasmids that express PLA2 genes under the control of the TMV promoter (cf. Methods for details). The transferred ORFs were previously identified in cell cultures of E. californica (data in upper panel) or C. roseus (data in middle panel). Leaves infected with the empty vector (bottom panel) were processed similarly and display the PLA2 activity of the host plant. Plasma membranes were isolated from the infected leaves and PLA2 activities assayed in the nonstimulated state as in Figure 8. Data are means ± se; n = 4 leaf samples. They are normalized to the mean PLA2 activity of similarly treated parallel samples, assayed in the absence of alkaloids, which is set to 100% (representing ∼5 pkat/mg membrane protein). Asterisks indicate significant (P < 0.05) inhibition of this activity by alkaloids. The experiment was done with one set of parallel treated greenhouse plants. Repetition with another set resulted in a similar relation of PLA2 activities and susceptibility to the alkaloids tested (cf. Supplemental Figure 5).
Figure 10.
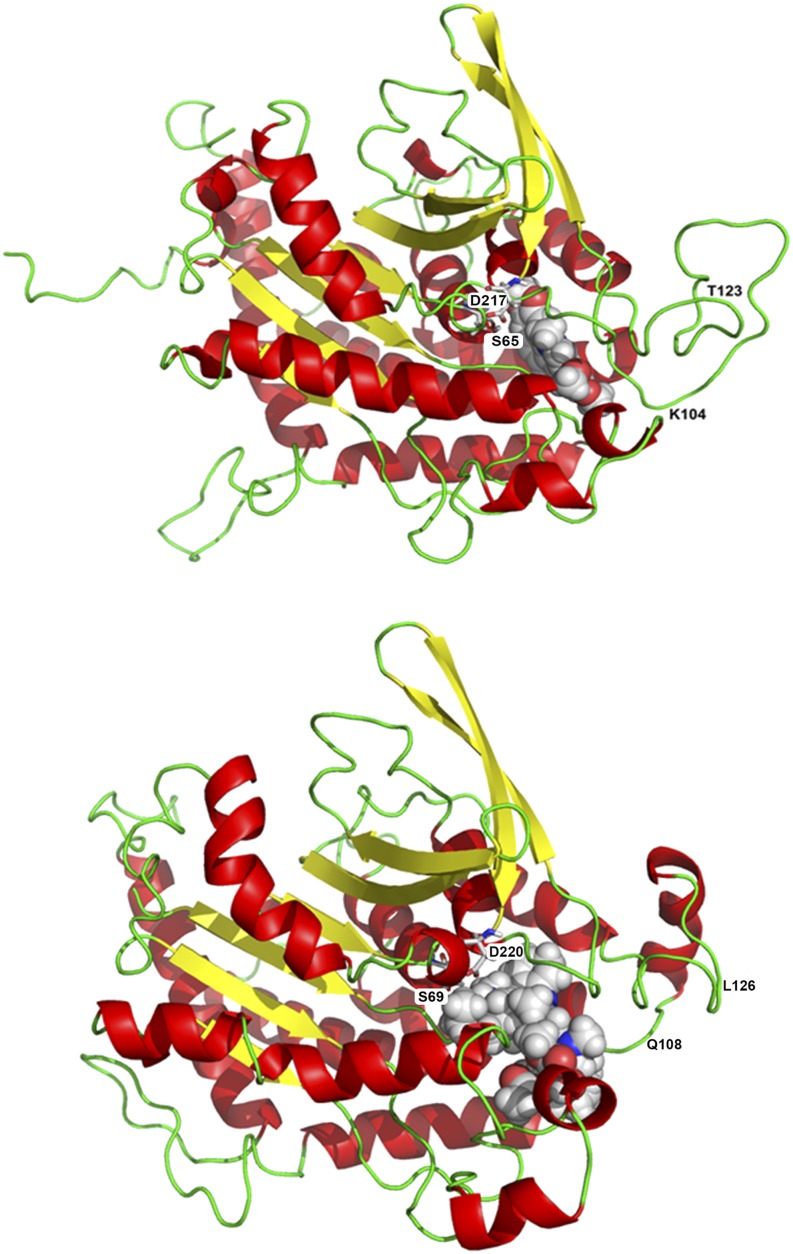
3D Homology Models of PLA2 from E. californica and C. roseus.
Peptide sequences of PLA2, derived from the used cell strains as described in Methods, were modeled to the template patatin 17 from S. cardiophyllum.
(A)Tertiary structure of E. californica PLA2, with docked 10-hydroxychelerythrine (space fill representation). The catalytic dyad formed by Ser-65 and Asp-217 represents the active site of the enzyme. Residues Thr-123 and Lys-104 represent the structure with the strongest deviation from the homologous enzyme of C. roseus.
(B) Tertiary structure of C. roseus PLA2 with docked vinblastine (space fill representation). The catalytic dyad formed by Ser-69 and Asp-220 represents the active site of the enzyme. Residues Gln-108 and Leu-126 represent the moiety with the strongest deviation from the homologous enzyme of E. californica.
In contrast, the plasma membranes of transformed N. benthamiana leaves display PLA2 activities that largely differ in their susceptibility to the test alkaloids: Transformation with the E. californica PLA2 gives rise to a plasma membrane enzyme that can be inhibited by the benzophenanthridines tested, mostly 10-OH-chelerythrine, while the monoterpenoid indole alkaloids cause little or no effect (Figure 9, top). On the contrary, transformation with the PLA2 gene of C. roseus results in a plasma membrane whose PLA2 activity is strongly inhibited by the dimeric MIAs vincristine and vinblastine but less by other MIAs and all tested benzophenanthridines (Figure 9, middle). This experiment also confirms that the transferred C. roseus gene (Caros024612.1) encodes a plasma membrane-bound PLA2.
