Loss of Arabidopsis retromer subunits suppresses immune receptor-mediated cell death and affects autophagy-related vacuolar processes, thus implicating retromer trafficking in cell death regulation.
Abstract
Membrane trafficking is required during plant immune responses, but its contribution to the hypersensitive response (HR), a form of programmed cell death (PCD) associated with effector-triggered immunity, is not well understood. HR is induced by nucleotide binding-leucine-rich repeat (NB-LRR) immune receptors and can involve vacuole-mediated processes, including autophagy. We previously isolated lazarus (laz) suppressors of autoimmunity-triggered PCD in the Arabidopsis thaliana mutant accelerated cell death11 (acd11) and demonstrated that the cell death phenotype is due to ectopic activation of the LAZ5 NB-LRR. We report here that laz4 is mutated in one of three VACUOLAR PROTEIN SORTING35 (VPS35) genes. We verify that LAZ4/VPS35B is part of the retromer complex, which functions in endosomal protein sorting and vacuolar trafficking. We show that VPS35B acts in an endosomal trafficking pathway and plays a role in LAZ5-dependent acd11 cell death. Furthermore, we find that VPS35 homologs contribute to certain forms of NB-LRR protein-mediated autoimmunity as well as pathogen-triggered HR. Finally, we demonstrate that retromer deficiency causes defects in late endocytic/lytic compartments and impairs autophagy-associated vacuolar processes. Our findings indicate important roles of retromer-mediated trafficking during the HR; these may include endosomal sorting of immune components and targeting of vacuolar cargo.
INTRODUCTION
Programmed cell death (PCD) plays a central role in many plant processes, most notably during development and pathogen-triggered disease or immunity (Hofius et al., 2007; Bozhkov and Lam, 2011; Coll et al., 2011). The hypersensitive response (HR) is a rapid, localized PCD reaction at the site of attempted pathogen invasion and a hallmark of effector-triggered immunity (ETI). This branch of the plant innate immune system relies on intracellular immune receptors, also known as disease resistance (R) proteins, which monitor the presence or activity of pathogen-derived effector proteins. In most cases, effectors function as virulence determinants of successful pathogens and are deployed into plants cells to manipulate host cell physiology and suppress basal immune responses (Bent and Mackey, 2007; Coll et al., 2011; Maekawa et al., 2011). These basal defenses are part of an ancient branch of the immune system and are activated by extracellular immune receptors upon recognition of pathogen-associated molecular patterns (PAMPs; Schwessinger and Ronald, 2012). In general, PAMP-triggered immunity and ETI share numerous downstream responses, but ETI exhibits higher amplitude and effectiveness (Jones and Dangl, 2006). Thus, ETI efficiently protects against adapted pathogens. However, it is still debated whether effector-triggered hypersensitive PCD is a cause or consequence of disease resistance. While some evidence supports the contribution of HR to growth restriction of strictly biotrophic pathogens (Wang et al., 2011), other evidence indicates that ETI can be separated from the HR (Bendahmane et al., 1999; Bulgarelli et al., 2010; Coll et al., 2010; Heidrich et al., 2011).
Most immune receptors controlling ETI are NB-LRR proteins, named after their central nucleotide binding (NB) and C-terminal leucine-rich repeat (LRR) domains (Caplan et al., 2008). The N-terminal regions include either a Toll/Interleukin-1 Receptor homology (TIR) or a predicted coiled-coil (CC) domain. The molecular mechanisms that regulate and execute ETI downstream of NB-LRR activation are not well known, and diversity in signaling mechanisms is likely (Eitas et al., 2008; Bonardi et al., 2011; Bonardi and Dangl, 2012). In particular, the mechanisms of ETI-associated HR remain ill defined. There is evidence that plants engage multiple routes to cellular demise in response to developmental and environmental cues (Bozhkov and Lam, 2011). A recent attempt to define these types of cell death focused on morphological criteria and proposed a classification into vacuolar cell death including autophagic mechanisms (see below) and necrosis (van Doorn et al., 2011). Importantly, plant HR could not be assigned to either type, since most cell death-associated morphologies show rather mixed and atypical features (van Doorn et al., 2011).
Distinct genetic components and morphological types of vacuole-mediated PCD are implicated in HR in response to activated NB-LRR proteins (Hara-Nishimura and Hatsugai, 2011). For instance, HR conditioned by the CC-NB-LRR proteins RESISTANCE TO P. SYRINGAE PV MACULICOLA1 (RPM1) and RESISTANCE TO P. SYRINGAE2 (RPS2) upon recognition of bacterial effectors relies on a fusion process between the tonoplast and plasma membrane, which results in the discharge of antimicrobial and death-inducing vacuolar content into the apoplast (Hatsugai et al., 2009). This membrane fusion system seems to be proteasome-dependent, as inhibition of the caspase-3-like activity of the 20S proteasome subunit PBA1 impaired HR induction (Hatsugai et al., 2009). By contrast, virus-induced HR triggered by the activated TIR-NB-LRR N protein requires caspase-1-like activity of the vacuolar protease VACUOLAR PROCESSING ENZYME and engages vacuolar membrane collapse and the release of hydrolytic enzymes into the cytosol (Hatsugai et al., 2004).
We have shown that autophagy components have death-promoting functions during the HR (Hofius et al., 2009). Autophagy is a conserved vacuolar pathway for the degradation and recycling of cell contents in eukaryotes and has been implicated in plant development, stress tolerance, and pathogen defense (Liu and Bassham, 2012). Using loss-of-function mutants of Arabidopsis thaliana AUTOPHAGY-RELATED (ATG) genes, we provided genetic evidence that HR conditioned by activated TIR-NB-LRR proteins (i.e., RPS4 and RECOGNITION OF PERONOSPORA PARASITICA1) largely depends on autophagy processes (Hofius et al., 2009). Autophagy components also contribute to HR mediated by RPM1, as do other PCD pathways involving metacaspases, cathepsins, and the proteasome (Hofius et al., 2009; Pajerowska-Mukhtar and Dong, 2009; Coll et al., 2010, 2014; Hackenberg et al., 2013).
The engagement of autophagy and vacuole-mediated execution steps during HR supports the importance of membrane trafficking in plant immunity (Teh and Hofius, 2014). Endocytosis, secretion, and vacuolar transport are implicated in immune receptor activation, signal transduction, and the targeting of defense compounds to sites of pathogen attack, mainly in association with PAMP-triggered immunity (Kwon et al., 2008; Beck et al., 2012; Inada and Ueda, 2014). However, much less is known of how trafficking regulators and endomembrane transport routes contribute to ETI-related HR and resistance responses (Nomura et al., 2011; Engelhardt et al., 2012).
Mutants expressing autoimmunity- and PCD-related phenotypes are good genetic models to identify components of defense and cell death pathways (Moeder and Yoshioka, 2008; Palma et al., 2010; Bonardi et al., 2011). One of these is the recessive accelerated cell death11 (acd11) Arabidopsis mutant, which exhibits constitutive activation of immune responses and PCD due to disruption of a ceramide-1-phosphate transfer protein (Brodersen et al., 2002, 2005; Simanshu et al., 2014). PCD in acd11 is initiated in seedlings at the two- to four-leaf stage and is dependent upon isochorismate-derived compounds, including the phytohormone salicylic acid. Such signaling compounds are metabolized in planta upon transgenic expression of the bacterial salicylate hydroxylase NahG (Heck et al., 2003; Brodersen et al., 2005), leading to full suppression of the lethal phenotype of acd11. In acd11 nahG, cell death is triggered upon application of the salicylic acid agonist benzothiadiazole-S-methyl ester (BTH). We previously used this to isolate acd11 suppressor mutants, termed lazarus (laz) (Malinovsky et al., 2010; Palma et al., 2010). These analyses showed that lethality in acd11 fully depends on the NB-LRR immune receptor LAZ5 and that LAZ2, a histone H3 lysine 36 methyltransferase, is required for LAZ5 expression (Palma et al., 2010). This indicates that loss of ACD11 results in inappropriate LAZ5 activation in the absence of pathogen effector recognition. Similarly, the PHOENIX21/ACTIVATED DISEASE RESISTANCE1-LIKE2 (ADR1-L2) CC-NB-LRR protein is required for autoimmunity in the lesion simulating disease resistance1 (lsd1) mutant (Bonardi et al., 2011). These results indicate that suppressors of at least some autoimmune mutants can define genes required for NB-LRR protein functions.
Here, we report that LAZ4 encodes VACUOLAR PROTEIN SORTING35B (VPS35B), one of three Arabidopsis VPS35 isoforms and a component of the multisubunit retromer complex. Plant retromer functions in endosomal protein sorting and vacuolar trafficking during development but has not been implicated in immunity before (Reyes et al., 2011; Robinson et al., 2012; Nodzynski et al., 2013). We demonstrate that VPS35-dependent trafficking pathways contribute to TIR-NB-LRR and CC-NB-LRR protein-mediated autoimmunity and HR cell death. We also show the specific involvement of retromer in disease resistance conditioned by a TIR-NB-LRR immune receptor. Finally, we provide evidence that retromer mutants are defective in HR-associated autophagic degradation, suggesting important functions of the retromer complex in vacuolar PCD.
RESULTS
Identification of LAZ4/VPS35B
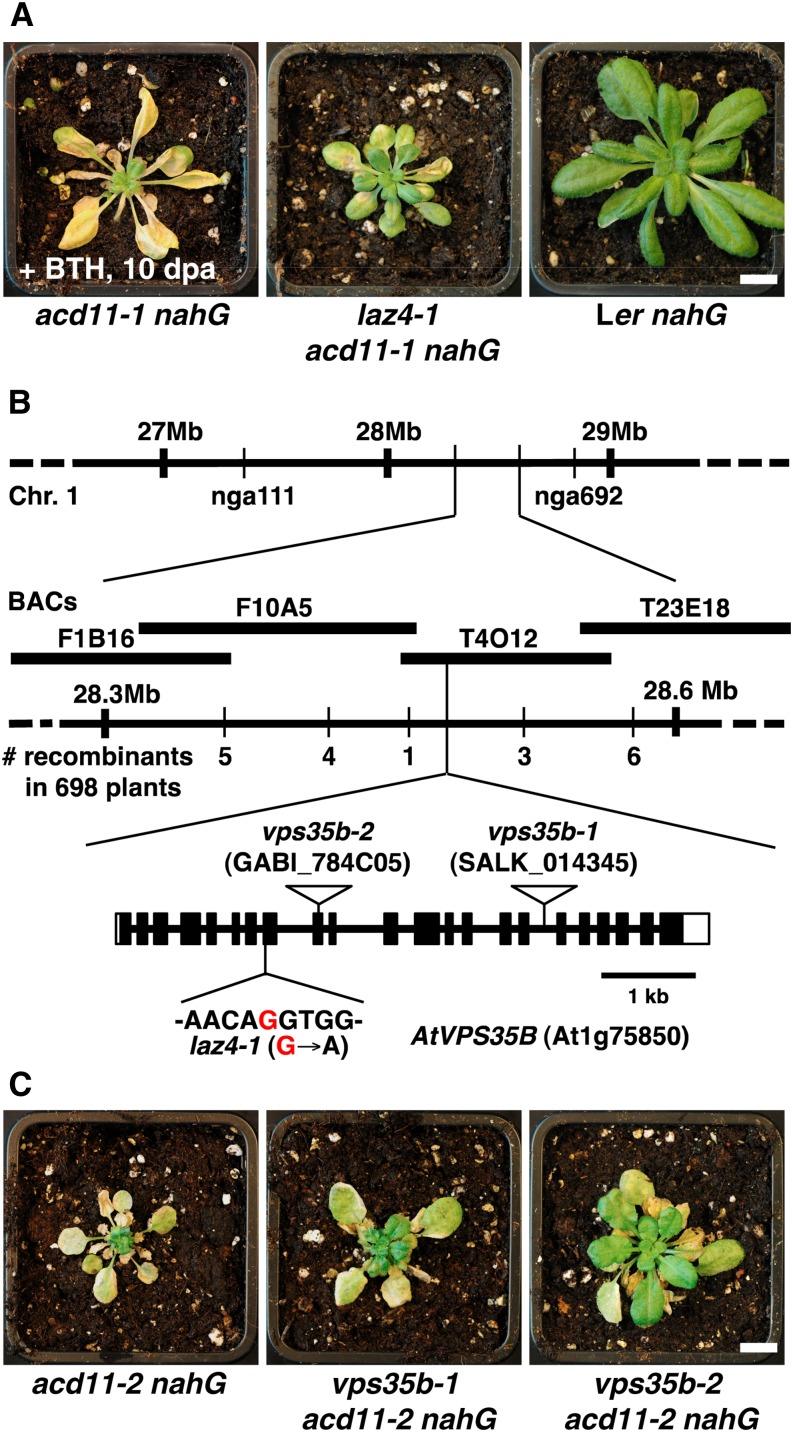
laz4-1 was isolated as an ethyl methanesulfonate (EMS)-induced, recessive suppressor of cell death in response to BTH in the Landsberg erecta (Ler) acd11-1 nahG background. Suppression appeared relatively weak compared with other laz mutants (Malinovsky et al., 2010; Palma et al., 2010), as cell death symptoms were visible in older leaf stages of laz4-1 acd11-1 nahG plants 5 d after BTH treatment (Figure 1A). However, cell death was more strongly attenuated in newly emerging leaves, which allowed laz4-1 acd11-1 nahG plants to survive throughout development to flower and set seed, in marked contrast with acd11-1 nahG plants (Supplemental Figure 1).