At the used concentrations of 10 µM, which is within the range measurable in the outer medium of elicited cultures, the most active alkaloid species of either class deactivate most or all of the available PLA2 activity (Figure 9, top and middle). In the alkaloid-producing cell cultures as well as in the transgenic leaves, the inhibition by species-specific alkaloids was generally more efficient than the unspecific inhibition by these toxins of the PLA2 from the other plant or the N. benthamiana host (compare Figures 8 and 9).
These data indicate that the selective inactivation of each PLA2 by alkaloids of the “own” species, as originally discovered in cultured cells of E. californica and C. roseus, can be established in a foreign host plant by the transfer of the encoding gene. Thus, the specific inhibition by distinct alkaloids is a property of the enzyme rather than of its membrane-associated activation mechanism.
In a search for the molecular background of the alkaloid effects, we compared the tertiary structures of PLA2 from E. californica and C. roseus. To this end, homology models were established with the template patatin (PDB: 1OXW_A, from Solanum), a PLA2 that harbors the typical structural elements of the patatin-like phospholipases (Rydel et al., 2003; Ryu, 2004). The PLA2 protein sequences to be assembled were derived from the cloned ORF of PLA2 in E. californica or the Caros024612.1 gene of C. roseus, which were both identified in the used cell cultures (see Methods). All protein sequences used for modeling are included in the alignment of Figure 2 (rows 1 to 3).
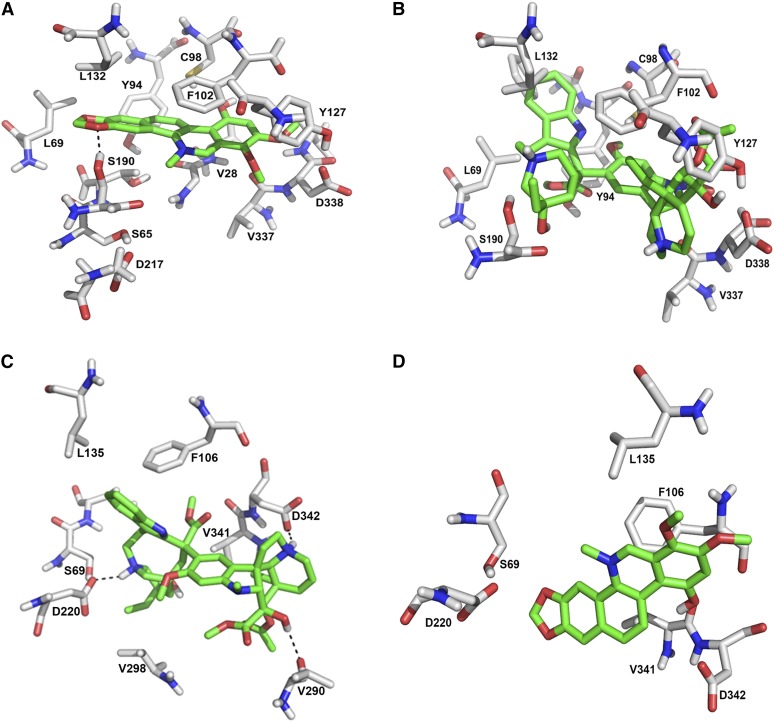
The resulting 3D structures, shown in Figure 10, display the expected similarities of two patatin-like phospholipases, but also substantial differences. These include binding areas that accommodate the tested alkaloids in close proximity to the catalytic center. A detailed view at the alkaloid binding sites (Figure 11), shows a high degree of complementarity to alkaloids of the same species: In the enzyme protein from E. californica, the strong inhibitor 10-hydroxychelerythrine fits well into a binding pocket, which fixes this compound by a hydrogen bond and three hydrophobic interactions. By contrast, the bulky structure of vincristine does not appear to find compatible binding sites. Vincristine fits well into an alkaloid binding pocket of the PLA2 from C. roseus, where it is precisely fixed by two salt bridges. However, the C. roseus enzyme model is hardly adaptable to the benzophenanthridine 10-hydroxychelerythrine, which might even cause repulsive interactions.
Figure 11.
Comparison of Alkaloid Binding Sites in E. californica and C. roseus PLA2.
The alkaloid binding pocket of either enzyme model is shown in two optimized docking arrangements: left, with the alkaloid that allows the highest interaction energy with this enzyme; right, with the alkaloid that allows the highest interaction energy with the enzyme of the other plant.
(A) Docking arrangement of 10-hydroxychelerythrine in the PLA2 of E. californica. The binding is stabilized by a hydrogen bond with Ser-190 and several hydrophobic interactions, e.g., with Val-28, Phe-102, and Tyr-94.
(B) Docking arrangement of vincristine at the PLA2 of E. californica. There are no favored interactions and the very bulky structure of vincristine tends to overlap with residues in the active site (Tyr-127 and Phe-102) and may cause reduced affinity.
(C) Docking arrangement of vincristine in the PLA2 of C. roseus. The binding is highly favored by the formation of two salt bridges between the protonated nitrogen atoms of the ligand with Asp-220 and Asp-342.
(D) Docking arrangement of 10-hydroxychelerythrine in the PLA2 of C. roseus. There are no favored interactions. The side chain of Asp-220 and backbone carbonyls of other amino acids undergo repulsive interactions with the ligand, which likely cause reduced affinity.
The visual impressions were confirmed by docking studies and a final comparison of the estimated interaction energies with the degree of inhibition caused by the same alkaloid (Table 1). The interaction data calculated from the model of E. californica PLA2 agree with the degree of inhibition of enzyme activity, with 10-hydroxychelerythrine being the most effective and sanguinarine the least effective benzophenanthridine in view of both inhibition and binding. With the exception of chelirubine, the effects of the other benzophenanthridines display a similar parallelity.
Table 1. Alkaloid Effects on PLA2 in E. californica and C. roseus.
| PLA2 of E. californica
|
PLA2 of C. roseus
|
|||
|---|---|---|---|---|
| Compound | Inhibition (%) by 10 µM | Interaction Energy (kcal/mol) | Inhibition (%) by 10 µM | Interaction Energy (kcal/mol) |
| Sanguinarine | 28 | −552 | 11 | −485 |
| Chelirubine | 80 | −590 | 18 | −542 |
| Macarpine | 65 | −608 | 20 | −605 |
| Chelerythrine | 61 | −583 | 16 | −550 |
| 10-OH-chelerythrine |
98 |
−630 |
14 |
−539 |
| Tabersonine | 14 | −311 | 33 | −415 |
| Catharanthine | 11 | −109 | 48 | −528 |
| Vindoline | 13 | −289 | 42 | −495 |
| Vincristine | 13 | Steric clash | 70 | −914 |
| Vinblastine | 4 | Steric clash | 65 | −913 |
For a collection of benzophenathridine and monoterpene indole alkaloids, the inhibition of both enzyme activities is compared to the proposed strength of interaction with each modeled enzyme protein. PLA2 activities were assayed experimentally with the nonstimulated enzyme (as in Figure 8); interaction data are calculated from docking studies. “Interaction energy” summarizes the interaction between ligand and enzyme plus the efforts caused by conformational changes in the enzyme structure (induced fit). Desolvation energy efforts for the ligands alone or for the enzyme are not considered. A horizontal line separates benzophenanthridines (top) and MIA (bottom). The highest value of each category appears in bold.
The monoterpenoid indole alkaloids all showed much lower interaction energies (vincristine and vinblastine did not even yield sufficient docking data at this pocket), which is accordant with their lacking effects on PLA2 activity.
Although generally higher than with the E. californica enzyme, the interaction energies obtained with the C. roseus PLA2 protein model confirm the high preference of the alkaloid binding pocket to vincristine or vinblastine over the other MIAs and the benzophenanthridines. In this respect, they correlate well with the pronounced inhibition of C. roseus PLA2 activity by these final, dimeric products of MIA biosynthesis.
DISCUSSION
Our data describe a far-reaching negative feedback control exerted by alkaloid phytoalexins over the expression of their own biosynthesis. It is based on the unique property of a plasma membrane-associated PLA2, i.e., its sensitivity to inhibition by self-made alkaloids. This mechanism prevents the elicitation process from continuing indefinitely and thus protects the alkaloid-producing cells from self-intoxication.
In E. californica, this PLA2 is clearly identified as an essential element of the signal transduction cascade acting between elicitor contact and the induction of biosynthetic enzymes (Viehweger et al., 2002, 2006, Heinze et al., 2007; this article). Here, the alkaloid feedback mechanism blocks the transfer of the elicitor-derived signal, consistent with the phenotype of low PLA2 mutants, established by specific gene silencing.
Although in C. roseus the role of PLA2 in the elicitor-triggered signal transfer was not yet investigated to the same depth, we see striking analogies to the E. californica system: C. roseus alkaloids inhibit the elicitor-triggered activation of both PLA2 and alkaloid production, and the PLA2 product LPC has an elicitor-like effect. They indicate a similar role of this enzyme in signaling and control of the overexpression of alkaloid biosynthesis.
The similarity of alkaloid effects on native and recombinant phospholipases of the plasma membrane leaves little doubt that PLA2 carries a selective susceptibility toward alkaloids of the plant of origin, with 10-OH chelerythrine and vincristine, respectively, as the most active inhibitors. They substantiate the existence of a specific alkaloid pocket in each PLA2, as suggested from the protein models. The relationship between calculated binding energies, inhibition of enzyme activity, and suppression of elicitor-triggered alkaloid biosynthesis all identified the same, most-active alkaloid of either group.
Given the basic divergence in alkaloid sensitivity between the PLA2 of E. californica and C. roseus, as demonstrated in this article, the inhibitory efficiency of individual alkaloids within the benzophenanthridine, or MIA, class requires more detailed investigation. Some minor differences between the PLA2s of E. californica or C. roseus and the ectopically expressed recombinant enzymes remain to be explained. An example is the different ranking of benzophenanthridines by their inhibitory power between Figures 8 and 9 (e.g., macarpine versus sanguinarine). More detailed insights into the alkaloid-PLA2 interactions would require templates for 3D modeling that reflect the structure of a membrane-associated PLA2 better than the actually available patatin, as well as high-throughput analyses of PLA2 activity that test more alkaloid structures, including sets of alkaloids produced in a distinct plant. Furthermore, the nonelicitable PLA2 of the E. californica plasma membrane is likewise inhibited by benzophenanthridines, as judged from the almost parallel decrease of PLA2 activity in elicited, elicitor-free, and nonelicitable cell strains (compare Figures 5B and 6). Attempts to characterize this enzyme, which was not in the scope of this paper, together with intracellular PLA2 might help to trace the evolutionary origin of the alkaloid pocket in the enzyme structure. Groups active in this field are invited to cooperate.
This study allows us to conclude that two evolutionarily distant plants, E. californica (Papaveraceae) and C. roseus (Apocynaceae); both harbor in their plasma membrane an elicitor-stimulated phospholipase with a molecular filter that discriminates self-made from foreign alkaloids. These alkaloid binding sites might thus be considered as molecular fingerprints that reflect evolutionary adaptations to the overexpression of species-specific phytoalexins. A selection pressure favoring such adaptations is most probably exerted by the cytotoxicity of the alkaloids: The overproduction of benzophenanthridines forced by increasing pathogen pressure, for instance, at high elicitor concentrations, causes severe intoxication of the producing cells (Weiss et al., 2006) comparable to those seen after silencing of the yield-controlling enzyme sanguinarine reductase (Müller et al., 2014). The precursor alkaloids, i.e., dihydrobenzophenanthridines, are less toxic to intact cells and less inhibitory to PLA2 (Müller et al., 2014; M. Heinze and W. Roos, unpublished data).
Similiarly, vincristine and vinblastine, the strongest inhibitors of PLA2 of C. roseus, are well known for their strong affinity to tubulin compared with their less affine monomeric precursors vindoline and catharanthine. The formation of dimeric alkaloids on demand is therefore considered a measure against self-intoxication (Sertel et al., 2011). In planta, dimerization to the cytotoxic dimers occurs in a final, oxygen-dependent step (Sottomayor et al., 2004) and thus parallels the formation of benzophenanthridines of E. californica at the ultimate enzyme dihydrobenzophenanthridine oxidase (Figure 1).