Figure 1.
laz4 Encodes the Retromer Component VPS35B.
(A) Twenty-eight-day-old acd11-1 nahG, laz4-1 acd11-1 nahG, and Ler nahG plants 10 d after treatment (dpa) with 100 μM BTH. Bar = 1 cm.
(B) The laz4 locus was mapped to a 65-kb interval between two markers on BAC T4O12 on chromosome 1. A G-to-A transition was found at the splice acceptor site of exon 8 in the At1g75850 gene, encoding the retromer component VPS35B. The structure of LAZ4/VPS35B (At1g75850) showing the laz4-1 mutation (red) and T-DNA insertions (open triangles) in vps35b-1 (SALK_014345) and vps35b-2 (GABI_784C05) is given. Closed boxes indicate exons, and lines between boxes indicate introns.
(C) Cell death phenotype of acd11-2 nahG harboring vps35b-1 and vps35b-2 in comparison with the acd11-2 nahG control. Photographs were taken 10 d after BTH treatment of 4-week-old plants. Bar = 1 cm.
To identify the LAZ4 locus, we mapped laz4 to a 65-kb interval at the bottom of chromosome 1 (Figure 1B). Candidate genes in this region with significantly induced expression in transcript profiles of acd11 nahG plants compared with wild-type and nahG plants upon BTH treatment were sequenced (Malinovsky et al., 2010). This identified a G-to-A transition in a splice acceptor site of At1g75850, which is annotated as VPS35B (Jaillais et al., 2007), encoding one of three VPS35 homologs in Arabidopsis (Figure 1B). VPS35 proteins are highly conserved in eukaryotes and form the large subunit of the retromer complex together with VPS26 and VPS29 (McGough and Cullen, 2011). Analysis of VPS35B transcripts in laz4 revealed that disruption of the intron splice site caused an in-frame 123-bp deletion corresponding to the loss of exon 8 (Supplemental Figure 2). Since only a single laz4 allele was identified, we introduced a 7.6-kb genomic fragment of the wild-type LAZ4/VPS35B locus into laz4-1 acd11-1 nahG to test for transgenic complementation. BTH treatment of independent T3 lines revealed growth arrest and cell death as for the parental acd11-1 nahG line in Ler (Supplemental Figure 1). In addition, introducing independent Columbia-0 (Col-0) ecotype knockout alleles of VPS35B into Col-0 acd11-2 nahG led to the suppression of cell death upon BTH treatment (Figure 1C). However, the effect of vps35b-1, a previously described Col-0 T-DNA insertion in intron 16 (the original designation vps35a-1 [Yamazaki et al., 2008] was changed to vps35b-1 according to TAIR nomenclature; see Methods), was considerably weaker compared with the Col-0 vps35b-2 (insertion in exon 9) (Figure 1C). Indeed, quantitative RT-PCR verified that vps35b-2 is a null mutant, whereas vps35b-1 still accumulated residual levels of VPS35B transcripts (Supplemental Figure 3). Together, these results demonstrate that laz4-mediated suppression of BTH-inducible PCD in acd11 nahG is caused by a mutation in VPS35B, and we refer to it as such hereafter.
VPS35B Is a Retromer Component
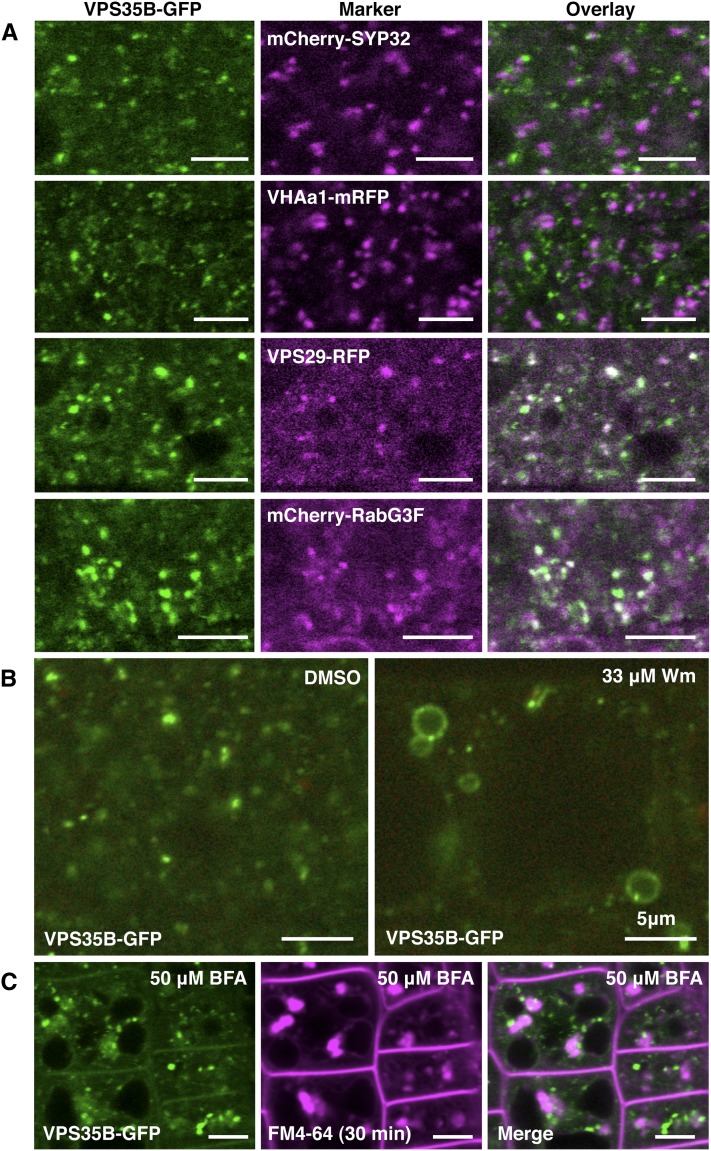
The retromer core complex in plants includes VPS29, VPS35, and VPS26 homologs (Jaillais et al., 2007; Yamazaki et al., 2008; Hashiguchi et al., 2010). These subunits localize to the prevacuolar compartment (PVC) and interact with each other (Oliviusson et al., 2006; Jaillais et al., 2007; Zelazny et al., 2013). However, in contrast with other VPS35 homologs, the subcellular localization and interaction capacity of VPS35B have not been studied in detail (Nodzynski et al., 2013; Zelazny et al., 2013). Therefore, we generated transgenic lines expressing a native promoter-driven VPS35B-GREEN FLUORESCENT PROTEIN (GFP) fusion in vps35b-2 and subsequently introduced subcellular compartment markers for colocalization studies. When analyzing root cells of transgenic seedlings, VPS35B-GFP appeared in punctate structures that did not overlap with markers for the Golgi (SYNTAXIN OF PLANTS32 [SYP32]) (Geldner et al., 2009) or the trans-Golgi network (TGN) (VACUOLAR H+ATPASE A1 [VHAa1]) (Dettmer et al., 2006). Instead, VPS35B-GFP colocalized with VPS29 and the Rab7 GTPase homolog RabG3F (Figure 2A) that physically interacts with VPS35A and recruits the retromer complex to endosomal membranes (Zelazny et al., 2013). Furthermore, treatment with the phosphatidylphosphate 3-kinase inhibitor wortmannin (Wm) caused ring-like structures labeled with VPS35B-GFP, indicating that VPS35B localizes to multivesicular bodies (MVBs)/PVCs, which are enlarged in the presence of Wm (Wang et al., 2009) (Figure 2B). Consistent with these findings, VPS35-GFP was hardly detectable in brefeldin A (BFA)-induced agglomerates, so-called BFA bodies, which consist of early secretory compartments (i.e., TGN/early endosomes) (Geldner et al., 2003) and are visualized by costaining with the plasma membrane-derived early endosome marker dye FM4-64 (Figure 2C).
Figure 2.
VPS35B Colocalizes with Retromer-Associated Proteins to MVBs/PVCs.
(A) VPS35B-GFP colocalizes with VPS29-mRFP and the late endosome marker and VPS35A-interacting protein mCherry-RabG3F but not with the Golgi marker mCherry-SYP32 or the TGN marker VHAa1-mRFP.
(B) VPS35B localization is Wm-sensitive. Punctate structures labeled by VPS35B-GFP show dilation after 90 min of treatment with 33 μM Wm, indicating an association of VPS35B-GFP with enlarged MVBs/PVCs.
(C) VPS35B localization is BFA-insensitive. VPS35B-GFP remained in punctate structures and was distinctively separated from FM4-64-labeled BFA bodies (magenta) after 1 h of treatment with 50 μM BFA.
Confocal images were taken in root tips of 6-d-old seedlings. Bars = 5 μm.