The far-reaching feedback seen in the two plants of our study is basically different from another mode of specific adaptation to self-made alkaloids, i.e., the positive selection of resistant mutants of an endangered molecular target. One of the rare examples is Camptotheca acuminata and related plants that produce camptothecin, an inhibitor of topoisomerase I. These plants have selectively enriched point mutations in topoisomerase I that confer camptothecin resistance to this enzyme (Sirikantaramas et al., 2008). Compared with this “resistance mode” of adaptation, the “cease induction mode” found in our study might have different consequences as it reduces the risk of self-intoxication and confines the flow of resources into secondary products at a stage prior to the accumulation of toxic levels.
Summarizing our findings, it appears that the evolution of secondary biosynthesis has left product-specific fingerprints in genes and enzymes of high regulatory importance. They may serve as markers in experiments aimed to reveal the embedding of secondary biosynthesis in the process of plant speciation and help to understand comparable adaptation mechanisms in human cells or organs, related to the medical use of plant-derived drugs or toxins. The hitherto underestimated in planta toxicity of specialized plant metabolites may thus become a more valuable source of knowledge to be shared between botanists and pharmacologists.
Even now, the ability of two valuable medicinal plants to discriminate between self-made and foreign secondary products opens new avenues of research in plant biotechnology, e.g., screening for self-regulatory product inhibitions of similar type in different plants and classes of phytoalexins or genetic engineering of cell-based production systems to remove bottlenecks resulting from the sensitivity of signaling enzymes to desired products, e.g., by manipulating the related inhibition sites.
METHODS
Plant Material and Elicitor Treatment
Cell suspensions of Eschscholzia californica and of Catharanthus roseus were cultured in modified Linsmayer-Skoog medium or M21 medium, respectively, on gyrotary shakers at 24°C, as described by Viehweger et al. (2002).
For elicitation experiments, 6- or 7-d-old cell suspensions were filtered without suction at a nylon mesh of 50-μm pore size, washed with 100 mM sorbitol, and resuspended in phosphate-free culture liquid diluted to 75% (50 mg fresh weight per mL).
Yeast elicitor is a glycoprotein fraction prepared from bakers' yeast according to Schumacher et al. (1987) and purified by ultrafiltration (30 kD), fast protein liquid chromatography (anion exchange and size exclusion), and SDS-PAGE. The active fractions are glycoproteins of 30 to 42 kD that contain ∼40% mannose (Roos et al., 1998). Dosage refers to the dry weight of the crude preparation and was usually set at 1 μg/mL, if not indicated otherwise.
The used culture of C. roseus was obtained as callus from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, and adopted to submerged growth as a cell suspension.
Identification and Cloning of PLA2 in E. californica
Based on tryptic peptides isolated from the plasma membrane proteins of E. californica cell cultures, two peptides typical of published PLA2 sequences were identified in an EST database of E. californica (MGIITKV and DLVQFYLDH). Primers based on these sequences (basic_for and basic_rev) and the codon usage of E. californica (www.kazusa.or.jp) amplified a 291-bp PCR fragment of the PLA2 gene. The full-length ORF was obtained via 5′ and 3′ RACE-PCR following the instructions of the Marathon cDNA amplification kit (Clontech). The gene-specific primers used to complete the 5′ end (race5′_re) and the 3′ end (race3′a_fo and race3′b_fo) are listed in Supplemental Table 1. The resulting complete sequences of PLA2 from E. californica are published at GenBank under accession numbers JQ886492 (mRNA) and AFJ92643 (protein). Supplemental Figure 1 provides the gene and amino acid sequence.
Silencing of PLA2 in Cultured Cells
A 622-bp cDNA fragment encoding the active site of PLA2 (positions −29 to 593 in Supplemental Figure 1) was amplified by PCR with primers designed to add flanking by BsaI sites and cloned in sense and antisense orientation into the vector pICH56022 (IconGenetics) following an approved protocol (Engler et al., 2009; Supplemental Figure 2). The vector was cloned in Escherichia coli using kanamycin resistance and extracted (GeneJET plasmid miniprep kit; Thermo Scientific) and the DNA used for biolistic bombardment (for details, see Heinze et al., 2013). The stably transformed cell lines were grown over three 8-d passages on paromomycin-containing selection agar.
The extent of silencing, i.e., the amount of remaining mRNA, was estimated by RT-PCR with PLA2-specific primers. To avoid the amplification of sequences from the incorporated RNAi vector, one primer (rtpcr_re; Supplemental Table 1) was designed not to bind PLA2-derived sequences of this vector. Expectedly, PCR products were obtained with the plant cDNA only, but not with vector DNA. The mRNA levels detected in this way were substantially below the wild-type levels (Supplemental Figure 3).
Identification of the PLA2 Gene of C. roseus
Genomic DNA was isolated from the used C. roseus culture and used as a template for a PCR-based search. Primers derived from EST_14671_iso_2 (patatin T5, C. roseus EST library of Michigan State University; http://www.medicinalplantgenomics.msu.edu) amplified a PCR product with typical PLA2 sequences that in turn allowed us to identify the full-length clone in the ORCAE gene library of the University of Gent (http://bioinformatics.psb.ugent.be/orcae/) under the annotation Caros024612.1. Figure 2, line 2, shows the complete ORF, which was amplified as an identical sequence from the DNA of the used C. roseus culture using the primers crcpla1_fo and crcpla2_re in Supplemental Table 1.
Heterologous Expression of PLA2 in Nicotiana benthamiana
The gene encoding the elicitor-responsive PLA2 protein of the E. californica plasma membrane (JQ886492, identified as described above) was cloned into the target vector pICH41308 using the BsaI-based Golden Gate cloning method (Engler et al., 2009; Weber et al., 2011). To this end, it was necessary to remove the restriction sites BsaI, BpiI, and Esp3I via silent mutations, using the PCR primer combinations plas1 to plas10 in Supplemental Table 1. A similar strategy was followed with the C. roseus gene Caros024612.1, which was identified in the used cell culture as described above. The complete gene was synthesized commercially (Life Technologies), and the restriction sites BsaI and Esp3I were then removed by silent mutations for subcloning in Golden Gate cloning vectors. Prior to infection, gene sequences lacking typeIIS restriction enzymes were confirmed by sequencing using primers moClo_fo and moClo_re (Supplemental Table 1).
Each gene was then subcloned in a TMV-based viral vector (pL1F-2/pICH75055) (Weber et al., 2011) and the resulting construct transformed in Agrobacterium tumefaciens strain GV3101:pMP90 by electroporation (Marillonnet et al., 2004). Leaves of 4-week-old greenhouse-grown N. benthamiana plants were infected by infiltration of transformed Agrobacterium strains using a syringe without needle. Infiltrated leaf areas were cut 4 d after infiltration and frozen in liquid nitrogen. Fifteen-gram portions were used for the preparation of plasma membranes as described below. RNA prepared from the same samples was used to prove the transformation by RT-PCR with gene specific primers, i.e., plas1_fo/plas8_re for PLA2 E. californica and cpla1_fo/cpla6_re for PLA2 C. roseus (Supplemental Table 1).
Cell Fractionation and Preparation of Solubilized Plasma Membrane Proteins
The plasma membrane of cultured cells of E. californica and C. roseus was isolated by cell fractionation and ultracentrifugation, followed by two-phase aqueous partitioning and solubilization by 0.5% sodium cholate (Heinze et al., 2013). Marker enzyme activities [NAD(P)H-cytochrome c oxidoreductase for endoplasmic reticulum, α-mannosidase for tonoplast, and erythrosin B-sensitive H+-ATPase for plasma membrane], assayed as described by Pönitz and Roos (1994), indicated that the preparation contained <10% endoplasmic reticulum and <2% tonoplast.
Assay of PLA2 Activity
Samples of the cholate-solubilized plasma membrane were incubated in black 96-well microplates with 400 nM of the artificial substrate bis-BODIPY-FL-C11-PC (Molecular Probes). Substrate hydrolysis was quantified by monitoring the initial rate of fluorescence increase at excitation of 485 nm and emission of 528 nm in a microplate fluorescence reader (FLX800; BioTek), following a previously published protocol (Heinze and Roos, 2013). If indicated, PLA2 activity was stimulated by adding 1 µg/mL of yeast elicitor plus 10 µM GTP at 10 min prior to the substrate. The PLA2 of C. roseus required 20 µg/mL of yeast elicitor preparation for full stimulation.
Alkaloids were added 10 min prior to the substrate, both to the nonstimulated and the stimulated enzyme. The PLA2 activity remaining after alkaloid treatment was estimated as a part of the maximum conversion rate assayed in parallel, alkaloid-free samples.
Assay of Alkaloid Production
Benzophenanthridine Alkaloids
The fluorimetric assay of the total benzophenanthridine alkaloid content was done as described in Angelova et al. (2010). In brief, aliquots of the cell suspension were incubated with methanol/KOH and centrifuged and the supernatant acidified with H2SO4. Alkaloid fluorescence was read at excitation of 360 nm and emission of 460 nm for dihydrobenzophenanthridines and excitation of 460 nm and emission of 570 nm (benzophenanthridines). Fluorescence intensities were calibrated to alkaloid concentrations using sanguinarine and dihydrosanguinarine as standard compounds. For HPLC-based analyses, alkaloids were extracted with ethylacetate, the solvent evaporated and the reminder dissolved in methanol. This solution was separated essentially as described by Weiss et al. (2006), except that a Dionex Ultimate 3000 HPLC system, equipped with a PDA-3000 diode array detector, was used at a flow rate of 1 mL/min. To confirm the identity of individual alkaloids, peak fractions were analyzed by electrospray ionization (ESI) mass spectrometry (MS) with an ESQUIRE-LC ion trap mass spectrometer (Bruker Daltoniks). The same benzophenantridines had also been identified in previous studies, e.g., Müller et al. (2014), using UPLC-ESI-MS/MS and ESI-FTICR-MS. The alkaloids routinely detected in elicited cultures are shown in Figure 5, top. Quantification was based on OD280 and calibrated using authentic sanguinarine and dihydrosanguinarine as references. Corrections for other alkaloids were derived from their UV spectra compared with the reference compounds.
MIAs
Alkaloids were extracted from 7-d-old C. roseus cultures with methanol/chloroform, speed vac-dried after solvent evaporation, and separated by HPLC with the same equipment as described above. The mobile phase gradient (phosphate/acetonitrile) was used as optimized by Tikhomiroff and Jolicoeur (2002), and the MIAs were eluted in the same order as published by these authors.
Alkaloid peaks were identified by UV spectra, which confirmed data of Hisiger and Jolicoeur (2007) and confirmed by ESI-MS of the peak fractions. The set of alkaloids routinely detected in elicited cultures is shown in Figure 7, top. The presence of the dimeric compounds vinblastine and vincristine is also supported by MS spectra. Vincristine was identified from a mass peak at m/z 825 (MW) and fragment ion peaks at m/z = 807, 783, 765, 733, 723, 687, and 353; mass peaks indicating vinblastine were recorded at m/z = 680, 569, 542, 524, 510, 508, 492, 490, 482, 406, and 381, concordantly with ESI-MS data published by Dubrovay et al. (2013) and Zhou et al. (2005). UV spectra of extracted vinblastine and vincristine are shown in Supplemental Figure 6.
Quantification of the peak fractions was based on OD280 and calibrated using authentic vinblastine as a reference.