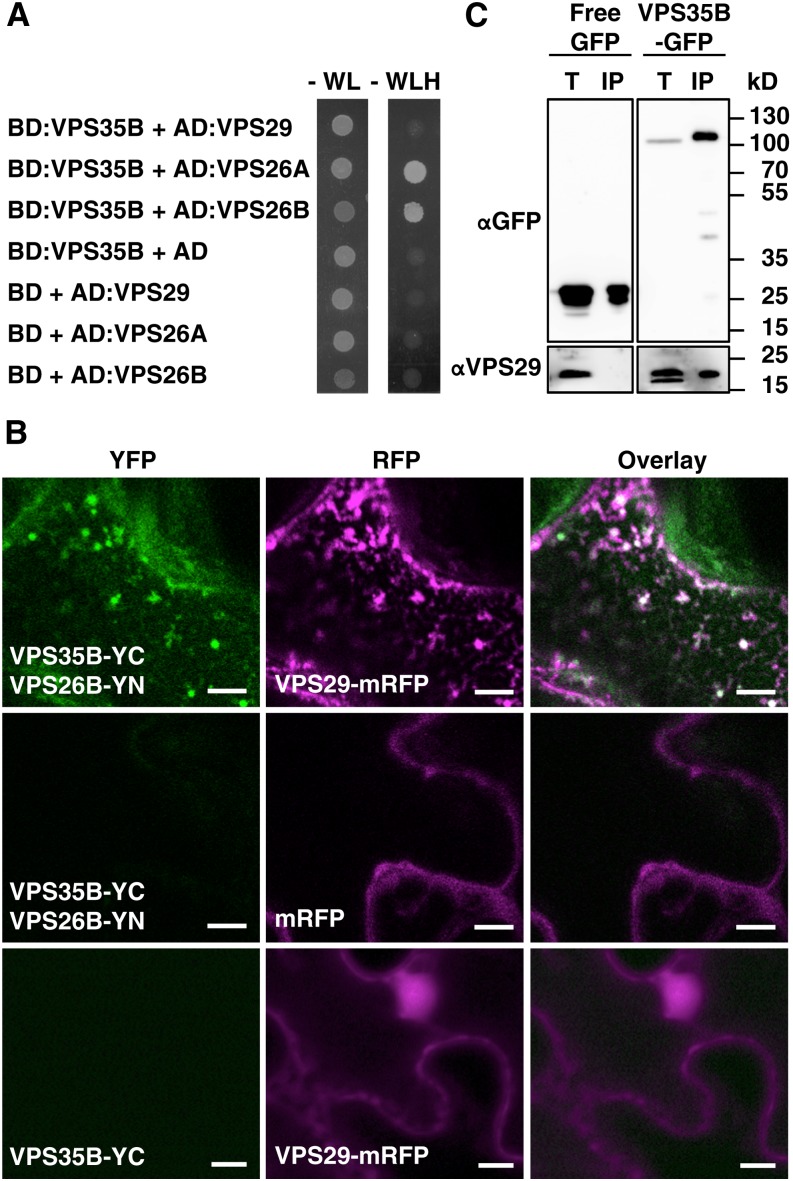
We then tested the ability of VPS35B to interact with VPS29 and VPS26. Yeast two-hybrid analysis verified the interaction with VPS26A and VPS26B, whereas direct binding to VPS29 was not clearly detectable (Figure 3A). Therefore, we investigated by bimolecular fluorescence complementation (BiFC) whether VPS35B is able to assemble into a retromer complex upon transient expression in leaves of Nicotiana benthamiana. In agreement with previous analysis of retromer assembly with VPS35A (Zelazny et al., 2013), the YELLOW FLUORESCENT PROTEIN (YFP)-derived fluorescence indicative of VPS35B-VPS26B interaction was detectable in the presence of RED FLUORESCENT PROTEIN (RFP)-tagged VPS29 but not of RFP alone (Figure 3B). This confirmed the capacity of VPS35B to reconstitute a stable retromer complex when coexpressed at comparable levels with other VPS subunits. In addition, we were able to detect VPS29 by immunoblot analysis in immunoprecipitates of native promoter-driven VPS35B-GFP in Arabidopsis seedlings (Figure 3C). Collectively, these results verified that VPS35B is a component of the MVB/PVC-localized retromer complex.
Figure 3.
VPS35B Interacts with Retromer Core Subunits.
(A) VPS35B interacts with VPS26A and VPS26B as indicated by the strong growth of yeast strains coexpressing VPS35B (bait) and VPS26A or VPS26B (prey) on selective medium (-WLH) after incubation for 3 d at 28°C. Only very weak growth is observed for VPS35B- and VPS29-expressing yeast cells. Control strains that expressed empty bait/prey vectors with the different retromer subunits show no growth under the same conditions. AD, activation domain; BD, binding domain.
(B) VPS29 is required for in vivo interaction of VPS35B and VPS26B. YFP fluorescence is detected in N. benthamiana leaves upon coexpression of VPS35B-YC and VPS26B-YN with VPS29-mRFP (top) but not with free mRFP (middle). No YFP signal is detected in infiltrated leaves expressing VPS35B-YC and VPS29-mRFP only (bottom). YC is the C-terminal portion of YFP, and YN is the N-terminal portion of YFP. Bars = 5 μm.
(C) Immunoblot analysis reveals the presence of VPS29 in VPS35B immunocomplexes. Total proteins from VPS35B-GFP- and free-GFP-expressing seedlings were extracted with salt-free buffer, and immunoprecipitation was performed with anti-GFP monoclonal antibody. Endogenous VPS29 (21 kD) was detected in total fractions (T) of both transgenic lines and coimmunoprecipitated (IP) with VPS35B-GFP (116 kD) but not with free GFP (27 kD). The total protein:immunoprecipitate ratio is 1:60. Immunoblots were probed with anti-GFP or anti-VPS29 antibody.
Genetic Functions of VPS35 Homologs in Autoimmunity-Triggered PCD
Despite the suppressive effect of VPS35B deficiency on BTH-inducible PCD in acd11 nahG, we were unable to obtain surviving acd11 vps35b double mutants in the absence of nahG in both the Ler and Col-0 backgrounds (Supplemental Figure 4). This indicated either a weak suppressor activity of mutated VPS35B or functional redundancy among the three VPS35 homologs. Indeed, previously described single knockouts of VPS35A (At2g17790) and VPS35C (At3g51310) (vps35a-1 and vps35c-1, designated according to TAIR nomenclature) (Yamazaki et al., 2008) showed no major effects on constitutive or BTH-inducible acd11-related death (Supplemental Figure 4). However, their respective combinations with the vps35b-1 allele permitted acd11-2 plants to survive in the absence of nahG (Figure 4A). Mutant growth under lower temperature (17°C) further enhanced the suppression phenotype and allowed reproductive development and seed production (Figure 4B). By contrast, vps35a-1 vps35c-1 double mutations did not suppress cell death in acd11 (Figure 4A) and acd11 nahG upon BTH treatment (Supplemental Figure 5A), indicating that VPS35B is predominantly required for cell death execution in acd11. Since triple mutant combinations of all VPS35 genes could only be generated previously in the presence of a leaky loss-of-function allele of VPS35A (At2g17790) (vps35a-2, designated according to TAIR) (Yamazaki et al., 2008), further genetic analysis of the VPS35 contribution to cell death in acd11 is prevented by the overlapping and essential roles of retromer in plant viability and development.
Figure 4.
Suppression of acd11- and lsd1-Triggered Autoimmunity by Loss-of-Function Mutations in VPS35 Genes.
(A) Col-0 wild-type, vps35a-1 b-1, vps35b-1 c-1, vps35a-1 c-1 (top) as well as acd11-2, acd11-2 vps35a-1 b-1, acd11-2 vps35b-1 c-1, and acd11-2 vps35a-1 c-1 (bottom) plants were grown for 4 weeks under short-day conditions at 21°C. Bars = 1 cm.
(B) acd11-2, acd11-2 vps35a-1 b-1, and acd11-2 vps35b-1 c-1 plants grown for 4 weeks under short-day conditions at 17°C. Bars = 1 cm.
(C) acd11-2 laz5-1 vps35b-1 and acd11 laz5-1 plants grown at ambient temperature (21°C) for 5 weeks under short-day conditions followed by 2 weeks under long-day conditions.
(D) BTH-triggered runaway cell death in lsd1-2 compared with lsd1-2 vps35a-1 b-1, lsd1-2 vps35b-1 c-1, and Col-0 wild-type plants. Plants were grown under short-day conditions for 25 d and sprayed with 100 μM BTH. Photographs were taken 8 d after treatment (dpa) in comparison with untreated controls. Bars = 1 cm.
acd11 plants carrying a null allele of LAZ5 due to a T-DNA insertion (acd11-2 laz5-1) (Palma et al., 2010) exhibited wild-type-like growth under short-day conditions but developed a distinct secondary cell death phenotype when transferred to a long-day photoperiod (Figure 4C). We speculate that this may be due to activation of an as yet uncharacterized NB-LRR protein, particularly since dominant-negative laz5 mutants fully suppress acd11 phenotypes (Palma et al., 2010). Notably, introducing the single vps35b-1 T-DNA mutation into acd11-2 laz5-1 considerably improved growth and reproductive performance under long-day conditions (Figure 4C). This indicates that VPS35B also contributes to LAZ5-independent forms of autoimmune cell death in acd11.
To analyze whether VPS35 functions in PCD control are limited to acd11, we introduced the VPS35 double mutant alleles into the lesion-mimic mutant lsd1. Runaway cell death conditioned by the CC-NB-LRR protein ADR1-L2 (Bonardi et al., 2011) was strongly suppressed by vps35b-1 c-1 but not by other mutant combinations (Figure 4D; Supplemental Figure 5B).
Collectively, these findings reveal that specific VPS35 proteins have important genetic functions in PCD triggered by both TIR-NB-LRR and CC-NB-LRR immune receptors.
VPS35B Is Required for Proper Localization of ACD11
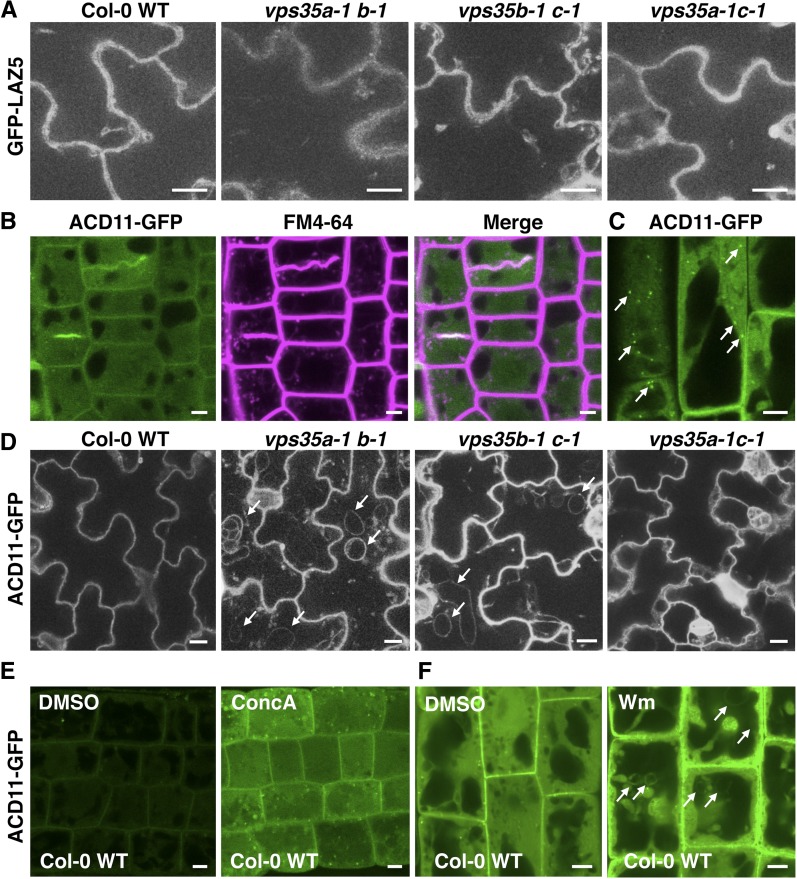
Since retromer is localized to endosomal compartments and functions in membrane trafficking (Robinson et al., 2012), VPS35 proteins may be involved in the subcellular targeting and localization of ACD11 and ACD11-related proteins. To examine this, we introduced GFP-LAZ5 and ACD11-GFP fusion constructs under the control of the constitutive UBQ10 promoter (Grefen et al., 2010) into the vps35 double mutant combinations and wild-type control. LAZ5-GFP showed cytoplasmic localization in cotyledon epidermal cells, which remained largely unaffected by vps35 mutations (Figure 5A). ACD11-GFP localized to the plasma membrane, cytoplasm, and some mobile punctate structures in wild-type seedling root cells (Figures 5B and 5C) but also appeared to be located in ring-like structures (with diameter up to 5 to 10 nm) in cotyledon epidermal cells of the acd11 cell death-suppressing double mutants vps35a-1 b-1 and vps35b-1 c-1 (Figure 5D). By contrast, ACD11-GFP localization was not altered in vps35a-1 c-1 compared with wild-type plants, indicating that VPS35B might be involved in proper intracellular trafficking of ACD11. We then applied chemical inhibitors to ACD11-GFP-expressing wild-type plants to further dissect the trafficking route of ACD11. Treatment of transgenic roots with the vacuolar type H+-ATPase inhibitor concanamycin A resulted in vacuolar accumulation of ACD11-GFP (Figure 5E). In addition, ACD11-GFP trafficking was sensitive to Wm treatment, leading to the aggregation of cytoplasmic ACD11 and relocalization to similar ring-like structures as observed in vps35a-1 b-1 and vps35b-1 c-1 mutants (Figure 5F). Colabeling with FM4-64 suggested that these spherical patterns might represent small vacuole-like structures (Supplemental Figure 6). These findings indicate that ACD11 is constitutively trafficked to the vacuole for degradation in a Wm-sensitive manner, which is also common to VPS35B-mediated trafficking. Therefore, Wm-induced vacuolation of ACD11-GFP mimics the misplacement of ACD11-GFP in vps35a-1 b-1 and vps35b-1 c-1. This suggests that proper localization of ACD11 requires functional VPS35B-containing retromer subcomplexes.