Homology Modeling and Docking Studies
3D protein models of PLA2 from E. californica and C. roseus were created and refined with the md-refinement option of YASARA (Krieger et al., 2009). YASARA identified the 3D structure of SeMet Patatin (1, PDB code: 1OXW) with a sequence identity of ∼43% to the query enzymes. After refinement, the quality of the models was checked for stereochemical quality by the PROCHECK software (Laskowski et al., 1993). This check confirmed that 90% of all amino acids lay in most favored areas of the Ramachandran plot, with three outliers from loop regions outside of the active sites. All general criteria of model quality were in the range predicted by PROCHECK or even better (overall G-factor). The putative native folding was analyzed by PROSA II (Sippl, 1990, 1993), and the graphical analyses showed nearly all residues in areas of negative energies except of a few loop regions. All these criteria strongly indicate a good model quality of the assumed native-like folding.
Thirty docking runs were performed for all ligands using GOLD (Verdonk et al., 2003; Hartshorn et al., 2007) with the Gold-Score fitness function and with standard settings. For the definition of the active site, in the PLA2 from E. californica the Cα atom of C98 and in the PLA2 from C. roseus the corresponding Cα of C102 was chosen as origin with a binding region of 18 Å. The following amino acid side chains were set to be flexible by applying the rotamer library included in GOLD: in the E. californica enzyme Cys-98, Phe-102, Tyr-127, Ser-190, and Thr-340, in the C. roseus enzyme Tyr-130, Tyr-208, Lys-129, Phe-106, Gln-108, and Asp-342.
The docking results were analyzed manually using Molecular Operating Environment, 2011.10 (MOE), and for the most favored poses from the Gold-score fitness values, the enzyme-ligand complexes were optimized using the Amber12-EHT force field (developed from Chemical Computing Group) to capture induced fit effects and to calculate more reasonable interaction energies between the ligands and the enzyme, compared with docking scores.
Energy efforts for desolvation and entropy effects of the ligands and the binding pocket were not included in the calculation. Due to the similarity of the alkaloids in each group, it is very likely that this correction does not change the relations between the interaction energies given in Table 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/NCBI and other databases under the following accession numbers:PLA2 of E. californica, GenBank JQ886492 (mRNA) and AFJ92643 (protein); PLA2 of C. roseus, Caros024612.1 (ORCAE gene library, University of Gent).
Supplemental Data
Supplemental Figure 1. Gene and Protein of PLA2 of E. californica with Sequences Relevant for Cloning and Silencing.
Supplemental Figure 2. DNA Map of the RNAi Vector Used for Silencing of PLA2.
Supplemental Figure 3. PLA2 mRNA Levels Assayed by RT-PCR in Wild-Type and Silenced Cultures.
Supplemental Figure 4. Coding Sequences of the PLA2 Genes of E. californica and C. roseus Used for Heterologous Expression in Nicotiana benthamiana.
Supplemental Figure 5. Heterologous Expression of PLA2 Genes from E. californica and C. roseus in Leaves of N. benthamiana (RT-PCR)
Supplemental Figure 6. Inhibition by Test Alkaloids of PLA2 Activities in Plasma Membranes of Transgenic N. benthamiana Leaves.
Supplemental Figure 7. UV Spectra of the Dimeric Alkaloids Vinblastine and Vincristine Extracted from C. roseus Cell Suspension Cultures.
Supplemental Table 1. Primers Used for the Identification and Modification of PLA2 Genes.
Supplementary Material
Acknowledgments
The work was supported by the German Research Council (Grant RO 889-12 to W.R.) and the Martin-Luckner Foundation (M.H.). We thank Carola Engler (Icon Genetics, Halle) for help with the construction of the RNAi vector, Angelika Schierhorn (Max-Planck-Unit for Enzymology of Protein Folding, Halle) for the ESI-MS of alkaloids, Ramona Grützner (IPB Halle) for cloning the constructs for PLA2 expression in N. benthamiana and plant infection, and Gabriele Danders (University of Halle) for careful cell cultivation and HPLC analyses. We thank J. Stöckigt, University of Mainz, for providing a set of C. roseus alkaloids.
AUTHOR CONTRIBUTIONS
M.H. did most laboratory experiments and contributed to the article. W.B. did the homology modeling. S.M. designed the Golden Gate cloning vectors for recombinant expression. W.R. coordinated the work and wrote the article.