Figure 5.
VPS35B Contributes to the Trafficking of ACD11-GFP but Not GFP-LAZ5.
(A) GFP-LAZ5 displays comparable cytoplasmic localization in cotyledons of the Col-0 wild-type and vps35 double mutant combinations.
(B) and (C) ACD11-GFP localizes to the cytoplasm and plasma membrane, as revealed by costaining (5 min) with the lipophilic dye FM4-64 (B), and to some mobile punctate structures (arrows; [C]) in wild-type plants.
(D) ACD11-GFP is mislocated to dilated ring-like structures in vps35a-1 b-1 and vps35b-1 c-1 double mutants (arrows) compared with wild-type and vps35a-1 c-1 plants.
(E) and (F) ACD11-GFP lies in a Wm-sensitive trafficking pathway en route to the vacuole for rapid turnover. Overnight treatment of ACD11-GFP-expressing seedlings with 1 μM concanamycin A (ConcA) causes ACD11-GFP accumulation in the vacuole of root epidermal cells (E). When treated with 33 μM Wm for 90 min, ACD11-GFP forms dilated ring-like structures (arrows), suggestive of a late endosomal trafficking pathway (F).
Imaging conditions are identical for each genotype in (A) and (D) as well as for DMSO control and concanamycin A or Wm treatment in (E) and (F). Bars in (A) and (D) = 10 μm; bars in (B), (C), (E), and (F) = 5 μm.
VPS35 Homologs Function in Effector-Triggered HR and Immunity
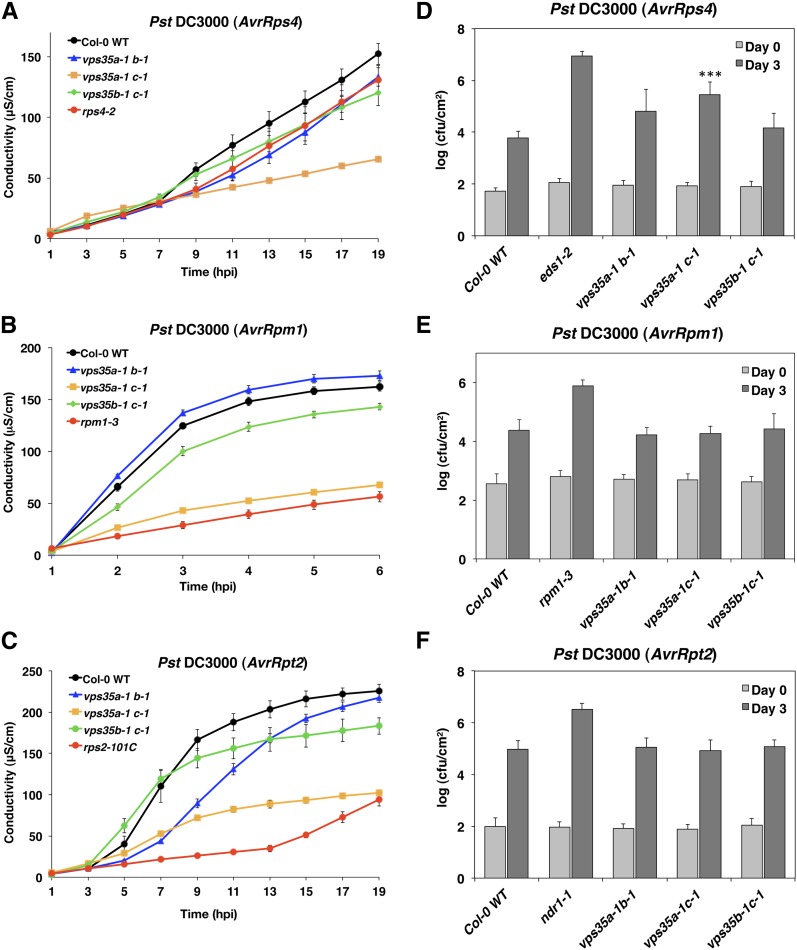
Since HR-like PCD in acd11 and lsd1 is mediated by inappropriate activation of NB-LRR immune receptors, suppressors of PCD may include regulators of NB-LRR proteins. Thus, VPS35B and its homologs also may be required for HR activated upon recognition of pathogen effectors. To investigate this, vps35 double mutant combinations were challenged with Pseudomonas syringae pv tomato (Pst) DC3000 strains expressing specific effectors to trigger ETI through the activation of specific NB-LRR proteins. In addition to the previously described double mutants vps35a-1 c-1 and vps35b-1 c-1 (Yamazaki et al., 2008), we used vps35a-1 b-1, which showed a similar dwarf phenotype to vps35a-1 c-1 but did not exhibit early leaf senescence (Supplemental Figure 7). To avoid potential interference of age-related processes with effects on HR and disease resistance measured by pathogen growth restriction, experiments were performed on leaves of short-day-grown plants (5 to 6 weeks) prior to the onset of senescence (Hofius et al., 2009, 2011). HR was monitored by ion leakage assays, as conductance increases upon electrolyte release from dying leaf tissue (Mackey et al., 2003; Hofius et al., 2009). These assays showed that the double mutants displayed marked differences in HR depending on the mutant combination and ETI event (Figure 6). Most strikingly, vps35a-1 c-1 showed strong suppression of HR as measured by ion leakage upon recognition of AvrRps4, AvrRpm1, and AvrRpt2 effector proteins (Figures 6A to 6C). Reductions in ion leakage observed in other vps35 mutant combinations were less consistent than in vps35a-1 c-1.
Figure 6.
Effect of VPS35 Deficiency on HR Cell Death and Disease Resistance.
(A) to (C) Ion leakage assays of 5- to 6-week-old Col-0 wild-type and vps35a-1 b-1, vps35a-1 c-1, and vps35b-1 c-1 double mutant plants after inoculation with avirulent strains of Pst DC3000 expressing AvrRps4 (A), AvrRpm1 (B), or AvrRpt2 (C). Loss-of-function mutants of the corresponding R genes RPS4 (rps4-2) (A), RPM1 (rpm1-3) (B), and RPS2 (rps2-101C) (C) served as additional controls. Means and se were calculated from four discs per treatment with three to four replicates within an experiment.
(D) to (F) Growth of avirulent strains of Pst DC3000 expressing AvrRps4 (D), AvrRpm1 (E), or AvrRpt2 (F) in 6-week-old Col-0 wild-type and vps35a-1 b-1, vps35a-1 c-1, and vps35b-1 c-1 double mutant plants 0 and 3 d after infiltration at OD600 = 0.00001. eds1-1 (D), rpm1-3 (E), and ndr1-1 (F) served as additional susceptible controls. Log-transformed values are means ± sd with n = 3 ([D] and [E]) or n = 4 to 6 (F). Asterisks indicate statistical significance (P < 0.001) determined by one-way ANOVA with posthoc Tukey’s test (compared with the wild type). cfu, colony-forming units.
To further analyze whether the observed changes in ion leakage resulted in altered disease resistance, bacterial titers were determined (Figures 6D to 6F). Notably, reduced levels of cell death triggered upon recognition of AvrRps4 were accompanied by up to 14-fold enhanced bacterial growth in vps35a-1 c-1 double mutants (Figure 6D). By contrast, RPM1- and RPS2-conditioned resistance remained largely unaffected, even in association with severe HR suppression in vps35a-1 c-1 (Figures 6E and 6F). Overall, these results indicate that VPS35A and VPS35C are generally required for HR mediated by activated NB-LRR proteins and have distinct functions in disease resistance conditioned by RPS4, a TIR-NB-LRR protein.
Finally, to assess whether VPS35 homologs are also required for basal defense, vps35 double mutants were infected with virulent Pst DC3000. Bacterial growth in the different vps35 mutant combinations was indistinguishable from that in wild-type controls (Supplemental Figure 8). This indicates that VPS35 deficiency does not compromise the induction of basal defense responses upon bacterial infection.
Retromer Dysfunction Suppresses HR
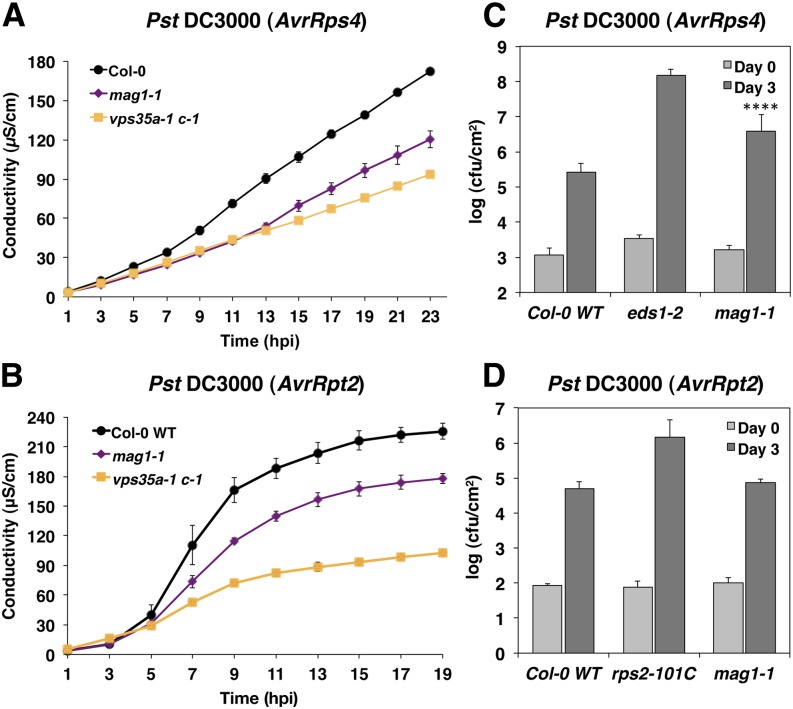
Suppression of HR by VPS35 loss-of-function mutations implicates retromer function in HR. Therefore, we assayed whether disruption of another retromer component similarly affects HR and disease resistance. VPS29 is encoded by a single gene in Arabidopsis, and vps29 mutants display severe dwarfism and morphological changes that may interfere with the analyses of immune system function. Therefore, we used the leaky T-DNA VPS29 allele maigo1-1 (mag1-1) (Shimada et al., 2006), which exhibits only moderate growth reduction and phenotypically resembled the vps35a-1 b-1 double mutant (Supplemental Figure 7). Ion leakage assays following infection with Pst DC3000 expressing AvrRps4 or AvrRpt2 indicated that mag1-1 mutants also were compromised in RPS4- and RPS2-conditioned HR (Figures 7A and 7B). Consistent with this, and similar to the vps35a-1 c-1 double mutant, mag1-1 supported increased growth of AvrRps4-expressing Pst DC3000 (P < 0.0001; Figure 7C), Together, these results indicate that retromer dysfunction, as a result of reduced VPS35 or VPS29 levels, is responsible for the suppression of HR upon NB-LRR activation.
Figure 7.
Effect of VPS29 Deficiency on HR Cell Death and Disease Resistance.
(A) and (B) Ion leakage assays of 6-week-old Col-0 wild-type, mag1-1, and vps35a-1 c-1 plants after inoculation with Pst DC3000 expressing AvrRps4 (A) or AvrRpt2 (B). Means and se were calculated from four discs per treatment with four (A) or three (B) replicates within an experiment.
(C) and (D) Growth of avirulent Pst DC3000 expressing AvrRps4 (C) or AvrRpt2 (D) in 6-week-old Col-0 wild-type, mag1-1, and eds1-2 (C) or ndr1-1 (D) plants 0 and 3 d after infiltration at OD600 = 0.0001 (C) or 0.00001 (D). Log-transformed values are means ± sd (n = 3). Asterisks indicate statistical significance (P < 0.0001) in mag1-1 determined by one-way ANOVA with posthoc Tukey’s test (compared with the wild type). cfu, colony forming units.
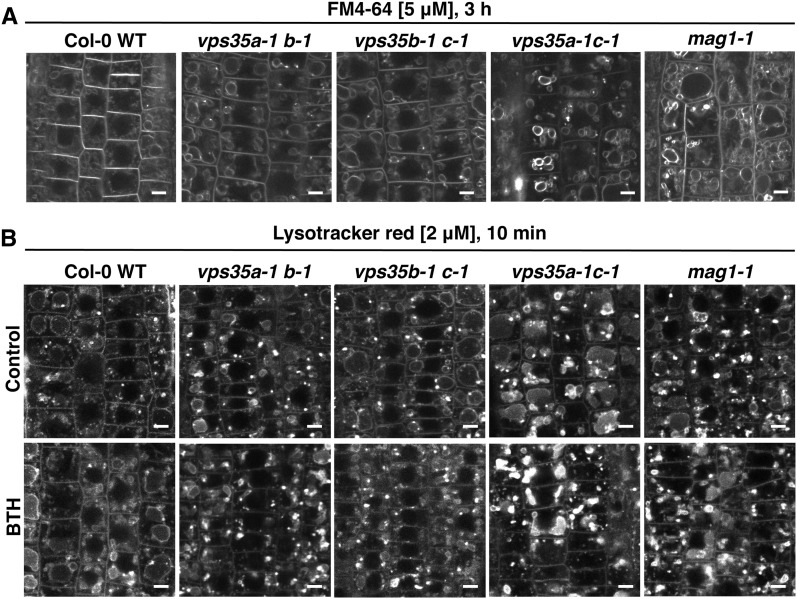
Retromer Deficiency Affects the Morphology of Late Endocytic/Lytic Compartments
Retromer is implicated in endomembrane trafficking and cargo delivery to the lytic vacuole, which plays essential roles in HR execution (Hara-Nishimura and Hatsugai, 2011; Robinson et al., 2012; Nodzynski et al., 2013). Thus, we investigated whether HR suppression in retromer mutants might be linked to alterations in the morphology and function of endosomal and vacuolar compartments. To this end, we performed uptake experiments with the endocytic tracer dye FM4-64 in roots to monitor endocytic trafficking events. Labeling of endosomes within 5 min revealed no obvious defects in early stages of endocytosis in mag1 and vps35 double mutant combinations compared with the wild-type control (Supplemental Figure 9). By contrast, detection of late endocytic and vacuolar compartments by prolonged FM4-64 staining (3 h) indicated morphological alterations in endomembrane structures of vps35a-1 c-1 and mag1-1 mutants, whereas vps35a-1 b-1 and vps35b-1 c-1 appeared comparable to the wild type (Figure 8A). To substantiate these findings, we stained lytic compartments with the acidophilic dye Lysotracker Red and frequently observed aberrant aggregation in vps35a-1 c-1 and mag1-1 (Figure 8B). Application of BTH further aggravated this phenotype. This indicates that defects in lytic compartments in response to retromer deficiency may be pronounced under HR-promoting conditions (Figure 8B).
Figure 8.
Retromer Deficiency Causes Defects in the Late Endocytic Pathway.
(A) Long-time uptake of FM4-64 reveals altered endomembrane structures and vacuolar morphology in vps35a-1 c-1 and mag1-1 mutants in comparison with Col-0 wild-type, vps35a-1 b-1, and vps35b-1 c-1 plants. Four-day-old seedlings were treated with 5 μM FM4-64 for 5 min and incubated in liquid half-strength MS medium at room temperature for 3 h before confocal imaging.
(B) Lysotracker Red staining indicates aberrant aggregation of lytic compartments in vps35a-1 c-1 and mag1-1 in comparison with Col-0 wild-type and other vps35 double mutant plants (top). The morphological phenotype was further aggravated in vps35a-1 c-1 and mag1-1 or mildly induced in vps35a-1 b-1 upon overnight treatment with 100 μM BTH (bottom). Control and BTH-treated 4-d-old seedlings were stained with 2 μM Lysotracker Red for 15 min before imaging.
Imaging conditions were identical across all genotypes. Bars = 5 μm.
Retromer Mutants Are Impaired in Autophagy Processes
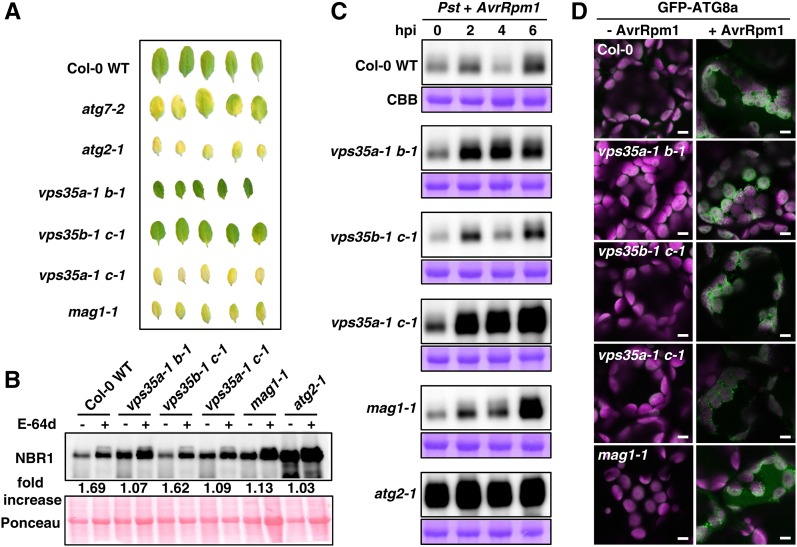
Lysotracker dyes are often used to label autolysosome-like structures that form after the fusion of autophagosomes with endosomes or vacuolar/lysosomal compartments to degrade autophagic cargo (Derrien et al., 2012; Kwon et al., 2013). Because autophagy contributes to vacuole-mediated forms of cell death condition by RPS4 and RPM1 (Hofius et al., 2009), we speculated that the abnormal morphology and potential dysfunction of late endocytic/lytic compartments may impact autophagy processes. To test this, we first compared the response to nutrient limitation of retromer versus atg mutants. Similar to atg2-1 and atg7-2, vps35a-1 c-1 and mag1-1 mutants responded with exaggerated senescence to leaf detachment and prolonged darkness, whereas vps35a-1 b-1 and vps35b-1 c-1 remained largely unaffected (Figure 9A). To further investigate whether autophagic activity is affected in retromer mutants, we monitored protein levels of the autophagic adaptor protein NEIGHBOR OF BRCA1 GENE1 (NBR1), whose vacuolar degradation is a marker for autophagic flux (Svenning et al., 2011; Minina et al., 2013). As expected, NBR1 levels remained low in untreated wild-type seedlings due to basal autophagy activity but were significantly enhanced upon inhibition of autophagic degradation by the cysteine protease inhibitor E-64d and in the atg2-1 mutant (Figure 9B). Importantly, vps35a-1 b-1, vps35a-1 c-1, and mag1-1 showed constitutively increased NBR1 amounts, which remained largely unaltered by E-64d treatment. This suggests that autophagic degradation cannot be completed in some retromer mutants (Figure 9B).
Figure 9.
Retromer Is Required for Autophagy Processes.
(A) Leaf detachment assay of Col-0 wild-type, atg7-2, and atg2-1 plants compared with vps35a-1 b-1, vps35a-1 c-1, and vps35b-1 c-1 double mutants and mag1-1. Detached leaves of 4-week-old plants were kept for 4 d on moist filter paper in darkness.
(B) Immunoblot analysis of NBR1 accumulation in vps35a-1 b-1, vps35b-1 c-1, vps35a-1 c-1, and mag1-1 mutant seedlings grown for 10 d on MS medium before treatment with DMSO (−) or the cysteine protease inhibitor E-64d (+) for 12 h. Equal amounts of crude extracts were separated by SDS-PAGE and probed on blots with anti-NBR1 antibody. Numbers correspond to the fold increase of NBR1 in E-64d-treated samples compared with the respective DMSO control. Ponceau staining of membrane-bound total proteins was used as a loading control and quantified for the normalization of NBR1 signal intensities.
(C) Immunoblot analysis of NBR1 accumulation upon infection with Pst DC3000 (AvrRpm1). Total proteins were extracted at the indicated time points from 5-week-old retromer mutants compared with Col-0 wild-type and atg2-1 controls and probed with anti-NBR1 antibody (top). Bottom panels indicate Coomassie Brilliant Blue (CBB) staining of Rubisco large subunit as a loading control.
(D) GFP-ATG8a accumulation is indicative of autophagosome formation in the Col-0 wild-type and retromer mutants upon infection with Pst DC3000 (AvrRpm1). Confocal images are representative of palisade parenchyma cells in leaves of 4-week-old plants before (−AvrRpm1) and 3 h after (+AvrRpm1) infection. GFP-ATG8a derived signals (green) are superimposed with chlorophyll fluorescence of chloroplasts (magenta). Bars = 5 μm.
We then analyzed the impact of retromer dysfunction on HR-associated autophagy, which is rapidly and strongly induced by Pst DC3000 expressing AvrRpm1 (Hofius et al., 2009). NBR1 levels in the wild type and vps35b-1 c-1 showed a characteristic biphasic response until 6 h after infection. By contrast, vps35a-1 b-1, vps35a-1 c-1, and mag1-1 mutants were altered in RPM1-dependent autophagic flux (Figure 9C). In particular, vps35a-1 c-1 plants displayed a dramatic increase of NBR1 protein within 6 h after infection, resulting in comparable levels to those in the constitutively NBR1-accumulating atg2-1 mutant. To further distinguish whether early or late autophagic steps are affected in retromer mutants, we monitored transgenically expressed GFP-ATG8a fusion protein. This fusion associates with autophagosomal membranes and is widely used as a marker for autophagosome formation (Yoshimoto et al., 2004). GFP-derived fluorescence before and 3 h after bacterial infection revealed normal accumulation of punctate autophagosome-like structures in leaves of vps35 and mag1 mutants, indicating that retromer deficiency does not impair autophagy initiation (Figure 9D). Overall, these findings suggest that the morphological abnormalities of late endocytic/lytic compartments in certain retromer mutants result in the disruption of vacuolar processes required for the execution of HR death-promoting autophagy.
DISCUSSION
A Novel Role for Retromer in Immune Receptor-Mediated HR
Retromer is a conserved multisubunit complex with functions in endosomal trafficking and the retrieval of transmembrane proteins (McGough and Cullen, 2011; Robinson et al., 2012). In yeast and mammalian cells, retromer binds specific peptide motifs in vacuolar and lysosomal acid hydrolase receptors, respectively, and mediates their recycling from endosomes back to the TGN (Nothwehr et al., 2000; Seaman, 2007; Bonifacino and Hurley, 2008). In addition, retromer-mediated protein sorting has been shown for a range of other transmembrane receptors and revealed retromer functions in processes such as Wnt-dependent signaling, apoptotic cell clearance, and the prevention of neurodegenerative diseases (Port et al., 2008; Chen et al., 2010; Lane et al., 2012). In plants, retromer components have been implicated in recycling vacuolar sorting receptors (Oliviusson et al., 2006; Kang et al., 2012) and in the vacuolar trafficking of plasma membrane proteins such as PIN-FORMED auxin efflux carriers (Kleine-Vehn et al., 2008; Nodzynski et al., 2013). These and other findings establish roles of the plant retromer in developmental processes including organogenesis, cell polarity, seed storage, and leaf senescence (Jaillais et al., 2007; Yamazaki et al., 2008; Hashiguchi et al., 2010; Pourcher et al., 2010).