Glossary
- LPC
lysophosphatidylcholine
- ORF
open reading frame
- RNAi
RNA interference
- MIA
monoterpenoid indole alkaloid
- TMV
Tobacco mosaic virus
- ESI
electrospray ionization
- MS
mass spectrometry
References
- Angelova S., Buchheim M., Frowitter D., Schierhorn A., Roos W. (2010). Overproduction of alkaloid phytoalexins in California poppy cells is associated with the co-expression of biosynthetic and stress-protective enzymes. Mol. Plant 3: 927–939. [DOI] [PubMed] [Google Scholar]
- Barták P., Šimánek V., Vlčková M., Ulrichová J., Vespalec R. (2003). Interactions of sanguinarine and chelerythrine with molecules containing a mercapto group. J. Phys. Org. Chem. 16: 803–810. [DOI] [PubMed] [Google Scholar]
- Blechert S., Brodschelm W., Hölder S., Kammerer L., Kutchan T.M., Mueller M.J., Xia Z.Q., Zenk M.H. (1995). The octadecanoic pathway: Signal molecules for the regulation of secondary pathways. Proc. Natl. Acad. Sci. USA 92: 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.-Y., Lee-Parsons C.W.T., Yoon S.-Y.H., Rhee H.S., Park J.M. (2007). Enhanced benzophenanthridine alkaloid production and protein expression with combined elicitor in Eschscholtzia californica suspension cultures. Biotechnol. Lett. 29: 2001–2005. [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix I., Khan M.F., Schriemer D.C., Cram D., Nowak J., Facchini P.J. (2010). Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 10: 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich H., Kutchan T.M. (1991). Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc. Natl. Acad. Sci. USA 88: 9969–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovay Z., Háda V., Béni Z., Szántay C. Jr. (2013). NMR and mass spectrometric characterization of vinblastine, vincristine and some new related impurities - part I. J. Pharm. Biomed. Anal. 84: 293–308. [DOI] [PubMed] [Google Scholar]
- Engler C., Gruetzner R., Kandzia R., Marillonnet S. (2009). Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini P.J., De Luca V. (2008). Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 54: 763–784. [DOI] [PubMed] [Google Scholar]
- Färber K., Schumann B., Miersch O., Roos W. (2003). Selective desensitization of jasmonate- and pH-dependent signaling in the induction of benzophenanthridine biosynthesis in cells of Eschscholzia californica. Phytochemistry 62: 491–500. [DOI] [PubMed] [Google Scholar]
- Gundlach H., Müller M.J., Kutchan T.M., Zenk M.H. (1992). Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. USA 89: 2389–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel J.M., Facchini P.J. (2013). Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 54: 647–672. [DOI] [PubMed] [Google Scholar]
- Haider G., von Schrader T., Füsslein M., Blechert S., Kutchan T.M. (2000). Structure-activity relationships of synthetic analogs of jasmonic acid and coronatine on induction of benzo[c]phenanthridine alkaloid accumulation in Eschscholzia californica cell cultures. Biol. Chem. 381: 741–748. [DOI] [PubMed] [Google Scholar]
- Hartshorn M.J., Verdonk M.L., Chessari G., Brewerton S.C., Mooij W.T., Mortenson P.N., Murray C.W. (2007). Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 50: 726–741. [DOI] [PubMed] [Google Scholar]
- Heinze M., Herre M., Massalski C., Hermann I., Conrad U., Roos W. (2013). Signal transfer in the plant plasma membrane: phospholipase A(2) is regulated via an inhibitory Gα protein and a cyclophilin. Biochem. J. 450: 497–509. [DOI] [PubMed] [Google Scholar]
- Heinze M., Roos W. (2013). Assay of phospholipase A activity. In Methods in Molecular Biology, Plant Lipid Signaling Protocols, Vol. 1009, Munnik T. and Heilmann I., eds (New York: Humana Press, Springer Science and Business Media; ), pp. 241–249. [DOI] [PubMed] [Google Scholar]
- Heinze M., Steighardt J., Gesell A., Schwartze W., Roos W. (2007). Regulatory interaction of the Galpha protein with phospholipase A2 in the plasma membrane of Eschscholzia californica. Plant J. 52: 1041–1051. [DOI] [PubMed] [Google Scholar]
- Hisiger S., Jolicoeur M. (2007). Analysis of Catharanthus roseus alkaloids by HPLC. Phytochem. Rev. 6: 207–234. [Google Scholar]
- Klein, M., and Roos, W. (2009). Handling dangerous molecules: transport and compartmentation of plant natural products. In Plant-Derived Natural Products: Synthesis, Function, and Application, A.E. Osbourn and V. Lanzotti, eds (Dordrecht, The Netherlands: Springer Science and Business Media; ), pp. 229–267. [Google Scholar]
- Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. (2009). Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77 (suppl. 9): 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. (1993). Procheck - A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Leonard E., Runguphan W., O’Connor S., Prather K.J. (2009). Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat. Chem. Biol. 5: 292–300. [DOI] [PubMed] [Google Scholar]
- Marillonnet S., Giritch A., Gils M., Kandzia R., Klimyuk V., Gleba Y. (2004). In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 101: 6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J., Verpoorte R., Kijne J.W. (2001). ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci. 6: 212–219. [DOI] [PubMed] [Google Scholar]
- Moreno P.R.H., Poulsen C., van der Heijden R., Verpoorte R. (1996). Effects of elicitation on different metabolic pathways in Catharanthus roseus (L.)G.Don cell suspension cultures. Enzyme Microb. Technol. 18: 99–107. [Google Scholar]
- Müller H., Heinze M., Heinke R., Schmidt J., Roos W. (2014). A non-biosynthetic enzyme controls alkaloid biosynthesis in cultured cells of Eschscholzia californica. Plant Cell Tissue Organ Cult. 119: 661–676. [Google Scholar]
- Mujib A., Ilah A., Aslam J., Fatima S., Siddiqui Z.H., Maqsood M. (2012). Catharanthus roseus alkaloids: application of biotechnology for improving yield. Plant Growth Regul. 68: 111–127. [Google Scholar]
- Pasquali G., Goddijn O.J.M., de Waal A., Verpoorte R., Schilperoort R.A., Hoge J.H.C., Memelink J. (1992). Coordinated regulation of two indole alkaloid biosynthetic genes from Catharanthus roseus by auxin and elicitors. Plant Mol. Biol. 18: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Pauw B., van Duijn B., Kijne J.W., Memelink J. (2004). Activation of the oxidative burst by yeast elicitor in Catharanthus roseus cells occurs independently of the activation of genes involved in alkaloid biosynthesis. Plant Mol. Biol. 55: 797–805. [DOI] [PubMed] [Google Scholar]