We provide several lines of evidence that the retromer complex has additional functions in plant immunity. First, we found that laz4, which partly suppresses BTH-induced PCD in acd11 nahG, encodes VPS35B, and we confirmed by localization and interaction studies that VPS35B functions similarly to the other Arabidopsis VPS35 homologs as a retromer component (Figures 2 and 3) (Oliviusson et al., 2006; Nodzynski et al., 2013). Second, distinct combinations of vps35 loss-of-function alleles significantly suppressed autoimmunity- and pathogen-triggered HR conditioned by different TIR-NB-LRR and CC-NB-LRR proteins. Third, reduced levels of the retromer component VPS29 in the weak mag1-1 mutant were sufficient to partially mimic the impact of VPS35 deficiency on HR development and disease resistance. Since immune responses could not be analyzed in retromer null mutants due to phenotypic constraints (vps29-5, vps26a-1 vps26b-1, or vps35 triple knockouts) (Jaillais et al., 2007; Yamazaki et al., 2008; Zelazny et al., 2013), we speculate that our data underestimate the contribution of retromer to PCD execution. Inducible and cell type-specific deletions of retromer components may circumvent pleiotropic developmental phenotypes and reveal the extent of effects of retromer loss on immune system function. Nonetheless, our analysis of single and double vps35 mutants indicates that the diversified roles of the three VPS35 homologs allow the genetic dissection of retromer-dependent trafficking in developmental and immunity-related contexts (Yamazaki et al., 2008; Hashiguchi et al., 2010; Nodzynski et al., 2013).
Role of VPS35B-Dependent Trafficking in acd11-Triggered Autoimmunity
VPS35B seems to be predominantly required for acd11-mediated autoimmune PCD (Figure 1; Supplemental Figure 4A). However, VPS35A and VPS35C can partially substitute the function of VPS35B, as constitutive acd11 cell death in the absence of nahG was attenuated only by vps35b-1 in combination with vps35a-1 or vps35c-1 (Figure 4A). Moderately reduced temperature (17°C) further enhanced the suppressive phenotype in acd11 vps35a-1 b-1 and acd11 vps35b-1 c-1 (Figure 4B), which may be due to an additional block of endosomal and/or retromer-associated trafficking (Kuismanen and Saraste, 1989; Gentzsch et al., 2004). Importantly, the acd11-suppressing double mutants vps35a-1 b-1 and vps35b-1 c-1 showed mislocalization of ACD11-GFP to membranes of small vacuole-like structures, which could be mimicked by Wm treatment in the wild type (Figure 5). Hence, VPS35B seems to be involved in late endosomal trafficking processes associated with ACD11-related PCD functions. ACD11 was recently identified as a ceramide-1-phosphate transfer protein and intermediary regulator of sphingolipid levels, in particular of death-promoting phytoceramides (Simanshu et al., 2014). It was proposed that subcellular imbalances of ceramide-1-phosphate and ceramide levels in acd11 might mimic pathogen effector targeting of ACD11 function in host sphingolipid metabolism, leading to inappropriate activation of HR via detection by LAZ5 (Simanshu et al., 2014). Such metabolic sensing of sphingolipid signals would not require the direct association of ACD11 with LAZ5 (Palma et al., 2010) and could explain why the disruption of VPS35B-dependent trafficking affects ACD11 but not LAZ5 localization (Figures 5A and 5B). Because our genetic data indicated that VPS35B function in acd11 particularly applies to LAZ5-independent PCD pathways (Figure 4C), it is also possible that another, yet unknown, immune receptor “guards” ACD11 and is corestricted to VPS35B-related endosomal compartments and trafficking. NB-LRR proteins are known to localize to multiple cellular compartments in unchallenged cells (Gao et al., 2011; Heidrich et al., 2011; Qi et al., 2012; Takemoto et al., 2012), and some can dynamically redistribute upon effector recognition (Qi and Innes, 2013; Teh and Hofius, 2014). Notably, the activated CC-NB-LRR protein R3a from potato (Solanum tuberosum) is targeted to late endosomes and initiates HR in a Wm-sensitive manner (Engelhardt et al., 2012). VPS35-containing retromer subcomplexes, therefore, might be required for endosomal trafficking or recycling of distinct NB-LRR proteins. Alternatively, altered localization of ACD11 in vps35b-containing double mutants and upon Wm treatment may reflect distinct morphological and functional changes of PVC (Nodzynski et al., 2013), which could block the trafficking of an important vacuolar component required for acd11-triggered PCD.
Diverse Functions of Retromer in Effector-Triggered HR
Although vps35a-1 c-1 double mutants did not suppress autoimmunity-triggered PCD (Figure 4A; Supplemental Figure 5), they strongly impacted HR cell death conditioned by RPM1 and RPS2 (Figures 6B and 6C) and ETI conditioned by RPS4 (Figure 6D). This suggests remarkable variation in the contribution of individual VPS35 proteins to plant immunity. Similar functional divergence between the VPS35 homologs has been observed in plant development, as loss of function of VPS35A, but not of VPS35B or VPS35C, suppressed the gravitropic phenotype of the zip1/vti11 mutant (Hashiguchi et al., 2010) and impaired PVC function and trafficking to lytic vacuoles (Nodzynski et al., 2013). Nonetheless, vps35c-1 and to a lesser extent vps35b-1 were able to enhance the suppressive effect of vps35a-1 on zip1/vti11 abnormalities (Hashiguchi et al., 2010), indicating again that VPS35 genes share common functions and can partially compensate for the loss of individual homologs. In support of this, we found that the previously observed morphological alterations of late endocytic and vacuolar compartments in VPS35A loss-of-function mutants (Nodzynski et al., 2013) were most pronounced in vps35a-1 c-1 but less evident in vps35a-1 b-1 double mutants (Figure 8). The aberrant endomembrane structures in vps35a-1 c-1 were further enhanced by BTH treatment and correlated well with the general impact of this double mutant on different forms of TIR-NB-LRR- and CC-NB-LRR-conditioned HR. Hence, we propose that the retromer-mediated integrity and function of lytic compartments are important for HR downstream of NB-LRR activation and signaling.
A common theme in HR is the requirement of vacuole-mediated execution steps (Hara-Nishimura and Hatsugai, 2011). RPS2-dependent HR involves fusion of the tonoplast with the plasma membrane and discharge of vacuolar hydrolytic enzymes into the extracellular matrix (Hatsugai et al., 2009). RPS4-dependent HR engages autophagy mechanisms (Hofius et al., 2009), which require intravacuolar breakdown of autophagosomes and their transported cargo. Late steps of autophagy-dependent cell death also may involve the collapse of vacuolar membranes and the release of hydrolytic enzymes into the cytosol (van Doorn et al., 2011). Interestingly, both forms of vacuolar cell death seem to be induced upon RPM1 activation (Hatsugai et al., 2009; Hofius et al., 2009). The normal formation of autophagosomes combined with the marked accumulation of the selective autophagy substrate NBR1 in vps35a-1 c-1 following RPM1 activation (Figures 9C and 9D) strongly suggest that retromer deficiency interferes with vacuolar processes and thus impairs autophagic flux during the HR. Due to the essential role of retromer in recycling vacuolar sorting receptors as well as in PVC morphology and function, it is expected that retromer dysfunction broadly disrupts the intracellular trafficking of vacuolar cargo (Oliviusson et al., 2006; Yamazaki et al., 2008; Kang et al., 2012; Nodzynski et al., 2013). Therefore, the execution steps of autophagy- and vacuole fusion-mediated cell death pathways may be affected likewise. This may explain the severe suppression of HR mediated by both TIR-NB-LRR and CC-NB-LRR proteins in vps35a-1 c-1 mutants.
Our observation that PCD suppression in vps35a-1 c-1 was not accompanied by an equally broad effect on bacterial growth restriction agrees with evidence that NB-LRR-conditioned disease resistance can be uncoupled from HR (Bendahmane et al., 1999; Coll et al., 2010; Heidrich et al., 2011). However, during RPS4-activated immunity, both cell death and pathogen growth restriction were compromised in vps35a-1 c-1 and vps29/mag1-1 (Figures 6 and 7). This may place retromer components upstream of HR and defense activation. Similar to the proposed role in acd11-triggered autoimmunity, retromer might be directly involved in the trafficking and proper localization of the immune receptor or associated complex components. Alternatively, impaired autophagic processes in vps35a-1 c-1 and vps29/mag1-1 may mimic the situation in autophagy-deficient mutants such as atg5-1, which exhibits compromised resistance to bacteria expressing AvrRps4 (Dong and Chen, 2013) but not AvrRpm1 or AvrRpt2 (Kwon et al., 2013).
A Direct Role of Retromer in Autophagy Regulation?
Our finding that retromer mutants exhibit additional defects in basal and starvation-induced autophagy (Figures 9A and 9B) further implicates important functions of retromer components and trafficking in autophagic mechanisms. In this regard, a recent proteomics analysis of autophagosome composition in mammalian cell cultures identified VPS35 as an associated protein (Dengjel et al., 2012). Subsequent genetic studies of the yeast orthologs revealed that vps35 knockouts are as defective in autophagy as atg mutants (Dengjel et al., 2012). Furthermore, a human Rab GTPase-activating protein interacted with the retromer component VPS29 and the autophagy protein ATG8/LC3 and was suggested to function as a molecular switch between retromer-decorated endosomes and autophagosomes (Popovic et al., 2012). Finally, a Parkinson’s disease-associated mutation in VPS35 impaired autophagy and trafficking of the transmembrane autophagy protein ATG9 (Zavodszky et al., 2014). Future investigations may reveal whether retromer subunits also are associated with autophagy components and compartments in plants.
In conclusion, this study provides a primary example of the involvement of retromer components in HR and disease resistance and highlights the emerging importance of specific membrane-trafficking routes in ETI (Nomura et al., 2011; Engelhardt et al., 2012; Teh and Hofius, 2014). Our finding that the retromer complex is genetically linked to NB-LRR-mediated HR signaling that also engages autophagy and/or other vacuole-mediated processes suggests a complex interrelationship between endosomal, autophagic, and vacuolar trafficking events. Our genetic models of autoimmunity- and pathogen-triggered HR are valuable tools to further dissect these pathways and characterize their regulatory interactions.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana acd11 and acd11 nahG in Ler (acd11-1) and Col-0 (acd11-2) as well as Col-0 nahG, lsd1-2, enhanced disease susceptibility1-1 (eds1-2; Ler eds1-2 introgressed into Col-0), non-race-specific disease resistance1-1 (ndr1-1), rps4-2, rpm1-3, rps2-101c, and acd11-2 laz5-1 have been described (Mindrinos et al., 1994; Aarts et al., 1998; Boyes et al., 1998; Brodersen et al., 2005; Bartsch et al., 2006; Kaminaka et al., 2006; Wirthmueller et al., 2007; Palma et al., 2010). The ATG-deficient T-DNA insertion mutants atg7-2 and atg2-1 were described before (Inoue et al., 2006; Hofius et al., 2009). Col-0 vps35a-1, vps35b-1, vps35c-1, and mag1-1 single mutants as well as vps35a-1 c-1 and vps35b-1 c-1 double mutants were characterized previously (Shimada et al., 2006; Yamazaki et al., 2008), and vps35a-1 and vps35b-1 mutant alleles were designated according to TAIR (www.arabidopsis.org) nomenclature for VPS35A (At2g17790) and VPS35B (At1g75850). The vps35b-2 T-DNA insertion line (GK-784C05) was obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info), and plants homozygous for the insertion were verified with T-DNA left border and gene-specific primers (5′-TTATGATTCAAGTATCAAACAGCCA-3′ and 5′-CCCTGGACGTGAATGTAGACAC-3′). Sequences of primers used to select the different mutant alleles in genetic crosses are available upon request. The following transgenic Arabidopsis marker lines have been described previously: mCherry-SYP32 (Geldner et al., 2009), VHAa1-mRFP (Dettmer et al., 2006), VPS29-RFP (Jaillais et al., 2007), and mCherry-RabG3F (Geldner et al., 2009).