- Pönitz J., Roos W. (1994). A glucose-activated electron transfer system in the plasma membrane stimulates the H(+)-ATPase in Penicillium cyclopium. J. Bacteriol. 176: 5429–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos W., Dordschbal B., Steighardt J., Hieke M., Weiss D., Saalbach G. (1999). A redox-dependent, G-protein-coupled phospholipase A of the plasma membrane is involved in the elicitation of alkaloid biosynthesis in Eschscholtzia californica. Biochim. Biophys. Acta 1448: 390–402. [DOI] [PubMed] [Google Scholar]
- Roos W., Evers S., Hieke M., Tschope M., Schumann B. (1998). Shifts of intracellular pH distribution as a part of the signal mechanism leading to the elicitation of benzophenanthridine alkaloids. Phytoalexin biosynthesis in cultured cells of Eschscholtzia californica. Plant Physiol. 118: 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos W., Viehweger K., Dordschbal B., Schumann B., Evers S., Steighardt J., Schwartze W. (2006). Intracellular pH signals in the induction of secondary pathways—the case of Eschscholzia californica. J. Plant Physiol. 163: 369–381. [DOI] [PubMed] [Google Scholar]
- Rydel T.J., Williams J.M., Krieger E., Moshiri F., Stallings W.C., Brown S.M., Pershing J.C., Purcell J.P., Alibhai M.F. (2003). The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42: 6696–6708. [DOI] [PubMed] [Google Scholar]
- Ryu S.B. (2004). Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9: 229–235. [DOI] [PubMed] [Google Scholar]
- Scherer G.F.E., Ryu S.B., Wang X., Matos A.R., Heitz T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15: 693–700. [DOI] [PubMed] [Google Scholar]
- Schmeller T., Latz-Brüning B., Wink M. (1997). Biochemical activities of berberine, palmatine and sanguinarine mediating chemical defence against microorganisms and herbivores. Phytochemistry 44: 257–266. [DOI] [PubMed] [Google Scholar]
- Schumacher H.M., Gundlach H., Fiedler F., Zenk M.H. (1987). Elicitation of benzophenanthridine alkaloid synthesis in Eschscholtzia cell cultures. Plant Cell Rep. 6: 410–413. [DOI] [PubMed] [Google Scholar]
- Schwartze W., Roos W. (2008). The signal molecule lysophosphatidylcholine in Eschscholzia californica is rapidly metabolized by reacylation. Planta 229: 183–191. [DOI] [PubMed] [Google Scholar]
- Sertel S., Fu Y., Zu Y., Rebacz B., Konkimalla B., Plinkert P.K., Krämer A., Gertsch J., Efferth T. (2011). Molecular docking and pharmacogenomics of vinca alkaloids and their monomeric precursors, vindoline and catharanthine. Biochem. Pharmacol. 81: 723–735. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. (1990). Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J. Mol. Biol. 213: 859–883. [DOI] [PubMed] [Google Scholar]
- Sippl M.J. (1993). Recognition of errors in three-dimensional structures of proteins. Proteins 17: 355–362. [DOI] [PubMed] [Google Scholar]
- Sirikantaramas S., Yamazaki M., Saito K. (2008). Mutations in topoisomerase I as a self-resistance mechanism coevolved with the production of the anticancer alkaloid camptothecin in plants. Proc. Natl. Acad. Sci. USA 105: 6782–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaninová I., Táborská E., Bochoráková H., Slanina J. (2001). Interaction of benzo[c]phenanthridine and protoberberine alkaloids with animal and yeast cells. Cell Biol. Toxicol. 17: 51–63. [DOI] [PubMed] [Google Scholar]
- Sottomayor M., Cardoso I.L., Pereira L.G., Barcel A.R. (2004). Peroxidase and the biosynthesis of terpenoid indole alkaloids in the medicinal plant Catharanthus roseus (L.) G Don. Phytochem. Rev. 3: 159–171. [Google Scholar]
- Tikhomiroff C., Jolicoeur M. (2002). Screening of Catharanthus roseus secondary metabolites by high-performance liquid chromatography. J. Chromatogr. A 955: 87–93. [DOI] [PubMed] [Google Scholar]
- van der Fits L., Memelink J. (2001). The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25: 43–53. [DOI] [PubMed] [Google Scholar]
- Vázquez-Flota F., De Luca V., Carrillo-Pech M., Canto-Flick A., de Lourdes Miranda-Ham M. (2002). Vindoline biosynthesis is transcriptionally blocked in Catharanthus roseus cell suspension cultures. Mol. Biotechnol. 22: 1–8. [DOI] [PubMed] [Google Scholar]
- Verdonk M.L., Cole J.C., Hartshorn M.J., Murray C.W., Taylor R.D. (2003). Improved protein-ligand docking using GOLD. Proteins 52: 609–623. [DOI] [PubMed] [Google Scholar]
- Viehweger K., Dordschbal B., Roos W. (2002). Elicitor-activated phospholipase A(2) generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signaling via the activation of Na+-dependent proton fluxes. Plant Cell 14: 1509–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viehweger K., Schwartze W., Schumann B., Lein W., Roos W. (2006). The Galpha protein controls a pH-dependent signal path to the induction of phytoalexin biosynthesis in Eschscholzia californica. Plant Cell 18: 1510–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D., Baumert A., Vogel M., Roos W. (2006). Sanguinarine reductase, a key enzyme of benzophenanthridine detoxification. Plant Cell Environ. 29: 291–302. [DOI] [PubMed] [Google Scholar]
- Wink M., Schmeller T., Latz-Bruning B. (1998). Modes of action of allelochemical alkaloids: Interaction with neuroreceptors, DNA, and other molecular targets. J. Chem. Ecol. 24: 1881–1937. [Google Scholar]
- Zenk M.H. (1994). The formation of benzophenanthridine alkaloids. Pure Appl. Chem. 66: 2023–2028. [Google Scholar]
- Zhou H., Tai Y., Sun C., Pan Y. (2005). Rapid identification of vinca alkaloids by direct-injection electrospray ionisation tandem mass spectrometry and confirmation by high-performance liquid chromatography-mass spectrometry. Phytochem. Anal. 16: 328–333. [DOI] [PubMed] [Google Scholar]
- Ziegler J., Facchini P.J. (2008). Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59: 735–769. [DOI] [PubMed] [Google Scholar]
- Zulak K.G., Cornish A., Daskalchuk T.E., Deyholos M.K., Goodenowe D.B., Gordon P.M.K., Klassen D., Pelcher L.E., Sensen C.W., Facchini P.J. (2007). Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta 225: 1085–1106. [DOI] [PubMed] [Google Scholar]
- Zulak K.G., Khan M.F., Alcantara J., Schriemer D.C., Facchini P.J. (2009). Plant defense responses in opium poppy cell cultures revealed by liquid chromatography-tandem mass spectrometry proteomics. Mol. Cell. Proteomics 8: 86–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.