Following seed surface sterilization and treatment at 4°C for 2 d, plants were grown in soil under short-day conditions (8/16-h light/dark cycles) in growth cabinets and under long-day conditions (16/8-h light/dark cycles) in a growth room at 150 μE m−2 s−1, 21°C, and ∼70% relative humidity. Sterile plants were grown on Murashige and Skoog (MS) agar plates with an 8- or 12-h photoperiod. The mutant screen and growth of F2 mapping populations were performed under controlled greenhouse conditions as described (Malinovsky et al., 2010; Palma et al., 2010).
Map-Based Cloning of the LAZ4 Locus
Ler laz4-1 acd11-1 nahG was isolated as a BTH-resistant suppressor in an M2 population of EMS-mutagenized acd11 nahG seeds (Malinovsky et al., 2010; Palma et al., 2010) and crossed with Col-0 acd11-2 nahG to generate a mapping population. Rough mapping was initiated on 30 to 40 F2 plants homozygous for laz4 using standard simple sequence length polymorphism markers (Zhang et al., 2007), and fine-mapping was performed on 698 F2 plants with simple sequence length polymorphism and cleaved-amplified polymorphic sequence markers designed from the Arabidopsis polymorphism and Landsberg sequence collection (http://www.arabidopsis.org/browse/Cereon/index.jsp). This mapped the LAZ4 locus to ∼65 kb on the bottom of chromosome 1. Microarray-derived expression profiles of genes in the interval between At1g75790 and At1g75970 were analyzed for increased expression in acd11 nahG relative to wild-type and nahG controls (Malinovsky et al., 2010), and candidates were sequenced.
BTH Treatment, Ion Leakage, and Bacterial Resistance Assays
PCD analysis of mapping populations, complemented lines, and mutant crosses was done after leaf spraying with 100 μM BTH. Ion leakage assays following syringe-infiltration of avirulent Pst DC3000 strains were performed with 2 × 108 colony-forming units/mL as described (Hofius et al., 2009). Resistance assays were performed with avirulent Pst DC3000 strains at OD600 = 0.00001 (unless stated otherwise) essentially as described (Mackey et al., 2003).
Plasmid Construction and Plant Transformation
For subcellular localization and immunocomplex analysis, a genomic fragment of VPS35B containing 1.5 kb of the predicted promoter region was amplified with the primer set VPS35B(-1500)-F/VPS35B-nostop-R (for primer sequences, see Supplemental Table 1), cloned into pENTR/D-TOPO (Life Technologies), and recombined via the Gateway LR reaction (Gateway Cloning Technology; Life Technologies) into the binary destination vector pGWB504 (Nakagawa et al., 2007) to generate the pVPS35B:VP35B-GFP construct. To obtain UBQ10:ACD11-GFP and UBQ10:GFP-LAZ5, genomic fragments of ACD11 (including the 5′ untranslated region) and LAZ5 were amplified using primers ACD11-5′UTR-F/ACD11-nostop-R or LAZ5-F/LAZ5-stop-R, ligated into pENTR/D-TOPO, and subsequently integrated into the binary pUBC-Dest and pUBN-Dest vectors, respectively (Grefen et al., 2010). For BiFC analysis, full-length cDNAs of VPS35B, VPS29, VPS26A, and VPS26B were amplified with primer pairs VPS35B-F/VPS35B-R, VPS29-TOPO-F/VPS29-TOPO-R, VPS26A-TOPO-F/VPS26A-TOPO-R, and VPS26B-TOPO-F/VPS26B-TOPO-R, respectively, subcloned into pENTR/D-TOPO, and recombined into pSITEII-3N1 (Martin et al., 2009) to produce 35S promoter-driven C-terminal fusions to either the N- or C-terminal half of YFP. Recombining the VPS29 cDNA into the binary vector pGWB554 resulted in 35S:VPS29-RFP. For labeling of autophagosomes, a full-length cDNA clone of ATG8a fused to the C terminus of GFP was recombined into the binary vector pMDC32 to obtain 2x35S:GFP-ATG8a (a gift from Elena Minina). Constructs were verified by sequencing, electroporated into Agrobacterium tumefaciens GV3101, and used for stable transformation of Arabidopsis by the floral dip method (Clough and Bent, 1998) or for transient expression in Nicotiana benthamiana.
Confocal Microscopy and Drug Treatment
Confocal images were acquired using a Zeiss LSM 780 and analyzed with ZEN imaging software (version 2011). Excitation/detection parameters for GFP and RFP/FM4-64/Lysotracker were 488 nm/490 to 552 nm and 561 nm/569 to 652 nm, respectively. Four- to 6-d-old seedlings grown on MS agar plates were used for drug treatments. To visualize BFA compartments, VPS35B-GFP-expressing seedlings were stained with FM4-64 (5 μM; Life Technologies) for 5 min followed by 1 h of incubation in 50 μM BFA (Sigma-Aldrich) at room temperature. To induce the dilation of MVBs, VPS35B-GFP- or ACD11-GFP-expressing seedlings were incubated in 33 μM Wm (Santa Cruz Biotechnology) for 90 min. To observe ACD11-GFP accumulation in the vacuole, ACD11-GFP-expressing seedlings were incubated overnight in 1 μM concanamycin A (Santa Cruz Biotechnology) before imaging. For vacuole visualization in roots, seedlings were stained either with 5 μM FM4-64 for 5 min, followed by 3 h of incubation at room temperature, or with 2 μM Lysotracker Red (Life Technologies) for 15 min before imaging.
Immunoprecipitation
For analysis of VPS35-GFP immunocomplexes, total proteins from 6- to 7-d-old seedlings were extracted with no-salt lysis buffer (50 mM Tris, pH 8.0, 0.1% Nonidet P-40, and EDTA-free protease inhibitor mixture [Roche]) at a fresh weight:buffer ratio of 1 g:1 mL. After centrifugation at 6000 rpm and 4°C for 5 min, 15 to 20 μL of anti-GFP microbeads (Miltenyi Biotec) was added to the resultant supernatant and incubated for 1 h at 4°C on a rotating wheel. Subsequent washing and elution steps were performed according to the manufacturer (μMACS GFP Isolation Kit; Miltenyi Biotec). Immunoblot analysis was done essentially as described below, and immunoprecipitates from transgenic lines expressing free GFP were used as controls. VPS35B-GFP and native VPS29 were detected by mouse anti-GFP (monoclonal antibody JL-8; Clontech) and rabbit anti-VPS29 (Agrisera) antibodies at final dilutions of 1:1000 and 1:5000, respectively.
Immunoblot Analysis
For NBR1 immunodetection, equal amounts of seedlings or leaf material were homogenized in protein extraction buffer (4 M urea, 100 mM DTT, and 1% [v/v] Triton X-100) and incubated on ice for 10 min. Protein samples were mixed 1:1 (v/v) with 2× Laemmli sample buffer, boiled for 10 min, and centrifuged at 13,000 rpm for 10 min. Total proteins of the supernatant were subjected to SDS-PAGE, and protein loading was verified by densitometry-based quantification of Coomassie Brilliant Blue-stained bands using ImageJ software. Proteins were separated, transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham, GE Healthcare), and blocked with 5% (w/v) nonfat milk powder in PBS containing 0.05% (v/v) Tween 20. Anti-Arabidopsis NBR1 (kindly provided by T. Johansen, Tromsø University) and secondary horseradish peroxidase-conjugated antibodies (Amersham, GE Healthcare) were diluted 1:2000 and 1:5000, respectively, in PBS containing 0.05% (v/v) Tween 20 with 1% (w/v) milk powder. The immunoreaction was developed using the ECL Prime kit (Amersham, GE Healthcare) and detected in a LAS-3000 Luminescent Image Analyzer (Fujifilm, Fuji Photo Film). Relative NBR1 amounts were quantified using ImageJ software and normalized to Ponceau-stained total protein levels.
Yeast Two-Hybrid Analysis
Yeast two-hybrid (Y2H) techniques were performed according to the Yeast Protocols Handbook (Clontech) using the Y2HGold yeast reporter strain (Clontech). Full-length cDNA fragments of VPS35B, VPS29, VPS26A, and VPS26B were amplified (see Supplemental Table 1 for yeast two-hybrid primer sequences) and cloned into pGBT9 and pGAD424 vectors (Clontech) via the restriction sites EcoRI/BamHI (VPS35B, VPS29, and VPS26) or SmaI/PstI (VPS26B). Yeast cells were cotransformed with the respective plasmid combinations, followed by selection of transformants on SD medium lacking Trp and Leu for 3 d at 28°C, and subsequent transfer to medium lacking Trp, Leu, and His for growth analysis.
BiFC
Cultures of Agrobacterium strain GV3101 harboring BiFC and control constructs were grown overnight at 28°C in Luria-Bertani medium, harvested and resuspended in infiltration buffer (0.5 mM MES, 10 mM MgCl2, and 100 μM acetosyringone), and incubated at room temperature for 2 h. Bacterial suspensions at OD600 = 0.05 were mixed in different combinations at equal ratio and infiltrated into abaxial sides of 4-week-old N. benthamiana leaves. Infiltrated leaves were imaged 36 to 40 h after infiltration with confocal microscopy.
Statistical Analysis
Statistical analysis was done using one-way ANOVA with posthoc Tukey’s test. Significance was accepted at the level of P < 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: VPS35A, AT2G17790; VPS35B, AT1G75850; VPS35C, AT3G51310; VPS29, AT3G47810; ACD11, AT2G34690; LAZ5, AT5G44870, LSD1, AT4G20380; RPS4, AT5G45250; RPM1, AT3G07040; RPS2, AT4G26090; EDS1, AT3G48090; NDR1, AT3G20600; SYP32, AT3G24350; VHAa1, AT2G28520; RabG3F, AT3G18820.
Supplemental Data
Supplemental Figure 1. Transgenic Complementation of laz4 Mutant.
Supplemental Figure 2. EMS-Induced Intron Splice Mutation in laz4 Results in Deletion of Exon 8 from VPS35B Transcripts.
Supplemental Figure 3. Analysis of VPS35B Transcript Levels in T-DNA Mutant Alleles vps35b-1 (SALK_014345) and vps35b-2 (GABI_784C05).
Supplemental Figure 4. Effect of Single Loss-of-Function Alleles in VPS35 Genes on PCD in acd11 and BTH-Treated acd11 nahG.
Supplemental Figure 5. Combined Loss-of-Function Mutations in VPS35A and VPS35C Do Not Suppress acd11 and lsd1 Autoimmune Cell Death.
Supplemental Figure 6. ACD11-GFP Localizes to Vacuole-Like Structures following Wortmannin (Wm) Treatment.
Supplemental Figure 7. Phenotypes of Retromer Deficient Mutants.
Supplemental Figure 8. VPS35 Deficiency Does Not Affect Basal Resistance to a Virulent Strain of Pst DC3000.
Supplemental Figure 9. Early Endocytosis Is Not Affected in Retromer Mutants.
Supplemental Table 1. Oligonucleotides.
Supplemental Table 2. Plasmids.
Supplementary Material
Acknowledgments
We thank Petra Epple (University of North Carolina) for technical and editorial contributions; Terje Johansen and Steingrim Svenning (University of Tromsø) for providing the NBR1 antibody and valuable comments on NBR1 analysis; Thierry Gaude (ENS Lyon) for fruitful discussions and seeds of the pVPS29:VPS29-RFP line; Stig U. Andersen (University of Aarhus) for seeds of atg2-1 and critical reading of the article; Noriyuki Hatsugai (Kyoto University) for sharing unpublished results; and Elena Minina (Swedish University of Agricultural Sciences) for the kind gift of the 2x35S:GFP-ATG8A plasmid. This work was supported by the Knut-and-Alice Wallenberg Foundation (to D.H.), by the Danish Research Councils (Grant 274-06-0460 to J.M.), by a German Research Foundation postdoctoral fellowship (Grant EL 734/1-1 to F.E.K.), by the Howard Hughes Medical Institute-Gordon and Betty Moore Foundation (to J.L.D.), and by the National Science Foundation (Arabidopsis 2010 Program Grant IOS-0929410 to J.L.D.).
AUTHOR CONTRIBUTIONS
D.M., O.-K.T., M.P., J.M., and D.H. designed the research. D.M., O.-K.T., F.G.M., Q.L., R.R.V., F.E.K., P.B., and D.H. performed the research. J.L.D. and I.H.-N. contributed materials and technical information. D.M., O.-K.T., Q.L., R.R.V., F.E.K., J.L.D., J.M., and D.H. analyzed the data. D.H. wrote the article with input from O.-K.T., F.E.K., P.B., M.P., J.L.D., and J.M.
Glossary
- PCD
programmed cell death
- HR
hypersensitive response
- ETI
effector-triggered immunity
- BTH
benzothiadiazole-S-methyl ester
- NB-LRR
nucleotide binding-leucine-rich repeat
- TIR-NB-LRR
Toll/Interleukin-1 Receptor-nucleotide binding-leucine-rich repeat
- CC-NB-LRR
coiled-coil-nucleotide binding-leucine-rich repeat
- EMS
ethyl methanesulfonate
- Ler
Landsberg erecta
- Col-0
Columbia-0
- PVC
prevacuolar compartment
- TGN
trans-Golgi network
- Wm
wortmannin
- MVB
multivesicular body
- BFA
brefeldin A
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- PAMP
pathogen-associated molecular pattern
- Pst
Pseudomonas syringae pv tomato
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Heard W., Mbengue M., Robatzek S. (2012). The INs and OUTs of pattern recognition receptors at the cell surface. Curr. Opin. Plant Biol. 15: 367–374. [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436. [DOI] [PubMed] [Google Scholar]
- Bonardi V., Dangl J.L. (2012). How complex are intracellular immune receptor signaling complexes? Front. Plant Sci. 3: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V., Tang S., Stallmann A., Roberts M., Cherkis K., Dangl J.L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 108: 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Hurley J.H. (2008). Retromer. Curr. Opin. Cell Biol. 20: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes D.C., Nam J., Dangl J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95: 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov P.V., Lam E. (2011). Green death: Revealing programmed cell death in plants. Cell Death Differ. 18: 1239–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Malinovsky F.G., Hématy K., Newman M.A., Mundy J. (2005). The role of salicylic acid in the induction of cell death in Arabidopsis acd11. Plant Physiol. 138: 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H.M., Olszak B., Skov S., Odum N., Jørgensen L.B., Brown R.E., Mundy J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Biselli C., Collins N.C., Consonni G., Stanca A.M., Schulze-Lefert P., Valè G. (2010). The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS ONE 5: e12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J., Padmanabhan M., Dinesh-Kumar S.P. (2008). Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135. [DOI] [PubMed] [Google Scholar]
- Chen D., Xiao H., Zhang K., Wang B., Gao Z., Jian Y., Qi X., Sun J., Miao L., Yang C. (2010). Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science 327: 1261–1264. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Smidler A., Puigvert M., Popa C., Valls M., Dangl J.L. (2014). The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 21: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Dengjel J., et al. (2012). Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics 11: M111.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Chen W. (2013). The role of autophagy in chloroplast degradation and chlorophagy in immune defenses during Pst DC3000 (AvrRps4) infection. PLoS ONE 8: e73091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas T.K., Nimchuk Z.L., Dangl J.L. (2008). Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 105: 6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt S., Boevink P.C., Armstrong M.R., Ramos M.B., Hein I., Birch P.R. (2012). Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell 24: 5142–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Chung E.H., Eitas T.K., Dangl J.L. (2011). Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 108: 7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M., Chang X.B., Cui L., Wu Y., Ozols V.V., Choudhury A., Pagano R.E., Riordan J.R. (2004). Endocytic trafficking routes of wild type and DeltaF508 cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 15: 2684–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Hackenberg T., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25: 4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Hatsugai N. (2011). The role of vacuole in plant cell death. Cell Death Differ. 18: 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y., Niihama M., Takahashi T., Saito C., Nakano A., Tasaka M., Morita M.T. (2010). Loss-of-function mutations of retromer large subunit genes suppress the phenotype of an Arabidopsis zig mutant that lacks Qb-SNARE VTI11. Plant Cell 22: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Iwasaki S., Tamura K., Kondo M., Fuji K., Ogasawara K., Nishimura M., Hara-Nishimura I. (2009). A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 23: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858. [DOI] [PubMed] [Google Scholar]
- Heck S., Grau T., Buchala A., Métraux J.P., Nawrath C. (2003). Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36: 342–352. [DOI] [PubMed] [Google Scholar]
- Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L., Parker J.E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404. [DOI] [PubMed] [Google Scholar]
- Hofius D., Munch D., Bressendorff S., Mundy J., Petersen M. (2011). Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 18: 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783. [DOI] [PubMed] [Google Scholar]
- Hofius D., Tsitsigiannis D.I., Jones J.D., Mundy J. (2007). Inducible cell death in plant immunity. Semin. Cancer Biol. 17: 166–187. [DOI] [PubMed] [Google Scholar]
- Inada N., Ueda T. (2014). Membrane trafficking pathways and their roles in plant-microbe interactions. Plant Cell Physiol. 55: 672–686. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Santambrogio M., Rozier F., Fobis-Loisy I., Miège C., Gaude T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kaminaka H., Näke C., Epple P., Dittgen J., Schütze K., Chaban C., Holt B.F. III, Merkle T., Schäfer E., Harter K., Dangl J.L. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25: 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Kim S.Y., Song K., Sohn E.J., Lee Y., Lee D.W., Hara-Nishimura I., Hwang I. (2012). Trafficking of vacuolar proteins: The crucial role of Arabidopsis vacuolar protein sorting 29 in recycling vacuolar sorting receptor. Plant Cell 24: 5058–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuismanen E., Saraste J. (1989). Low temperature-induced transport blocks as tools to manipulate membrane traffic. Methods Cell Biol. 32: 257–274. [DOI] [PubMed] [Google Scholar]
- Kwon C., Bednarek P., Schulze-Lefert P. (2008). Secretory pathways in plant immune responses. Plant Physiol. 147: 1575–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Kim S.R., Park O.K. (2013). The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161: 1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane R.F., St. George-Hyslop P., Hempstead B.L., Small S.A., Strittmatter S.M., Gandy S. (2012). Vps10 family proteins and the retromer complex in aging-related neurodegeneration and diabetes. J. Neurosci. 32: 14080–14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kufer T.A., Schulze-Lefert P. (2011). NLR functions in plant and animal immune systems: So far and yet so close. Nat. Immunol. 12: 817–826. [DOI] [PubMed] [Google Scholar]
- Malinovsky F.G., et al. (2010). Lazarus1, a DUF300 protein, contributes to programmed cell death associated with Arabidopsis acd11 and the hypersensitive response. PLoS ONE 5: e12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- McGough I.J., Cullen P.J. (2011). Recent advances in retromer biology. Traffic 12: 963–971. [DOI] [PubMed] [Google Scholar]
- Mindrinos M., Katagiri F., Yu G.L., Ausubel F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Minina E.A., Sanchez-Vera V., Moschou P.N., Suarez M.F., Sundberg E., Weih M., Bozhkov P.V. (2013). Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging Cell 12: 327–329. [DOI] [PubMed] [Google Scholar]
- Moeder W., Yoshioka K. (2008). Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal. Behav. 3: 764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nodzynski T., Feraru M.I., Hirsch S., De Rycke R., Niculaes C., Boerjan W., Van Leene J., De Jaeger G., Vanneste S., Friml J. (2013). Retromer subunits VPS35A and VPS29 mediate prevacuolar compartment (PVC) function in Arabidopsis. Mol. Plant 6: 1849–1862. [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Ha S.A., Bruinsma P. (2000). Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 151: 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviusson P., Heinzerling O., Hillmer S., Hinz G., Tse Y.C., Jiang L., Robinson D.G. (2006). Plant retromer, localized to the prevacuolar compartment and microvesicles in Arabidopsis, may interact with vacuolar sorting receptors. Plant Cell 18: 1239–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K., Dong X. (2009). A kiss of death—Proteasome-mediated membrane fusion and programmed cell death in plant defense against bacterial infection. Genes Dev. 23: 2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6: e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D., Akutsu M., Novak I., Harper J.W., Behrends C., Dikic I. (2012). Rab GTPase-activating proteins in autophagy: Regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32: 1733–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Kuster M., Herr P., Furger E., Bänziger C., Hausmann G., Basler K. (2008). Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat. Cell Biol. 10: 178–185. [DOI] [PubMed] [Google Scholar]
- Pourcher M., Santambrogio M., Thazar N., Thierry A.M., Fobis-Loisy I., Miège C., Jaillais Y., Gaude T. (2010). Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 22: 3980–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D., Innes R.W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D., DeYoung B.J., Innes R.W. (2012). Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673. [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Pimpl P., Scheuring D., Stierhof Y.D., Sturm S., Viotti C. (2012). Trying to make sense of retromer. Trends Plant Sci. 17: 431–439. [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Ronald P.C. (2012). Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63: 451–482. [DOI] [PubMed] [Google Scholar]
- Seaman M.N. (2007). Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 120: 2378–2389. [DOI] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Simanshu D.K., Zhai X., Munch D., Hofius D., Markham J.E., Bielawski J., Bielawska A., Malinina L., Molotkovsky J.G., Mundy J.W., Patel D.J., Brown R.E. (2014). Arabidopsis accelerated cell death 11, ACD11, is a ceramide-1-phosphate transfer protein and intermediary regulator of phytoceramide levels. Cell Rep. 6: 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D., Rafiqi M., Hurley U., Lawrence G.J., Bernoux M., Hardham A.R., Ellis J.G., Dodds P.N., Jones D.A. (2012). N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol. Plant Microbe Interact. 25: 379–392. [DOI] [PubMed] [Google Scholar]
- Teh O.K., Hofius D. (2014). Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 65: 1297–1312. [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., et al. (2011). Morphological classification of plant cell deaths. Cell Death Differ. 18: 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Barnaby J.Y., Tada Y., Li H., Tör M., Caldelari D., Lee D.U., Fu X.D., Dong X. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029. [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M.N., Moreau K., Jimenez-Sanchez M., Breusegem S.Y., Harbour M.E., Rubinsztein D.C. (2014). Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5: 3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E., Santambrogio M., Pourcher M., Chambrier P., Berne-Dedieu A., Fobis-Loisy I., Miège C., Jaillais Y., Gaude T. (2013). Mechanisms governing the endosomal membrane recruitment of the core retromer in Arabidopsis. J. Biol. Chem. 288: 8815–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Glazebrook J., Li X. (2007). Identification of components in disease-resistance signaling in Arabidopsis by map-based cloning. Methods Mol. Biol. 354: 69–78. [DOI] [PubMed] [Google Scholar]
- In a recent study, Saucet et al. (2015) reported that RPS4 loss-of-function phenotypes are partial because of the continued presence of the RPS4B/RRS1B NB-LRR protein pair. This finding may explain the significantly enhanced suppression of AvrRps4-triggered HR in vps35a-1 c-1 compared with the rps4-2 control (Figure 6A). [Google Scholar]
- Saucet S.B, Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G. (2015). Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun., doi/10.1038/ncomms7338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.